Abstract
Impaired ethanol metabolism can lead to various alcohol-related health problems. Key enzymes in ethanol metabolism are alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH); however, neuroendocrine pathways that regulate the activities of these enzymes are largely unexplored. Here we identified a neuroendocrine system involving Corazonin (Crz) neuropeptide and its receptor (CrzR) as important physiological regulators of ethanol metabolism in Drosophila. Crz-cell deficient (Crz-CD) flies displayed significantly delayed recovery from ethanol-induced sedation that we refer to as hangover-like phenotype. Newly generated mutant lacking Crz Receptor (CrzR01) and CrzR-knockdown flies showed even more severe hangover-like phenotype, which is causally associated with fast accumulation of acetaldehyde in the CrzR01 mutant following ethanol exposure. Higher levels of acetaldehyde are likely due to 30% reduced ALDH activity in the mutants. Moreover, increased ADH activity was found in the CrzR01 mutant, but not in the Crz-CD flies. Quantitative RT-PCR revealed transcriptional upregulation of Adh gene in the CrzR01. Transgenic inhibition of cyclic AMP-dependent protein kinase (PKA) also results in significantly increased ADH activity and Adh mRNA levels, indicating PKA-dependent transcriptional regulation of Adh by CrzR. Furthermore, inhibition of PKA or cAMP response element binding protein (CREB) in CrzR cells leads to comparable hangover-like phenotype to the CrzR01 mutant. These findings suggest that CrzR-associated signaling pathway is critical for ethanol detoxification via Crz-dependent regulation of ALDH activity and Crz-independent transcriptional regulation of ADH. Our study provides new insights into the neuroendocrine-associated ethanol-related behavior and metabolism.
Introduction
Chronic ethanol consumption causes serious health problems such as liver cirrhosis and various types of cancer [1]. Detoxification of the ethanol involves a sequence of reactions, in which ethanol is first oxidized to acetaldehyde by ADH, and then further oxidized to acetate by mitochondrial ALDH. Acetaldehyde promotes adduct formation, leading to the dysfunction of various key proteins and DNA damage [2]. Thus, the accumulation of acetaldehyde introduces significant toxic effects, causing the ethanol-associated health problems. Therefore, the regulatory mechanisms of ethanol metabolism are important to understand pathophysiological effects of ethanol.
The rate of ethanol metabolism in individuals is greatly influenced by genetic polymorphisms, which gives rise to enzyme variants with different catalytic properties [3], [4]. In addition to the genetic factors, transcriptional regulation and post-translational modifications affect ADH/ALDH activity [5]–[8]. Interestingly, hormones, such as thyroid and growth hormone, modulate ADH activity and its expression, indicating that hormonal factors could play a part in the ethanol detoxification [9]–[13]. However, little is known about neuroendocrine regulation of ethanol metabolism.
The genetic basis of ethanol-induced behaviors has been investigated in the fruit fly, Drosophila melanogaster, since its progressive behavioral patterns in response to acute ethanol exposure are quite similar to those of humans [14]. Moreover, genetic and transgenic toolkits available for this species make it an attractive model system to investigate the molecular mechanisms underlying pathophysiological and behavioral outcome of the ethanol consumption in vertebrates. Various studies have reported that several neuropeptides are involved in the regulation of ethanol-related behavior in Drosophila [15]–[18]. These studies suggest that peptidergic networks play an important role in modulating sensitivity to ethanol.
The neuropeptide Corazonin (Crz) was first found in the American cockroach [19]. Although the sequence and structure of Crz is highly conserved among different insect species [20], it has been shown to affect diverse physiological functions in a species-specific manner; cardio-acceleration in the cockroach [19], induction of cuticular pigmentation in the migratory locust [21], reduction of the spinning rate and pupal development in the silkworm [22], and induction of ecdysis in a moth [23]. In Drosophila adult, Crz is produced by a major group of neurosecretory cells in the brain and abdominal ganglion [24], [25]. Given the complexity of the Crz neuronal architecture, Crz is predicted to deliver multiple biological functions: Crz neurons are shown to be associated with sperm transfer and copulation duration [26], regulation of trehalose levels [25], and responses to physiological and nutritional stresses [27], [28].
Drosophila CrzR is a member of G-protein coupled receptor (GPCR) family and structurally homologous to the mammalian GnRH (Gonadotropin releasing hormone) receptor [29]. Physiological roles of the CrzR have not been explored. Using mutant flies lacking Crz neurons or CrzR, we investigated biological functions of the Crz signaling system in D. melanogaster. Both types of mutant flies displayed significantly delayed recovery from ethanol-induced sedation. We further show that such ‘hangover-like’ phenotype of the CrzR mutant likely results from fast acetaldehyde accumulation due to higher ADH production as a result of transcriptional up-regulation and lower ALDH activity. Only the latter event was observed in the Crz-CD mutant, suggesting a complicated signaling mechanism associated with the CrzR.
Results
Hangover-like phenotype mediated by Crz neurons
To explore the role of the Crz signaling pathway in ethanol-induced behavior, we employed Crz-cell deficient (Crz-CD) D. melanogaster that was produced by transgenic expression of a cell death gene hid. When flies were exposed to the vapor derived from 30–70% ethanol, we did not see a significant difference in the rates of sedation between controls and Crz-CD [30]. This is in contrast to a recent report that showed that Crz-CD is resistant to ethanol-induced sedation [16]. We do not know the cause of such discrepancy, except for the experimental setting.
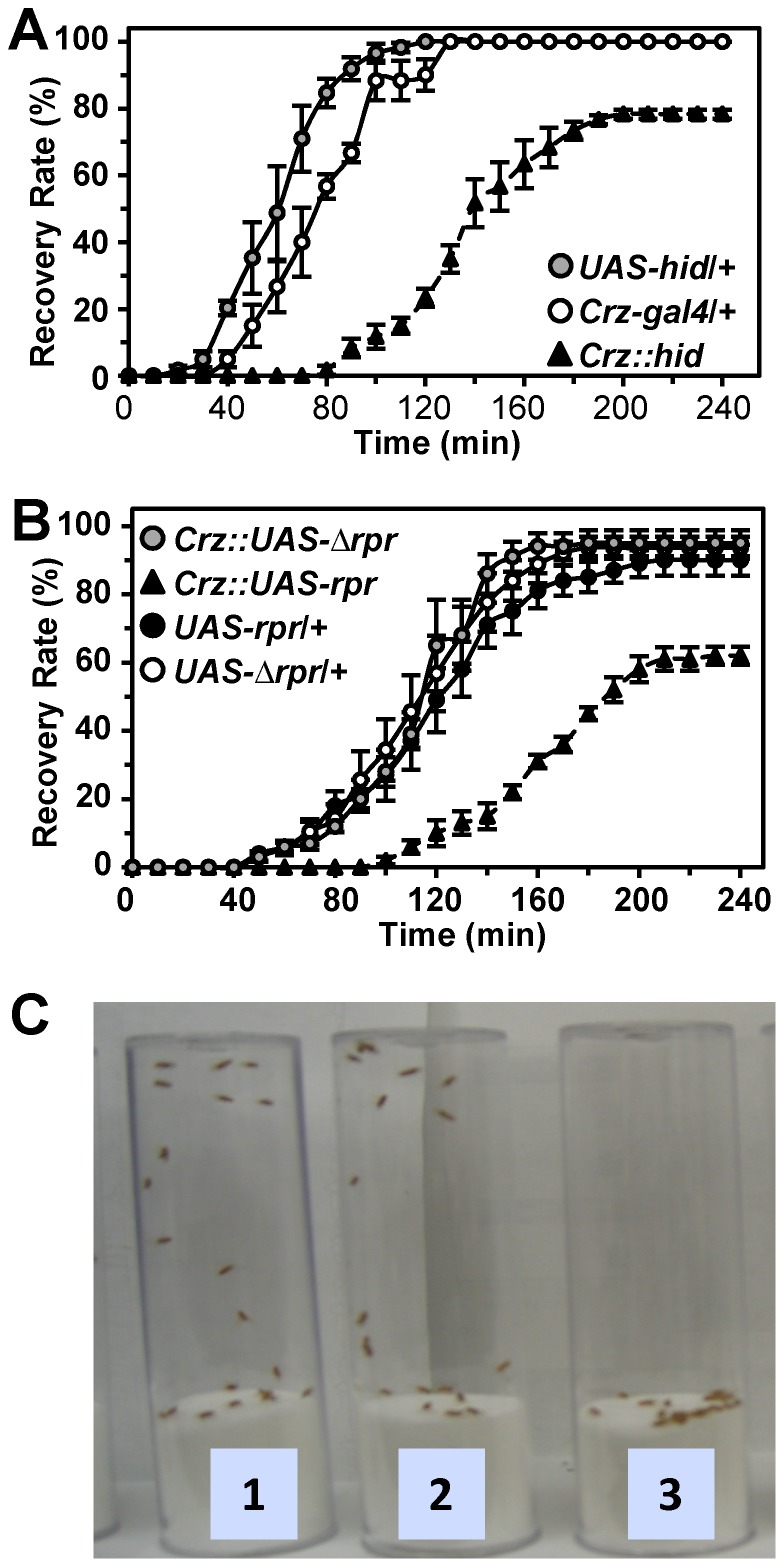
During this study, however, we noticed that Crz-CD flies showed significantly delayed recovery from ethanol-induced sedation. To characterize this phenotype further, flies that were completely sedated with vapor from 100% ethanol for 17∼18 min were allowed to recover in an ethanol-free environment. Control flies began to assume normal standing posture and mobile around 40 min and most of them recovered after 2 h. In contrast, the recovery of Crz-CD flies (Crz::hid) was significantly delayed; it was first observed at 80 min and maximum 80% of flies recovered by 4 h while the remaining flies did not survive (Figure 1, A and C). Crz-CD flies generated by ectopic expression of a different cell death gene reaper (rpr) produced results similar to the Crz::hid, whereas expression of a mutant rpr (▵rpr, an rpr lacking IBM death domain) showed no difference from controls (Figure 1B). These results suggest that Crz neurons are required for the recovery from ethanol-induced sedation. We refer to such delayed recovery from ethanol-induced sedation as “hangover-like” phenotype.
