Abstract
Background and Purpose
Visit-to-visit variability in BP is associated with ischemic stroke. We sought to determine whether such variability has a genetic aetiology and whether genetic variants associated with BP variability are also associated with ischemic stroke.
Methods
A GWAS for loci influencing BP variability was undertaken in 3,802 individuals from the Anglo-Scandinavian Cardiac Outcome Trial (ASCOT) study where long-term visit-to-visit and within visit BP measures were available. Since BP variability is strongly associated with ischemic stroke, we genotyped the sentinel SNP in an independent ischemic stroke population comprising of 8,624 cases and 12,722 controls and in 3,900 additional (Scandinavian) participants from the ASCOT study in order to replicate our findings.
Results
The ASCOT discovery GWAS identified a cluster of 17 correlated SNPs within the NLGN1 gene (3q26.31) associated with BP variability. The strongest association was with rs976683 (p=1.4×10−8). Conditional analysis on rs976683 provided no evidence of additional independent associations at the locus. Analysis of rs976683 in ischemic stroke patients found no association for overall stroke (OR 1.02; 95% CI 0.97-1.07; p=0.52) or its sub-types: CE (OR 1.07; 95% CI 0.97-1.16; p=0.17), LVD (OR 0.98; 95% 0.89-1.07; p=0.60) and SVD (OR 1.07; 95% CI 0.97-1.17; p=0.19). No evidence for association was found between rs976683 and BP variability in the additional (Scandinavian) ASCOT participants (p=0.18).
Conclusions
We identified a cluster of SNPs at the NLGN1 locus showing significant association with BP variability. Follow up analyses did not support an association with risk of ischemic stroke and its subtypes.
Keywords: Blood pressure variability, stroke, GWAS, gene, polymorphism
Introduction
Familial studies have long provided evidence of heritability (31%-68%) of blood pressure (BP)1. In recent years substantial progress has also been made in our understanding of the genetics of various measures of BP (Systolic BP, Diastolic BP, Mean Arterial Pressure and Pulse Pressure)2-7. However, episodic hypertension or variability in BP remains understudied despite evidence supporting their role as risk factors in vascular events8. Visit-to-visit variability in systolic BP is a strong predictor of ischemic stroke independent of mean BP9, with hypertensives showing the most BP variability over a series of visits having the greatest risk of a cardiovascular event8, 10.
Determining whether BP variability has a genetic basis is difficult given the lack of prospective cohorts with visit-to-visit BPs recorded and accompanying GWAS data. The Anglo-Scandinavian Cardiac Outcome Trial (ASCOT) study is a longitudinal study investigating the impact of a calcium channel blocker against a beta-blocker regime in hypertensive individuals at moderate risk of a cardiovascular (CV) outcome recruited in the United Kingdom, Ireland and Nordic countries from 1998-200011. Long-term BP variability measurements spread over 5 years and genome wide genotyping were available for a ASCOT study subset, the ASCOT United Kingdom-Ireland cohort (ASCOT-UK-IR) allowing a genome wide analysis to be conducted for genetic risk variants of BP variability.
We hypothesized that since visit-to-visit BP variability is associated with the risk of ischemic stroke more than haemorrhagic stroke8 and hypertension is a major modifiable risk factor, any genetic variants which are associated with BP variability may also be associated with ischemic stroke. Based on recently published GWAS studies12, 13 which show the genetic risk of stroke to be subtype specific, we tested the genetic variant in ischemic stroke subtypes. In an effort to replicate our findings, we also tested the genetic variant for association with BP variability in an independent set of individuals from the ASCOT Scandinavian (ASCOT-DK-FI-NO-SE) cohort.
Methods
Study populations
ASCOT
The ASCOT Blood Pressure Lowering Arm (ASCOT-BPLA) is an investigator-led multi-centre trial which included over 19,000 hypertensive patients, aged 40-79 years at baseline, with an average SBP of 140/90 mmHg on-treatment and 160/100 mmHg off-treatment 11. Patients had no history of CHD but had at least three other risk factors for cardiovascular disease such as LVH, type II diabetes mellitus, peripheral artery disease, previous stroke/TIA, male, ≥ 55 years of age or cigarette smoking. The study tested the impact of a contemporary calcium channel blocker based regimen against an older beta blocker based regime in hypertensives at moderate risk of a CV outcome. The primary objective of the blood pressure-lowering arm was to assess and compare the long-term effects of two blood-pressure-lowering regimens on the combined endpoint of non-fatal myocardial infarction (including silent myocardial infarction) and fatal CHD. Blood pressure was measured in a seated position by a uniform automated device (Omron HEM705CP) in all participants over an average of 13 visits across 5.5 years.
