Summary
The biogenic amine transporters (BATs) regulate endogenous neurotransmitter concentrations and are targets for a broad range of therapeutic agents that include selective serotonin reuptake inhibitors (SSRIs), serotonin-norepinephrine reuptake inhibitors (SNRIs) and tricyclic antidepressants (TCAs)1, 2. Because eukaryotic BATs are recalcitrant to crystallographic analysis, our understanding of the mechanism of these inhibitors and antidepressants is limited. LeuT is a bacterial homolog of BATs and has proven a valuable paradigm for understanding relationships between structure and function in BATs3. However, because LeuT has only ~20% amino acid sequence identity to BATs and is a promiscuous amino acid transporter4, it does not recapitulate the pharmacological properties of BATs. Indeed, SSRIs and TCAs bind in the extracellular vestibule of LeuT5-7 and act as non-competitive inhibitors of transport5. In contrast, multiple studies demonstrate that both TCAs and SSRIs are competitive inhibitors for eukaryotic BATs and bind to the primary binding pocket8-16. Here, we engineered LeuT to harbor human BAT-like pharmacology by mutating key residues around the primary binding pocket. The final LeuBAT mutant binds the SSRI sertraline with a binding constant of 18 nM and displays high affinity binding to a range of SSRIs, SNRIs and a TCA. We determined 12 crystal structures of LeuBAT in complex with four classes of antidepressants. The chemically diverse inhibitors have a remarkably similar mode of binding in which they straddle TM3, wedge between TM3/TM8 and TM1/TM6, and lock the transporter in a sodium and chloride-bound outward facing open conformation. Together, these studies define common and simple principles for the action of SSRIs, SNRIs and TCAs on BATs.
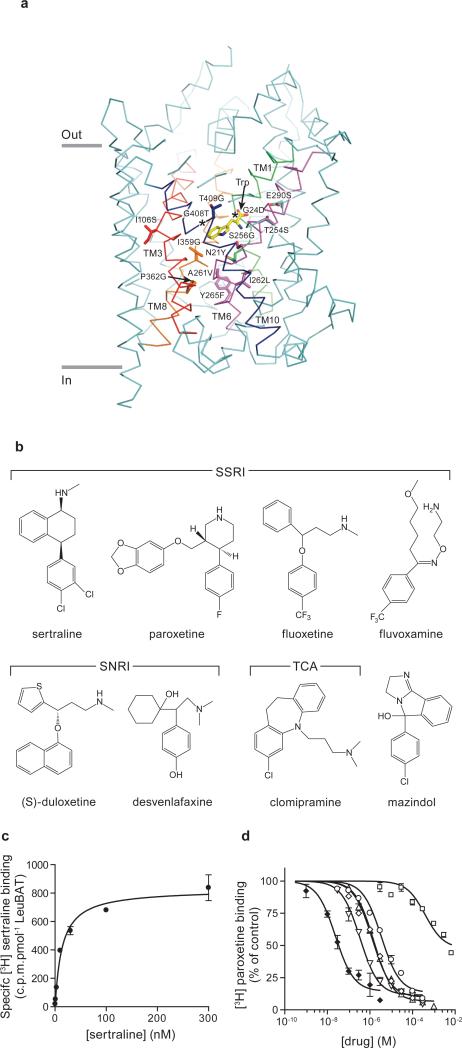
We used the structure of wild-type LeuT in complex with the competitive inhibitor tryptophan (PDB code 3F3A)4 as a template for mutant design (Fig. 1a). We analyzed residues within a 10 Å-radius of the primary binding pocket of the LeuT-Trp complex (Fig. 1a) together with a LeuT/human serotonin transporter (hSERT) amino acid sequence alignment to identify about 20 residues which point toward the primary binding pocket and are divergent from hSERT (Supplementary Fig. 1). These residues are located in both bundle and scaffold domains17, sodium binding sites3, the chloride binding site18, 19 and the extracellular vestibule. Previous studies have demonstrated the importance of many of these residues in hSERT pharmacology9-12, 15, 20, 21. By tracking the binding constant (Kd) of [3H]-paroxetine, we introduced these mutations into LeuT, focusing initially on ‘first shell’ residues predicted to interact directly with inhibitors and next on ‘second shell’ residues (Supplementary Table I). The Kd values for paroxetine and mazindol binding to the final LeuBAT mutant, deemed Δ13 LeuBAT (Supplementary Table I), are 431±24 nM and 112±18 nM, respectively (Supplementary Fig. 2). Strikingly, the Kd of Δ13 for mazindol is similar to that of hSERT (103±4.7 nM)9. Because uptake experiments using the Δ6 or Δ13 variants reconstituted into liposomes show that the constructs are not active in transporting either serotonin or dopamine (Supplementary Fig. 3), further experiments are required to engineer a variant of LeuBAT that possesses both high affinity inhibitor binding and transport activity.
