Abstract
An important issue in nanopore sensing is to construct stable and versatile sensors that can discriminate analytes with minute differences. Here we report a means of creating nanopores that comprise ultrashort single-walled carbon nanotubes inserted into a lipid bilayer. We investigate the ion transport and DNA translocation through single-walled carbon nanotube nanopores and find that our results are fundamentally different from previous studies using much longer single-walled carbon nanotubes. Furthermore, we utilize the new single-walled carbon nanotube nanopores to selectively detect modified 5-hydroxymethylcytosine in single-stranded DNA, which may have implications in screening specific genomic DNA sequences. This new nanopore platform can be integrated with many unique properties of carbon nanotubes and might be useful in molecular sensing such as DNA-damage detection, nanopore DNA sequencing and other nanopore-based applications.
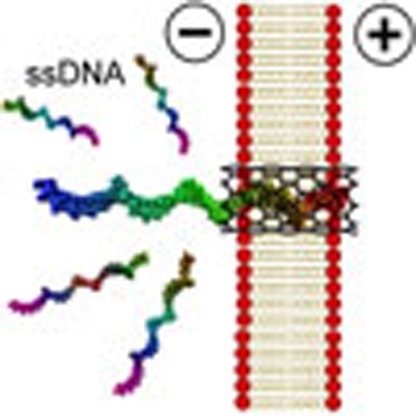 Nanopore sensors are a promising tool for the controlled detection of a range of possible substrates. Here the authors describe a nanopore sensor based on short single-walled carbon nanotubes inserted into a lipid bilayer, with modified sensing properties compared to longer nanotubes.
Nanopore sensors are a promising tool for the controlled detection of a range of possible substrates. Here the authors describe a nanopore sensor based on short single-walled carbon nanotubes inserted into a lipid bilayer, with modified sensing properties compared to longer nanotubes.
A nanopore sensor can be regarded as a Coulter counter at the single-molecule scale1. The first major breakthrough in nanopore sensing occurred in the mid 1990s when α-haemolysin (αHL) was used as a nanopore sensor. Kasianowicz et al.2 reported the investigation of the movement of polynucleotides through an αHL nanopore, which marked the beginning of the era of nanopore DNA sequencing. At about the same time, Bayley and coworkers3,4,5 started to develop stochastic sensing of a wide variety of analytes based on αHL and mutants. Another breakthrough came from the successful fabrication of solid-state (SS) nanopores with subnanometre control. Those pores are mostly drilled on silicon-based materials with ion-beam sculpting6 or electron-beam lithography7. The precise control of nanopore diameters and their inherent high durability have greatly expanded the scope of sensing substrates8. Recently, carbon nanotubes of micron length9, drilled graphene membranes10,11,12 and designed DNA nanostructures13 were also used as nanopores for DNA analysis. Despite the advances over the last two decades, to develop new sensors with unique functions such as high sensitivity and selectivity is still the central task in nanopore technology.
Here we report a means of creating nanopores that comprise ultrashort single-walled carbon nanotubes (SWCNTs) inserted into a lipid bilayer. We study the ion transport and single-stranded DNA (ssDNA) translocation through SWCNT nanopores and find that our results are fundamentally different from previous studies conducted with much longer SWCNTs9,14. The occurrence of exceptionally large conductance changes during ssDNA traversing SWCNT nanopores reveals that the ion mobilities inside SWCNTs are two to four times higher than the bulk mobility. Furthermore, we utilize the new SWCNT nanopores to selectively detect modified 5-hydroxymethylcytosine (5hmC) in ssDNA, which may have implications for high-throughput screening of genomic DNA fragments that contain 5hmC15. Our findings provide an effective and reliable strategy for constructing a new type of carbon-based sensors and demonstrate its potential for nucleic acid analysis. This new nanopore platform, integrated with many special properties of SWCNTs, is expected to be useful in molecular sensing and many other nanopore-based applications, such as ion channels16 and nanopore DNA sequencing17,18,19.
Results
Construction of SWCNT nanopores
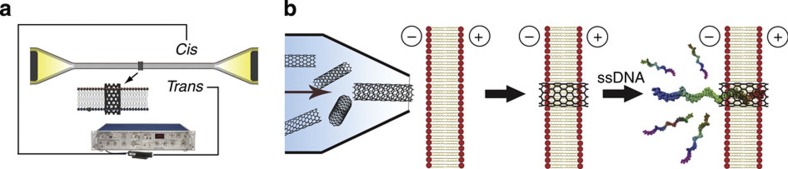
The ultrashort SWCNTs with diameter of 0.8–2.0 nm and length of 5–10 nm were obtained by cutting long SWCNTs (Supplementary Fig. S1) with sonication in concentrated sulphuric acid/nitric acid (3/1) and subsequent size-exclusion separation on high-performance liquid chromatography (HPLC). The setup for protein nanopore research was adopted for the current study (Fig. 1a). The thickness of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPc) bilayer is ∼5 nm; therefore, the ideal length of the SWCNTs for our nanopore system is approximately between 5 and 10 nm, which was proved by atomic force microscopy characterization and the insertion experiments with different HPLC fractions (Supplementary Figs S2–S5). However, there is still a remaining challenge for the nanopore system to be viable, that is, how to effectively insert a single SWCNT into the lipid bilayer. After several unsuccessful attempts, we eventually designed a micro-injection probe that could reliably insert sub-10 nm SWCNTs into lipid bilayer (Fig. 1b, Supplementary Fig. S6 and Supplementary Movies 1 and 2). On insertion, the tip of the probe with an injection rate of 1.0 μl min−1 was brought in close proximity to lipid bilayer but without touching it. After a current jump caused by the insertion occurred, the probe was slowly taken away from the bilayer to avoid any further interference. About two-thirds of the insertions afforded stable current signals straightway and the rest underwent a stabilizing period varying from several seconds to a few minutes (Supplementary Fig. S7). Less than 5% of the inserted SWCNTs showed persistent unstable current levels and were thus abandoned.