The ASCOT United Kingdom-Ireland (ASCOT-UK-IR) GWA study population included 3802 individuals extracted from the original cohort of 19,342 hypertensives. Visit-to-visit BP variability measurements were recorded prospectively for within visit and between visit BP variability over 5.5 years. Blood samples for DNA isolation were collected of which 3,802 individuals of European ancestry from UK and Ireland were genotyped allowing a genome wide analysis to be conducted for risk variants of BP variability. A subset of 3,900 individuals from the ASCOT study recruited in Denmark, Finland, Norway and Sweden (ASCOT-DK-FI-NO-SE) for whom DNA was available were utilised for replication analyses. The recruitment criterion for the Scandinavian ASCOT participants was identical to the UK and Irish participants, and all had BP measurements taken at similar time-points to calculate BP variability.
Details of ASCOT-UK-IR study population are tabulated in Table I of data supplement methods (Please see http://stroke.ahajournals.org).
Ischemic Stroke
The stroke population included 8,624 cases and 12,722 controls from 7 different cohorts (see Supplemental Material): Australian Stroke Genetics Collaborative (ASGC) 13, 14, Bio-Repository of DNA in Stroke (BRAINS)15, 16, Genetics of Early Onset Stroke (GEOS) 17, 18, Ischemic Stroke Genetics Study and Siblings with Ischemic Stroke Study (ISGS 19/ SWISS 20), Welcome Trust Case Control Consortium 2 United Kingdom (WTCCC2-UK) 21, Welcome Trust Case Control Consortium 2 Germany (WTCCC2-Germany) 21 and Vitamin Intervention for Stroke Prevention trial (VISP) 22.All participating cohorts received institutional ethical clearance and signed consent from each participating study subject. ISGS/SWISS, GEOS and VISP used gender and age matched stroke-free controls recruited from the local population. BRAINS and WTCCC2-UK used the WTCCC 1958 British Birth cohort and National Blood Service (NBS) controls. WTCCC2-Germany derived controls of German Caucasian origin from the KORAgen study (www.gsf.de/kora).
TOAST classification23 was performed by an in-house neurologist and all stroke cases were classified into 3 categories: cardioembolic stroke, large artery disease and small vessel disease. All cohorts except VISP provided stroke subtype data.
Details of stroke cohort study populations are tabulated in Table II of data supplement methods (Please see http://stroke.ahajournals.org).
Genotyping and Imputation
The genotyping, imputation and quality control for the ASCOT GWAS has been described previously24. A detailed description of genotyping, imputation and quality control methods for each participating study in the ischemic stroke analysis is given in the supplement methods section and Table III (Please see http://stroke.ahajournals.org). Single SNP genotyping of rs976683 in 3,900 Scandinavian ASCOT samples was performed using the KASPAR assay at Bart's and the London Genome Centre. Image processing and genotype calling was using SDS (Applied Biosystems) and Autocaller (Applied Biosystems). Any genotypes discrepant between the two calling algorithms was manually inspected and corrected.
Data Analysis
In the ASCOT study BP was measured in all participants over an average of 13 visits across 5.5 years. Measurements for the first 6 months after starting therapy were excluded because this was a period of forced medication titration and any differential medication effects could have acted as a confounder. Data simulations demonstrated that the combination of within visit BP variability and visit-to-visit BP variability allowed the use of more BP measurements. Within-individual visit-to-visit BP variability phenotype was expressed as mean (±SD) and coefficient of variation (SD/mean) using the 2nd and 3rd readings for every visit for ASCOT-BPLA cohort. The Variance Independent of Mean (VIM) transformation was applied if there was a correlation between the mean SBP and coefficient of variation10. The SBP-VIM was derived for all on-treatment SBP values, analysing total variability (within visit and between visit variability) using a coefficient of variation (SD/meank) where k was determined from curve fitting10. Analysis also included use of Residual Standard Deviation (RSD) for effect size estimates which is the square root of the total squared deviation of data points from a linear regression of blood pressure values against time, divided by (n–2), where n is the number of readings10. All analyses were adjusted for age, gender, gender-age [gender*age where gender is coded as 1 (male) or 2 (female)], SBP mean, and the first 10 principal components (from decomposition of the genotype matrix).