Fig. 1. LeuBAT design and pharmacology.
(a) The representation of mutation positions around the primary binding pocket in wild-type LeuT-Trp structure (PDB 3F3A). Bound tryptophan (yellow) and the mutated residues are in sticks. The transmembrane helices TM1, TM3, TM6, TM8 and TM10 around the pocket are highlighted as green, red, purple, orange and blue, respectively. Asterisks depict the glycine residue positions. (b) Chemical structures of four SSRIs, two SNRIs, one tricyclic antidepressant (clomipramine) and one stimulant (mazindol); (c) Measurement of [3H] sertraline binding (filled circles) to Δ13 LeuBAT; (d) Dose-response curves for inhibition of [3H] paroxetine binding to Δ13 LeuBAT by sertraline (filled diamonds), fluvoxamine (empty circles), fluoxetine (empty diamonds), duloxetine (empty inverted triangles), clomipramine (empty triangles), desvenlafaxine (empty squares). Error bars, s.e.m, n = 3.
For the Δ13 LeuBAT construct we performed competition experiments using [3H] paroxetine and multiple cold SSRIs, SNRIs and a TCA (Fig.1; Supplementary Table II). Strikingly, sertraline possesses the highest affinity (Ki=14±2 nM; Kd=18±2 nM; Fig. 1), thus approaching the reported value for sertraline binding to hSERT (0.3 nM)22. To demonstrate that the Δ6 and Δ13 variants possess increased affinities for inhibitors relative to wild-type LeuT, we determined that the Kd values for sertraline and mazindol binding to wild-type LeuT are 308±63 nM and 22.3±5.4 μM, respectively, while the binding of paroxetine could not be fit to an isotherm because of low affinity (Supplementary Fig. 2). The substrate alanine, which binds to the primary pocket of wild-type LeuT4, could not suppress the binding of sertraline to wild-type LeuT, consistent with the conclusion that these drugs bind within the extracellular vestibule of wild-type LeuT5-7.
We determined crystal structures of LeuBAT in complex with a panel of SSRIs, SNRIs and a TCA using the Δ5, Δ6 and Δ13 variants (Supplementary Table III). For the Δ5 and Δ6 mutants, we determined structures for the Δ5-mazindol, Δ6-sertraline, Δ6-desvenlafaxine, Δ6-duloxetine, and Δ6-mazindol complexes at resolutions of 2.3 Å- 2.7 Å. For the Δ13 variant, we determined seven structures with sertraline, paroxetine, fluoxetine, fluvoxamine, duloxetine, desvenlafaxine, and clomipramine (CMI) at resolutions of 2.85 Å-3.31Å (Supplementary Fig. 4; Supplementary Table III). Because the binding of inhibitors is similar between the Δ6 and Δ13 constructs (Supplementary Fig. 5), we used the higher resolution structures of the Δ6 complexes for analysis and validation of the drug binding sites in Δ13.
All LeuBAT structures adopt an outward-facing open conformation (Figs. 2 and 3), similar to that of wild-type LeuT in complex with tryptophan (PDB 3F3A)4 (r.m.s.d. of 0.48 Å for Cα atoms). All drugs bind to the primary binding pocket3, interact with both the bundle (TM1, TM6) and scaffold (TM3, TM8) domains, and are lodged between the extracellular gate residues Arg30 and Asp4043. These observations invalidate the notion that SSRIs and TCAs elicit their effects on hSERT by binding in the extracellular vestibule6, 7, 23.