Figure 1. Schematic illustration of the setup and workflow.
(a) A patch-clamp amplifier was adopted to measure the currents through SWCNT nanopores. (b) Illustration of the insertion of a SWCNT into lipid bilayer with a micro-injection probe and ssDNA translocation through the inserted SWCNT.
Ion transport inside different SWCNT nanopores
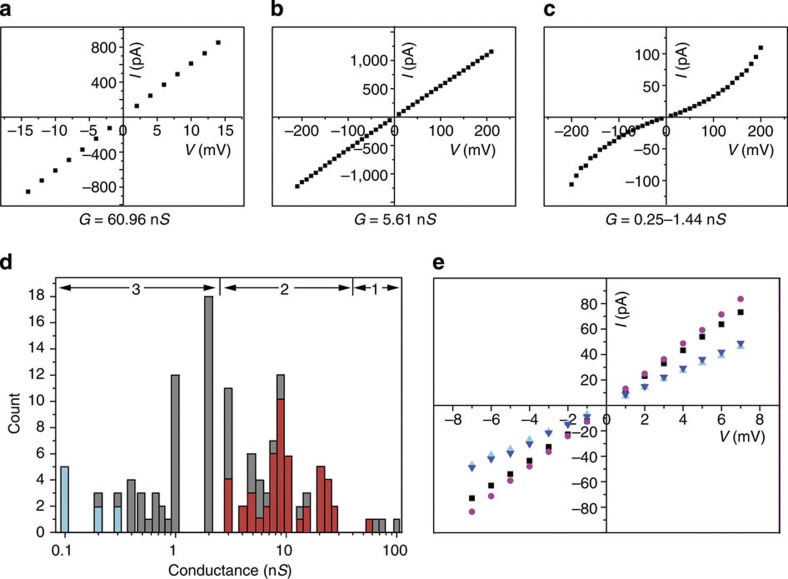
We first measured the conductance of SWCNT-based nanopores. Among the 180 nanopores that have been recorded, all the current traces show stable baseline and high signal-to-noise ratio (Supplementary Fig. S8). Three typical current–voltage (I–V) curves and their corresponding conductance values are shown in Fig. 2a–c. We did not observe any stochastic pore blocking by ions as reported by Strano and coworkers14, probably because the SWCNTs used in our experiments are ∼100,000 times shorter than theirs and the voltage applied in our SWCNT nanopores is also much lower than the threshold voltage in their measurements. The conductance of all the SWCNT nanopores we measured spans four orders of magnitude and can be roughly categorized into three groups (Fig. 2d). Group 1 has enormously large conductance of 60–100 nS, and group 3 has relatively small conductance of 0.1–2 nS, whereas group 2 of intermediate values (3–24 nS) is sandwiched in the middle (standard buffer conditions: 1 M KCl, 10 mM Tris, pH 8.0). The large conductance group contains all metallic SWCNTs, as suggested in a similar scenario in ref. 9. Conductance measurements using ultrashort pure metallic SWCNTs (IsoNanotubes-M) also afforded conductance values in the similar ranges to group 1.
Figure 2. Categorization of SWCNT nanopores.
(a–c) Representation of I–V curves and conductance value of a SWCNT nanopores of group 1–3, respectively. Data were acquired in the buffer of 1 M KCl and 10 mM Tris, pH 8.0. (d) Conductance distribution of SWCNT nanopores categorized in three groups 1–3. The SWCNTs that passed ssDNA molecules are marked in red (the unmarked tubes in group 2 were not tested for DNA translocation). The SWCNTs that showed nonlinear I–V curves are marked in turquoise. (e) I–V curves of a SWCNT nanopore in group 2 under four different pH conditions (pH 10.0, circle; pH 8.0, square; pH 3.44, inverted triangle; pH 2.89, triangle). The experiments were repeated in six different SWCNT nanopores.
The SWCNTs in group 2 and 3 are semiconductors, and the ones in group 2 have larger diameters that enable them to pass ssDNA, which we observed experimentally (Fig. 2d, red histogram). For electrolyte inside a nanotube with diameter of 1–2 nm and length of 5–10 nm, the calculated ionic conductance is between 1.18 and 9.40 nS without the consideration of free-energy barrier (Supplementary Methods). However, many measured conductance values in group 2 exceed this predicted range (Fig. 2d). This has prompted us to investigate whether there is a different ion transport mechanism in this new nanopore system, as will be discussed vide infra. Similarly, according to the conductance calculation, it appears that SWCNTs in group 3 have diameters <1 nm. This has been confirmed by Raman spectroscopy characterization (Supplementary Fig. S9 and Supplementary Table S1) and ssDNA translocation experiments, as none of the SWCNTs in group 3 could pass ssDNA. Another notable result obtained in group 3 is when the conductance of SWCNT nanopores decreases to 0.1–0.3 nS, the I–V curves become nonlinear but remain symmetrical for positive and negative potentials (Fig. 2c,d, turquoise histograms). A plausible explanation could be found in a simulation study by Peter and Hummer20, which shows that when ions transport through a nanopore with diameter <1 nm, the hydration layers around the ions need to be stripped off due to the spatial constrains, creating a large energetic penalty. It was previously shown that the ionic conductance through 2-μm-long SWCNTs was dependent on the pH of buffer solutions9. We also measured the ionic conductance of ultrashort SWCNT nanopores under different pH values (Fig. 2e). The results confirmed that the charged end groups (carboxylic acid residues) played a vital role in ion conduction through nanotubes.