For the stroke meta-analysis, the candidate SNPs were extracted from the genome wide data and site-specific logistic regression analysis was performed to test association of top SNP with overall ischemic stroke and its major sub-types (large artery disease, cardioembolic stroke and small vessel disease) under an additive genetic model. Age and gender were used as covariates. β coefficients, standard errors and p values from different studies were pooled via inverse variance meta-analysis using a fixed effects model. Meta-analysis was carried out for over-all ischemic stroke and its sub-types based on the TOAST criteria23 using METAL software25. Pooled ORs were calculated using estimated effect size of the SNP and standard error of the effect size estimate. 95% confidence intervals were calculated using odd ratios and standard error. A detailed description of the statistical analysis methods for each participating study is given in the Table IV of data supplement methods (Please see http://stroke.ahajournals.org).
Power for the stroke meta-analysis was calculated using the CATS genetic power calculator26. The following parameters were used to calculate the power for the replication of SNPs rs976683 in the ischemic stroke population using an additive genetic model: N (cases): 8624, N (controls): 12,722, stroke prevalence: 7.2% 27, rs976683 MAF: 0.25 and significance level: 0.05. The sample size provided sufficient power to detect modest effect sizes ranging from 1.1-1.4 for overall ischemic stroke but had reduced power for subtypes.
Results
ASCOT GWAS
The ASCOT GWA study population consisting of 3802 subjects was primarily male (82.3%) with a mean age of 63.7 (±8.1) years. Mean baseline SBP, mean baseline DBP and mean VIM were 161.6. (±17.6) mm Hg, 92.4 (±9.9) mm Hg and 0.004 (±0.001) mm Hg respectively. Details of ASCOT-UK-IR study population are tabulated in Table I of data supplement methods (Please see http://stroke.ahajournals.org).
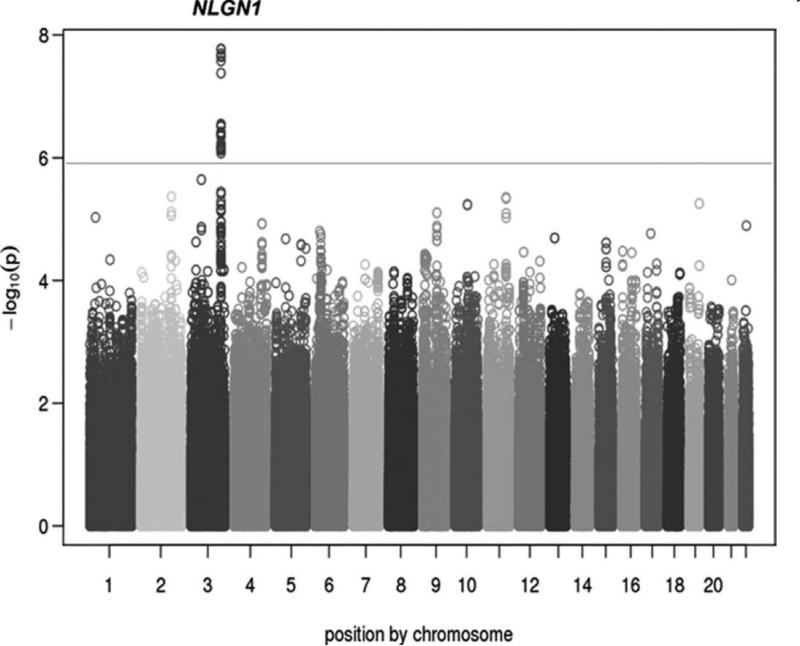
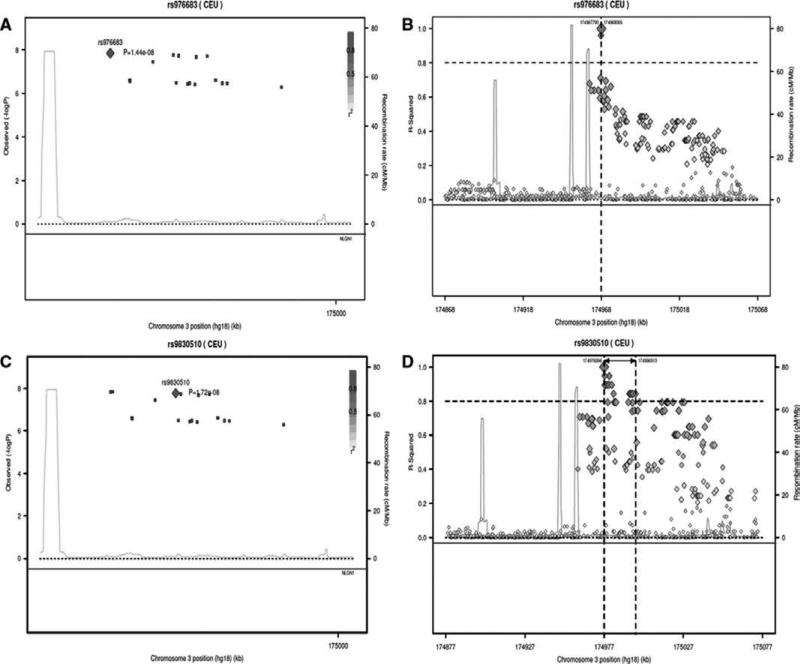
GWAS for BP variability identified a cluster of 17 correlated SNPs within the Neuroligin-1 (NLGN1) gene on 3q26.31) (ENCODE ID: ENSG00000169760.13) (Figure 1 and Table V of data supplement methods, http://stroke.ahajournals.org). Within the cluster, 12 SNPs were directly genotyped and 5 were imputed. Seven SNPs (3 imputed and 4 genotyped) reached genome wide significance (p ≤ 5 × 10−8) with the strongest association at the imputed SNP rs976683 (p=1.4 × 10−8) (Figure 2A and 2B). The effect size for SNP rs976683 association was small (β=0.000179) corresponding to a 0.01% mm Hg change in BP variability per copy of the risk allele. Conditional analysis using rs976683 provided no evidence of an independent signal at this locus (p=0.18).
