Abstract
BEL1-type proteins are ubiquitous plant transcription factors in the three-amino-acid-loop-extension superfamily. They interact with KNOTTED1-like proteins, and function as heterodimers in both floral and vegetative development. Using the yeast two-hybrid system with POTATO HOMEOBOX1 (POTH1) as the bait, seven BEL1-type proteins were originally identified. One of these genes, designated StBEL5, has transcripts that move long distances in the plant and enhance tuberization and root growth. Using the potato genome database, 13 active BEL1-like genes were identified that contain the conserved homeobox domain and the BELL domain, both of which are essential for the function of BEL1-type proteins. Phylogenetic analysis of the StBEL family demonstrated a degree of orthology with the 13 BEL1-like genes of Arabidopsis. A profile of the gene structure of the family revealed conservation of the length and splicing patterns of internal exons that encode key functional domains. Yeast two-hybrid experiments with KNOTTED1-like proteins and the new StBELs confirmed the interactive network between these two families. Analyses of RNA abundance patterns clearly showed that three StBEL genes, BEL5, -11, and -29, make up approximately two-thirds of the total transcript values for the entire family. Among the 10 organs evaluated here, these three genes exhibited the 12 greatest transcript abundance values. Using a phloem-transport induction system and gel-shift assays, transcriptional cross-regulation within the StBEL family was confirmed. Making use of the potato genome and current experimental data, a comprehensive profile of the StBEL family is presented in this study.
Key words: BELL1, KNOTTED1, mobile RNA, Solanum tuberosum, TALE, tuberization.
Introduction
The BEL1-like family (BELL) of transcription factors is ubiquitous among plant species and they interact with KNOTTED1-like proteins to regulate a range of developmental processes (Müller et al., 2001; Chen et al., 2003; Smith and Hake 2003; Kanrar et al., 2008; Lal et al., 2011; Li et al., 2012). These BEL1-like homeodomain (BLH) proteins have significant roles in meristem and floral development, and their functions are often overlapping and redundant. ARABIDOPSIS THALIANA HOMEOBOX 1 (ATH1), PENNYWISE (PNY), and POUNDFOOLISH (PNF) are BLH proteins of Arabidopsis that are critical for the initiation, maintenance, and development of the shoot apical meristem (Rutjens et al., 2009; Ung et al., 2011) and inflorescence architecture (Smith and Hake, 2003; Ragni et al., 2008; Khan et al., 2012). SAW1 (BLH2) and SAW2 (BLH4) are negative regulators of BREVIPEDICELLUS (BP), an important class I KNOX protein and positive regulator for growth. In the saw1 saw2 double mutant, BP can be expressed on the margin of the leaf, and the leaf will develop with a serrated and revolute shape (Kumar et al., 2007). Misexpression of BLH1 in the embryo sac will switch one of the synergid cells into an egg cell (Pagnussat et al., 2007), and loss of function of the AtBEL1 gene blocks the development of integuments (Brambilla et al., 2007). In Arabidopsis, there are 13 BEL1-like family members, all of which can form heterodimers with KNOX proteins (Kumar et al., 2007).
BEL1- and KNOTTED1-type proteins interact in a tandem complex to regulate transcription of target genes (Bellaoui et al., 2001; Chen et al., 2004; Hake et al., 2004; Lin et al., 2013). BEL1 also interacts with MADS-box transcription factors and SPOROCYTELESS to support ovule development in Arabidopsis (Brambilla et al., 2007; Bencivenga et al., 2012). POTH1 (potato homeobox 1) is a member of the class I KNOTTED-like homeobox proteins of potato. Using POTH1 as bait in the yeast two-hybrid system, seven BEL1-like proteins, designated StBEL5, -11, -13, -14, -22, -29, and -30, were isolated from stolon and leaf libraries of potato (Chen et al., 2003). The heterodimer of StBEL5 and POTH1 exhibits a strong binding affinity to the promoter of GA20ox1 and negatively regulates its expression (Chen et al., 2004). DNase footprinting experiments identified the binding site of the POTH1–StBEL5 dimer in the GA20ox1 promoter as a TTGAC double tandem motif. The TTGAC motif can be recognized by either POTH1 or StBEL5, but only when both TTGAC motifs are intact can the POTH1–StBEL5 heterodimer function (Chen et al., 2004).
Several studies have demonstrated the role of StBEL5 and POTH1 in tuber development (Chen et al., 2003; Rosin et al., 2003; Banerjee et al., 2009). Overexpression of each of these genes in transgenic potato lines produced plants that exhibited enhanced tuber yields. Heterografting experiments showed that the mRNA of StBEL5 is mobile in both a downward and upward direction (Hannapel, 2013). Movement from leaves to stolon tips was enhanced under short-day (SD) conditions and mediated by the untranslated regions (Banerjee et al., 2006, 2009). The mobility of StBEL5 mRNA was dramatically reduced without the untranslated regions (UTRs), whereas a non-mobile mRNA exhibited increased mobility upon fusion with the StBEL5 UTRs (Banerjee et al., 2009). Besides enhancing movement of the mRNA, the UTRs also suppressed translation of a β-glucuronidase (GUS) marker in a transient expression system (Banerjee et al., 2009). Recent mobility studies have also demonstrated movement of StBEL5 into roots that impacts growth (Lin et al., 2013).
From the recently published potato genome (Xu et al., 2011), 14 BEL1-like loci have been identified including the seven original StBEL proteins isolated from the yeast two-hybrid screen (Chen et al., 2003). Except for StBEL5, however, very little information is available on the other StBEL1-like family members. Transcripts of all seven of the original BEL1-like proteins were detected in RNA from phloem-enriched exudate or laser-captured microdissected phloem cells (Yu et al., 2007; Campbell et al., 2008). Making use of the reference potato genome (Xu et al., 2011) and current experimental data, an extended analysis of the BEL1-like family of potato is presented in this study. Because of their functional relationship with KN1-like proteins and the potential for long-distance trafficking of their mRNAs, the BEL1 genes of potato represent a valuable model for assessing the dynamic role these transcription factors play in plant development.
Materials and methods
Phylogenetic and gene structure analysis
A phylogenetic tree was constructed using the neighbour-joining method (Saitou and Nei, 1987) available in the MEGA 4.0.2 software package (Tamura et al., 2007). Full-length proteins of the StBEL family were aligned using the ClustalW algorithm (Thompson et al., 1994) included in the BioEdit software package (Hall, 1999). Gene expression data of StBELs for both the RH and DM genotypes was downloaded from Potato Genome Sequencing Consortium website (http://potatogenome.net). The StBEL genes were drawn to scale and assigned to potato chromosomes based on their positions shown in the PTGS (Release: Annotation v3.4, Assembly v3, Pseudomolecules v2.1.11).
Real-time quantitative reverse transcription-PCR (qRT-PCR) for StBEL expression analysis
Solanum tuberosum ssp. andigena plants were soil grown for 4 weeks under long-day (LD) conditions and then transferred to either SD or maintained under LD conditions for a further 10 d. Leaf and stolon tip samples were harvested, frozen in liquid nitrogen and stored at –80 °C. RNA preparation and qRT-PCR were performed as described previously (Lin et al., 2013). The relative gene quantification (comparative threshold cycle) method (Livak and Schmittgen, 2001) was used to calculate the expression levels of the StBEL RNAs. StACT8 (accession number GQ339765) was used as an internal control. Products ranged from 98 to 160bp and were mostly designed spanning the introns in order to detect any genomic DNA contamination (Supplementary Table S1 available at JXB online). The specificity of primers was determined by melting curve analyses and agarose gel (3%) electrophoresis performed following the qRT-PCR experiments. A standard curve was generated based on six-point (10-fold) serial dilutions of cDNA to calculate the gene-specific PCR efficiency. PCR efficiencies of primers ranged from 97 to 110 %.