Figure 1. Hangover-like phenotype of Crz-CD.
Crz neuron ablation induced by hid (A) and rpr (B) transgene expression (triangles) leads to delayed and incomplete recovery compared to wild-type (circles). Each data point is a mean ± sem (n = 3–5). All genotypes are in y w background. (C) Flies recovered from ethanol-induced sedation at 2 hours after exposure. (1, Crz-gal4/+; 2, UAS-hid/+; 3, Crz-gal4/UAS-hid).
Severe hangover-like phenotype of a CrzR null mutant
To investigate whether the foregoing result is due to the lack of Crz signaling, we generated a null mutant lacking CrzR (a.k.a. GRHRII). A putative hypomorphic mutant allele, Mi{ET1}GRHRIIMB00838 (for short, MB00838), carries a Minos transposable element bearing eye-specific GFP (green fluorescence protein) marker inserted in the fifth intron (Flybase; Figure 2A). To generate a CrzR-null mutant, we mobilized MB00838 using heat-shock-induced expression of Minos transposase [31]. The mobilization was confirmed by a mosaic pattern of GFP expression in the eyes. Subsequently, we established one hundred GFP-negative lines, one of which was identified to be a deletion mutant lacking CrzR by PCR. Further refined PCR confirmed an 8-kb deletion including exon 2–5 (Figure 2). In addition to this deletion, PCR revealed a footprint of the gal4-coding region derived from the MB00838 element at its original insertion site. RT-PCR targeting the 5' region of CrzR (–59∼+328) confirmed the absence of CrzR transcription, suggesting that this is a bona fide null mutant (Figure 2C). Thus we designate this mutant as CrzR01 for the first amorphic allele of this gene.
Figure 2. Generation of CrzR-null allele.
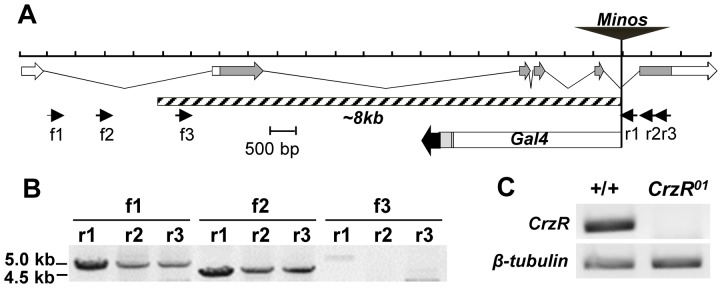
(A) Diagram of CrzR encoding exons and approximate locations of PCR primers. Exons and introns are shown as arrows and solid lines, respectively. Coding exons are shown in grey, and UTRs in white. An 8-kb deletion was indicated by the hatched box. A footprint of gal4 coding region and flanking sequence derived from the Minos element was found at the original insertion site. (B) PCR products derived from the designated primer sets and mutant genomic DNA. No specific PCR product was produced from f3 primer. (C) RT-PCR. CrzR01 did not produce PCR product, while the wild-type did it with expected size.
Homozygous CrzR01 mutants were viable and fertile and had no discernible morphological or developmental aberrations. Mutant females are reproductively normal as their fecundity is comparable to wild-type (Figure S1). In addition, circadian locomotor activity rhythms are indistinguishable between wild-type and CrzR01 mutant flies, indicating that CrzR01 mutants are as healthy as wild-type (Figure S2).
Next we measured hangover-like phenotype of the CrzR01 mutant, as done for Crz-CD. When flies were exposed to ethanol for 17.5 min, the recovery of CrzR01 flies was severely retarded; only 20% recovered fully after 4 h, whereas 70% of control flies did (Figure 3A). To confirm whether this is caused by the deletion of the CrzR locus, we performed similar assays using hemizygous CrzR01 (CrzR01/Df) or control (+/Df) with w1118 background. After 17.5-min exposure, most Df/+ flies recovered after 2 h, while CrzR01/Df did after 4 h (Figure 3B). The recovery of Df/+ flies was largely unaffected when the exposure duration was increased to 18 min, while only 70% of CrzR01/Df recovered by 4 h (Figure 3C). Similar hangover-like phenotype was observed with RNA interference (RNAi)-mediated CrzR knock-down flies (CrzR-KD). CrzRRNAi was constructed as described in the supporting protocol (Protocol S1). In response to an actin-gal4, two independent lines, UAS-CrzRRNAiS3S and UAS-CrzRRNAiT17, showed severe reduction of CrzR mRNA (Figure S3A). These CrzR-KD flies displayed hangover-like phenotype, with around 20% and 35% of the flies recovered by 4 h, which is in contrast to 80–90% recovery of the heterozygous transgenic controls (Figure S3B). Taking together with the data obtained with Crz-CD flies suggests that Crz signaling is causally associated with the hangover-like phenotype. This phenotype is unlikely to represent a general response to sedative agents, as neither Crz-CD nor a CrzR01 mutation affected the recovery rates from ethyl ether-induced sedation (Figure S4).
Figure 3. Hangover-like phynotype of the CrzR01 mutant flies.