ssDNA translocation through SWCNT nanopores
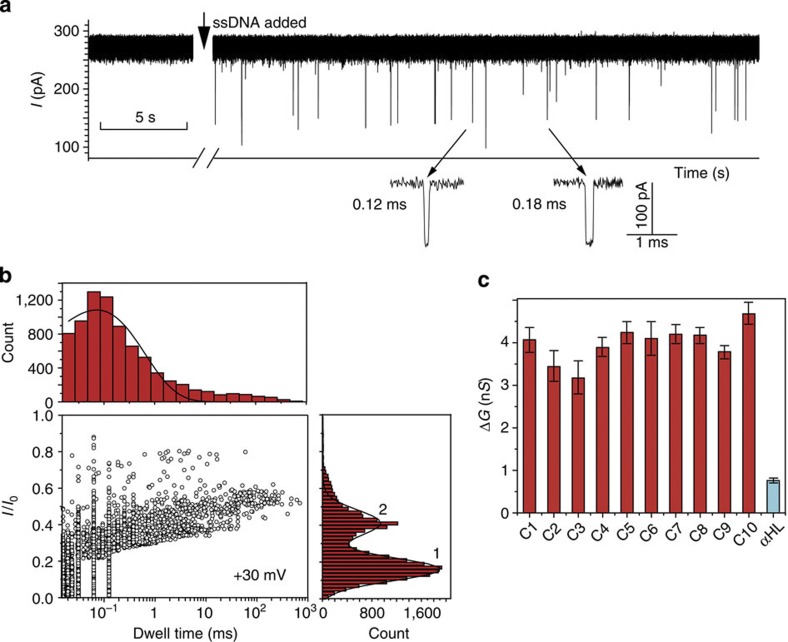
Next we examined ssDNA translocation through SWCNT nanopores, taking advantage of the ultrahigh sensitivity of PCR for detecting small numbers of molecules. We used 120-nucleotide oligomers (DNA1 in Supplementary Table S2. PCR protocol shown in Supplementary Table S3) with sequences predicted to have minimal secondary structures2. DNA solution was added in the cis compartment (final concentration 1.0 μM), which was connected to ground, and DNA molecules were driven through SWCNT nanopores under positive potentials. Three different scenarios could be observed when DNA oligomers were added for translocation through various SWCNT nanopores. First, the current trace remained quiescent if the conductance of the SWCNT nanopore was relatively small (≤2.0 nS). Second, transient current blockades of ∼20 pA started to occur frequently (Supplementary Fig. S10). Third, remarkably large current blockades appeared immediately after the addition of DNA (Fig. 3a). We identified these large-amplitude blockades as DNA translocation events by quantitative PCR analysis of the translocated DNA molecules (Supplementary Fig. S11). The short-lived 20-pA blockades are ascribed to DNA colliding with the ends of SWCNTs, as PCR showed negative results.
Figure 3. Translocation of ssDNA through SWCNT nanopores.
(a) Typical ionic current signal recorded when DNA1 (final concentration 1.0 μM) was added to the cis side of a SWCNT nanopore (GCNT=8.97 nS). The capture rate is ∼2,300 events per h. Data were acquired in the buffer of 1 M KCl and 10 mM Tris, pH 8.0, with the transmembrane potential held at +30 mV. (b) Scatter plot of the events (normalized current blockage I/I0 versus event durations) in a. The dwell-time histogram was constructed using the data points with I/I0>0.3. The solid line in dwell-time histogram is a mono-exponential fit to the data. Peak 1 in I/I0 histogram represents SWCNT–DNA colliding events and peak 2 represents DNA translocation events. The lines on peaks 1 and 2 are Gaussian fit to the histogram. (c) Comparison of conductance change values (ΔG) of DNA translocation through SWCNT nanopores (C1–10) and αHL. The error bars are defined as s.d. of at least three experiments. The conductance values of the nanopores are as follows: GCNT (1–10) 8.30, 8.39, 8.54, 8.65, 9.63, 16.41, 16.75, 24.59, 25.28 and 60.75 nS; GαHL=0.93 nS.
It is important to note that our observation of DNA translocation through ultrashort SWCNTs (5–10 nm) in 1 M KCl is entirely different from an earlier report9, in which DNA translocation through 2-μm-long SWCNTs was shown to be current-increasing spikes rather than current blockades. The mechanism accounting for this drastic difference requires further investigation. For comparison with other nanopore systems, we performed detailed studies of ssDNA translocation through different SWCNT nanopores (Fig. 3b, and Supplementary Figs S12 and S13). Two distinctive features are observed and discussed below.
First, the voltage required to drive DNA through SWCNT nanopores (10–40 mV) is much lower than that for αHL (≥80 mV) (refs 21, 22) and SS-nanopores (≥100 mV when dpore≈10 nm) (ref. 23). The diameter of SWCNTs in our experiments is close to or even smaller than that of the other two types of nanopores, which means the entropic barrier for DNA translocation in SWCNT nanopore should be similar to or higher than that of other nanopores. The difference in driving voltage must arise from DNA–nanopore interactions, which involve multiple weak force interactions within αHL and SS-nanopores, whereas in SWCNT nanopores DNA molecules are significantly attracted into SWCNTs through favourable interactions of the bases with the SWCNT pore wall24,25.