Yeast two-hybrid system
The Matchmaker two-hybrid system (Clontech) was used for the yeast (Saccharomyces cerevisiae) two-hybrid screen with yeast strain pJ69-2A. The StBEL constructs were amplified by PCR and cloned into the vector pACT-AD (Supplementary Table S1 available at JXB online), in frame with the GAL4 activation domain. The tobacco Knox cDNA constructs were amplified by PCR and cloned into pBridge (Clontech) in frame with the GAL4-binding domain. Sequencing of selected cDNAs and constructs was performed at the Iowa State University DNA Facility, Ames, IA, USA. Positive interactions were confirmed by co-transforming into pJ69-2A with each purified pAD and pBridge plasmid and plating on –Leu/–Trp (transformation control) and –Leu/–Trp/–His/–Ade (selection) nutrient medium. Knox/StBEL interactions were quantified for lacZ induction using a β-galactosidase assay (Pierce Chemical). The Knox cDNA clones from tobacco (NTH1, -15, -20, and -22) were graciously provided by M. Matsuoka (Nishimura et al., 2000).
RT-PCR for StBEL RNAs in GAS:BEL5 plants
Production and characterization of the GAS:BEL5 transgenic line and verification of StBEL5 RNA movement has been described previously by Banerjee et al. (2006, 2009). Transgenic or wild-type (WT) S. tuberosum ssp. andigena plants were grown under LD conditions for 3 weeks and then transferred to SD conditions for 10 d. RNA was extracted from leaves and roots and one-step RT-PCR was performed using 200–250ng of total RNA, a non-plant-sequence primer fused to the transgenic RNA, and a gene-specific primer for the StBEL5 transcripts and a pair of gene-specific primers for StBEL6, -34, -22, and -14 (Supplementary Table S1 available at JXB online). All PCRs were standardized and optimized to yield a product in the linear range. Homogenous PCR products were quantified using ImageJ software (Abramoff et al., 2004) and normalized using 18S rRNA values.
Gel-shift assays
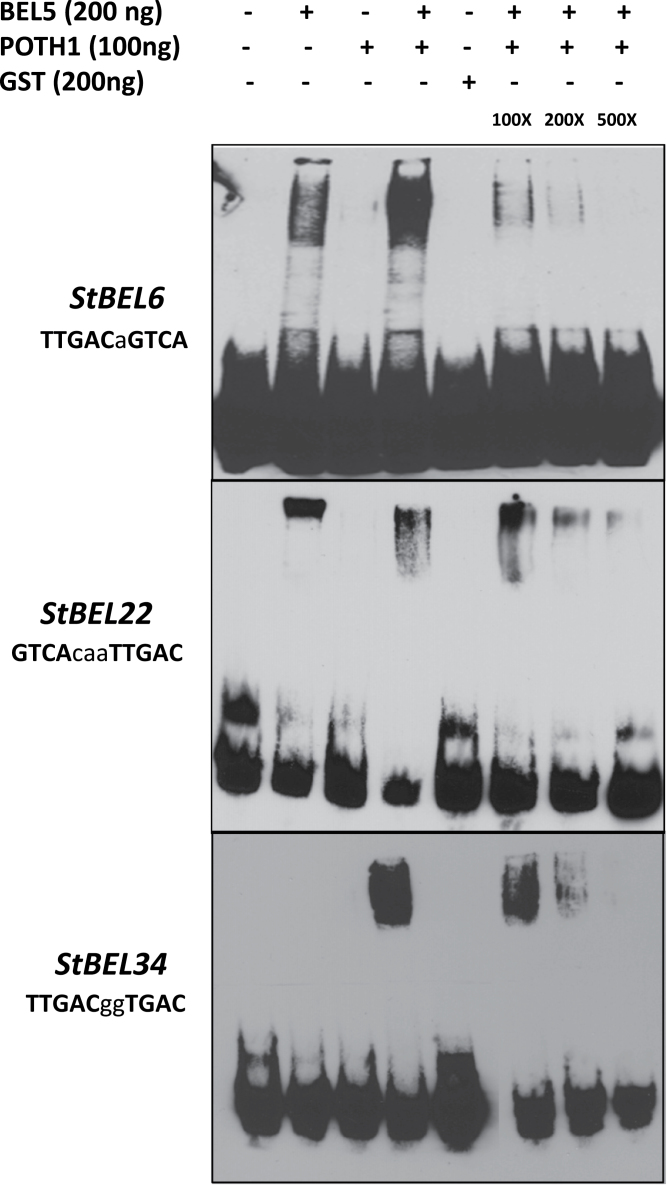
Oligonucleotides with 3′ biotin labelling were synthesized at the DNA Facility, Iowa State University, Ames, IA, USA. dsDNA was prepared by hybridization of complementary synthetic oligonucleotides (Supplementary Table S1 available at JXB online). Gel-shift assays were performed using a LightShift Chemiluminescent EMSA kit from Thermo Scientific according to the manufacturer’s protocol with the following modifications. Twenty microliters of binding reactions were set up on ice containing 20mM HEPES (pH 7.5), 10% glycerol (v/v), 0.5% Triton X-100 (v/v), 0.5mM EDTA (pH 8.0), 50mM KCl, 2mM MgCl2, 20ng μl–1 of BSA, 1mM dithiothreitol, and 50ng μl–1 of poly(dI-dC) as a non-specific competitor. Ten femtomoles of labelled DNA was used for all assays. Two hundred nanograms of StBEL5–GST, 100ng of POTH1–GST or 200ng of glutathione S-transferase (GST) proteins were used (see Fig. 9). The binding mix was incubated on ice for 60min before electrophoresis. For the competition assays, unlabelled dsDNA fragments (100×, 200×, and 500×) were incubated with the recombinant protein on ice for 30min before addition of the labelled probe. Both the unlabelled and labelled DNA fragments used here were the same sequence.
Fig. 9.
Gel-shift assays of various tandem TGAC core motifs (bold, upper-case nucleotides) in three putative target genes of StBEL5 and POTH1 with a range of linker sequence (lower-case nucleotides) between motifs. Upstream sequences of StBEL6 contain the tandem motifs in a tail-to-tail orientation on opposite DNA strands and StBEL22 motifs exhibit a head-to-head orientation, whereas StBEL34 contains the tandem motifs in a tail-to-head orientation on the same (+) DNA strand. The StBEL5 and POTH1 proteins were expressed and purified with a C-terminal GST fusion tag. Each DNA bait was tested for binding with StBEL5–GST, POTH1–GST, or GST alone or with StBEL5–GST and POTH1–GST together. Ten femtomoles of synthesized DNA probes of 30 (StBEL6) or 50 nt labelled with biotin were used in the binding reaction. The amounts of StBEL5 and POTH1 proteins used in these assays were adjusted to achieve equivalent molarity. Unlabelled DNA bait at 100×, 200×, and 500× concentrations relative to the labelled probe was used in the competition assays.
Results
Phylogeny of the StBEL family
Originally, seven BEL1-like proteins were identified using the yeast two-hybrid system (Chen et al., 2003). Additional sequences for BEL1-like genes in potato were retrieved by querying 269 nt sequence runs, covering the conserved BELL domain and the homeodomain of StBEL5, against the PGSC_DM_v3.4_gene.fasta file from the Potato Genome Sequencing Consortium website (http://potatogenomics.plantbiology.msu.edu). Based on genomic and expressed RNA sequence data, six new active StBEL genes, StBEL6, -31, -32, -33, -34, and -35, and one pseudogene were identified (Table 1). The open reading frames for these new BELs were predicted with FGENESH (http://linux1.softberry.com/berry.phtml) using the most closely related gene orthologue in tomato as a reference. Based on these amino acid sequences, a phylogenetic tree was constructed for the StBEL family (Fig. 1). BEL1-like proteins are characterized by four conserved regions: the SKY-box located in the N-terminal region, the BELL domain, the homeodomain, and the VSLTLGL motif in the C terminus (Supplementary Fig. S1 available at JXB online). The TALE (three-amino acid loop extension) is the proline-tyrosine-proline (PYP) link located between helices I and II.
Table 1.