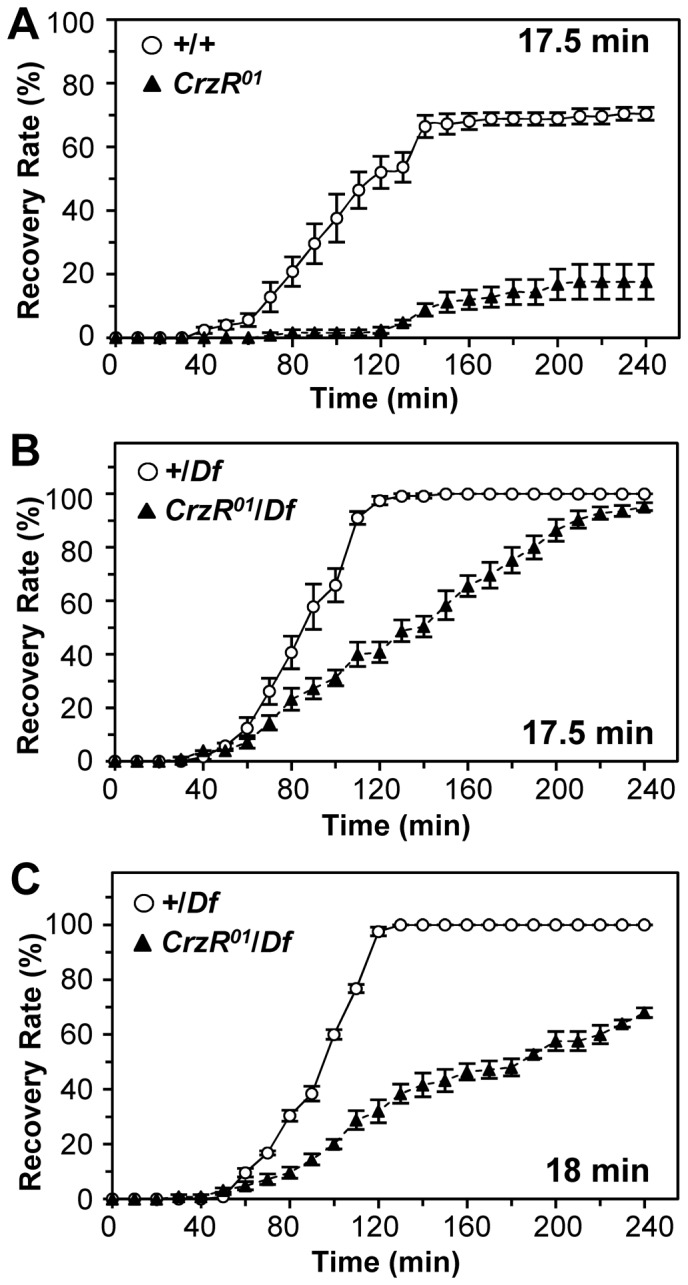
Flies were exposed to 100% ethanol as indicated. (A) Homozygous CrzR01. (genotypes: y w;; CrzR01) (B) CrzR01/Df. (genotypes: w1118;; +/Df: w1118;; CrzR01/Df). (C) Same as in (B), except for 18-min exposure period. Each data point represents mean ± sem (n = 5).
Defective acetaldehyde metabolism in CrzR01 mutant
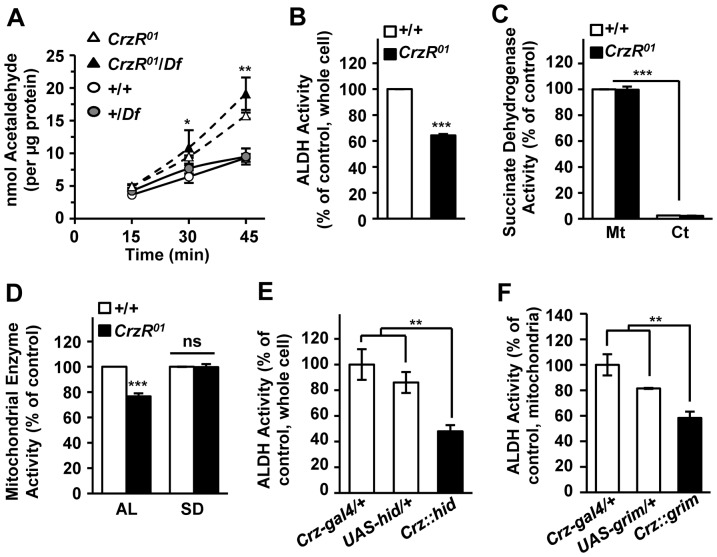
Ethanol is first oxidized to acetaldehyde, which has been considered a major cause of hangover symptoms in humans [32]. To test this possibility for CrzR01 flies, we used gas chromatography to monitor the contents of acetaldehyde following exposure to ethanol. Remarkably acetaldehyde contents in both CrzR01 and CrzR01/Df flies were up to 100% higher than those of control flies 45 min after ethanol exposure (Figure 4A).
Figure 4. Lack of Crz/CrzR leads to reduced ALDH activity.
(A) Levels of acetaldehyde in adult males exposed to 100% ethanol. (n = 4). (B) CrzR01 mutation resulted in the reduction of whole cell ALDH activity (n = 3). (C) Succinate dehydrogenase activity in mitochondria (Mt) and cytosol (Ct). The activity was detected almost exclusively in the mitochondrial fraction (n = 3). (D) Reduction of mitochondrial ALDH activity (AL) in CrzR01 mutant. Succinate dehydrogenase activity (SD) is similar between control and mutant. (n = 3). (E, F) Crz-CD leads to reduced ALDH activity. (E) Whole cell ALDH activities of Crz::hid are significantly lower than those of the transgenic controls (n = 8). (F) grim-induced Crz-CD produced significant reduction of mitochondrial ALDH activity (n = 3). (*P<0.05; **P<0.01; ***P<0.001; ns, not significant). Each data point represents mean ± sem for the indicated replicates. All genotypes are in y w background.
The accumulation of acetaldehyde in CrzR01 mutants is possibly due to subnormal acetaldehyde oxidation as a result from lower ALDH activity. Indeed, ALDH activity in whole fly extract of CrzR01 was only 63% of wild-type (Figure 4B). Since acetaldehyde oxidation occurs mainly inside the mitochondria, the ALDH activity was measured with isolated mitochondria. Succinate dehydrogenase activity, a mitochondrial marker, was observed almost exclusively in the mitochondrial fraction in both genotypes, verifying successful separation of the mitochondria from the cytosol (Figure 4C). Around 30% reduced ALDH activity was observed in the CrzR01 mutant, while succinate dehydrogenase activity was indistinguishable between wild-type and CrzR01 (Figure 4D). Reduction of ALDH activity was also observed in whole cell extracts of Crz-CD flies (Crz::hid) or in the mitochondrial fraction of Crz-CD induced by ectopic grim expression (Figure 4, E and F). In aggregate, these results suggest that Crz signaling plays an important role in the regulation of ALDH activity.
Loss of CrzR results in enhanced ADH activity
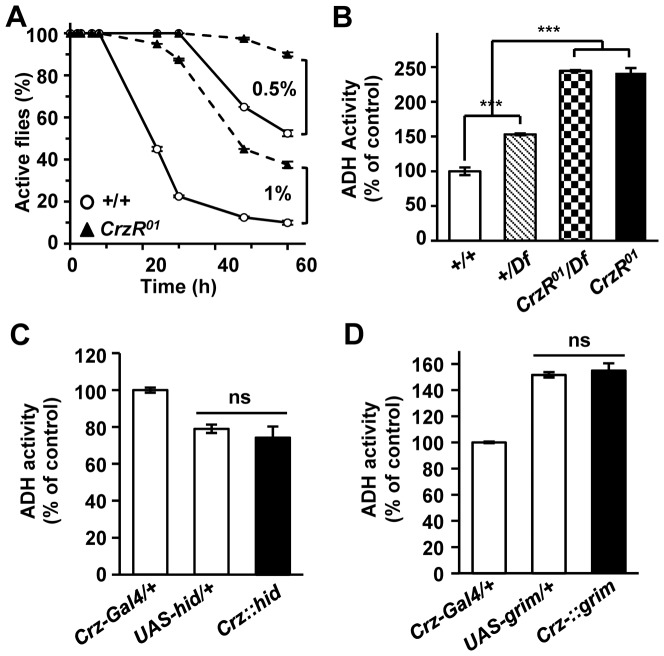
Since CrzR01 mutants have lower ALDH activity, they could be more sensitive to exogenously provided acetaldehyde than wild-type. To test this, flies were fed low concentrations of acetaldehyde without causing acute toxicity [33]. Surprisingly, the CrzR01 flies were more resistant to acetaldehyde-mediated intoxication than the wild-type. With 0.5% (v/v) acetaldehyde, ∼ 50% of wild-type flies became immobilized after 60 h, while only 10% of CrzR01 did so (Figure 5A). Significant differences between the two genotypes were also apparent in response to 1% acetaldehyde.