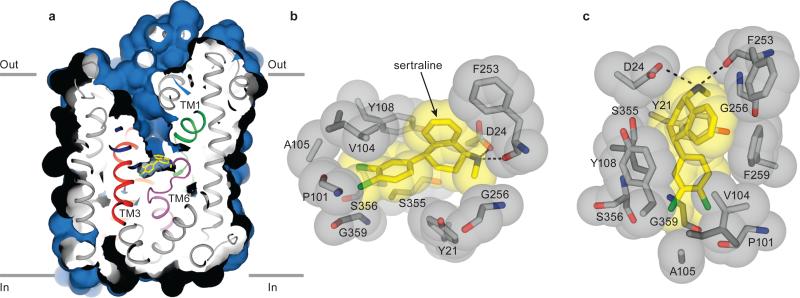
Fig. 2. LeuBAT Δ13-sertraline complex adopts an outward-facing open conformation.
(a) Cross-sections of the crystal structure of Δ13-sertraline showing the solvent accessible surface area (blue). Bound sertraline is shown in yellow sticks. (b-c) Zoom into the sertraline binding pocket viewed within the membrane plane (b) and from the extracellular side (c). Sertraline and key residues in the pocket are depicted in both sticks and spheres showing the van der Waals radius. Salt bridges and hydrogen bonds are in dashed lines. Phe259 in (b) is omitted for clarity.
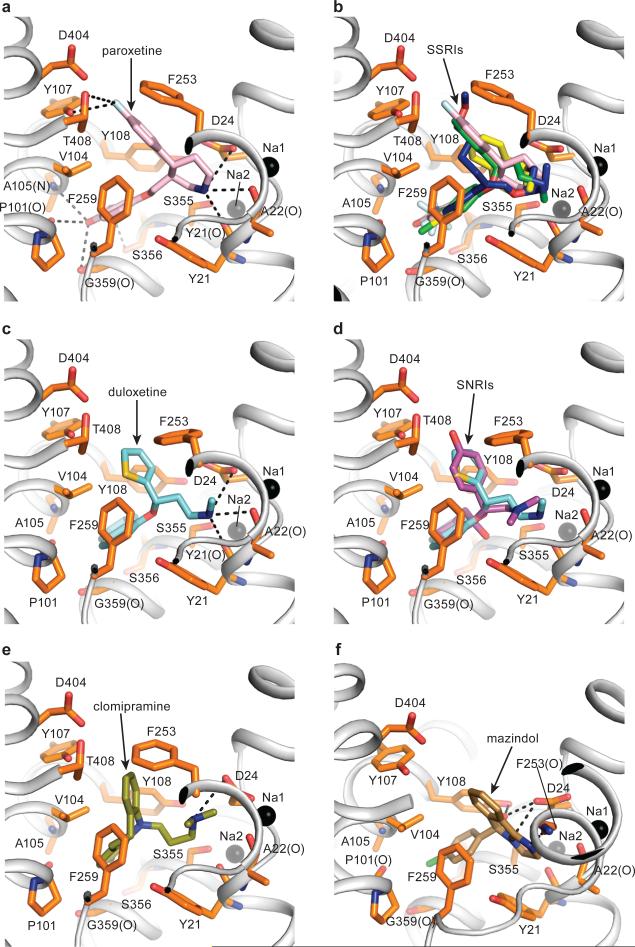
Fig. 3. SSRIs, SNRIs, TCA and mazindol share similar binding features.
(a) Paroxetine binding site in Δ13 LeuBAT mutant viewed within the membrane plane with paroxetine shown as pink sticks. Sodium ions and key residues are shown as black spheres and orange sticks, respectively. Hydrogen bonds, salt bridges and polar interactions are in dashed lines. (b) Superimposition of paroxetine (pink), (R)-fluoxetine (green), fluvoxamine (blue) and sertraline (yellow) in the primary binding pocket of the Δ13 LeuBAT; (c) (S)-Duloxetine binding site in the Δ13 LeuBAT with duloxetine shown as cyan sticks; (d) Superimposition of (S)-duloxetine (cyan) and (S)-desvenlafaxine (magenta) in the primary drug-binding pocket; (e) Clomipramine (CMI) binding site in Δ13 LeuBAT with CMI shown as olive sticks; (f) Mazindol binding site in LeuBAT Δ6 variant with mazindol molecule shown as sand-colored sticks.