Sequence structure of 14 genes in the BEL1 family of potatoAn asterisk indicates the presence of a tandem TGAC-core motif in the promoter with no more than a 3 nt linker between the TGAC cores. CDS, coding sequence.
| PGSC locus no. | gene | 5′ UTR (nt) | Intron in 5′ UTR (nt) | CDS (aa) | 3′ UTR (nt) |
|---|---|---|---|---|---|
| PGSC0003DMG400005930 | BEL5* | 149 | 203 | 2067 (688) | 503 |
| PGSC0003DMG400021323 | BEL29 | 259 | 1893 | 2130 (710) | 491 |
| PGSC0003DMG400019635 | BEL11 | 268 | 177 | 2130 (710) | 317 |
| PGSC0003DMG400010086 | BEL13 | 388 | 470 | 2217 (738) | 111 |
| PGSC0003DMG400012329 | BEL14 | 307 | 832 | 1902 (633) | 76 |
| PGSC0003DMG400022011 | BEL22* | 362 | 195 | 2088 (695) | 74 |
| PGSC0003DMG400030961 | BEL30 | 466 | 966 | 1938 (645) | 57 |
| PGSC0003DMG400003751 | BEL32 | 777 | 467 | 1986 (661) | 414 |
| PGSC0003DMG400024267 | BEL33 | 234 | None | 1518 (505) | 209 |
| PGSC0003DMG400008057 | BEL34* | 54 | None | 2079 (692) | 233 |
| PGSC0003DMG400019142 | BEL35 | 69 | None | 1728 (575) | 109 |
| PGSC0003DMG400029946 | BEL6* | 359 | 1090 | 1725 (574) | 155 |
| PGSC0003DMG400003750 | BEL31 | 130 | None | 1272 (423) | 175 |
| PGSC0003DMS000003755 | BEL15 | pseudogene |
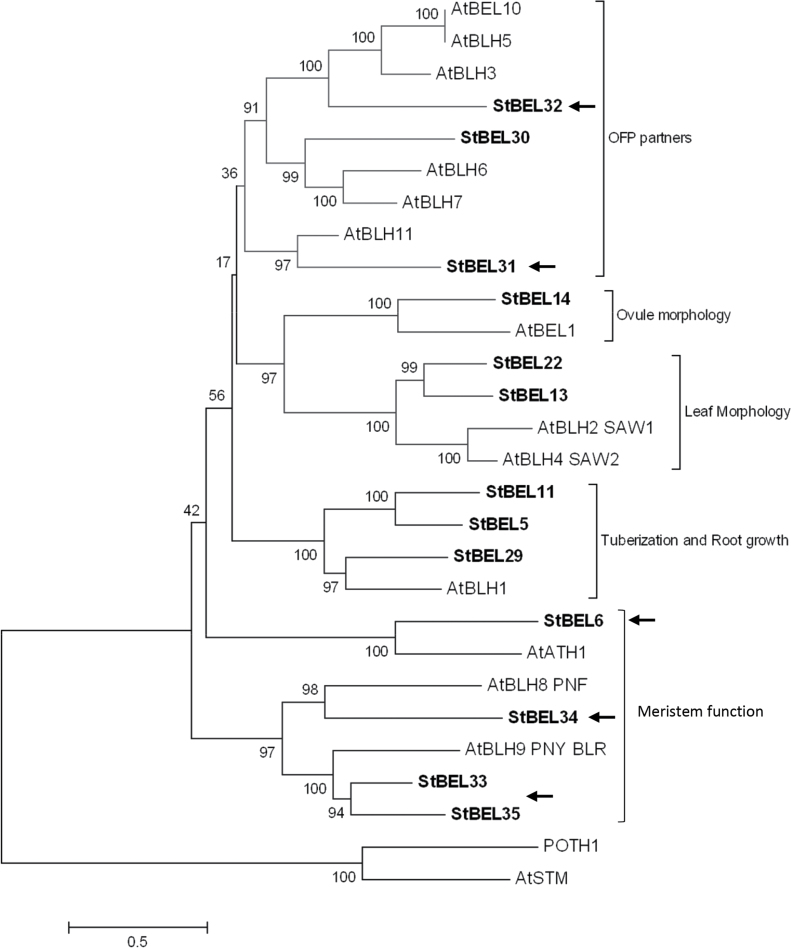
Fig. 1.
Phylogenetic relationship of the BEL1-like proteins of Arabidopsis and potato. The amino acid sequences of the 13 known potato BEL1-like proteins were analysed and compared with BEL1 proteins of Arabidopsis. These data were organized into a phylogenetic tree with the MEGA4.0.2 package and the neighbour-joining program. The numbers listed at the branching points are boot-strapping values that indicate the level of significance (percentage) for the separation of two branches. The length of the branch line indicates the extent of difference according to the scale at the lower left-hand side. StBELs are represented in bold letters. Putative functions are listed for each group. Arrows designate the six new StBEL proteins.
Phylogenetic analysis was structured on alignment of the StBEL1-like proteins with the 13 known members of the Arabidopsis BEL1 family (Rutjens et al., 2009; Fig. 1). Overall the StBEL proteins clustered into five main clades that further branched into subclades. The six new BELs delineated into five independent branches of the phylogenetic tree with the closely related BEL33 and -35 clustered on the same branch (Fig. 1, arrows). In general, the BELs of potato matched very closely with their Arabidopsis orthologues.
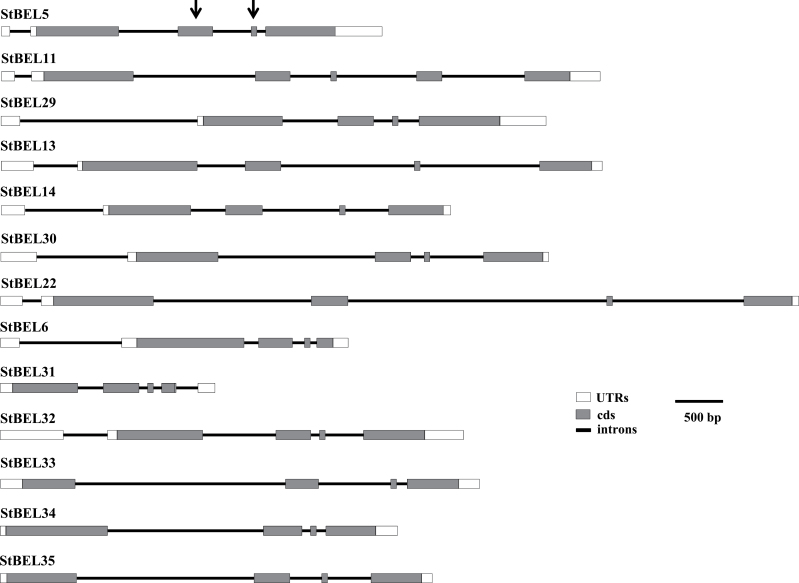
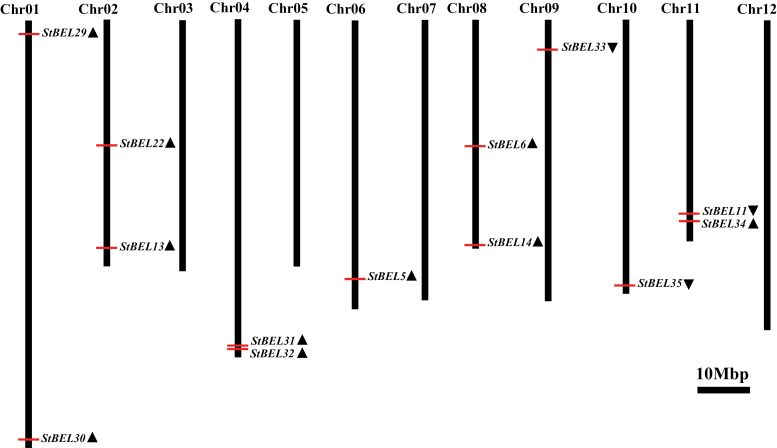
The amino acid sequences of the StBEL proteins range from 423 for BEL31 to 738 for BEL13 (Table 1) and displayed a range of divergence outside the four conserved regions (Fig. 2). Although conserved sequence motifs like LSLSL and DFV were evident towards the N and C termini, respectively, their functional significance is unknown (Fig. 2). BEL6 and -31 were relatively short BEL1-like proteins at 574 and 423 aa, respectively, and did not contain either the VSLTLGL or the DFV C-terminal motifs (Fig. 2). One other StBEL gene (BEL15, locus no. PGSC0003DMS000003755) was phylogenetically related to StBEL14 and -22, but its ORF encoded a truncated protein structure and no expressed sequence tags were identified for it, suggesting it is inactive. The overall gene structure of this family was highly conserved. Twelve of the 13 active StBEL genes contained three or four introns. StBEL11 contained five (Fig. 3). Twelve of the 13 also had four exons. Again, BEL11 was the exception with five. Scoring UTRs, exons, and introns, 10 of the 13 genes ranged in length from approximately 4.0kb (BEL5 and -34) to 6.3kb (BEL11 and -13). BEL31 and -6 were 2.3 and 3.65kb, respectively. BEL22 was approximately 8.4kb. The length of the second and third coding sequence exons were conserved throughout the family (Fig. 3, arrows). Splicing appeared to be consistent at these four internal exonic junctions to produce sequences ranging from 353 to 411 nt for exon 2 and 61 nt for exon 3. For all potato BELs, exon 2 encoded an amino acid sequence that spanned a portion of both the BELL domain and the homeodomain. The third exon encoded a sequence in the homeodomain. The 3′ end of exon 2 contained 2 nt (CC) of the codon that encodes the first proline in the PYP TALE, whereas the 5′ end of exon 3 contained the other nucleotide (any of the four bases) of this proline codon. BEL34 was the lone exception with the complete proline codon present at the end of exon 2 and only 60 nt in exon 3. This exonic splicing pattern in the middle of the PYP TALE is conserved in tomato, rice, and Arabidopsis BEL1 genes. The 13 active BEL genes were distributed over eight of the 12 potato chromosomes (Fig. 4). No more than two genes were located on any one chromosome. The close proximity of BEL31 and -32, only 2021bp apart on chromosome 4 (Supplementary Table S2 available at JXB online), suggested a recent tandem duplication event.
