Summary
In search of the ancestors of Native American mitochondrial DNA (mtDNA) haplogroups, we analyzed the mtDNA of 531 individuals from nine indigenous populations in Siberia. All mtDNAs were subjected to high-resolution RFLP analysis, sequencing of the control-region hypervariable segment I (HVS-I), and surveyed for additional polymorphic markers in the coding region. Furthermore, the mtDNAs selected according to haplogroup/subhaplogroup status were completely sequenced. Phylogenetic analyses of the resulting data, combined with those from previously published Siberian arctic and sub-arctic populations, revealed that remnants of the ancient Siberian gene pool are still evident in Siberian populations, suggesting that the founding haplotypes of the Native American A–D branches originated in different parts of Siberia. Thus, lineage A complete sequences revealed in the Mansi of the Lower Ob and the Ket of the Lower Yenisei belong to A1, suggesting that A1 mtDNAs occasionally found in the remnants of hunting-gathering populations of northwestern and northern Siberia belonged to a common gene pool of the Siberian progenitors of Paleoindians. Moreover, lineage B1, which is the most closely related to the American B2, occurred in the Tubalar and Tuvan inhabiting the territory between the upper reaches of the Ob River in the west, to the Upper Yenisei region in the east. Finally, the sequence variants of haplogroups C and D, which are most similar to Native American C1 and D1, were detected in the Ulchi of the Lower Amur. Overall, our data suggest that the immediate ancestors of the Siberian/Beringian migrants who gave rise to ancient (pre-Clovis) Paleoindians have a common origin with aboriginal people of the area now designated the Altai-Sayan Upland, as well as the Lower Amur/Sea of Okhotsk region.
Keywords: mtDNA variation, native Siberians, Native Americans
Introduction
The diffusion of the first modern humans in Siberia appears to have been restricted to the regions south of the 55°N parallel, where multiple Upper Paleolithic sites have been found and dated to 43,000-39,000 YBP (reviewed by Vasil’ev et al. 2002). Environmental conditions permitting human entry into the New World apparently existed shortly before and after 30,000 YBP and 13,000 YBP, when many areas in Siberia/Beringia remained ice-free and may have been periodically connected to the North American interior through an ice-free corridor, which was repeatedly buried under continental glaciers (Wright, 1991; West, 1996; Goebel et al. 2003; Pitulko et al. 2004). In addition, the first Americans could have spread along the Pacific Coast among the islands and bays of Alaska and Canada, at a time when the North American interior was an inhospitable, ice-covered wasteland (Dalton, 2003)
The affinity of southern Siberians with Native Americans is supported by anthropological (Kozintsev et al. 1999), dental (Turner, 1994) and genetic evidence, including paternally inherited Y chromosome polymorphisms (Lell et al. 2002) and autosomal HLA class II gene(s) variation (Uinuk-ool et al. 2002; Volodko et al. 2003). Intriguingly, the HLA class II gene frequencies separate Siberian/Asian/Native American populations into two clusters, one of which encompasses nearly all of the Siberian and all of the Native American populations, while the other consistes of central, eastern and southeastern Asians (Uinuk-ool et al. 2002). Thus, the debate continues about precisely where in Siberia the ancestors of Native Americans arose, and when and how they spread into the non-glaciated interior of Alaska, subsequently migrating southward into the American West.
Early studies of Native American mtDNA variation have shown that all Native American mtDNAs belong to haplogroups A, B, C, D and X, and that some of these haplogroups are also common along the northern Pacific Rim (Torroni et al. 1993a, b; Ward et al. 1993; Forster et al. 1996; Starikovskaya et al. 1998; Brown et al. 1998; Schurr et al. 1999). Specifically, the analysis of mtDNA diversity in the Chukchi and Siberian Eskimos of extreme northeastern Siberia revealed haplogroups A, C, and D. In contrast, the Koriak and Itelmen of the adjacent Kamchatka Peninsula, who speak a language from the same language phylum (Chukchi-Kamchatkan), harbour the east Eurasian haplogroups G, Z and Y, which are completely absent in Native Americans. Interestingly, the Aleuts of the Commander Islands, adjacent to the Kamchatka Peninsula, were founded by a single lineage of haplogroup D2 (Derbeneva et al. 2002a). While haplogroup B is absent in aboriginal populations of northwestern and northern Siberia (Derbeneva et al. 2002b, c), it has been found in populations restricted to the south-western and south-central periphery of the subcontinent (Sukernik et al. 1996; Derenko et al. 2000, 2003;).