Figure 5. CrzR mutation causes significant increase of ADH activity.
(A) Survival rates of CrzR01 (dashed lines) and wild-type (solid line) in response to acetaldehyde as indicated (n = 3). (B) Histogram showing ADH activity (n = 4). (C,D) ADH activity in Crz-CD. (C) hid- and (D) grim-induced Crz cell ablation did not increase the whole cell ADH activity (P > 0.05, n = 3). (*** P<0.001). Each data point represents mean ± sem for the indicated replicates. All genotypes are in y w background.
Since D. melanogaster ADH is known to mediate the detoxification of the acetaldehyde by converting acetaldehyde to ethanol [33], we wondered if greater resistance to acetaldehyde by CrzR01 is due to higher ADH activity in the mutant. Indeed, around 2.4-fold increased ADH activity was measured in both homozygous CrzR01 and CrzR01/Df as compared to wild-type (Figure 5B). Df/+ flies showed a 1.5-fold increase of ADH activity, suggesting a dosage-dependent regulation of the ADH by CrzR.
In contrast to CrzR01, Crz-CD did not elevate ADH activity (Figure 5, C and D), indicating that Crz neurons are required only for the regulation of ALDH activity. The reasons for such difference are currently unclear but may be related to an unknown ligand that controls CrzR activity or intrinsic activity of CrzR (see discussion).
CrzR regulates Adh transcription in PKA-dependent manner
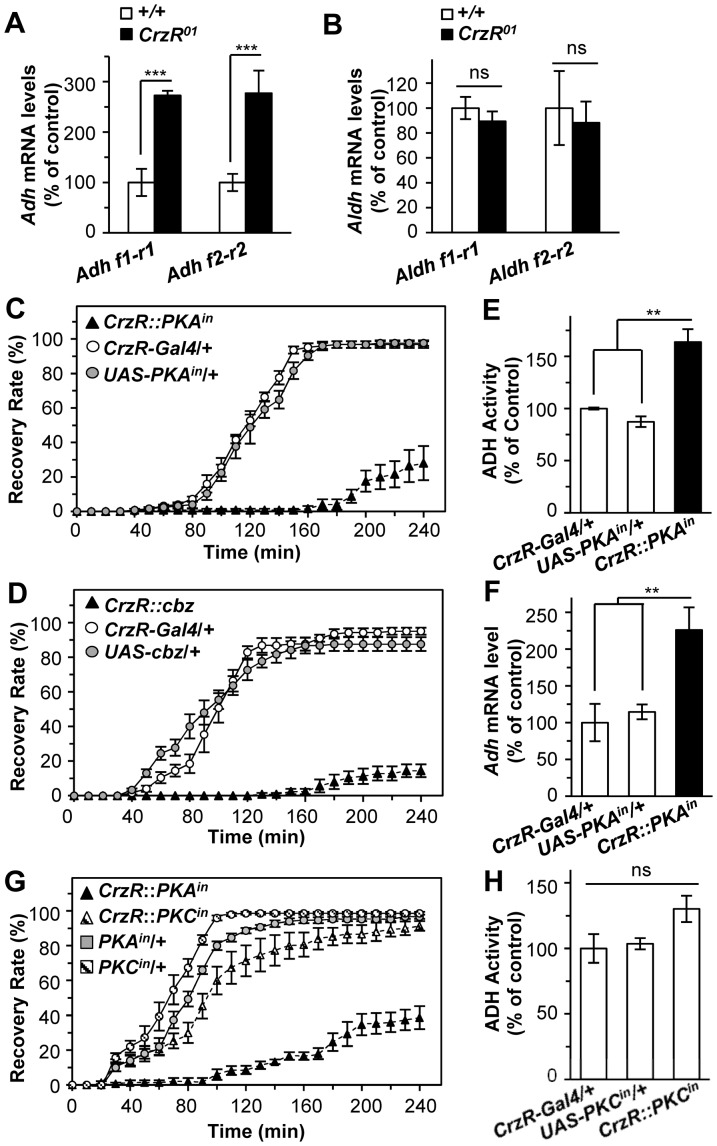
To further delve into the mechanisms as to how Crz/CrzR regulates ADH and ALDH activities, mRNA levels of Adh and Aldh were analyzed by real-time RT-PCR. An average of 2.7-fold increase of Adh transcript levels were observed from the CrzR01 using two different primer sets (Figure 6A) while no difference was found for Aldh (Figure 6B). These results indicate that CrzR is involved in the transcriptional regulation of Adh, but not of Aldh.
Figure 6. CrzR regulates Adh mRNA levels through PKA-dependent pathway.
(A) RT-qPCR using two different sets of primers revealed 2.7-fold increase of Adh transcript level in CrzR01 compared to the wild-type (n = 4). (B) No significant change of Aldh transcript level in CrzR01 (n = 4). (C,D) Expression of PKA inhibitor (C) or dominant negative CREB (D) using CrzR-gal4 induced severe hangover-like phenotype (n = 5). (E) PKA inhibition in CrzR cells increased ADH activity by 50% compared to the controls (n = 3). (F) PKA inhibition in CrzR cells leads to significant increase of Adh transcript level (n = 3). (G) No delayed recovery was observed from flies expressing PKC inhibitor from CrzR-Gal4 (P > 0.05, n = 5). (H) Inhibition of PKC did not affect ADH activity (p > 0.05, n = 3). (**P<0.005; *** P<0.001). Each data point represents mean ± sem for the indicated replicates. All genotypes are in y w background.
To elucidate signaling mechanisms underlying CrzR-regulated Adh transcription, transgenic manipulations were performed in the CrzR cells using a CrzR-gal4 line. Although we have been unsuccessful to obtain endogenous CrzR expression patterns using in situ hybridization, reporter gene expression patterns driven by the CrzR-gal4 lines were consistent with high throughput expression data from FlyAtlas (Flybase: flybase.org; see also ref. 28). Simultaneous expression of membrane-bound GFP (mCD8GFP) and nuclear RFP (Redstinger) showed respective florescent signals in the larval central nerve system (CNS) and salivary glands, but not in the larval fat body (Figure 7 A-C). Interestingly, unlike in the larval fat body, strong fluorescent signals were detected in the adult fat body, indicating developmental regulation of CrzR expression in this metabolic tissue (Figure 7D). Such spatial and developmental expression patterns agree with the FlyAtlas data. Moreover, since the fat body is the major target tissue for the neuroendocrine regulation of hemolymph trehalose levels [34] and Crz is likely to be involved in such physiological event [25], CrzR expression in the adult fat body was expected.
Figure 7. CrzR-gal4 driven reporter gene expression.