Figure 1.
Genome-wide Manhattan plot for the ASCOT Anglo IR-UK GWAS showing a cluster of 17 SNPs in/near NLGN1 associated with BP variability (P < 5 × 10−7). Individual –log10 p values are plotted against their genomic position by chromosome. The dotted line at 10−6 marks the threshold for promising SNPs and the solid line at 10−8 marks the genome-wide significance threshold.
Figure 2.
Regional association and LD plots for the 17 correlated SNP's within the Neuroligin-1 (NLGN1) gene (3q26.31). The plots A & B are conditioned on the imputed sentinel SNP rs976683 and C & D are conditioned on the top genotyped SNP rs9830510. In plot 2A & C, each coloured square represents a SNP p value, with the colour scale correlating the r2 values for that SNP to the target SNP (red diamond) taken from the HapMap phase 2 CEU panel. In plot 2B & C, the target SNP (orange diamond) is represented in linkage disequilibrium with the cluster of 16 SNPs and other SNPs in the HapMap phase 2 CEU panel.
The top genotyped SNP to reach genome wide significance (p=1.72×10−8) was rs9830510 (Figure 2C and 2D). The direction of effect was in concordance with rs976683 however the SNPs were not highly correlated (r2=0.5, D=0.93).
Ischemic Stroke population demographics
8624 cases and 12722 controls of European descent from 7 studies spread across Europe, America and Australia: ASGC, BRAINS (European arm), GEOS, ISGS/SWISS, VISP, WTCCC2-UK and WTCCC2-Germany were available. The mean age of study participants ranged from 41.0 ± 7.0 years to 72.87 ± 13.16 years for stroke cases and 39.5 ± 6.7 to 66.28 ± 7.54 for controls. The male: female ratio was approximately 50:50. The three main ischemic stroke subtypes; cardioembolic, large vessel disease and small vessel disease accounted for 1523, 1639 and 1254 cases, respectively. The demographic data such as age, gender distribution and stroke sub-type frequencies for each population are summarized in Table II of data supplement methods (Please see http://stroke.ahajournals.org).
Association with overall ischemic stroke and sub-types
SNP rs976683 was directly genotyped in all 7 cohorts with an average MAF of 0.26 (Table VI of data supplement methods (Please see http://stroke.ahajournals.org) and was not significantly associated (at p≤0.05) with the increased risk of ischemic stroke or its subtypes. Pooled odds ratios were as follows; overall ischemic stroke (OR 1.02; 95% CI 0.97-1.07; p=0.52), cardioembolic (OR 1.07; 95% CI 0.97-1.16; p=0.17), large vessel disease (OR 0.98; 95% 0.89-1.07; p=0.60) and small vessel disease (OR 1.07; 95% CI 0.97-1.17; p=0.19). There was no significant heterogeneity between studies (Table VII of data supplement methods (Please see http://stroke.ahajournals.org).