Second, the conductance change (ΔG) caused by DNA traversing SWCNT nanopores are exceptionally large (Fig. 3a) compared with that in αHL. The ΔG values of DNA translocation of SWCNT nanopores (ten examples) αHL under identical buffer conditions are listed in Fig. 3c. When a DNA molecule enters the nanopore, it causes two competing effects. On one hand, the conductance is decreased because of the steric exclusion of both cations and anions by DNA26; on the other hand, the counterions shielding the phosphate groups on the DNA backbone are introduced into the nanopore and thus enhance the conductance. According to a previously established model27, the conductance change due to the DNA translocation through SWCNT nanopores, ΔGCNT, can be expressed as:
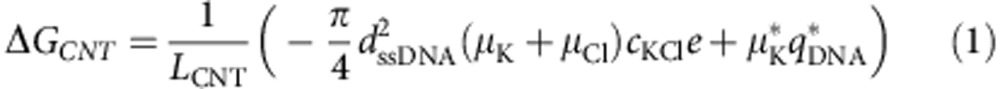 |
where LCNT represents the length of the SWCNT, dssDNA (∼1.1 nm) is the diameter of ssDNA, μK* is the effective electrophoretic mobility of potassium ions moving along the DNA and qDNA* is the effective charge on the DNA per unit length, which is assumed to be constant. For the ten SWCNT nanopores, we obtained the mean ΔGCNT=3.98±0.43 nS and LCNT=5–10 nm, whereas for αHL nanopore ΔGαHL=0.75 nS and LαHL≈10 nm. In high-ionic-strength electrolyte solution inside nanopores (cKCl=1 M, Dpore>1.1 nm), μK≈μCl. If we take an approximation that μK* equals the ion mobility inside nanopores μK (ref. 27), we can obtain ΔGpore∝μK/Lpore, that is, μK∝ΔGporeLpore. By comparing these two groups of data, we can conclude that the ion mobilities in SWCNT nanopores are enhanced roughly two- to four times higher than that in αHL, where ion mobilities are shown to be the same as bulk mobility. This conclusion is in qualitative agreement with a molecular dynamic simulation study by Peter and Hummer20, which showed an ∼50% increase in ion mobilities inside SWCNTs. Moreover, our deduced value seems to nicely coincide with the measurements of ion mobilities through membranes that contain a large number of SWCNT pores in a recent report16. However, caution must be taken in the comparison because of the distinct differences between the two systems, including SWCNT diameter (1.1–1.8 nm versus 0.9 nm) and length (5–10 nm versus 5 μm), salt concentration (1 M versus 6 mM) and single-channel versus ensemble measurements and so on.
Identification of chemically modified 5hmC in ssDNA
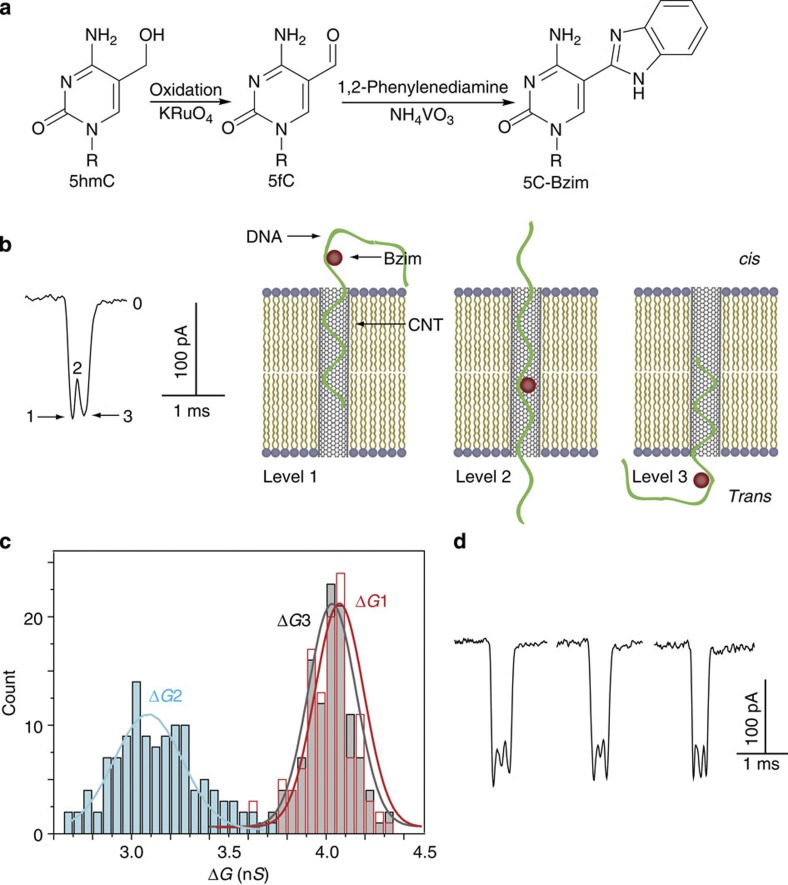
To explore the potential advantages of SWCNT nanopores, we carried out DNA-sensing experiments for identifying 5hmC in the sequence. 5hmC, now considered the sixth base, has been increasingly recognized not only as an intermediate in DNA demethylation but also for having important roles in epigenetic reprogramming and regulation of tissue-specific gene expression15. However, conventional bisulphite sequencing cannot discriminate 5hmC from 5-methylcytosine (5mC). Thus, various methods have been and are being developed to differentiate these two bases in DNA sequences28,29,30,31. Here we employed a strategy of a combination of chemical modification and nanopore sensing to selectively detect 5hmC in a short ssDNA. The two-step modification is bio-orthogonal and highly specific to 5hmC (Fig. 4a and Supplementary Fig. S14). We then used SWCNT nanopores to screen the modified DNAs (Supplementary Table S2), hoping to exploit their intrinsic high sensitivity. A characteristic type of current signature was observed when benzoimidazole (Bzim)-modified 5hmC-containing DNA2 was translocated through SWCNT nanopores (Fig. 4b). In this multilevel blockade, level 1 state represented either 3′- or 5′-end of DNA2–Bzim entering SWCNT nanopore, whereas the Bzim-modified part had not yet reached the nanopore. Level 2 with a shape of current-increasing spike immediately followed level 1 and represented the Bzim part entering and sliding through the nanopore. Finally, the current dropped to level 3, which was caused by the translocation of the rest of DNA2–Bzim, before returning to the full open-pore level (Fig. 4b). We plotted the histograms of the conductance difference between the three levels (ΔG1–3) and the open-pore level (Fig. 4c). It was seen that ΔG2 varied in a wider range than ΔG1 and ΔG3, while the latter two always remained at about the same height (for examples, see Supplementary Fig. S15). The frequency of signature events (fsig) was found to be proportional to the DNA2–Bzim concentration range from 10 to 100 nM (Supplementary Fig. S16a). The detection limit might be lowered for several orders of magnitude when asymmetric electrolyte conditions are used32,33,34. It should be noted that the proportion of the characteristic events is ∼10% of total events, probably because DNA degradation during the 5hmC–DNA oxidation step28 generated unmodified DNA fragments that retained in the final solution and contributed to the total number of DNA translocation events.