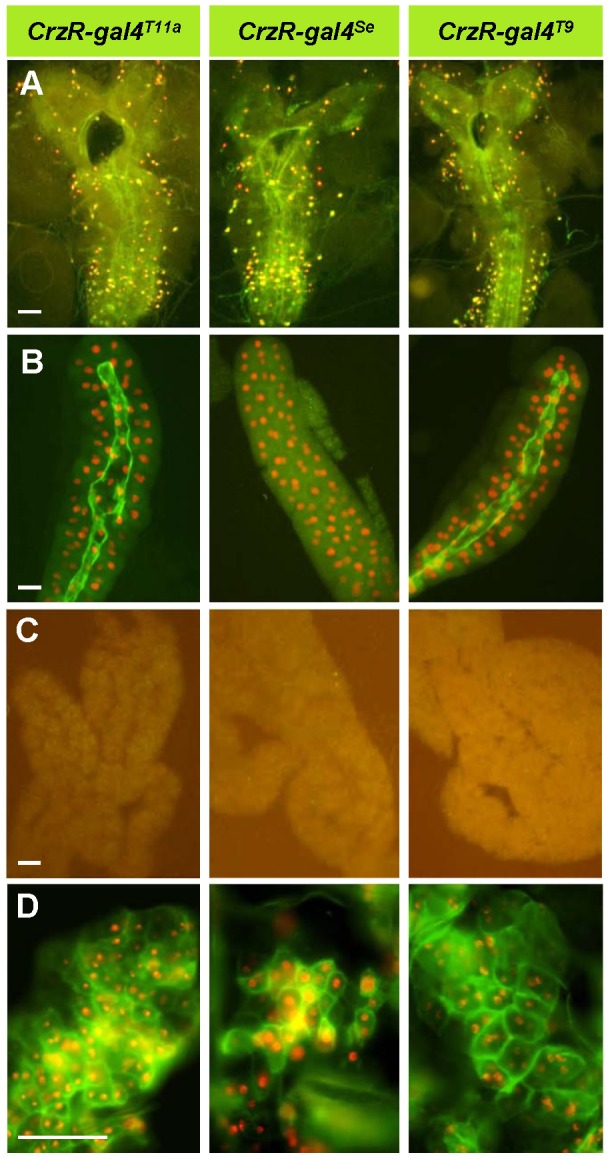
UAS-mCD8GFP;; UAS-Redstinger was expressed under the control of three CrzR-gal4 drivers as indicated. (A-C) Third instar larva. (A) CNS. (B) salivary gland. (C) fat body. (D) Adult fat tissue. Scale bar = 50 µm. (green, membrane bound GFP; red, nuclear RFP).
CrzR is a member of Class-A GPCR family [20], [29], [35]. A signaling pathway typical of these receptors involves PKA, which regulates activity of a transcription factor CREB via phosphorylation. Thus we explored whether this is true for the CrzR-regulated expression of Adh. To do this, PKA signaling was disrupted by ectopic expression of a PKA inhibitor (PKAin), a dominant-negative form of the regulatory subunit of a Drosophila PKA [36], or Cbz, a dominant negative CREB [37] using CrzR-gal4. Remarkably, hangover-like phenotypes of these flies were comparable to that of the CrzR01 mutant (Figure 6, C and D). In addition, the inhibition of PKA also increased ADH activity and Adh mRNA levels significantly (Figure 6, E and F), suggesting that CrzR-regulated Adh expression requires PKA-dependent pathway. In contrast, ectopic expression of the protein kinase C (PKC) inhibitor [38] showed a wild-type pattern of the hangover-like phenotype and ADH activity (Figure 6, G and H), indicating no positive role for PKC.
Discussion
Drinking alcohol is one of the oldest habits of humankind and has some positive effects on human society, but uncontrolled consumption causes serious psychopathic symptoms and other health problems. Excessive consumption of alcoholic beverage and associated hangovers cost $223.5 billion per year in the U.S. alone, due to losses in workplace productivity, health care expenses, costs associated with law enforcement and criminal justice, and accidents (http://www.cdc.gov/Features/AlcoholConsumption/).
Depending on the concentrations, effects of ethanol consumption in humans include euphoria, impaired motor function and speech, followed by vomiting, coma and even death in certain cases. Upon cessation of drinking, hangover is characterized by unpleasant physical pains such as headache, sensory problems such as vertigo, gastrointestinal symptoms such as nausea and vomiting, and disruption of sleep and biological rhythms [39]. These symptoms are mainly from acetaldehyde accumulation, as supported by high incidence of ethanol intoxication in Eastern Asian populations due to the polymorphic deficiency of functional ALDH2 allele [40], [41] and ALDH2-knockout mice [42]. In line with this, drugs inhibiting ALDH, such as disulfiram, have been used to treat chronic alcoholism by causing adverse symptoms from ethanol intake [43], [44]. Despite these reports, relatively little is known about the regulation of ethanol metabolism by physiological factors.
Like mammals, D. melanogaster metabolizes ethanol to acetaldehyde and acetate catalyzed by ADH and ALDH, respectively [45], and Adh and Aldh mutant flies showed dramatically reduced tolerance to ethanol [46]. Intriguingly, our present data suggest that a neuroendocrine system involving Crz plays an important role in regulating ADH and ALDH activities. CrzR activation leads to the PKA-dependent transcriptional repression of Adh and to post-transcriptional activation of ALDH via unknown pathways, which together suppress acetaldehyde accumulation.
In contrast to CrzR01, Crz-CD did not affect ADH activity levels, but reduced ALDH activity. Such a difference could explain that the Crz-CD flies displayed milder hangover-like phenotype than CrzR01 did. One might argue that Crz-CD still has residual Crz function. Although this is a possibility, undetectable Crz neurosecretory cells in the Crz-CD brains indicate near lack of Crz function. This is much more severe than RNAi-induced Crz knockdown with respect to the level of Crz expression [16]. Another caveat is that Crz-CD phenotype is due to an elimination of other co-existing transmitters. In fact, a subset of Crz neurons was shown to co-express small neuropeptide F (sNPF) [47]. However, since sNPF-producing neurons are so widely distributed in the brain, ablation of a few neurons co-expressing Crz and sNPF is unlikely to affect sNPF functions. Although more definitive evidence for the Crz await Crz-null mutant, our data with CrzR-null mutant support a role for Crz in the hangover-like phenotype.
Assuming that Crz-CD eliminates Crz function nearly entirely, we propose that CrzR mediates two separate signaling pathways; Crz-dependent up-regulation of ALDH activity and Crz-independent down-regulation of Adh transcription. The latter pathway might involve a distinct ligand that activates CrzR. One possible candidate is adipokinetic hormone (AKH), as the AKH receptor is structurally related to the CrzR with 56% amino acid sequence similarity [29]. However, CrzR showed very little affinity for AKH [35], indicating that AKH is unlikely to be a natural ligand for CrzR. Alternatively, CrzR might have an intrinsic activity that is not required for ligand binding. It is not uncommon that some GPCRs have intrinsic (or spontaneous) activities in the absence of ligand biding [48].
Although the identity of the second ligand for CrzR is speculative, a growing body of evidence suggests that a GPCR couples to different G-proteins in response to different agonists; such molecular flexibility is often referred to as “functional selectivity” of GPCR [49], [50]. According to this hypothesis, binding properties of different ligands induce and stabilize a unique conformational status of GPCR, which in turn shifts coupling preference to different G-proteins. Recently, CrzR isolated from the silkworm, Bombyx mori, was shown to couple dually to the Gq and Gs proteins in cell-based assays [51]. However, our results indicate that Gq-associated PKC activation is unlikely to be involved in the regulation of Adh transcription in Drosophila. Thus we propose that Gs-led PKA activation is the major in vivo signaling pathway of the CrzR at least for the Adh regulation. Nevertheless, it seems that CrzR is an excellent model system to unravel the physiological dynamics of the GPCR.
The International Agency for Research on Cancer (IARC) classified ethanol as a Group 1 carcinogen (http://monographs.iarc.fr/ENG/Classification/ClassificationsAlphaOrder.pdf). Accumulation of acetaldehyde as a result of defective ALDH2 is a major culprit of the carcinogenesis [52], [53]. In line with this, many cancer cells have disproportional activities of ADH and ALDH, making them less efficient in removing acetaldehyde compared to normal tissues [54]. In this regard, CrzR01 flies might be prone to cancer development, and perhaps can serve as an interesting model system to elucidate the mechanisms of ethanol-induced carcinogenesis.
Materials and Methods
Fly strains
Flies were reared on standard cornmeal/yeast medium at 25°C. The following strains were used: y w; Mi{ET1}GRHRIIMB00838 (Bloomington stock no. 22910), w1118; snaSco/SM6a, P{hsILMiT}2.4 (Bloomington stock no. 24613), w1118; UAS-PKAin (gifted from Ben White at NIH), w1118; UAS-PKCin (Bloomington stock no. 4589), y w;; UAS-cbz/TM3, Sb, Ser (Bloomington stock no. 7222). w1118;; Df(3L)BSC380/TM6C, Sb1 (for short Df; Bloomington stock no. 24404). The following transgenic lines were used to induce targeted ablation of Crz neurons: y w;; Crz-gal4 [25], y w; UAS-rpr, y w;; UAS-▵rpr, y w;; UAS-grim, and y w; UAS-hid/CyO, y+ [55].