The headwaters of the Ob, Yenisei, Lena and Amur rivers, the largest rivers in Eurasia, served as major routes out of Inner Asia where migrations initiated northward to the northern and eastern perimeters of former Beringia. Hence, populations from these areas might be expected to harbour the founding mtDNA lineages present in Native Americans. The occurrence of haplogroups A, B, C and D in southern Siberia and adjacent areas of Mongolia (Kolman et al. 1996; Sukernik et al. 1996; Derenko et al. 2000, 2003; Keyser-Tracqui et al. 2003), and the traces of haplogroup X mtDNAs revealed in the mountainous Altai (Derenko et al. 2001), supports this hypothesis. To further clarify the relationship between Siberian and Native American mtDNA haplogroups and subhaplogroups, we have undertaken a broad survey of mtDNA variation by high-resolution RFLP analysis, sequencing of the HVS-I, and surveying some additional diagnostic markers, in nine distinct populations that have evolved in the southern extent of Siberia. Finally, mtDNAs selected according to haplogroup/subhaplogroup status were then completely sequenced. The data obtained were integrated with similar data sets representing Siberian arctic and sub-arctic populations: the Mansi of the Lower Ob River basin, the Ket of the Lower Yenisei and the Nganasan of the Taimir Peninsula (Derbeneva et al. 2002b, c), the Chukchi and Siberian Eskimos of Chukotka (Starikovskaya et al. 1998) and the Itelmen and Koriak of Kamchatka (Schurr et al. 1999). Together, these data encompass all of the linguistic groups of indigenous Siberian populations, and support a dual Siberian origin for Native Americans: a migration derived from the Altai-Sayan Upland and a migration from the Lower Amur River/Sea of Okhotsk region.
Subjects and Methods
Populations and Samples
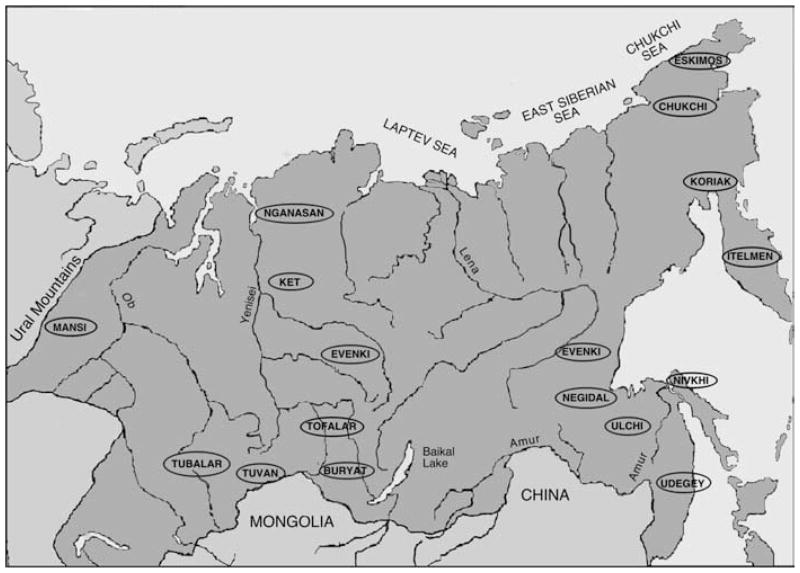
Blood samples were collected from nine indigenous populations with appropriate informed consent during multiple field expeditions conducted by Rem I. Sukernik, Elena B. Starikovskaya, and Natalia V. Volodko (Figure 1). The individuals who participated in these studies were interviewed and verified their family histories prior to blood being drawn, only from those subjects who were unrelated through at least three generations and lacked non-native maternal ancestors. A brief description of each population follows. Note that the Nivkhi, the Udegey and most of the Evenki samples are those previously surveyed by Torroni et al. (1993a). In that study, RFLP analysis with 14 restriction endonucleases and sequence analysis of a few selected HVS-I regions representing haplogroups A, C and D were performed.
Figure 1.
Approximate location of Siberian populations analysed for mtDNA variation.