Figure 4. Detection of 5hmC in ssDNA using SWCNT nanopore.
(a) 5hmC modification scheme: oxidation28 of 5hmC to 5fC followed by oxidative condensation with 1,2-phenylenediamine to afford 5C–Bzim. The R group is DNA. (b) A typical multilevel current signature and diagram showing molecular mechanism of DNA2–Bzim translocation through SWCNT nanopore. Level 1: DNA2–Bzim entering the nanopore before Bzim-modified part reaches the pore; level 2: the Bzim part entering and sliding through the nanopore; level 3: translocation of the rest of DNA2–Bzim. The mean duration of 118 single-spike events is 406±111 μs. (c) Histograms of the conductance differences between level 1–3 and the open-pore level G0 in b (118 events in total). (d) New type of current signature caused by the translocation of DNA5–Bzim, which contains double 5hmC sites. The mean duration of 110 double-spike events is 536±161 μs. All the data were acquired in the buffer of 1 M KCl and 10 mM Tris, pH 8.0, with the transmembrane potential held at +40 mV.
To confirm that the presence of current signature was due to the modified 5hmC, we carried out control experiments in which 5hmC in DNA2 was replaced with 5mC or cytosine (C). The results showed that 5mC/C-containing DNAs were inert under the modification conditions and no characteristic current signature was observed in those control groups. In addition, we also examined DNA5, which contains two 5hmC sites (Supplementary Table S2). Translocation of DNA5–Bzim afforded a new type of current signature with double current-increasing spikes and its frequency was proportional to the DNA5–Bzim concentration between 10 and 100 nM (Fig. 4d and Supplementary Fig. S16b). This result further substantiated the correlation of the spikes in current signature with the modified 5hmCs in DNA.
Discussion
It has been demonstrated in literature that modified ssDNA could produce characteristic current signals when passing through αHL35. However, those modifiers were generally very bulky groups, which caused a further current blockade on top of the DNA-occupying level during translocation. To reveal whether Bzim-modified DNA can generate any kind of ‘signature events’ when translocated through αHL, we carried out DNA2–Bzim traversing αHL experiments under otherwise identical conditions for SWCNT nanopores. The results are shown in Supplementary Fig. S15b. It is interesting to see that DNA2–Bzim did produce a small fraction of ‘unusual current events’ (<10% of total events) during translocation through αHL. Those events all have a transient medium-level blockade before the DNA translocation level. Previous studies by Butler et al.36 had shown that this medium-conductance state was caused by DNA occupancy in the cis vestibule of αHL. It seems that the Bzim modification is not bulky enough to cause any conductance change during DNA translocation, but it somehow delays the process of DNA lingering in the cis vestibule and thus produces a marginally discernible conductance state in the events. However, the minor difference between these current events is not determinative enough for the identification of 5hmC in ssDNA using αHL. Interestingly, a very recent report also described the detection of chemically modified 5hmC in immobilized ssDNA with αHL nanopore37. Although modification of 5hmC with different chemical groups could be discriminated by residual current levels in nanopore analysis, this strategy suffered from the fact that the 5hmC has to be at a specific position in the ssDNA to be detected.
In comparison with αHL, identification of modified 5hmC in ssDNA through SWCNT nanopores can be achieved with high confidence owing to the characteristic current signals. Although the locations of 5hmC cannot be quantitatively determined from the signature events due to the exceedingly fast DNA translocation through SWCNT nanopores, this new sensor offers a simple and powerful tool for: (i) high-throughput screening of genomic DNA fragments that contain 5hmC and (ii) differentiating 5mC and 5hmC in bisulphite sequencing. This strategy can detect any 5hmC site in the DNA strand regardless of the local sequence context and might be extended to many other useful applications in chemistry and biology.