Fig. 2.
Amino acid sequence alignment of the six new StBELs with StBEL5. Black- and grey-boxed letters represent identical or similar residues, respectively. The conserved BELL domain (starting Leu272), homeodomain (starting Trp393) and the N-terminal SKY and C-terminal VSLTLGL boxes have been underlined. The amino acids for conserved domains are aligned in relation to the BEL5 protein.
Fig. 3.
The structure of StBEL genes drawn to scale, according to the alignment of cDNA sequences against the corresponding genomic sequences. The cDNA sequence for the new StBELs was obtained by RT-PCR with gene-specific primers in combination with both 5′ and 3′ RACE and by utilizing genome sequence as needed. All genomic sequence for the StBEL family was obtained from sequence data from the Potato Genome Sequencing Consortium website (http://solanaceae.plantbiology.msu.edu). The conserved internal second and third exons representing coding sequence are indicated by arrows on the BEL5 gene structure.
Fig. 4.
Genomic distribution of the StBEL genes on potato chromosomes. Chromosome numbers are shown at the top of each bar. The triangles following the gene names indicate the direction of transcription. The position (bp) of each StBEL gene on PGSC potato chromosome pseudomolecules (Release: Annotation v3.4, Assembly v3, Pseudomolecules v2.1.11) is specified in Supplementary Table S1 available at JXB online.
Because of their importance in regulating RNA mobility (Banerjee et al., 2006, 2009), UTRs were scored for length by using the longest sequence obtained either by rapid amplification of cDNA ends (RACE) or from available web-based RNA sequence data. BEL5, -11, -29 and -32 had 3′ UTRs ranging from 317 to 503 nt, the four longest in the family (Table 1). Common sequence motifs were observed in the 3′ UTR sequences of BEL5, -11, and -29 (Supplementary Fig. S2 available at JXB online). BEL6, -13, -22, -30 and -32 contained the longest 5′ UTR sequences, ranging from 359 to 777 nt. Several polypyrimidine clusters of at least 3 nt were identified in the 3′ UTRs of BEL5, -11, and -29 (Supplementary Fig. S2 available at JXB online) and in the 5′ UTRs of BEL13, -14, and -30. These motifs are recognized by the polypyrimidine tract-binding proteins, an important class of RNA-binding proteins (Auweter and Allain, 2008; Ham et al., 2009; Mahajan et al., 2012). Intronic sequences ranging from 177 to 1893 nt interrupted the 5′ UTRs of all the BELs except BEL31, -33, -34, and -35 (Table 1).
Expression patterns of potato BEL1-like genes
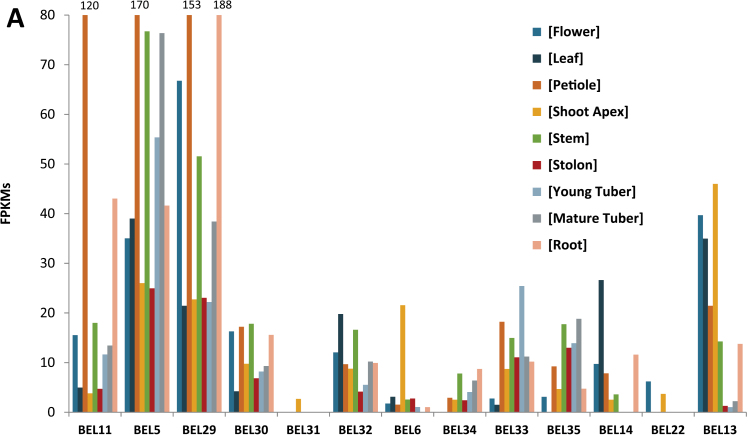
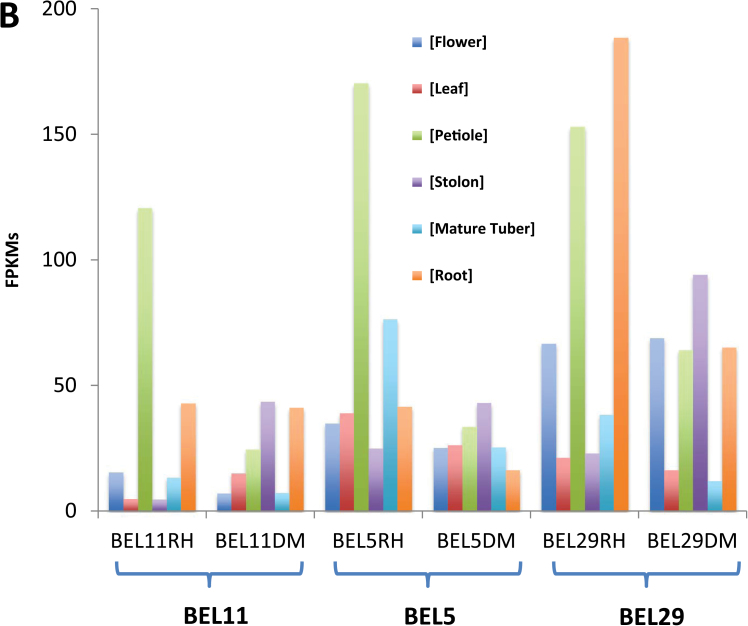
Using RNA-Seq data of S. tuberosum group Tuberosum RH89-039-16 from the recently published potato genome (Xu et al., 2011) and RT-PCR, widespread ubiquitous accumulation of most StBEL transcripts was generally observed (Fig. 5A and Supplementary Fig. S3 available at JXB online). With the RNA-Seq data, however, detectable fragment counts were observed in only two organs for StBEL31 (sprouts and shoot apices) and -22 (flowers and shoot apices). The most striking feature of this overall RNA expression profile, however, was that three of the StBEL genes (StBEL5, StBEL11, and StBEL29) exhibited a very high proportion, greater than two-thirds, of the overall RNA accumulation values for the entire StBEL family (Fig. 5A). BEL5 was the most abundant RNA in six of the 10 organs and placed in the top three ranking for all organs (Supplementary Table S3 available at JXB online). BEL29 was most abundant in three others. The one exception was in the shoot apex where StBEL13 was most abundant. Overall, the 12 highest fragments per kb per million mapped reads (FPKMs) values compiled for all organs were for BEL5, -11, and -29,and these three accounted for 22 out of 30 of the top three abundance values in each organ category (Supplementary Table S3 available at JXB online). All three registered relatively high transcript values for petioles (170, 121, and 153, respectively) and stems (77, 17, and 52, respectively), both prominent organs involved in the transport of mobile signals (Banerjee et al., 2006; Ham et al., 2009). As an example, StBEL5 mRNA is transcribed in leaf veins and petioles and moves into sieve elements of the phloem of both and is transported via the stem to stolon tips and roots to regulate growth (Banerjee et al., 2006; Lin et al., 2013). No transcription was observed for StBEL5 in stems despite the high accumulation of its RNA detected in this organ (Banerjee et al., 2006). The abundant and ubiquitous nature of StBEL5, -11, and -29 RNAs, particularly in the petiole, and their phylogenetic similarity, suggest that they may act in a network of mobile RNA signals that regulates development throughout the plant. In roots, values were greatest for StBEL5, -11, and -29 at 42, 43, and 188 FPKMs, respectively (Supplementary Table S3 available at JXB online). In tuber sprouts, their values topped out at 60, 46, and 62, respectively. The value of 188 for BEL29 in roots was the greatest observed among all potato BELs in any organ. In the less robust (smaller tubers, smaller plants) S. tuberosum group Phureja DM1-3 516 R44 haplotype (Xu et al., 2011), total transcript values for BEL5, -11, and -29 were much less than in the RH haplotype, making up only 55% of total transcript values compared with 71% of the RH total abundance values (Fig. 5B). Abundance values of the other 10 StBEL RNAs were essentially the same in the two genotypes (Supplementary Fig. S4 available at JXB online).