Despite no evidence of an additional signal from the conditional analysis, the genotyped SNP rs9830510 was also tested for association in the ischemic stroke cohort to ensure that the association result of imputed SNP rs976683 was not an imputation artefact. rs9830510 was directly genotyped in all 7 cohorts with an average MAF of 0.15 (Table VI of data supplement methods (Please see http://stroke.ahajournals.org)). Association with increased risk of ischemic stroke or its subtypes was not significant (at p≤0.05) with pooled odds ratios as follows; overall ischemic stroke (OR 0.96; 95% CI 0.90-1.02; p=0.54), cardioembolic (OR 1.03; 95% CI 0.91-1.15; p=0.83), LVD (OR 0.76; 95% CI 0.66-0.80; p=0.03) and small vessel disease (OR 1.01; 95% CI 0.89-1.14; p=0.92). There was no significant heterogeneity between studies (Table VIII of data supplement methods (Please see http://stroke.ahajournals.org).
ASCOT BP variability follow up
Association testing of rs976683 with BP variability in the ASCOT Scandinavian arm provided no evidence of association (p = 0.18).
Discussion
We provide evidence supporting a role of genetic variants at the Neuroligin-1 (NLGN1) locus with BP variability but were unable to demonstrate association between this locus and ischemic stroke and BP variability in an independent Scandinavian sample. A GWAS for BP variability in the UK-IR discovery cohort identified a cluster of 17 correlated SNPs within the NLGN1 gene which encode a neuronal cell surface protein implicated in the growth and remodelling of the vascular system28. The strongest association reaching genome wide significance, was at imputed SNP rs976683 (p=1.4×10−8) and a correlated genotyped SNP rs9830510 (p=1.7×10−8), which represents a novel locus for BP variability in hypertensives and has not been detected in any of the previously published BP GWA studies. The effect size for the sentinel association was extremely small (β=0.000179) corresponding to a 0.01% unit change in BP variability per copy of the risk allele. Similar observations have been made in genome wide analysis of other measures of BP where effect sizes were also very small (1mmHg SBP and 0.5mmHg DBP) but could have the potential to significantly alter the outcomes at a population level. This evidence lead us to believe that the observed effect (albeit small) may be part of a battery of unrelated and common gene loci that exert independent but small effects which compound to cause the disease. However this hypothesis can only be confirmed via large prospective GWAS studies.
We attempted to replicate our findings with BP variability in two ways, firstly testing the top SNPs for association with ischemic stroke in an independent population comprising 8,624 cases and 12,722 controls from 7 cohorts. This is a common exploratory approach used to study candidate genes that maybe associated with different vascular disorders such as MI and stroke through their effect on shared risk factors such as hypertension, diabetes and smoking29. Our sample size provided sufficient power to detect modest effect sizes ranging from 1.1-1.4 for overall ischemic stroke but as with other studies, had reduced power for subtypes due to small sample size. SNPs rs976683 and rs9830510 were not significantly associated (p≤0.05) with the risk of overall stroke or its subtypes with the estimated pooled ORs ranging from 1.02-0.96 for overall ischemic stroke, 1.07-1.03 for cardio embolic, 0.98-0.76 for large vessel disease and 1.07-1.01 for small vessel disease.
The failure to detect an association with overall stroke could be due to several reasons. Genes affecting multi-factorial diseases such as stroke usually have small effect sizes and are difficult to identify in modestly sized study populations. Insufficient statistical power, given the small observed effect size for rs976683 on BP variability, is the most likely cause of an undetectable association with ischemic stroke. Another reason could be the clinical phenotypic heterogeneity introduced due to the diverse aetiology of ischemic stroke which makes it difficult to differentiate true signals from noise. Large studies such as the recent METASTROKE30 meta-analysis which included 15 stroke cohorts comprising of 12,000 cases and 60,000 controls also failed to identify any new genetic risk variants and only validated previous findings of variants within genes PITX2, ZFHX3, and HDAC9. Despite the large study population, the observed effect sizes were small (OR 1.39 - 0.96) suggesting that a combined burden of risk alleles carried by an individual is the likely cause, as shown in haemorrhagic stroke31. These studies have also highlighted the subtype-specific nature of the risk which lends support to the fact that true associations may be hidden under the multi-factorial pathogenesis of stroke. Our work also has a number of limitations which include possible inaccuracy of the TOAST classification into stroke sub-types. The case-control study design of our meta-analysis may be another limitation as some studies can induce survival bias by including recurrent stroke, thus allowing selection of milder forms of strokes.