Tubalar
The present study includes 72 Tubular, 27 individuals of whom were collected from the Tubalar admixed with Chelkan in the villages of Suronash, Tuloi and Artybash (Sukernik et al. 1996), while 45 are new samples drawn from the Tubalar in the villages of Pyzha, Tunzha, Paspaul, Salganda, Ynyrga, Kara-Koksha and Urlu-Aspak (Turochak and Choiski Districts, Altai Republic). The Tubalar and Chelkan are recent descendants of small hunting-gathering bands who a century ago occupied the coniferous forest zone (taiga) of northeastern Altai. They differ from numerous southern Altaians in culture, language, and physical appearance (Levin & Potapov, 1964).
Tuvan
The Tuvan are largely Turkic-speakers, and their total population size exceeds 100,000. A total of 95 Tuvan blood samples were collected across the Tuva Republic. Much of the Tuva Republic is situated in the south extreme of Siberia, with the taiga zone of Altai-Sayan ranges to the northwest, north and northeast, and the Mongolian arid sandy wastes to the south. Many families still perform nomadic/pastoral subsistence activities (Levin & Potapov, 1964).
Buryat
The Buryat are Mongolic-speakers who are regarded as a northern extension of the ethnic groups from Mongolia and former Manchuria (Levin & Potapov, 1964). At present, the total population size of the Russian Buryat is over 200,000. Geographically and culturally they are close to the Tuvan. Our sample consisted of 25 non-related individuals currently residing in Kushun village (Nizhneudinsk District, Irkutsk Region), and represents the Buryat of the western Baikal Upland. The family history of a few of the individuals sampled indicated Tofalar ancestry through the maternal line.
Tofalar (former Karagas)
The Tofalar are a small tribe of hunters and reindeer breeders inhabiting the northern slopes of the Sayan mountain range. The current population, which totals less than four hundred individuals, is subdivided into two territorial groups, one residing in the western (the village of Upper Gutara) and the other in the eastern part of mountainous Sayan (the villages of Nerkha and Alygdzher). Originally, the Tofalar, as well as the Tubalar and Chelkan tribes of the northeastern Altai, spoke a Samoyed language of the Uralic language family, but later adopted a Turkic language (Levin & Potapov, 1964). Our sample included 46 individuals collected from Upper Gutara and Nerkha. Several current residents of Nerkha were born in the adjoining village of Alygdzher in the same Nizhneudinsk District, Irkutsk Region.
Evenki
The present study includes 71 Evenki mtDNAs, of which 53 were collected from the Evenki living in the villages of Poligus and Surinda, located in the middle of the Stony Tunguska River basin (Torroni et al. 1993a), and 18 are new samples obtained from the Evenki inhabiting the Sea of Okhotsk region (Lell et al. 2002; Uinuk-ool et al. 2002). Most of the Evenki are hunters and reindeer breeders, with a total population of approximately 20,000. They speak the northern Tungusic language of the Altaic linguistic family, and inhabit the vast expanses of boreal forest extending from the Lower Yenisei in the west to the coast of the Okhotsk Sea in the east (Levin & Potapov, 1964).
Negidal
Thirty-three Negidal samples were analysed. They were collected from the village of Vladimirovka (Polina Osipenko District, Khabarovsk Region), and from the villages of Takhta, Tir, and Mago (Nikolaevskiy District, Khabarovsk Region). Genetically, the Negidals may represent the last remnants in the southern Okhotsk region of a culture based on the hunting of small sea mammals, which was first described by Middendorf (1869). In the late 1800s, several hundred Negidal lived in small settlements spread along the course of the Amgun, the left tributary of the Lower Amur, and the adjacent Tugur, which flow into the Sea of Okhotsk. Subsequently, they have been admixed and influenced by the expanding Tungusic Evenki and Even tribes, and now speak a dialect belonging to the Tungusic language group (Levin & Potapov, 1964).
Ulchi
We analysed 87 samples, obtained from elderly Ulchi residing in Old and New Bulava, two neighbouring villages (Ulchi District, Khabarovsk Region). Until recently, the Ulchi were a well-defined tribe of hunters and fishermen dispersed along the lakes and the reaches of the Lower Amur. They speak a language of the Tungusic-Manchurian group. The Ulchi sampled are representative of the few hundred individuals who are left from several small Ulchi villages (Levin & Potapov, 1964; Black, 1988) that are no longer in existence.