Fig. 5.
(A) Expression profile of StBEL family members mined using the RNA-seq data from the publically available Potato Genome Database from the Tuberosum RH89-039-16 haplotype (Xu et al., 2011). Nine organs are presented and abundance values are shown in FPKMs (fragments per kb per million mapped reads). (B) A comparison of expression profiles of StBEL11, -5, and -29 mined using the RNA-seq data from the publically available Potato Genome Database from both the RH and the DM1-3516-R44 haplotypes (Xu et al., 2011). Six organs available from the DM database are presented for comparison and abundance values are shown in FPKMs.
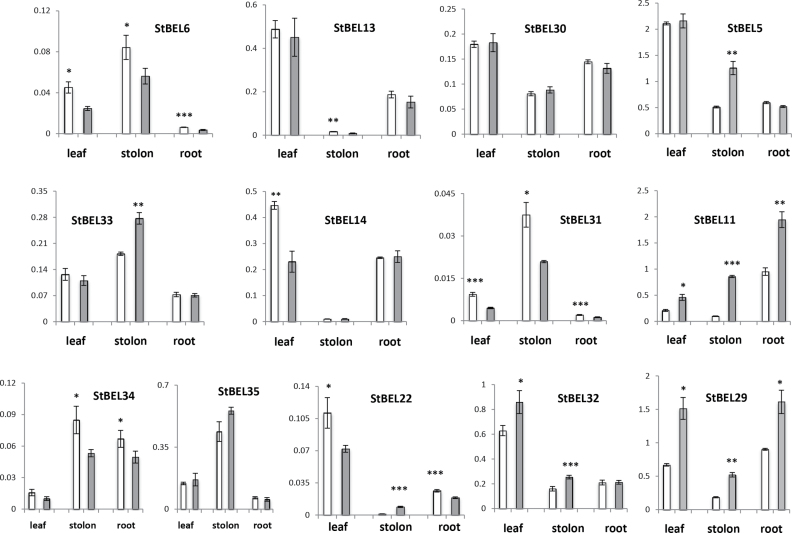
Previous work on the accumulation of StBEL5 RNA showed that its abundance and mobility was regulated by photoperiod but that its transcription was induced by SDs only in stolons (Chen et al., 2003; Banerjee et al., 2006; Chatterjee et al., 2007). Real-time qRT-PCR was performed on all members of the StBEL family to determine their RNA accumulation patterns in leaves, stolons, and roots in response to day length in the photoperiod-responsive S. tuberosum ssp. andigena (Fig. 6). StBEL5, -11, -22, -29, -32, and -33 displayed upregulation in one or more organs from SD plants, whereas StBEL6 and -31 exhibited increased levels in all three organs under LDs. StBEL34 RNA levels increased in both stolons and roots under LDs. No photoperiod effect was observed for either StBEL30 or -35. StBEL5, -11, and -29 exhibited the strongest induction in RNA accumulation in stolons in response to SDs. StBEL6, -31, -33, -34, and -35 exhibited proportionately more RNA in stolons than leaves, whereas only a trace of RNA for StBEL13, -14, and -22 was detected in stolons (Fig. 6). The relative abundance levels of these RNAs were generally consistent with the RNA-Seq data (Fig. 5A) with StBEL6, -22, -31, and -34 being the least abundant and StBEL5, -11, and -29 being the most abundant among the StBEL genes. Clearly, a diverse range of transcript concentrations was evident in these StBEL family members. For example, the relative abundance difference between transcript levels in SD leaves for StBEL5 and -31 was approximately 480-fold.
Fig. 6.
Effect of photoperiod on StBEL RNA accumulation in the photoperiod-responsive potato species, S. tuberosum ssp. andigena in leaves, stolon tips, and roots grown for 4 weeks. Relative levels of StBEL transcripts are presented on the y-axes and were quantified using total RNA extracted from new leaves (leaf), 0.5cm samples from the tip of stolons (stolon), or secondary roots (root) from plants grown under long (open bars) or short (grey shaded bars) days. SD plants were harvested after 10 d of SD conditions (8h light, 16h dark). Real-time qRT-PCR with gene-specific primers was used to calculate the relative amounts of RNA for each StBEL gene. The expression of each BEL gene was calculated as the 2−ΔCt value and normalized to the endogenous reference gene, StAct8. The StBELs are organized phylogenetically by columns into four groups (see Fig. 1). Standard errors of the means of three biological replicates are shown with asterisks indicating significant differences (*P<0.05; **P<0.01; ***P<0.001, respectively) using Student’s t-test.
Interaction with KNOX partners
The BEL1-like homeodomain proteins interact physically with their KNOX homeodomain protein partners to regulate gene expression by controlling the transcription of target genes (Bellaoui et al., 2001; Müller et al., 2001; Smith et al., 2002; Chen et al., 2003, 2004). The seven originally identified members of the StBEL family displayed selective interaction with the Knotted1-like protein, POTH1 (Chen et al., 2003), and four other tobacco class I-type KNOX (NTH1, -15, -20 and -22) proteins (Supplementary Table S4 available at JXB online). To test for the interaction of the new BEL1-like gene family members with Knotted1-type proteins, all of the six new BELs and three other previously identified StBELs (StBEL5, -13, and -30) representative of a wide phylogenetic range across the StBEL family, were evaluated for protein interaction in the yeast two-hybrid system.
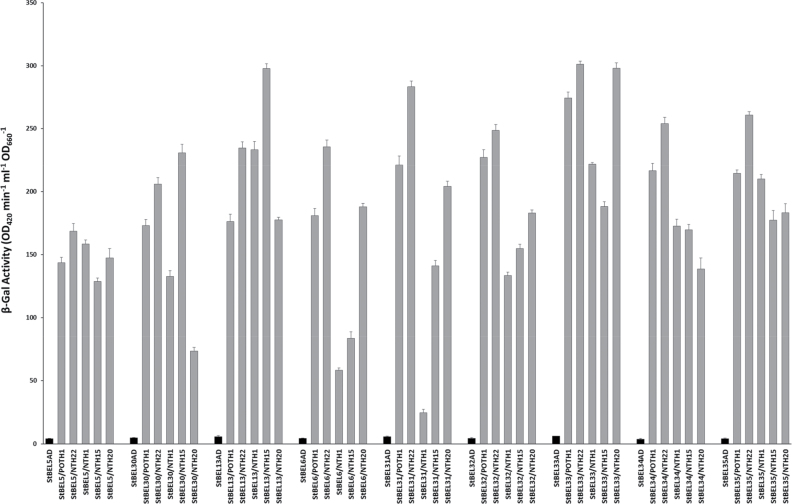
Interaction was tested with POTH1 and the four tobacco KNOX types and quantified using β-galactosidase activity (Fig. 7). NTH22 is the tobacco orthologue of POTH1. All of the potato BEL1-like proteins displayed an interaction with all five KNOX proteins but their binding affinities varied considerably. The NTH22 interaction with StBEL33, for example, based on β-galactosidase activity was the strongest among all the StBEL proteins tested (301 Miller units), whereas the interactions between NTH1 and StBEL31 and -6 were the weakest (25 and 58 Miller units, respectively). Overall, StBEL interactions with NTH1 exhibited some of the lowest levels of β-galactosidase activity with an average of 149 Miller units per interaction. Among the KNOX types, NTH22 exhibited the greatest activity levels with an average of 243 units per interaction. Interactions with StBEL13, -33, and -35 had the strongest overall interactions with the five KNOX types. Among the StBEL proteins, the most robust interactions with NTH1 and -15 (SHOOTMERISTEMLESS orthologue) were with StBEL13 (233 and 298 units, respectively), whereas the strongest interactions with POTH1, NTH22, and NTH20 were with StBEL33 (274, 301, and 297 units, respectively). StBEL5, -6, -30, and -31 displayed the weakest interactions overall with these KNOX partners.