The second attempt to replicate our findings included testing for association of rs976683 with BP variability in 3900 individuals from the Scandinavian arm consisting of the ASCOT study. Although not an ideal resource for follow up of our original observation, it was the only available replication population where individuals were selected using identical recruitment criteria as the ASCOT-UK-IR cohort and BP measurements were taken at the same time points allowing identical analysis of BP variability. However, replication analysis in this population provided no evidence of association between NLGN1 and BP variability (p = 0.18). Failure to replicate this association may in part be due to population stratification induced by Anglo-Scandinavian differences such as admixture of Finnish and central European ancestry 32 and recruitment of the ‘ASCOT-SE’ samples in Sweden. There are considerable genetic differences amongst Europeans and studies have demonstrated autosomal substructure in the Finnish and Swedish populations, warning researchers against making assumptions of genetic homogeneity in isolated European populations33-35. These studies have also shown that the British population is genetically less differentiated as compared to the Scandinavian populations33. Such findings have an impact on the choice of study participants for a GWAS because undetected population substructure is known to introduce bias in GWA studies36. Further, it is also possible that the genetic effect is confined to specific sub-populations of smokers, alcohol consumers and furosemide-exposed individuals within the ASCOT-UK-IR cohort. Identified SNPs from the ASCOT-UK-IR GWAS could also be artifactual.
The power to detect the effect size of a genetic risk variant is dependent on its minor allele frequency37. It is interesting that the minor allele frequency of SNPs rs976683 and rs9830510 in both study populations were similar (0.25 in ASCOT-UK-IR and 0.15 in ischemic stroke cohorts). However, even though the point estimates of the effect sizes observed for stroke were larger than BP variability, no comparative conclusions could be drawn from this as neither SNP was significantly associated with the increased risk of stroke. SNPs rs976683 and rs9830510 are intronic and an in silico regulatory SNP detection framework38 predicts that these SNPs alter transcription factor binding sites for over 150 cellular transcription factors. Due to the lack of association with any phenotypic trait at genome wide significance, information on eQTL, tissue specific expression and histone marks remains scarce through conventional data mining resources. NLGN1 gene may play a role in BP variability via processes involving the growth and remodelling of the vascular system28 . The NLGN1 protein in ubiquitously produced outside the central nervous system and expression of its α and β protein isoforms in the blood vessel walls and pancreatic beta-cells39 supports roles in atherosclerosis and insulin regulation respectively, cellular processes that may play a role in stroke. The wide spread impact of the misfiring NLGN1 gene is demonstrated in its association with type II granular corneal dystrophy 40 and autism 41 which suggests that its effects can be mediated through varied cellular processes. So far SNP rs976683 has only been suggestively implicated in Parkinson's disease42.
Our findings implicate SNPs at the NLGN1 locus are associated with BP variability but not ischemic stroke, although a suitable replication cohort could not be found to confirm our results. In order to understand the true relationship between visit-to-visit BP variability and risk of stroke, large prospective longitudinal studies following healthy cohorts for stroke occurrence are required. There is a need for international guidelines for clinical monitoring of BP variability that advocate diagnosis and assessment of treatment response in hypertension to be based upon the average of a series of blood pressure measures. Calibration of measuring devices is also needed to avoid phenotypic bias.
Supplementary Material
Acknowledgments
Sources of Funding
ASCOT was supported by Pfizer (NY, USA), Servier Research Group (Paris, France) and Leo Laboratories (Copenhagen, Denmark). ASGC was funded through grants from the Hunter Medical Research Institute and the National health, Medical Research Council, University of Newcastle and the Vincent Fairfax Family Foundation. BRAINS received support from Department of Health (United Kingdom), Henry Smith Charity, British Council, United Kingdom-India Education and Research Institute (UKIERI) and the Qatar National Research Foundation (www.BrainsGenetics.com). GEOS was supported by the following grants: NIH Genes, Environment and Health Initiative (GEI) Grant U01 HG004436, NIH grant HHSN268200782096C, GENEVA Coordinating Centre grant U01 HG 004446, Office of Research and Development, Medical Research Service, Baltimore Geriatrics Research, Education, and Clinical Centre of the Department of Veterans Affairs, National Institute of Neurological Disorders and Stroke (NINDS), NIH Office of Research on Women's Health (R01 NS45012, U01 NS069208-01) and Department of Veterans Affairs BLR&D Career Development Award (CDA-2). ISGS/SWISS was supported in part by the Intramural Research Program of the NIA (NIH project Z01 AG-000954-06 and NIH project Z01 AG-000015-50), NIH-NINDS Grant R01 NS-42733 (ISGS, J F Meschia) and NIH-NINDS Grant R01 NS-39987 (SWISS, J F Meschia). VISP was funded by the NINDS/NIH (R01 NS34447) and National Human Genome Research Institute (NHGRI) (Grant U01 HG005160). WTCCC-Ger and WTCCC-UK were funded by the Wellcome Trust as part of the WTCCC2 project (085475/B/08/Z, 085475/Z/08/Z and WT084724MA).