The successful fabrication of ultrashort SWCNT-based nanopores is another exciting advance in the development of nanopore technology. Currently, the major obstacle for SWCNT nanopore development is the complex composition of the as-grown CNT and precise manipulation of individual SWCNTs. The recent successes in the controlled growth of SWCNTs with single chirality38 or structure-specific separation of CNT mixtures39 might significantly improve the efficiency and reliability of the fabrication of SWCNT nanopores. Although the current device is based on lipid bilayer membrane, which is intrinsically fragile yet convenient to manipulate, the technology of embedding SWCNTs in solid support is readily available40. Many of the peculiar properties of CNTs can be integrated into nanopore to achieve special goals unobtainable from other nanopore systems. For instance, SWCNTs of different diameters with subangstrom precision provide a large repertoire of candidates for constructing various artificial ion channels, which nicely complements a recently reported approach that forms transmembrane channels in lipid bilayers with DNA-based nanostructures13. For another example, SWCNT nanopores of different chiralities may have special recognitions for different small molecules such as 20 amino acids, which provides the basis of protein sequencing. More generally, the electronic structure41 and ferromagnetic anisotropy of SWCNTs42 would allow the incorporation of gate voltage and magnetic interactions into the platform of SWCNT nanopores and investigate the controlled mass transport at the single-molecule level. The simplicity of the environment inside SWCNT nanopores and the perfect uniformity of the graphitic wall of SWCNTs would provide a great platform for developing many other nanopore-related applications, especially for nanopore DNA sequencing and DNA-damage sensing.
Methods
Materials
DPhPc was purchased from Avanti Polar Lipids (Beijing distributor). Wild type D8H6 αHL was produced by expression in BL21 (DE3) pLysS Escherichia coli cells and self-assembled into heptamers34. DNA1, DNA6-8 and (GT)29 oligomer were purchased from Sangon Biotech, Shanghai, and DNA2-5 oligomers were purchased from Takara Bio Inc. (Dalian). SWCNTs (90% purity) were purchased from Timesnano (Chengdu) and further purified in our laboratory. The SWCNTs were synthesized by chemical vapour deposition method, with the length of 5–30 μm and diameter of 1–2 nm. IsoNanotubes-M (metallic SWCNTs) was purchased from Nanointegris.
Preparation of short SWCNTs
The short SWCNTs were prepared in a two-step process. One hundred milligrams of the purified SWCNTs were suspended in 24 ml mixture of concentrated H2SO4/HNO3 (3/1) in a 250-ml conical flask and sonicated in a water bath for 48 h at 35–40 °C (ref. 43). The suspension was then diluted with 200 ml of water and neutralized with 6 M NaOH solution. The relatively long SWCNTs (>300 nm by scanning electron microscopy, CNT1) were collected on a 100-nm pore size filter membrane (PTFE; Millipore) and the shorter SWCNTs (CNT2) were collected in the filtrate. The solution was taken for centrifugal ultrafiltration (3 kDa UFC900308; Millipore) for 40 min and then Milli-Q water was added to dilute the solution for further centrifugal ultrafiltration. The washing and ultrafiltration were repeated three times before water was completely removed to afford a black powder as the product.
Length separation of short SWCNTs
We followed the protocol developed by Zheng and coworkers44 to conduct the length separation of short SWCNTs on HPLC. Both SWCNT samples of CNT1 and CNT2 were dispersed with DNA oligomer (GT)29 before HPLC separation. In a typical procedure, 0.8 mg of SWCNTs was suspended with 0.8 mg (GT)29 in 1 ml aqueous buffer solution (40 mM Tris, pH 7.0, 0.5 mM EDTA and 0.2 M NaCl). The mixture was kept in an ice-water bath and sonicated (Sonics, vibra cell) for 90 min. After sonication, the mixture was then centrifuged (Sigma 3K15) for 90 min at 16,000g to remove insoluble material, resulting in a DNA-dispersed SWCNTs solution. Length separation by HPLC was carried out using size-exclusion columns (COSMOSIL CNT, Nacalai Tesque Inc). We used two types of columns (dimension: 7.5 mm × 300 mm, pore size: 300 and 1,000 Å) connected in series to separate both CNT1 and CNT2 samples. Dispersed DNA–SWCNT solutions were filtrated through a 0.22-μm membrane and loaded (∼0.5 ml) onto the column series. The hybrid was eluted with pH 7.0 buffer containing 40 mM Tris, 0.5 mM EDTA and 0.2 M NaCl. Each fraction was collected at every 1-min interval.
Micro-injection probe
We attempted several approaches to solve the SWCNT insertion problem, including continuous stirring (up to 2 h), agarose gel probe45 and glass probe46, but none of them worked. Eventually, we designed a micro-injection probe that could reliably insert ultrashort SWCNTs into lipid bilayer. The micro-injection probe is controlled with a micromanipulator (Narishige Scientific Instrument Lab., Japan). The probe is composed of two main parts: a homemade micro-injector and a glass needle. The needle is made from a taper glass capillary tube stretched by a capillary puller. The opening of the needle has outer diameter of ∼20 μm and an inner diameter of 3–5 μm.