We find two sodium ions, Na1 and Na2, bound to sites similar to those in the LeuT-Trp complex with the following distinction. In the wild-type LeuT structures3, 4 the α-carboxyl group of Leu or Trp participates in the coordination of Na1, while in the LeuBAT structures a water molecule is found at the equivalent position, bridging the carboxylate of Asp24 and Na1 (Supplementary Fig. 6). We detected electron density for a chloride ion in the LeuBAT-paroxetine structure ~4.5 Å from Na1, coordinated by Tyr47, Ser254, Asn286 and Ser290 as previously predicted (Supplementary Fig. 6)18, 19, 24.
We solved structures of four LeuBAT-SSRI complexes: sertraline, paroxetine, fluoxetine and fluvoxamine. Sertraline occupies the primary pocket surrounded by TM1, TM3, TM6 and TM8 and buries 438.6 Å2 or 93% of its surface area (Fig. 2). The amine group forms a salt bridge with the carboxyl group of Asp24 while the tetrahydronaphthalene ring participates in hydrophobic interactions with Tyr21 and is sandwiched between Val104, Tyr108 and Phe259 (Fig. 2). The two chlorine atoms on the dichlorophenyl ring insert into a groove formed by Pro101, Val104, Ala105, Ser356 and Gly359. We suggest that these extensive hydrophobic and van der Waals interactions contribute to the high affinity of sertraline for LeuBAT and hSERT.
The other SSRIs - paroxetine, fluoxetine and fluvoxamine - also bind to the primary binding pocket in an orientation similar to that of sertraline. Consistent with previous hSERT-paroxetine and hSERT-fluoxetine models13, the amine groups are proximal to the carboxyl group of Asp24 (Fig. 3a,b). The amine groups of the inhibitors also form direct hydrogen bonds with main chain carbonyl groups of Tyr21, Ala22 and/or Phe253. The benzodioxol group of paroxetine and trifluoromethylphenyl rings from fluoxetine and fluvoxamine insert to the same groove as does the chlorophenyl ring of sertraline, forming hydrophobic interactions with Val104, Tyr108 and Phe259, and/or van der Waals interactions with the main chain carbonyl groups of Pro101, Ala105, Ser356 and Gly359. In addition, the fluorophenyl ring of paroxetine, the phenyl ring of fluoxetine and the ether chain of fluvoxamine extend into the extracellular vestibule, forming hydrophobic interactions, van der Waals contacts and hydrogen bonding interactions with Tyr107, Phe253, Asp404 and/or Thr408.
In the SNRI complexes with duloxetine and desvenlafaxine, the inhibitors sit in the primary pocket with the amine groups interacting with the carboxyl group of Asp24 and, in the case of duloxetine, also with the main chain carbonyl of Tyr21 (Fig. 3c, d). The naphthalene ring from duloxetine and the cyclohexanol ring from desvenlafaxine are sandwiched by hydrophobic groups Tyr21, Val104, Phe259, and Tyr108. The thiophene ring from duloxetine and the phenol ring from desvenlafaxine protrude into the extracellular vestibule and interact with Phe253 by π-stacking interactions. For desvenlafaxine, the hydroxyl group in the cyclohexanol moiety makes a hydrogen bond with the main chain carbonyl group of Ser355 while the phenol hydroxyl interacts with Asp404 and the phenol group of Tyr107.