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ehret GB. Genome-wide association studies: Contribution of genomics to understanding blood pressure and essential hypertension. Current hypertension reports. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nature genetics. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nature genetics. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Consortium for Blood Pressure Genome-Wide Association S. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east asians. Nature genetics. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, et al. Blood pressure loci identified with a gene-centric array. American journal of human genetics. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothwell PM, Howard SC, Dolan E, O'Brien E, Dobson JE, Dahlof B, et al. Prognostic significance of visit-to-visit variability, maximum systolic blood pressure, and episodic hypertension. Lancet. 2010;375:895–905. doi: 10.1016/S0140-6736(10)60308-X. [DOI] [PubMed] [Google Scholar]
- 9.Rothwell PM. Limitations of the usual blood-pressure hypothesis and importance of variability, instability, and episodic hypertension. Lancet. 2010;375:938–948. doi: 10.1016/S0140-6736(10)60309-1. [DOI] [PubMed] [Google Scholar]
- 10.Rothwell PM, Howard SC, Dolan E, O'Brien E, Dobson JE, Dahlof B, et al. Effects of beta blockers and calcium-channel blockers on within-individual variability in blood pressure and risk of stroke. Lancet neurology. 2010;9:469–480. doi: 10.1016/S1474-4422(10)70066-1. [DOI] [PubMed] [Google Scholar]
- 11.Sever PS, Dahlof B, Poulter NR, Wedel H, Beevers G, Caulfield M, et al. Rationale, design, methods and baseline demography of participants of the anglo-scandinavian cardiac outcomes trial. Ascot investigators. Journal of hypertension. 2001;19:1139–1147. doi: 10.1097/00004872-200106000-00020. [DOI] [PubMed] [Google Scholar]
- 12.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nature genetics. 2012;44:1147–1151. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEvoy M, Smith W, D'Este C, Duke J, Peel R, Schofield P, et al. Cohort profile: The hunter community study. International journal of epidemiology. 2010;39:1452–1463. doi: 10.1093/ije/dyp343. [DOI] [PubMed] [Google Scholar]
- 15.Yadav S, Schanz R, Maheshwari A, Khan MS, Slark J, de Silva R, et al. Bio-repository of DNA in stroke (brains): A study protocol. BMC medical genetics. 2011;12:34. doi: 10.1186/1471-2350-12-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotlarciuc I, Khan MS, Maheshwari A, Yadav S, Khan FY, Al-Hail H, et al. Bio-repository of DNA in stroke: A study protocol of three ancestral populations. J R Soc Med Cardiovasc Dis. 2012:1. doi: 10.1258/cvd.2012.012019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacClellan LR, Mitchell BD, Cole JW, Wozniak MA, Stern BJ, Giles WH, et al. Familial aggregation of ischemic stroke in young women: The stroke prevention in young women study. Genet Epidemiol. 2006;30:602–608. doi: 10.1002/gepi.20171. [DOI] [PubMed] [Google Scholar]
- 18.Kittner SJ, Stern BJ, Wozniak M, Buchholz DW, Earley CJ, Feeser BR, et al. Cerebral infarction in young adults: The baltimore-washington cooperative young stroke study. Neurology. 1998;50:890–894. doi: 10.1212/wnl.50.4.890. [DOI] [PubMed] [Google Scholar]
- 19.Meschia JF, Brott TG, Brown RD, Jr., Crook RJ, Frankel M, Hardy J, et al. The ischemic stroke genetics study (isgs) protocol. BMC neurology. 2003;3:4. doi: 10.1186/1471-2377-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meschia JF, Kissela BM, Brott TG, Brown RD, Jr., Worrall BB, Beck J, et al. The siblings with ischemic stroke study (swiss): A progress report. Clinical medicine & research. 2006;4:12–21. doi: 10.3121/cmr.4.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Stroke Genetics C, Wellcome Trust Case Control C. Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, et al. Genome-wide association study identifies a variant in hdac9 associated with large vessel ischemic stroke. Nature genetics. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spence JD, Howard VJ, Chambless LE, Malinow MR, Pettigrew LC, Stampfer M, et al. Vitamin intervention for stroke prevention (visp) trial: Rationale and design. Neuroepidemiology. 2001;20:16–25. doi: 10.1159/000054753. [DOI] [PubMed] [Google Scholar]