Fig. 7.
Specific interaction of POTH1 and four KNOTTED1-type proteins of tobacco with nine BEL1-like proteins, BEL5, -30, -13, -6, -31, -32, -33, -34 and -35, of potato using a β-galactosidase assay to assess the strength of interaction. The new potato BELs were cloned into the pACT-AD vector and the KNOTTED1-types were expressed in pBridge. BEL5/POTH1-BD was used as a reference and the new BEL proteins in the pACT-AD vector transformed into yeast are shown as negative controls. LacZ induction in the yeast strain pJ69-2A was assayed in transformed yeast cultures using a quantitative yeast β-galactosidase assay method. Standard errors of the means of three replicate samples are shown for each combination.
Tandem TTGAC motifs in the StBEL gene family
StBEL5 functions in tandem with its KNOX partner, POTH1, to bind specifically to a 10bp sequence consisting of twin TTGAC core motifs to regulate developmental processes in potato (Chen et al., 2003, 2004). Examination of the StBEL5 promoter revealed inverted tandem TTGAC motifs spaced 3 nt apart in a head-to-head orientation on opposite stands 820 nt upstream from the start of its 5′ UTR. Using a mutated promoter driving GUS expression, this double motif was confirmed to be involved in mediating autoregulation of StBEL5 in stolons and roots (Lin et al., 2013). To check for the possibility of cross-regulation among the StBELs, upstream sequences up to 3kb for each StBEL gene were screened for TGAC motifs. Four of the 13 StBEL members, including StBEL5, harboured the TGAC core motif in tandem separated, at most, by a 3 nt linker (Fig. 8). StBEL6 contained the motif in a tail-to-tail orientation on opposite stands (TTGACaGTCA, 520 nt upstream from the start of the 5′ UTR). StBEL22 had the motif in a head-to-head orientation again on opposite stands (GTCAcaaTTGAC, 1471 nt upstream from the start of the 5′ UTR), whereas the motifs are present in a tail-to-head direction on the same (+) strand in StBEL34 promoter sequence (TTGACggTGAC, 1459 nt upstream from the start of the 5′ UTR).
Fig. 8.
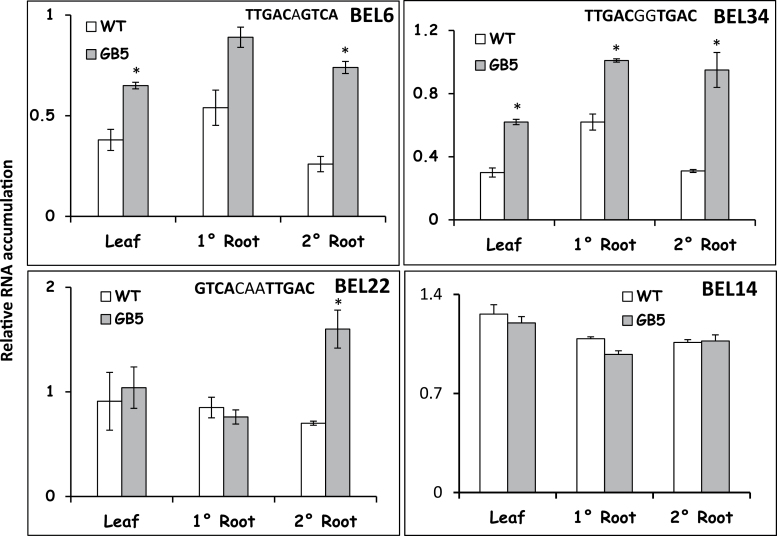
Cross-regulation of endogenous StBEL6, -34, and -22 in GAS:BEL5 overexpression lines. The movement of transgenic StBEL5 mRNA from leaf to primary and secondary roots was confirmed previously using transgenic lines expressing full-length StBEL5 RNA driven by the GAS promoter of melon (Cucumis melo) grown under SD conditions (Fig. 1B; Lin et al., 2013). This promoter is expressed in the minor veins of leaf mesophyll (Ayre et al., 2003; Banerjee et al., 2009). Substantial amounts of transgenic BEL5 RNA moved into primary and secondary roots and activated accumulation of WT StBEL5 transcripts (Fig. 7; Lin et al., 2013). This same RNA was used to assess levels of endogenous RNA for StBEL6, -34, -22, and -14 in both the WT (open bars) and transgenic BEL5 line (grey shaded bars) in leaves, primary (1° Root) and secondary (2° Root) roots. The existing upstream double TGAC core motifs are shown for each gene. The BEL14 upstream sequence contains no tandem TGAC motif and was included as a negative control. One-step RT-PCR was performed using 200–250ng of total RNA and gene-specific primers for StBEL6, -34, -22, and -14. All PCRs were standardized and optimized to yield product in the linear range. Homogenous PCR products were quantified using ImageJ software (Abramoff et al., 2004) and normalized using 18S rRNA values. Standard errors of the means of three replicate samples are shown. The asterisk indicates a significant difference (P<0.05) using Student’s t-test.
Using gel-shift assays, tandem head-to-head and tail-to-head TGAC motifs present on the StBEL5 and StGA20ox1 promoters, respectively, were confirmed as binding targets to the StBEL5/POTH1 heterodimer (Chen et al., 2004; Lin et al., 2013). Double palindromic tail-to-tail TGAC core motifs present in upstream sequence of StGA2 oxidase1 also bound to this tandem protein complex (Lin et al., 2013). In all three of these examples, specificity of binding to the tandem TGAC element was confirmed through mutagenesis. To further study the significance of these motifs on the promoter activity of StBEL6, -22, and -34, RNA accumulation for these genes was assayed in a transgenic line that couples leaf-specific overexpression of StBEL5 with the capacity to transport BEL5 transcripts into stolons and roots (Banerjee et al., 2006). Using the leaf-specific galactinol synthase (GAS) promoter (Ayre et al., 2003), movement of StBEL5 RNA from leaves to stolons and roots was readily observed with the greatest level of accumulation occurring in secondary roots (Lin et al., 2013). In theory, any RNA driven by the GAS promoter (in this case, StBEL5) that is detected in organs other than the leaf is the result of long-distance transport. In this way, this system monitors the induction of a target gene by a mobile RNA signal. The relative expression patterns of StBEL6, -22, and -34 were assayed in leaves, primary and secondary roots of the GAS:BEL5 transgenic line relative to non-transformed controls. RNA levels were enhanced 2- to 3-fold for all three BEL genes in secondary roots in correlation with transgenic StBEL5 RNA accumulation (Fig. 8). Induction was also observed in leaves and primary roots for StBEL34 and in leaves for StBEL6. No induction was observed in the GAS:BEL5 transgenic plants in the levels of mRNA for StBEL14, a StBEL gene without a tandem TGAC motif present in its upstream sequences. DNA-binding assays confirmed the interaction of the StBEL5/POTH1 protein heterodimer with the double TGAC core motifs present in all three of these StBEL genes (Fig. 9). In the case of the StBEL6 and -22 motifs, a strong interaction with the StBEL5 protein alone was also observed. The tightly resolved band in the upper portion of lane two (BEL5 protein alone) for StBEL22 suggested the presence of a homodimer. Three of the four 5′→3′ DNA strand orientations for the two core motifs are represented in this StBEL group: tail-to-tail, head-to-head, and tail-to-head on the (+)-strand (Table 2 and Fig. 9). In previous work, mutated forms of the identical tandem motifs present in StBEL6, -22 and -34 exhibited diminished binding to the StBEL5/POTH1 complex (Chen et al., 2004; Lin et al., 2013). As reported previously (Fig. 6), these three putative targets of the StBEL5 complex are among the rarest of the StBEL transcripts.