The tricyclic antidepressant CMI binds to the primary binding pocket of Δ13 LeuBAT (Fig. 3e) in agreement with a hSERT-TCA model11, 12 and in contrast to wild-type LeuT-TCA structures5, 6. The tricyclic ring is surrounded by hydrophobic residues that include Tyr21, Val104, Tyr108, Phe253 and Phe259. The chlorine atom in the tricyclic ring extends to the pocket formed by Ala105, Ser356 and Gly359, similar to the chorine positions in sertraline or the trifluoromethyl moiety in fluoxetine. Not only are previously proposed interactions12 observed in our LeuBAT-CMI structure, such as the salt bridge between Asp24 (Asp98 in hSERT) and the tertiary aliphatic amine of the TCA, the structure is consistent with interactions between Ala105 (Ala173 in hSERT) and the TCA 3-position, and with Phe253 (Phe335 in hSERT) being near the TCA 7-position. Our structure is in harmony with the conclusion that Ser438 in hSERT (Ser355 in LeuBAT) is vicinal to the aminopropyl chain of TCA and that the S438T mutation affects the binding affinity because of steric clash11.
The stimulant mazindol binds to the primary site and is surrounded by a hydrophobic pocket formed by Tyr21, Val104, Tyr108 and Phe259 (Fig.3f). The amine nitrogen forms a salt bridge with the carboxyl group of Asp24. The hydroxyl group hydrogen bonds with the carboxyl group of Asp24 and the phenol group of Tyr108. The chlorophenyl ring inserts into the pocket formed by Ala105, Ser356 and Gly359, similar to sertraline. The importance of these interactions is supported by the fact that removal of the chlorophenyl ring or changes in the substitution on the phenyl ring decreases the affinity of mazindol to hSERT9.
By soaking Δ6 mutant crystals in 20 mM desvenlafaxine, we identified a second desvenlafaxine molecule in the extracellular vestibule (Supplementary Fig. 7). The second molecule occupies a similar position as the TCAs, sertraline and fluoxetine occupy in wild-type LeuT structures reported previously5-7 (Supplementary Fig. 7). These results demonstrate that the n-octyl-β-d-glucoside (β-OG) molecule bound to the extracellular vestibule of wild-type LeuT25, 26 is readily substituted by a drug molecule and that the extracellular vestibule is a site for the low affinity, non-specific binding of small molecules.
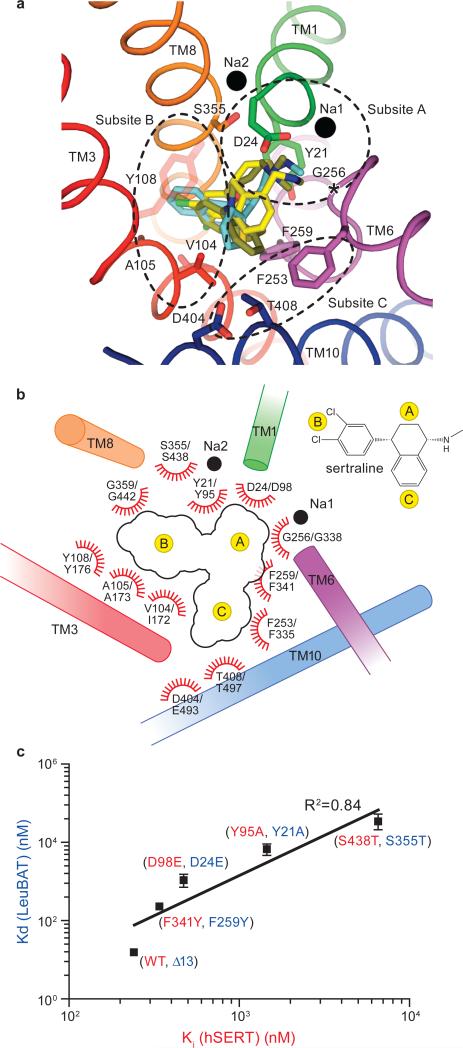
Analysis of the sertraline (SSRI), duloxetine (SNRI) and CMI (TCA) complexes with LeuBAT allows us to identify three subsites within the primary binding site to which the pharmacophores of these chemically diverse inhibitors bind (Fig.4a, b). Subsite A is defined by Asp24, Tyr21, Gly256 and Ser355 from TM1, TM6 and TM8 and this subsite accommodates the polar, amine moiety of the inhibitors27. Asp24 interacts with the amine groups of most drugs by salt bridge, which echoes the suggestion that Asp98 in hSERT forms a similar interaction with the amine groups of SSRIs, TCAs and serotonin9, 12, 13, 20, 21.
Fig 4. Implication for drug binding in hSERT and validation by mutational studies.