- 23.Adams HP, Jr., Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. Toast. Trial of org 10172 in acute stroke treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 24.Deshmukh HA, Colhoun HM, Johnson T, McKeigue PM, Betteridge DJ, Durrington PN, et al. Genome-wide association study of genetic determinants of ldl-c response to atorvastatin therapy: Importance of lp(a). Journal of lipid research. 2012;53:1000–1011. doi: 10.1194/jlr.P021113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nature genetics. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 27.Lee S, Shafe AC, Cowie MR. Uk stroke incidence, mortality and cardiovascular risk management 1999-2008: Time-trend analysis from the general practice research database. BMJ open. 2011;1:e000269. doi: 10.1136/bmjopen-2011-000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottos A, Destro E, Rissone A, Graziano S, Cordara G, Assenzio B, et al. The synaptic proteins neurexins and neuroligins are widely expressed in the vascular system and contribute to its functions. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20782–20787. doi: 10.1073/pnas.0809510106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng YC, Anderson CD, Bione S, Keene K, Maguire JM, Nalls M, et al. Are myocardial infarction--associated single-nucleotide polymorphisms associated with ischemic stroke? Stroke. 2012;43:980–986. doi: 10.1161/STROKEAHA.111.632075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, et al. Genetic risk factors for ischaemic stroke and its subtypes (the metastroke collaboration): A meta-analysis of genome-wide association studies. Lancet neurology. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falcone GJ, Biffi A, Devan WJ, Jagiella JM, Schmidt H, Kissela B, et al. Burden of risk alleles for hypertension increases risk of intracerebral hemorrhage. Stroke. 2012;43:2877–2883. doi: 10.1161/STROKEAHA.112.659755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, et al. Correlation between genetic and geographic structure in europe. Current biology : CB. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 33.Salmela E, Lappalainen T, Fransson I, Andersen PM, Dahlman-Wright K, Fiebig A, et al. Genome-wide analysis of single nucleotide polymorphisms uncovers population structure in northern europe. PLoS One. 2008;3:e3519. doi: 10.1371/journal.pone.0003519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelis M, Esko T, Magi R, Zimprich F, Zimprich A, Toncheva D, et al. Genetic structure of europeans: A view from the north-east. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphreys K, Grankvist A, Leu M, Hall P, Liu J, Ripatti S, et al. The genetic structure of the swedish population. PLoS One. 2011;6:e22547. doi: 10.1371/journal.pone.0022547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nature genetics. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 37.Tabangin ME, Woo JG, Martin LJ. The effect of minor allele frequency on the likelihood of obtaining false positives. BMC proceedings. 2009;3(Suppl 7):S41. doi: 10.1186/1753-6561-3-S7-S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macintyre G, Bailey J, Haviv I, Kowalczyk A. Is-rsnp: A novel technique for in silico regulatory snp detection. Bioinformatics. 2010;26:i524–530. doi: 10.1093/bioinformatics/btq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suckow AT, Comoletti D, Waldrop MA, Mosedale M, Egodage S, Taylor P, et al. Expression of neurexin, neuroligin, and their cytoplasmic binding partners in the pancreatic beta-cells and the involvement of neuroligin in insulin secretion. Endocrinology. 2008;149:6006–6017. doi: 10.1210/en.2008-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eun-Ju Lee KJK, Kim Han-Na, Bok Jeong, Jung Sung-Chul, Kweon Kim Eung, Lee Jong-Young, et al. Genome-wide scan of granular corneal dystrophy, type ii: Confirmation of chromosome 5q31 and identification of new co-segregated loci on chromosome 3q26.3. Experimental and Molecular Medicine. 2011 Jul;43:393–400. doi: 10.3858/emm.2011.43.7.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, et al. Genome-wide association study confirms snps in snca and the mapt region as common risk factors for parkinson disease. Annals of human genetics. 2010;74:97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.