General procedure for 5hmC–DNA modification
Oxidation
About 20 μg of synthetic 5hmC–DNA (DNA2 or DNA5) was made up to 24 μl with NaOH (0.05 M final concentration) on ice, 1 μl of a KRuO4 (Alfa Aesar) solution (15 mM in 0.05 M NaOH final concentration) was added and the reaction was held on ice for 1 h, with occasional vortexing28. Next, 100 μl buffer (100 mM HEPES, 1 mM EDTA, pH 7.7) was added to the mixture. The reaction mixture was purified with a Micro Bio-SpinP6 column (Bio-Rad).
Condensation
Fifteen microlitres ammonium metavanadate (5.31 mM) (Alfa Aesar, Tianjin) and o-phenylenediamine (91.15 mM) containing buffer solution (100 mM HEPES, 1 mM EDTA, pH 7.7) was added to 5-formylcytosine DNA solution from the oxidation step. The reaction was held at room temperature for 2 h, with vortexing. Finally, the mixture was purified with a Micro Bio-Spin P6 column. The recovery rate of the modified 5hmC–DNA is ∼50% based on the comparison of the DNA concentrations before and after modification (the reaction yields are assumed to be 100%).
Single-channel current recording
A bilayer of DPhPc was formed across an aperture 100–150 μm in diameter in a 20-μm-thick polycarbonate film (Goodfellow, Malvern, PA) that divided a planar bilayer chamber into two compartments, cis and trans. Both compartments contained 1 ml of buffer. SWCNTs and DNA were added to the cis compartment, which was connected to ground. The trans compartment was connected to the head-stage of the amplifier. All experiments were carried out in 1 M KCl, 10 mM Tris, pH 8.0, at 22.5±2 °C, unless otherwise stated. Currents were recorded by connecting a patch-clamp amplifier (Axopatch 200B; Axon instruments, Foster City, CA) to an external eight-pole Bessel low-pass filter (type 901, Frequency Devices) operating at 29.9 kHz, and sampled at 100 kHz by a computer equipped with a Digidata 1440A/D converter (Axon instruments). For 5hmC–DNA-sensing experiments, data were acquired at 100 kHz with a 10-kHz low-pass Bessel filter.
Data analysis
Current traces were analysed with Clampfit 10.2 software (Axon Instruments). Events were detected using the Event Detection feature and used to construct amplitude and dwell-time histograms. Origin (Microcal, Northampton, MA) and Clampfit were used for histogram construction, curve fitting and graph presentation. Adobe Illustrator was used for making figures.
Author contributions
L.L. and C.Y. performed SWCNT preparation and characterization experiments. C.Y. performed DNA modification experiments. L.L., K.Z. and H.-C.W. performed single-channel recording experiments. L.L., J.L. and H.-C.W. performed data analysis. L.L. and H.-C.W. conceived the project, designed the experiments and wrote the paper.
Additional information
How to cite this article: Liu, L. et al. Ultrashort single-walled carbon nanotubes in a lipid bilayer as a new nanopore sensor. Nat. Commun. 4:2989 doi: 10.1038/ncomms3989 (2013).
Supplementary Material
Supplementary Figures S1-S16, Supplementary Tables S1-S3, Supplementary Methods and Supplementary References
Micro-injection probe interaction with the aperture.
SWCNT insertion into lipid bilayer.
Acknowledgments
We thank M. Zheng, M. Chen and T. Zeng for helpful discussions, T.S. Lin, Q. Yuan for assistance with quantitative PCR experiments and T. Li and Y.R. Li for assistance with SWCNT preparation and data analysis. This work was funded by the National Natural Science Foundation of China (numbers 21175135 and 21205119), National Basic Research Program of China (973 programme, numbers 2010CB933600, 2013CB932800 and 2013CB933704) and the ‘100 Talents’ programme of the Chinese Academy of Sciences.
References
- Coulter W. H. Means for counting particles suspended in a fluid. U.S. Patent 2,656,508 (1953).
- Kasianowicz J. J., Brandin E., Branton D. & Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L. Q., Braha O., Conlan S., Cheley S. & Bayley H. Stochastic sensing of organic analytes by a pore-forming protein containing a molecular adapter. Nature 398, 686–690 (1999). [DOI] [PubMed] [Google Scholar]
- Braha O. et al. Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 18, 1005–1007 (2000). [DOI] [PubMed] [Google Scholar]
- Movileanu L., Howorka S., Braha O. & Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 18, 1091–1095 (2000). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Ion-beam sculpting at nanometre length scales. Nature 412, 166–169 (2001). [DOI] [PubMed] [Google Scholar]
- Storm A. J., Chen J. H., Ling X. S., Zandbergen H. W. & Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2, 537–540 (2003). [DOI] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat. Nanotech. 2, 209–215 (2007). [DOI] [PubMed] [Google Scholar]
- Liu H. et al. Translocation of single-stranded DNA through single-walled carbon nanotubes. Science 327, 64–67 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaj S. et al. Graphene as a subnanometre trans-electrode membrane. Nature 467, 190–193 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant C. A. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 2915–2921 (2010). [DOI] [PubMed] [Google Scholar]
- Schneider G. F. et al. DNA translocation through graphene nanopores. Nano Lett. 10, 3163–3167 (2010). [DOI] [PubMed] [Google Scholar]
- Langecker M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Choi W., Han J.-H. & Strano M. S. Coherence resonance in a single-walled carbon nanotube ion channel. Science 329, 1320–1324 (2010). [DOI] [PubMed] [Google Scholar]