Table 2.
Eight target genes of StBEL5The TGAC core motifs running 5′→3′ are in bold letters. Linker sequence between the motifs is shown in lower-case letters. The location of the motif is designated upstream from either the transcription (TSS) or the translation (AUG) start site.
| Gene | Motif a | Orientationb | Location of motif (nt upstream) | RNA levels regulated by StBEL5 | Binding confirmed by EMSAc | Reference |
|---|---|---|---|---|---|---|
| StBEL5 | GTCAAtgcTTGAC | HtH | 820 (TSS) | Yes | Yes | Lin et al., 2013 |
| StBEL6 | TTGACaGTCA | TtT | 520 (TSS) | Yes | Yes | Figs 8 and 9 |
| StBEL22 | GTCAcaaTTGAC | HtH | 1471 (TSS) | Yes | Yes | Figs 8 and 9 |
| StBEL34 | TTGACggTGAC | TtH (+) | 1459 (TSS) | Yes | Yes | Figs 8 and 9 |
| StGA20ox1 | TTGACTTGAC | TtH (+) | 700 (TSS) | Yes | Yes | Chen et al., 2004 |
| StGA2ox1 | TTGACaaGTCA | TtT | 1768 (AUG) | Yes | Yes | Lin et al., 2013 |
| StIPT | TTGACaaGTCA | TtT | 1408 (AUG) | Yes | Yes | Hannapel et al., 2013 |
| YUCCA1a | TTGACcttaTTGAC | TtH (+) | 641 (AUG) | Yes | Yes | Lin et al., 2013 |
a The criteria for these motifs was the inclusion of at least one TTGAC and one TGAC on either strand of the DNA with a linker sequence of no more than four nt.
b Three 5′→3′ orientations were observed: head-to-head (HtH), tail-to-tail (TtT), or tail-to-head on the plus strand (TtH(+)). No double motifs aligned tail-to-head on the (–) strand were identified in this initial screen.
c Verified binding to the BEL5/POTH1 complex via EMSA. Stronger binding with the BEL5/POTH1 complex was observed than with either protein alone.
Discussion
Targets of the StBEL transcription factors
To date, members of the BEL1-like family of transcription factors have been identified in every plant species that has so far been studied. With the advent of full-genome sequences, the breadth and potential functions of this key family of DNA-binding proteins may now be fully understood. Clear evidence has established the role of the BELs in both floral and vegetative development. A catalogue of the known target genes for BEL1 transcription factors supports this premise. Most prominent in this currently minute collection are GA20 oxidase1, GA2 oxidase1, YUCCA1a, isopentenyl transferase, StBEL5, PIN1 and -2 (Chen et al., 2004; Lin et al., 2013; Hannapel et al., 2013), and of course, as shown here, other BEL1-like genes (Table 2). All of these targets contribute to important aspects of plant growth, including meristem maintenance, tuberization, and leaf and root development. BEL1 proteins may also play important roles in response to biotic stress and pathogen challenge (Luo et al., 2005) and in regulating lignin biosynthesis (Mele et al., 2003). The strong wound response exhibited by the promoter of StBEL5 (Chatterjee et al., 2007) suggests that BEL1 proteins may function in defence against abiotic stress.
The StBEL5/11/29 clade
These three StBEL types group phylogenetically in a unique cluster with AtBLH1 of Arabidopsis. AtBLH1 functions with KNAT3 to affect establishment of cell fates in the mature embryo sac (Pagnussat et al., 2007). Each of these three StBEL proteins was among the largest proteins in the potato group (688 aa for BEL5 and 710 for both BEL11 and -29) and contained conserved amino acid sequence domains outside the canonical motifs. Their overall transcript abundance levels were consistent and unique. All three exhibited relatively high levels in petioles, stolons, roots, and tuber sprouts, whereas StBEL5 and -29 exhibited high levels in flowers, shoot apices, and young and mature tubers (Supplementary Table S3 available at JXB online). All three exhibited enhanced levels of RNA accumulation in stolons from SD plants (Fig. 6). Together, these observations suggested that StBEL5, -11, and -29 are relatively stable RNAs that play pivotal roles in regulating development in actively growing organs. Within this group, StBEL5 functions as a mobile RNA that impacts growth in both tubers and roots (Banerjee et al., 2006; Lin et al., 2013). As discussed previously, the RNA metabolism of StBEL5 is mediated by its 3′ UTR (Banerjee et al., 2006, 2009), and there are sequences within this region that are common to both StBEL11 and -29 (Supplementary Fig. S2 available at JXB online). It is conceivable that StBEL members of this subgroup are functionally redundant and share a similar long-distance, non-cell-autonomous delivery system.
Levels of regulation controlling StBEL gene activity
Perhaps the most intriguing aspect of the StBEL family is its complex mode of regulating expression and activity at both the transcriptional and post-transcriptional levels. StBEL5 regulates activity of its own promoter (Lin et al., 2013), and in the current study, movement and accumulation of transgenic BEL5 RNA were also correlated with an increase in steady-state levels of three other StBEL RNAs. This increase was observed only with genes containing the tandem core TGAC motif recognized by the BEL/KNOX complex (Smith et al., 2002; Chen et al., 2004; Lin et al., 2013). Auto- and cross-regulation among plant transcription factors in the same family is now known to be quite common. MADS genes are regulated by MADS-box proteins in a wide network of protein–DNA interaction. SEPALLATA3 binds to cis-regulatory elements of other MADS-box genes and is a key component in the transcriptional network regulating the formation of floral organs (Kaufmann et al., 2009). Positive autoregulation of Knox genes in rice was essential for shoot apical meristem development (Tsuda et al., 2011). In this study, OSH1 directly bound to five KNOX loci (including itself) to upregulate expression. Using ChIP-seq in maize, Bolduc et al. (2012) showed that KN1 directly targets upstream sequence of numerous transcription factors, including its own gene, nine other KNOX types, and five BEL1-like genes. Two of the target maize BEL1-like genes are orthologues of StBEL6 and -34. As a mechanism for enhancing specificity, it is very likely that many of these maize DNA interactions are mediated by BEL/KNOX tandem complexes. Gel-shift assays with native DNA sequences of potato showed that binding of BEL/KNOX complexes was consistently stronger than with either protein alone (Fig. 9; Chen et al., 2004; Lin et al., 2013; Hannapel et al., 2013).
At the post-transcriptional level, BEL1-like proteins exhibit several potential mechanisms for the control of activity or expression. These include, first, the availability and binding affinity of protein partners. These partners may include KNOX proteins, ovate family proteins (OFPs), or a MADS-box homeodomain protein complex that contains the SEPALLATA MADS-box proteins (Brambilla et al., 2007). Interaction with KNOX proteins facilitates selective transport of the tandem complex into the nucleus (Bhatt et al., 2004). In a similar manner, the ovate family proteins, AtOFP1 and AtOFP5, associate with the cytoskeleton and interact with both BEL and KNOX proteins to regulate their subcellular localization to the cytoplasm (Hackbusch et al., 2005). By preventing nuclear localization, the OFPs essentially block BEL/KNOX activity. In another example of partner interaction, a truncated form of a KNOX protein of Arabidopsis, designated KNATM-B, encodes a MEINOX domain but not the homeodomain (Magnani and Hake, 2008). This new class of KNOX proteins is conserved in eudicots, including both tomato (Magnani and Hake, 2008) and potato (PGSC0003DMP400031538) and selectively interacts with BEL proteins through the MEINOX domain. These results suggest that KNATM-B may prevent specific BEL proteins from taking part in transcriptional complexes by sequestering them in an inactive dimer or by localization in the cytoplasm.