CrzR null mutant and CrzR-gal4 line
Mobilization of the y w; Mi{ET1}GRHRIIMB00838 (Bloomington stock no. 22910) was used to induce CrzR mutation. Homozygous GRHRIIMB00838 females were crossed to males carrying a genomic source of heat-shock inducible Minos transposase (hsLMit2.4). Progenies were incubated at 37°C water bath for 1 h per day until pupariation. Fifty single male progeny with eyes showing mosaic pattern of GFP were crossed individually to a balancer y w;; Ly/TM6C, Sb, Tb females. Two GFP-negative progeny derived from each cross were randomly selected and crossed individually to the balancer to establish a total of 100 excision lines. Genomic DNA from homozygous flies was used for PCR-based screening to detect the deletion of the CrzR locus. For CrzR-gal4, 3.5-kb sequence upstream of the CrzR (–3130 to +331, +1 is transcription start site) was PCR amplified (primer sequences are shown in Table S1). This fragment was cloned into the pPTGAL vector at Xba I/Not I sites for germline transformation.
Behavior assays
Groups of 25 males (1–3 days old) were maintained in a food vial for 1–2 days and used for the following behavior assays.
Recovery from ethanol sedation. Flies were transferred into an empty plastic vial (O.D.×H: 25×95 mm) and then sealed with cotton plug. One ml of 100% ethanol was applied onto the cotton plug. Following 17–18 min of exposure, the cottons were replaced with fresh buzz plugs and then the vials were placed upside down. Flies capable of climbing up were considered “recovered” and the numbers of recovered flies were recorded every 10 min.
Toxicity assay. Assay was performed according to Barbancho et al [56], with slight changes. Groups of 20 males were placed in hermetically sealed plastic vials (O.D.× H: 25×95 mm) containing 3% sucrose medium supplemented with acetaldehyde. The toxicity of acetaldehyde was represented by the number of knocked-down flies as a function of exposure time.
Procedures of fecundity, circadian rhythm and ethyl ether recovery assays are provided in supporting protocol (Protocol S1).
Activity assays of ADH and ALDH
A hundred flies were homogenized in 1 ml of isolation buffer [50 mM sodium phosphate (pH 7.4), 0.24 M sucrose, 0.5 mM EDTA, 0.5 mM dithiothreitol, 0.001% (w/v) phenylthiourea], as described [57]. The homogenate was centrifuged for 10 min at 1000 rpm and then the supernatant was saved. The pellet was resuspended in 0.5 ml of the isolation buffer. Following centrifugation as before, the supernatant was combined with the first one. The pooled supernatant was centrifuged for 45 min at 14,000 rpm to separate cytosolic and mitochondrial fractions [58]. The pellet was washed with isolation buffer twice and resuspended in 250 µl of the same buffer supplemented with 1% (v/v) Triton-X-100. The resuspension was allowed to stand for 15 min and then centrifuged at 14,000 rpm for 20 min. The supernatant was used for mitochondrial fraction. All operations were performed on ice except centrifugation at 4°C. The protein content was determined by the BCA (Bicinchoninic acid) method. Three independent extracts per genotype were prepared. ALDH and ADH activities were assayed spectrophotometrically by measuring reduced β-NAD+ at 340 nm. ALDH activity assay was done following the method of Moxon et al. [59]. Briefly, the assay was started by adding 25 µl fly extract into 500 µl of 50 mM Tris/HCl (pH 8.6) including 3.6 mM acetaldehyde, 1 mM β-NAD+, and 20 mM pyrazole. The absorbance was recorded every min for up to 20 min by using Smartspect™ 3000 kinetics program (BioRad). ADH activity was assayed, as described in Barbancho et al. [56], by adding 20 µl extract into 500 µl of 50 mM Tris/HCl (pH 8.6) buffer containing 100 mM 2-propanol, 1 mM β-NAD+ and 1.6 mM cyanamide. The absorbance was recorded every 10 sec for up to 3 min. Succinate dehydrogenase activity was assayed by following the reduction of INT [2-(p-indophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium] [60]. The reaction was initiated by adding 2 µl of mitochondrial or 20 µl of cytosolic fraction into 200 µl substrate solution (0.11% w/v INT, 55 mM sodium succinate, 25 mM sucrose in 100 mM sodium phosphate, PH 7.4), followed by incubation in 37°C water bath for 15 min. The reaction was stopped by adding 200 µl of 3% ice-cold HCl. The chromogen was extracted with 600 µl ethyl acetate by centrifugation at 14,000 rpm for 5 min, and then its absorption was determined at 490 nm.
Measurement of acetaldehyde content
Forty flies were homogenized in 1 ml sterilized ice-cold water with 20 mM pyrazole and 1.6 mM cyanamide to inhibit ADH and ALDH activities. The homogenates were centrifuged at 14,000 rpm for 20 min at 4°C and the supernatant was kept in ice. Standard solutions were prepared from pure acetaldehyde in the same solution. Acetaldehyde content was measured using an Agilent 7890 gas chromatograph equipped with Agilent model G1888 headspace auto sampler, a flame ionization detector and a DB-624 capillary column (60 m x 0.32 mm x 1.8 µm). Separation of acetaldehyde was complete under the following conditions; injection volume 0.2 µl; split ratio 50:1; injection port temperature 200°C; oven temperature program: 60°C for 2 min, 25°C min−1 to 200°C for 1 min, FID temperature 300°C; carrier gas N2; and gas flow 3.0 ml/min (helium). For the auto sampler: auto sampler oven temperature 70°C; transfer line temperature 125°C; loop temperature 125°C; vial equilibration time 15 min; high shaking (mixing) speed; loop fill time 0.03 min; inject time 0.50 min; vial pressure 10 psig; pressurization time 0.5 min. All determinations were carried out in triplicates.
Quantitative RT-PCR
Total RNA was purified from 20 adult males using TRIzol reagent (Invitrogen) according to the manufacture’s protocol. About 5–10 µg of total RNA was added to 40 µl of Go Superscript reverse transcription mix (Promega) to generate cDNA using the oligo-dT-Ad primer (GACTCGAGTCGACATCGAT20). cDNA was then purified and eluted with 40-μl water using a PCR purification kit (Qiagen). SYBR Green reaction kit (Bio-Rad) was used for real-time PCR, with 0.5 µl cDNA included in 25 µl reaction volume. The PCR was carried out using an iQ™5 Multicolor Real-Time PCR Detection System (Bio-Rad). Amplifications were initiated with a 10-min denaturation at 95°C, followed by 40 cycles of 95°C, 15sec 55°C, 30 sec 72°C, 30 sec. Reactions were run in triplicates. Primers (Table S1) were designed to flank at least one intron to ascertain that the PCR products were derived from the cDNA. β-tubulin was used as an internal control to normalize expression levels of the target genes. Expression data were obtained using the 2−ΔΔCT method, as described by Livak and Schmittgen [61] where ΔΔCT equals the normalized cycle threshold (ΔCT) of Aldh or Adh in test genotypes minus the ΔCT of the same gene in control flies.
Histology
CrzR-gal4 line was crossed to UAS-mCD8GFP;; UAS-Redstinger. The resulting male offspring were dissected and incubated in fixative (4% paraformaldehyde in PBS) for 30 min at room temperature. Tissues were washed in PBS (3×5 min) and mounted in a quenching medium (0.5% n-propyl gallate in 90% glycerol and 10 mM phosphate buffer, pH 7.4). Differential fluorescent signals were captured by Olympus BX-61 microscope equipped with a CCD camera and then merged.
Statistics
Statistical analyses were done using Instat 2.0 (GraphPad Software). Student’s unpaired t-test was used to determine significant differences between two means. Difference among multiple means was tested by one-way ANOVA followed by Student-Newman-Keuls (SNK) multiple comparisons test.