- Branco M. R., Ficz G. & Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13 (2012). [DOI] [PubMed] [Google Scholar]
- Wu J., Gerstandt K., Zhang H., Liu J. & Hinds B. J. Electrophoretically induced aqueous flow through single-walled carbon nanotube membranes. Nat. Nanotech. 7, 133–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotech. 4, 265–270 (2009). [DOI] [PubMed] [Google Scholar]
- Cherf G. M. et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-angstrom precision. Nat. Biotechnol. 30, 344–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C. & Hummer G. Ion transport through membrane-spanning nanopores studied by molecular dynamics simulations and continuum electrostatics calculations. Biophys. J. 89, 2222–2234 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson S. E., Misakian M., Robertson B. & Kasianowicz J. J. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 85, 3057–3060 (2000). [DOI] [PubMed] [Google Scholar]
- Meller A., Nivon L. & Branton D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86, 3435–3438 (2001). [DOI] [PubMed] [Google Scholar]
- Skinner G. M., van den Hout M., Broekmans O., Dekker C. & Dekker N. H. Distinguishing single- and double-stranded nucleic acid molecules using solid-state nanopores. Nano Lett. 9, 2953–2960 (2009). [DOI] [PubMed] [Google Scholar]
- Yeh I. C. & Hummer G. Nucleic acid transport through carbon nanotube membranes. Proc. Natl Acad. Sci. USA 101, 12177–12182 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulevich V., Kim S., Grigoropoulos C. P. & Noy A. Frictionless sliding of single-stranded DNA in a carbon nanotube pore observed by single molecule force spectroscopy. Nano. Lett. 11, 1171–1176 (2011). [DOI] [PubMed] [Google Scholar]
- Rabin Y. & Tanaka M. DNA in nanopores: Counterion condensation and coion depletion. Phys. Rev. Lett. 94, 148103 (2005). [DOI] [PubMed] [Google Scholar]
- Smeets R. M. M. et al. Salt dependence of ion transport and DNA translocation through solid-state nanopores. Nano Lett. 6, 89–95 (2006). [DOI] [PubMed] [Google Scholar]
- Booth M. J. et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336, 934–937 (2012). [DOI] [PubMed] [Google Scholar]
- Song C.-X. et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat. Methods 9, 75–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laszlo A. H. et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethyl-cytosine with nanopore MspA. Proc. Natl Acad. Sci. USA 110, 18904–18909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M., Morrison W., Rabin Y., Grosberg A. Y. & Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotech. 5, 160–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zheng D. L., Tan Q. L., Wang M. X. & Gu L. Q. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nat. Nanotech. 6, 668–674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen S. et al. Highly sensitive and selective DNA-based detection of mercury(II) with α-hemolysin nanopore. J. Am. Chem. Soc. 133, 18312–18317 (2011). [DOI] [PubMed] [Google Scholar]
- Mitchell N. & Howorka S. Chemical tags facilitate the sensing of individual DNA strands with nanopores. Angew. Chem. Int. Ed. 47, 5565–5568 (2008). [DOI] [PubMed] [Google Scholar]
- Butler T. Z., Gundlach J. H. & Troll M. Ionic current blockades from DNA and RNA molecules in the α-hemolysin nanopore. Biophys. J. 93, 3229–3240 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.-W., Gong L. & Bayley H. Single-molecule detection of 5-hydroxymethylcytosine in DNA through chemical modification and nanopore analysis. Angew. Chem. Int. Ed. 52, 4350–4355 (2013). [DOI] [PubMed] [Google Scholar]
- Li X. et al. Selective synthesis combined with chemical separation of single-walled carbon nanotubes for chirality selection. J. Am. Chem. Soc. 129, 15770–15771 (2007). [DOI] [PubMed] [Google Scholar]
- Tu X., Manohar S., Jagota A. & Zheng M. DNA sequence motifs for structure-specific recognition and separation of carbon nanotubes. Nature 460, 250–253 (2009). [DOI] [PubMed] [Google Scholar]
- Holt J. K. et al. Fast mass transport through sub-2-nanometer carbon nanotubes. Science 312, 1034–1037 (2006). [DOI] [PubMed] [Google Scholar]
- Iijima S. & Ichihashi T. Single-shell carbon nanotubes of 1-nm diameter. Nature 363, 603–605 (1993). [Google Scholar]
- Islam M. F., Milkie D. E., Torrens O. N., Yodh A. G. & Kikkawa J. M. Magnetic heterogeneity and alignment of single wall carbon nanotubes. Phys. Rev. B 71, 201401 (2005). [Google Scholar]
- Liu J. et al. Fullerene pipes. Science 280, 1253–1256 (1998). [DOI] [PubMed] [Google Scholar]
- Huang X. Y., McLean R. S. & Zheng M. High-resolution length sorting and purification of DNA-wrapped carbon nanotubes by size-exclusion chromatography. Anal. Chem. 77, 6225–6228 (2005). [DOI] [PubMed] [Google Scholar]
- Holden M. A. & Bayley H. Direct introduction of single protein channels and pores into lipid bilayers. J. Am. Chem. Soc. 127, 6502–6503 (2005). [DOI] [PubMed] [Google Scholar]
- Holden M. A., Jayasinghe L., Daltrop O., Mason A. & Bayley H. Direct transfer of membrane proteins from bacteria to planar bilayers for rapid screening by single-channel recording. Nat. Chem. Biol. 2, 314–318 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1-S16, Supplementary Tables S1-S3, Supplementary Methods and Supplementary References
Micro-injection probe interaction with the aperture.
SWCNT insertion into lipid bilayer.