(a) Superposition of SSRI (sertraline, yellow), SNRI (duloxetine, cyan) and TCA (CMI, olive) in the primary binding pocket of LeuBAT, viewed from extracellular side. The key residues in the pockets and the two sodium ions are shown as sticks and black spheres, respectively. The regions enclosed by dashed lines define subsites A, B and C in the primary drug binding pocket. (b) Schematic representation of drug interactions in the primary binding pockets of LeuBAT/hSERT. The transmembrane helices are shown as cylinders. Residue numbering follows LeuBAT and hSERT, respectively. (c) Plot of sertraline binding constants for Y21A, D24E, F259Y and S355T mutants of the Δ13 LeuBAT against the inhibition constants for the corresponding mutants Y95A, D98E, F341Y and S438T in hSERT, respectively. Error bars, s.e.m, n = 3
Subsite B includes residues from TM3, TM6 and TM8 and involves two types of interactions. First, non polar residues form hydrophobic interactions with the hydrophobic rings of the drugs. Phe259 (Phe 341 in hSERT) together with Val104 (Ile172 in hSERT or Val148 in hNET) define a non polar ridge that accommodates the hydrophobic groups of the drugs27,28. Previous studies showed that the F341Y mutation in hSERT reduces the potency of paroxetine and escitalopram27. Here we suggest that the F341Y mutation leads to a clash with the fluorophenyl ring of paroxetine. The second type of interaction in subsite B is the groove delineated by Pro101, Ala105, Gly359 and Ser356. This groove accommodates the polar groups in the drugs’ rings, such as the chloro, dichloro, trifluoromethyl and benzodioxol groups of CMI, sertraline, fluoxetine and paroxetine, respectively. An hSERT- imipramine model suggests that the imipramine 3-position is vicinal to Ala173 (Ala105 in LeuBAT)12, and here we show that the chlorine atom in the 3-position of CMI forms a direct contact with this ridge.
Subsite C, distal to the primary, orthosteric binding site, is located in the extracellular vestibule, and is comprised of residues in TM6 and TM10, including Phe253, Asp404 and Thr408. Subsite C interacts with bulky drugs such as paroxetine, desvenlafaxine, fluoxetine and likely plays a role in enhancing inhibitor affinity and specificity. Indeed, previous studies determined that mutation of E493Q in hSERT (Asp404 in LeuBAT) attenuates the potency of fluoxetine7.
To buttress the conclusion that the LeuBAT complexes represent reliable models for BAT-inhibitor complexes, we prepared and analyzed the individual Y21A, D24E, F259Y and S355T mutations in the context of the Δ13 LeuBAT construct. Y21A, D24E and S355T are in subsite A and F259Y is in subsite B. We next measured their [3H]-sertraline Kd values and plotted logKd of the LeuBAT mutants against logKi of the homologous hSERT mutants27(Fig. 4c). The resulting linear relationship suggests that the mutations in LeuBAT have similar effects to those in hSERT, and suggests that LeuBAT represents a framework for understanding the pharmacology of hSERT. We further compared the pharmacological selectivity of LeuBAT with all human BATS by plotting logKi (LeuBAT) vs. logKi (hDAT, hNET or hSERT) (Supplementary Fig. 8). Inspection of these plots suggests that the pharmacological properties of LeuBAT is a hybrid of human BATs. Moreover, we compared the TCA binding site of the LeuBAT-CMI complex with the recently determined structure of the Drosophila dopamine transporter (dDAT)-nortriptyline complex29 (Supplementary Fig. 9). We note that the overall outward-open conformation and the essential elements of inhibitor binding are shared between the two structures. Finally, we reverted residues Asp24, Gly256 and Gly359 in LeuBAT to their LeuT identities and investigated their effects on the drug binding. These mutations profoundly diminish drug binding (Supplementary Fig. 10), thus supporting the conclusion that the LeuBAT-inhibitor crystal structures represent specifically bound ligand-transporter complexes.
Taken together, the LeuBAT complexes allow us to map crucial subsites within the primary, orthosteric binding site that are responsible for binding the pharmacophores of a chemically diverse group of SSRIs, SNRIs and TCAs, they show that these inhibitors act by binding to the outward-open conformation of the transporter and, perhaps most importantly, they provide molecular guideposts for the development of new therapeutic agents.