Supporting Information
Fecundity assay. Numbers of eggs laid per fly were recorded per day (n = 8). No difference was found between CrzR01 and wild type females.
(PDF)
Actograms showing circadian locomotor activity rhythms. Flies were entrained for 3 days of 12:12 LD followed by 7 days of DD. A majority of CrzR01/Df (23/25; 92%) and Df/+(26/27; 96%) flies showed normal circadian rhythmic activities, with the mean period length (± sem) of 23.9 h (± 0.05) for CrzR01/Df and 23.7 h (± 0.07) for Df/+.
(PDF)
Knocking down of CrzR mRNA leads to severe hangover-like phenotype. (A) RT-PCR showed significant reduction of CrzR mRNA levels using two UAS-CrzRRNAi lines, S3S and T8. (B) Inducing CrzRRNAi with actin-gal4 driver (triangles) reuslted in severe hangover-like phynotype, compared to transgenic controls (circles). Each data point is a mean ± sem (n = 5). All genotypes are in y w background.
(PDF)
Recovery test from ethyl ether-induced sedation. (A) No obvious difference of the recovery rate was observed between Crz-CD flies (triangles) and controls (squares). Each data point represents mean ± sem (n = 3). (B) No delayed recovery was observed for CrzR01 mutant (n = 4).
(PDF)
Primers used.
(PPTX)
Supporting protocol.
(DOCX)
Acknowledgments
We would like to thank K. LeClair for her assistance on fecundity assay and Z. Yu for the CrzR-gal4 construct. We also thank J. May for DNA sequencing.
Funding Statement
This work was supported by National Science Foundation (NSF) grants (IOS-0919797, IBN-0133538) (http://www.nsf.gov/) and a Fite fellowship from BCMB (K.S.) (http://web.bio.utk.edu/bcmb/grad/financial_support/advanced.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bagnardi V, Blangiardo M, La Vecchia C, Corrao G (2001) Alcohol consumption and the risk of cancer: a meta-analysis. Alcohol Res Health 25: 263–270. [PMC free article] [PubMed] [Google Scholar]
- 2. Setshedi M, Wands JR, Monte SM (2010) Acetaldehyde adducts in alcoholic liver disease. Oxid Med Cell Longev 3: 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen YC, Peng GS, Wang MF, Tsao TP, Yin SJ (2009) Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chem Biol Interact 178: 2–7. [DOI] [PubMed] [Google Scholar]
- 4. Edenberg HJ (2007) The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health 30: 5–13. [PMC free article] [PubMed] [Google Scholar]
- 5. Crabb DW, Pinaire J, Chou WY, Sissom S, Peters JM, et al. (2001) Peroxisome proliferator-activated receptors (PPAR) and the mitochondrial aldehyde dehydrogenase (ALDH2) promoter in vitro and in vivo. Alcohol Clin Exp Res 25: 945–952. [PubMed] [Google Scholar]
- 6. Dannenberg LO, Chen HJ, Edenberg HJ (2005) GATA-2 and HNF-3beta regulate the human alcohol dehydrogenase 1A (ADH1A) gene. DNA Cell Biol 24: 543–552. [DOI] [PubMed] [Google Scholar]
- 7. Song BJ, Abdelmegeed MA, Yoo SH, Kim BJ, Jo SA, et al. (2011) Post-translational modifications of mitochondrial aldehyde dehydrogenase and biomedical implications. J Proteomics 74: 2691–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu M, Jerome RE, Edenberg HJ (1994) A negative regulatory element upstream from the mouse Adh-1 gene can down regulate a heterologous promoter. Gene 141: 249–254. [DOI] [PubMed] [Google Scholar]
- 9. Hillbom ME, Pikkarainen PH (1970) Liver alcohol and sorbitol dehydrogenase activities in hypo- and hyperthyroid rats. Biochem Pharmacol 19: 2097–2103. [DOI] [PubMed] [Google Scholar]
- 10. Israel Y, Videla L, Fernandez-Videla V, Bernstein J (1975) Effects of chronic ethanol treatment and thyroxine administration on ethanol metabolism and liver oxidative capacity. J Pharmacol Exp Ther 192: 565–574. [PubMed] [Google Scholar]
- 11. Mezey E, Potter JJ (1979) Rat liver alcohol dehydrogenase activity: effects of growth hormone and hypophysectomy. Endocrinology 104: 1667–1673. [DOI] [PubMed] [Google Scholar]
- 12. Potter JJ, Yang VW, Mezey E (1989) Influence of growth hormone on the synthesis of rat liver alcohol dehydrogenase in primary hepatocyte culture. Arch Biochem Biophys 274: 548–555. [DOI] [PubMed] [Google Scholar]
- 13. Simon FR, Fortune J, Iwahashi M, Sutherland E (2002) Sexual dimorphic expression of ADH in rat liver: importance of the hypothalamic-pituitary-liver axis. Am J Physiol Gastrointest Liver Physiol 283: G646–G655. [DOI] [PubMed] [Google Scholar]
- 14. Singh CM, Heberlein U (2000) Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol Clin Exp Res 24: 1127–1136. [PubMed] [Google Scholar]
- 15. Corl AB, Rodan AR, Heberlein U (2005) Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster . Nat Neurosci 8: 18–19. [DOI] [PubMed] [Google Scholar]
- 16. McClure KD, Heberlein U (2013) A small group of neurosecretory cells expressing the transcriptional regulator apontic and the neuropeptide corazonin mediate ethanol sedation in Drosophila. J Neurosci 33: 4044–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, et al. (1998) Ethanol intoxication in Drosophila: Genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell 93: 997–1007. [DOI] [PubMed] [Google Scholar]
- 18. Wen T, Parrish CA, Xu D, Wu Q, Shen P (2005) Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A 102: 2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veenstra JA (1989) Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett 250: 231–234. [DOI] [PubMed] [Google Scholar]
- 20. Sha K, Conner WC, Choi DY, Park JH (2012) Characterization, expression, and evolutionary aspects of Corazonin neuropeptide and its receptor from the House Fly, Musca domestica (Diptera: Muscidae). Gene 497: 191–199. [DOI] [PubMed] [Google Scholar]
- 21. Tawfik AI, Tanaka S, De Loof A, Schoofs L, Baggerman G, et al. (1999) Identification of the gregarization-associated dark-pigmentotropin in locusts through an albino mutant. Proc Natl Acad Sci U S A 96: 7083–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanaka Y, Hua Y, Roller L, Tanaka S (2002) Corazonin reduces the spinning rate in the silkworm, Bombyx mori. J Insect Physiol 48: 707–714. [DOI] [PubMed] [Google Scholar]
- 23. Kim YJ, Spalovska-Valachova I, Cho KH, Zitnanova I, Park Y, et al. (2004) Corazonin receptor signaling in ecdysis initiation. Proc Natl Acad Sci U S A 101: 6704–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi YJ, Lee G, Hall JC, Park JH (2005) Comparative analysis of Corazonin-encoding genes (Crz's) in Drosophila species and functional insights into Crz-expressing neurons. J Comp Neurol 482: 372–385. [DOI] [PubMed] [Google Scholar]
- 25. Lee G, Kim KM, Kikuno K, Wang Z, Choi YJ, et al. (2008) Developmental regulation and functions of the expression of the neuropeptide corazonin in Drosophila melanogaster. . Cell Tissue Res 331: 659–673. [DOI] [PubMed] [Google Scholar]
- 26. Tayler TD, Pacheco DA, Hergarden AC, Murthy M, Anderson DJ (2012) A neuropeptide circuit that coordinates sperm transfer and copulation duration in Drosophila. Proc Natl Acad Sci U S A 109: 20697–20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao Y, Bretz CA, Hawksworth SA, Hirsh J, Johnson EC (2010) Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PLoS One 5: e9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Veenstra JA (2009) Does corazonin signal nutrional stress in insects? Insect Biochem Molec Biol 39: 755–762. [DOI] [PubMed] [Google Scholar]
- 29. Cazzamali G, Saxild N, Grimmelikhuijzen C (2002) Molecular cloning and functional expression of a Drosophila corazonin receptor. Biochem Biophys Res Commun 298: 31–36. [DOI] [PubMed] [Google Scholar]
- 30.Choi SH (2009) The regulation of neuropeptide Corazonin and its functional analyses in Drosophila melanogaster. Tennessee Research and Creative Exchange Doctoral Dissertations.