Secondly, the binding affinity of the BELL/KNOX complex for the various tandem TGAC motifs can regulate activity. As shown previously, binding may occur to double elements with tail-to-tail, head-to-head, or (+)- and (–)-strand tail-to-head orientations (Table 2; Chen et al., 2004; Hannapel et al., 2013; Lin et al., 2013). Very little is known about how these various configurations affect the interaction of the BEL/KNOX or KNOX/KNOX complexes with the upstream target cis-element. Such differences in binding affinity could certainly impact the results on cross-regulation of StBEL6, -22, and -34 presented here (Figs 8 and 9) as each of these promoters contain a unique configuration of the tandem core TGAC element.
The third post-transcriptional mechanism that can significantly affect BEL1 activity is the non-cell-autonomous nature of BEL1-like mRNAs. Specific StBEL RNAs are transcribed in one organ and have the capacity to move long distances via the phloem to target organs. The best example of a mobile RNA in the BEL1 family is StBEL5. RNA movement assays demonstrated that StBEL5 transcripts move through the phloem to stolon tips to regulate tuber formation. StBEL5 mRNA originates in the leaf and its movement to stolons is induced by a SD photoperiod (Banerjee et al., 2006). Movement of StBEL5 RNA into roots correlated with increased growth and the accumulation of several transcripts associated with hormone metabolism has also been reported (Hannapel et al., 2013; Lin et al., 2013). Regulated long-distance transport of full-length mRNAs is a unique signalling process and represents a dynamic mechanism to separate transcription and translation, and in this case, to control both the temporal and spatial activity of a pivotal transcription factor. Based on RNA profiling in phloem cells (Yu et al., 2007; Campbell et al., 2008), it is very likely that other StBEL genes (like StBEL11 and -29) may function in a similar manner.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Schematic protein structure of StBELs.
Supplementary Fig. S2. Alignment of the 3′ UTRs of StBEL11 and -29 to StBEL5.
Supplementary Fig. S3. RT-PCR for select StBEL genes.
Supplementary Fig. S4. Expression profile for StBEL family members using RNA-Seq data.
Supplementary Table S1. List of primers and oligonucleotides.
Supplementary Table S2. Position of StBEL genes on PGSC chromosome pseudomolecules.
Supplementary Table S3. RNA-Seq data for 13 StBEL genes.
Supplementary Table S4. Interaction of tobacco KNOX and potato BEL1 proteins.
Acknowledgements
Thanks to Anjan Banerjee and Hao Chen for their contributions to our understanding of StBEL5 biology. This research was supported by the NSF Plant Genome Research Program award no. DBI-0820659 and National Research Initiative grant no. 2008–02806 from the USDA National Institute of Food and Agriculture.
Glossary
Abbreviations:
- FPKM
fragments per kb per million mapped reads
- GAS
galactinol synthase
- GUS
β-glucuronidase
- GST
glutathione S-transferase
- LD
long day
- NTH
Nicotiana tabacum homeobox
- qRT-PCR
quantitative reverse transcription-PCR
- RACE
rapid amplification of cDNA ends
- SD
short day
- UTR
untranslated region
- WT
wild type.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–42 [Google Scholar]
- Auweter SD, Allain FHT. 2008. Structure-function relationships of the polypyrimidine tract binding protein. Cellular and Molecular Life Sciences 65, 516–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayre BG, Blair JE, Turgeon R. 2003. Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiology 133, 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Chatterjee M, Yu Y, Suh SG, Miller WA, Hannapel DJ. 2006. Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee AK, Lin T, Hannapel DJ. 2009. Untranslated regions of a mobile transcript mediate RNA metabolism. Plant Physiology 151, 1831–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellaoui M, Pidkowich MS, Samach A, Kushalappa K, Kohalmi SE, Modrusan Z, Crosby WL, Haughn GW. 2001. The Arabidopsis BELL1 and KNOX TALE homeodomain proteins interact through a domain conserved between plants and animals. Plant Cell 13, 2455–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S, Simonini S, Benková E, Colombo L. 2012. The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis . Plant Cell 24, 2886–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt AM, Etchells JP, Canales C, Lagodienko A, Dickinson H. 2004. VAAMANA, A BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis . Gene 328, 103–111 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. 2012. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes and Development 26, 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla V, Battaglia R, Colombo M, Masiero S, Bencivenga S, Kater MM, Colombo L. 2007. Genetic and molecular interactions between BELL1 and MADS box factors support ovule development in Arabidopsis . Plant Cell 19, 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BA, Hallengren J, Hannapel DJ. 2008. Accumulation of BEL1-like transcripts in Solanaceous species. Planta 228, 897–906 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Banerjee AK, Hannapel DJ. 2007. A BELL1-like gene of potato is light activated and wound inducible. Plant Physiology 145, 1435–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Banerjee AK, Hannapel DJ. 2004. The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1 . The Plant Journal 38, 276–284 [DOI] [PubMed] [Google Scholar]
- Chen H, Rosin FM, Prat S, Hannapel DJ. 2003. Interacting transcription factors from the TALE superclass regulate tuber formation. Plant Physiology 132, 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF. 2005. A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proceedings of the National Academy of Sciences, USA 102, 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. 2004. The role of knox genes in plant development. Annual Review of Cell and Developmental Biology 20, 125–151 [DOI] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98 [Google Scholar]
- Ham BK, Brandom JL, Xoconostle-Cázares B, Ringgold V, Lough TJ, Lucas WJ. 2009. A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21, 197–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel DJ, Sharma P, Lin T. 2013. Phloem-mobile messenger RNAs and root development. Frontiers in Plant Science 4, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannapel DJ. 2013. Long-distance signaling via mobile RNAs. In: Baluška F, ed. Long-distance systemic signaling and communication. Berlin: Springer-Verlag [Google Scholar]
- Kanrar S, Bhattacharya M, Arthur B, Courtier J, Smith HMS. 2008. Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis . The Plant Journal 54, 924–937 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. 2009. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biology 7, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M, Tabb P, Hepworth SR. 2012. BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal Behavior 7, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Kushalappa K, Godt D, Pidkowich MS, Pastorelli S, Hepworth SR, Haughn GW. 2007. The Arabidopsis BEL1-LIKE HOMEODOMAIN proteins SAW1 and SAW2 act redundantly to regulate KNOX expression spatially in leaf margins. The Plant Cell 19, 2719–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal S, Pacis LB, Smith HM. 2011. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis . Molecular Plant 4, 1123–1132 [DOI] [PubMed] [Google Scholar]
- Li Y, Pi L, Huang H, Xu L. 2012. ATH1 and KNAT2 proteins act together in regulation of plant inflorescence architecture. Journal of Experimental Botany 63, 1423–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Sharma P, Gonzalez DH, Viola IL, Hannapel DJ. 2013. The impact of the long-distance transport of a BEL1-like mRNA on development. Plant Physiology 161, 760–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Luo H, Song F, Goodman RM, Zheng Z. 2005. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biology 7, 459–468 [DOI] [PubMed] [Google Scholar]
- Magnani E, Hake S. 2008. KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20, 875–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Bhogale S, Kang IH, Hannapel DJ, Banerjee AK. 2012. The mRNA of a Knotted1-like transcription factor of potato is phloem mobile. Plant Molecular Biology 79, 595–608 [DOI] [PubMed] [Google Scholar]
- Mele G, Ori N, Sato Y, Hake H. 2003. The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes and Development 17, 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Wang Y, Franzen R, Santi L, Salamini F, Rohde W. 2001. In vitro interactions between barley TALE proteins suggest a role for protein-protein associations in the regulation of Knox gene function. The Plant Journal 27, 13–23 [DOI] [PubMed] [Google Scholar]
- Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M. 2000. Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant and Cell Physiology 41, 583–590 [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu H-J, Sundaresana V. 2007. Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1 . Plant Cell 19, 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V. 2008. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20, 888–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ. 2003. Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiology 132, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. 2009. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. The Plant Journal 58, 641–654 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Smith HM, Boschke I, Hake S. 2002. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proceedings of the National Academy of Sciences, USA 99, 9579–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HMS, Hake S. 2003. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. The Plant Cell 15, 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N. 2011. Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung N, Lal S, Smith HM. 2011. The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical meristem in Arabidopsis . Plant Physiology 156, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Pan S, Cheng S, et al. 2011. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 [DOI] [PubMed] [Google Scholar]
- Yu YY, Lashbrook CC, Hannapel DJ. 2007. Tissue integrity and RNA quality of laser microdissected phloem of potato. Planta 226, 797–803 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.