METHODS SUMMARY
The LeuBAT mutants were expressed and purified as previously described3 except that lauryl maltose neopentyl glycol was used for solubilization and purification. After final purification by size-exclusion chromatography in β-OG, LeuBAT was concentrated to 2.5 mg/ml and supplemented with saturated serotonin or mazindol. All LeuBAT-drug complexes except mazindol were formed by soaking LeuBAT crystals in crystallization solutions containing 3-20 mM of each drug. The structures were solved by molecular replacement using LeuT-Trp structure4 as a search probe and then subjected to crystallographic refinement. Functions of LeuBAT mutants were examined by [3H]-paroxetine, [3H]-sertraline and [3H]-mazindol saturation and competition binding assays.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Michel for help in binding experiments, D. Claxton for comments, L. Vaskalis for assistance with illustrations. We also thank the beamline staff at the Advanced Light Source (beamlines 8.2.1 and 5.0.2) and Advanced Photon Source (Argonne National Laboratory, beamlines 24-ID-C and 24-ID-E). H.W. also thanks the presenters in 2012 CCP4/APS summer school for useful lectures and tutorials. This work was supported by the NIH. E.G is an Investigator with the Howard Hughes Medical Institute.
Footnotes
AUTHOR CONTRIBUTION
H.W. and E.G. designed the research; H.W., A.G., and R.R. performed protein expression and purification; H.W., A.G., K.H.W. and A.P. carried out ligand-binding and flux experiments; H.W. conducted crystallization and structure determination; H.W. and E.G. wrote the manuscript together with comments from all authors.
ACCESSION CODES
Coordinates and structure factors for the LeuBAT Δ13-paroxetine, Δ13-sertraline, Δ13-duloxetine, Δ13-desvenlafaxine, Δ13-fluoxetine, Δ13-fluvoxamine, Δ13-clomipramine, Δ6-sertraline, Δ6-desvenlafaxine, Δ6-duloxetine, Δ6-mazindol, Δ5-mazindol cocrystal structures have been deposited in the Protein Data Bank with codes 4MM4, 4MM5, 4MM6, 4MM7, 4MM8, 4MM9, 4MMA, 4MMB, 4MMC, 4MMD, 4MME, 4MMF, respectively.
REFERENCES
- 1.Bröer S, Gether U. The solute carrier 6 family of transporters. Br. J. Pharmacol. 2012;167:256–278. doi: 10.1111/j.1476-5381.2012.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kristensen AS, et al. SLC6 Neurotransmitter transporters: Structure, function, and regulation. Pharmacol. Rev. 2011;63:585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 2005;437:215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448:952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z, et al. LeuT-desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science. 2007;317:1390–1393. doi: 10.1126/science.1147614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, et al. Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures. Nat. Struct. Mol. Biol. 2009;16:652–657. doi: 10.1038/nsmb.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Talvenheimo J, Nelson PJ, Rudnick G. Mechanism of imipramine inhibition of platelet 5-hydroxytryptamine transport. J. Biol. Chem. 1979;254:4631–5. [PubMed] [Google Scholar]
- 9.Barker EL, et al. High affinity recognition of serotonin transporter antagonists defined by species-scanning mutagenesis: An aromatic residue in transmembrane domain I dictates species-selective recognition of citalopram and mazindol. J. Biol. Chem. 1998;273:19459–19468. doi: 10.1074/jbc.273.31.19459. [DOI] [PubMed] [Google Scholar]
- 10.Henry LK, et al. Tyr-95 and Ile-172 in transmembrane segments 1 and 3 of human serotonin transporters interact to establish high affinity recognition of antidepressants. J. Biol. Chem. 2006;281:2012–2023. doi: 10.1074/jbc.M505055200. [DOI] [PubMed] [Google Scholar]
- 11.Andersen J, et al. Location of the antidepressant binding site in the serotonin transporter: importance of Ser-438 in recognition of citalopram and tricyclic antidepressants. J. Biol. Chem. 2009;284:10276–10284. doi: 10.1074/jbc.M806907200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinning S, et al. Binding and orientation of tricyclic antidepressants within the central substrate site of the human serotonin transporter. J. Biol. Chem. 2010;285:8363–8374. doi: 10.1074/jbc.M109.045401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tavoulari S, Forrest LR, Rudnick G. Fluoxetine (Prozac) Binding to serotonin transporter is modulated by chloride and conformational changes. J. Neurosci. 2009;29:9635–9643. doi: 10.1523/JNEUROSCI.0440-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koldso H, et al. The two enantiomers of citalopram bind to the human serotonin transporter in reversed orientations. J. Am. Chem. Soc. 2010;132:1311–1322. doi: 10.1021/ja906923j. [DOI] [PubMed] [Google Scholar]