- 31. Metaxakis A, Oehler S, Klinakis A, Savakis C (2005) Minos as a genetic and genomic tool in Drosophila melanogaster . Genetics 171: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yokoyama M, Yokoyama A, Yokoyama T, Funazu K, Hamana G, et al. (2005) Hangover susceptibility in relation to aldehyde dehydrogenase-2 genotype, alcohol flushing, and mean corpuscular volume in Japanese workers. Alcohol Clin Exp Res 29: 1165–1171. [DOI] [PubMed] [Google Scholar]
- 33. Leal JFM, Barbancho M (1992) Acetaldehyde detoxification mechanisms in Drosophila melanogaster adults involving aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH) enzymes. Insect Biochem Mol Biol 22: 885–892. [Google Scholar]
- 34. Lee G, Park JH (2004) Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster . Genetics 167: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Park Y, Kim YJ, Adams ME (2002) Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci U S A 99: 11423–11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodan AR, Kiger JA Jr, Heberlein U (2002) Functional dissection of neuroanatomical loci regulating ethanol sensitivity in Drosophila. J Neurosci 22: 9490–9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eresh S, Riese J, Jackson DB, Bohmann D, Bienz M (1997) A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J 16: 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Broughton SJ, Kane NS, Arthur B, Yoder M, Greenspan RJ, et al. (1996) Endogenously inhibited protein kinase C in transgenic Drosophila embryonic neuroblasts down regulates the outgrowth of type I and II processes of cultured mature neurons. J Cell Biochem 60: 584–599. [DOI] [PubMed] [Google Scholar]
- 39. Swift R, Davidson D (1998) Alcohol hangover: mechanisms and mediators. Alcohol Health Res World 22: 54–60. [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshida A, Huang IY, Ikawa M (1984) Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in orientals. Proc Natl Acad Sci U S A 81: 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crabb DW, Edenberg HJ, Bosron WF, Li TK (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 83: 314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Isse T, Oyama T, Kitagawa K, Matsuno K, Matsumoto A, et al. (2002) Diminished alcohol preference in transgenic mice lacking aldehyde dehydrogenase activity. Pharmacogenetics 12: 621–626. [DOI] [PubMed] [Google Scholar]
- 43. Chick J, Gough K, Falkowski W, Kershaw P, Hore B, et al. (1992) Disulfiram treatment of alcoholism. Br J Psychiatry 161: 84–89. [DOI] [PubMed] [Google Scholar]
- 44. Wright C, Moore RD (1990) Disulfiram treatment of alcoholism. Am J Med 88: 647–655. [DOI] [PubMed] [Google Scholar]
- 45. Gibson JB, May TW, Wilks AV (1981) Genetic variation at the alcohol dehydrogenase locus in Drosophila melanogaster in relation to environmental variation - ethanol levels in breeding sites and allozyme frequencies. Oecologia 51: 191–198. [DOI] [PubMed] [Google Scholar]
- 46. Fry JD, Saweikis M (2006) Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet Res 87: 87–92. [DOI] [PubMed] [Google Scholar]
- 47. Nässel DR, Enell LE, Santos JG, Wegener C, Johard HA (2008) A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. De Ligt RAF, Kourrunakis AP, Izerman AP (2000) Inverse agonism at G protein-coupled receptors: (patho)physiological relevance and implications for drug discovery. Brit J Pharmacol 130: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deupi X, Kobilka B (2007) Activation of G protein-coupled receptors. Adv Protein Chem 74: 137–166. [DOI] [PubMed] [Google Scholar]
- 50. Seifert R, Dove S (2009) Functional selectivity of GPCR ligand stereoisomers: new pharmacological opportunities. Mol Pharmacol 75: 13–18. [DOI] [PubMed] [Google Scholar]
- 51. Yang J, Huang H, Yang H, He X, Jiang X, et al. (2013) Specific activation of the G protein-coupled receptor BNGR-A21 by the neuropeptide corazonin from the silkworm, Bombyx mori, dually couples to the G(q) and G(s) signaling cascades. J Biol Chem 288: 11662–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boccia S, Hashibe M, Galli P, De Feo E, Asakage T, et al. (2009) Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev 18: 248–254. [DOI] [PubMed] [Google Scholar]
- 53. Seitz HK, Stickel F (2007) Molecular mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer 7: 599–612. [DOI] [PubMed] [Google Scholar]
- 54. Jelski W, Szmitkowski M (2008) Alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) in the cancer diseases. Clin Chim Acta 395: 1–5. [DOI] [PubMed] [Google Scholar]
- 55. Lee G, Kikuno K, Nair S, Park JH (2013) Mechanism of postecdysis-associated programmed cell death of peptidergic neurons in Drosophila melanogaster. . J Comp Neurol 521: 3972–3991. [DOI] [PubMed] [Google Scholar]
- 56. Barbancho M, Sanchez-Canete FJ, Dorado G, Pineda M (1987) Relation between tolerance to ethanol and alcohol dehydrogenase (ADH) activity in Drosophila melanogaster: selection, genotype and sex effects. Heredity (Edinb) 58 (Pt 3): 443–450. [DOI] [PubMed] [Google Scholar]
- 57. Heinstra PW, Geer BW, Seykens D, Langevin M (1989) The metabolism of ethanol-derived acetaldehyde by alcohol dehydrogenase (EC 1.1.1.1) and aldehyde dehydrogenase (EC 1.2.1.3) in Drosophila melanogaster larvae. Biochem J 259: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Leal JFM, Barbancho M (1993) Aldehyde dehydrogenase (Aldh) activity in Drosophila melanogaster adults - Evidence for cytosolic localization. Insect Biochem Mol Biol 23: 543–547. [DOI] [PubMed] [Google Scholar]
- 59. Moxon LN, Holmes RS, Parsons PA, Irving MG, Doddrell DM (1985) Purification and molecular properties of alcohol dehydrogenase from Drosophila melanogaster - Evidence from NMR and kinetic studies for function as an aldehyde dehydrogenase. Comp Biochem Physiol B Biochem Mol Biol 80: 525–535. [Google Scholar]
- 60. Pennington RJ (1961) Biochemistry of dystrophic muscle. Mitochondrial succinate-tetrazolium reductase and adenosine triphosphatase. Biochem J 80: 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fecundity assay. Numbers of eggs laid per fly were recorded per day (n = 8). No difference was found between CrzR01 and wild type females.
(PDF)
Actograms showing circadian locomotor activity rhythms. Flies were entrained for 3 days of 12:12 LD followed by 7 days of DD. A majority of CrzR01/Df (23/25; 92%) and Df/+(26/27; 96%) flies showed normal circadian rhythmic activities, with the mean period length (± sem) of 23.9 h (± 0.05) for CrzR01/Df and 23.7 h (± 0.07) for Df/+.
(PDF)
Knocking down of CrzR mRNA leads to severe hangover-like phenotype. (A) RT-PCR showed significant reduction of CrzR mRNA levels using two UAS-CrzRRNAi lines, S3S and T8. (B) Inducing CrzRRNAi with actin-gal4 driver (triangles) reuslted in severe hangover-like phynotype, compared to transgenic controls (circles). Each data point is a mean ± sem (n = 5). All genotypes are in y w background.
(PDF)
Recovery test from ethyl ether-induced sedation. (A) No obvious difference of the recovery rate was observed between Crz-CD flies (triangles) and controls (squares). Each data point represents mean ± sem (n = 3). (B) No delayed recovery was observed for CrzR01 mutant (n = 4).
(PDF)
Primers used.
(PPTX)
Supporting protocol.
(DOCX)