- 15.Andersen J, et al. Mutational mapping and modeling of the binding site for (S)-Citalopram in the human serotonin transporter. J. Biol. Chem. 2010;285:2051–2063. doi: 10.1074/jbc.M109.072587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jørgensen AM, et al. Homology modeling of the serotonin transporter: insights into the primary escitalopram-binding Site. ChemMedChem. 2007;2:815–826. doi: 10.1002/cmdc.200600242. [DOI] [PubMed] [Google Scholar]
- 17.Forrest LR, Rudnick G. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology. 2009;24:377–386. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zomot E, et al. Mechanism of chloride interaction with neurotransmitter:sodium symporters. Nature. 2007;449:726–730. doi: 10.1038/nature06133. [DOI] [PubMed] [Google Scholar]
- 19.Forrest LR, Tavoulari S, Zhang Y-W, Rudnick G, Honig B. Identification of a chloride ion binding site in Na+/Cl− dependent transporters. Proc. Natl. Acad. Sci. 2007;104:12761–12766. doi: 10.1073/pnas.0705600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Celik L, et al. Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation . J. Am. Chem. Soc. 2008;130:3853–3865. doi: 10.1021/ja076403h. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann KW, et al. Structural determinants of species-selective substrate recognition in human and Drosophila serotonin transporters revealed through computational docking studies. Proteins. 2009;74:630–642. doi: 10.1002/prot.22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatsumi M, Groshan K, Blakely RD, Richelson E. Pharmacological profile of antidepressants and related compounds at human monoamine transporters. Eur. J. Pharmacol. 1997;340:249–258. doi: 10.1016/s0014-2999(97)01393-9. [DOI] [PubMed] [Google Scholar]
- 23.Sarker S, Weissensteiner R, Steiner I, Sitte HH, Ecker GF, Freissmuth M, Sucic S. The high-affinity binding site for tricyclic antidepressants resides in the outer vestibule of the serotonin transporter. Mol Pharmacol. 2010;78:1026–35. doi: 10.1124/mol.110.067538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavoulari S, Rizwan AN, Forrest LR, Rudnick G. Reconstructing a chloride-binding site in a bacterial neurotransmitter transporter homologue. J. Biol. Chem. 2011;286:2834–2842. doi: 10.1074/jbc.M110.186064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat. Struct. Mol. Biol. 2012;19:212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc. Natl. Acad. Sci. 2009;106:5563–5568. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen L, et al. Interaction of antidepressants with the serotonin and norepinephrine transporters: mutational studies of the S1 substrate binding pocket. J. Biol. Chem. 2012;287:43694–43707. doi: 10.1074/jbc.M112.342212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersen J, et al. Molecular determinants for selective recognition of antidepressants in the human serotonin and norepinephrine transporters. Proc. Natl. Acad. Sci. 2011;108:12137–12142. doi: 10.1073/pnas.1103060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penmatsa A, Wang KH, Gouaux E. X-ray structure of a dopamine transporter in complex with a tricyclic antidepressant. Nature. 2013 doi: 10.1038/nature12533. doi:10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 31.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collaborative Computing Project The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, et al. Phenix: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen VB, et al. Molprobity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Gouaux E. Substrate binds in the S1 site of the F253A mutant of LeuT, a neurotransmitter sodium symporter homologue. EMBO Rep. 2012;13:861–866. doi: 10.1038/embor.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.