Nivkhi
The 56 Nivkhi were collected from the village of Nekrasovka and from the tiny fishing settlements located in the northwestern part of Sakhalin Island (Torroni et al. 1993a). In traditional times, the Nivkhi were small sea mammal hunters of the Lower Amur/Southern Okhotsk region and numbered several thousands. The Nivkhi is a language isolate with no known affiliation to existing language families (Levin & Potapov, 1964; Black, 1988). At present, this population is subdivided into two adjoining groups, one in the lower-most Amur River and Amur delta, and the other in the northern part of Sakhalin.
Udegey
We extended the analysis of 46 Udegey mtDNAs from the Gvasiugi village located in the central part of the Sikhote-Alin Range, Khabarovsk Region (Torroni et al. 1993a). This hunting-fishing group historically inhabited both slopes of the Sikhote-Alin Range, adjacent to the Sea of Japan, but are now on the brink of extinction. The Udegey language belongs to the southern branch of the Tungusic language group (Levin & Potapov, 1964; Krauss, 1988).
mtDNA Analysis
Genomic DNAs were extracted either from buffy coats or lymphoblast cell lines using standard procedures. To determine RFLP haplotypes, the entire mtDNA of each sample was amplified in ten overlapping PCR fragments (Torroni et al. 1993a; 1996). Each of the ten PCR segments was then digested with 14 restriction endonucleases (AluI, AvaII, BamHI, DdeI, HaeII, HaeIII, HhaI, HincII, HinfI, HpaI, MboI, MspI, RsaI, and TaqI). In addition, all mtDNAs were screened for the presence/absence of the BstNI site at nucleotide position (np) 13704, the AccI sites at nps 14465 and 15254, the BfaI site at np 4914, the NlaIII sites at nps 4216 and 4577, and the MseI sites at nps 14766 and 16297. The polymorphism at np 12308 was also surveyed by using a mismatched primer that generates a HinfI site when the A12308G mutation is present (Torroni et al. 1996). RFLP analysis was supplemented by sequencing of HVS-I, which was performed as described in Starikovskaya et al. (1998). In addition, selected mtDNAs representing major lineages and sub-lineages in Siberia were completely sequenced using cycle sequencing, “Big Dye Terminators” (ABI/Perkin-Elmer Cetus) dideoxy nucleotide terminators and an ABI Prism 3100 DNA Analyzer. Trace files were analysed using the Sequencher (v.4.0.5 GeneCode Corp.) software. During the course of the present and related studies 67 mtDNAs collected throughout Siberia and the adjacent Commander Islands were subjected to complete sequencing (Derbeneva et al. 2002a; Mishmar et al. 2003). Twenty of these 67 mtDNA sequences are new, and have been submitted to GenBank under accession codes AY519484–AY519497 and AY570524–AY570526, AY615359–615361.
Categorization of Haplotypes and Haplogroups
The Siberian mtDNA types were categorized into haplogroups denoting monophyletic clusters of mitochondrial sequences that arose on the ancestral haplotype background (Torroni et al. 1993a, 1994a, 1996; Forster et al. 1996; Macaulay et al. 1999; Finnila et al. 2001; Derbeneva et al. 2002a, b, c; Herrnstadt et al. 2002; Mishmar et al. 2003). “Founder” or “ancestral” haplotypes (versus “derived” haplotypes) are those from which all other related haplotypes were “derived” through acquisition of new mutations. Haplogroups (subhaplogroups) are also referred to as mtDNA lineages of related mtDNA haplotypes. “Candidate” founders for Native American mtDNA haplogroups were identified by comparing the polymorphisms of similar haplotypes from Eurasia and the Americas. Relationships were confirmed and extended using complete mtDNA sequences.
Phylogenetic analysis
Phylogenetic relationships between 16 Siberian and two adjacent Native American populations of the North Pacific Rim (Aleut and Haida) were determined by the neighbor-joining algorithm using DA distances:
where m is the number of subhaplogroups across populations and xi and yi are frequencies of the i-th mtDNA subhaplogroups in populations X and Y, respectively. DA distance is more efficient in obtaining a correct tree topology for a small number of loci (Takezaki & Nei, 1996).
Results
mtDNA Diversity
The mtDNAs of 531 maternally unrelated individuals from nine native Siberian populations were characterized by RFLP analysis and HVS-I sequencing (Table 1). The populations studied extend from the Northeastern Altai in the west to the Lower Amur/Sea of Okhotsk region in eastern Siberia. The overall distribution of mtDNA subhaplogroups across Siberia is shown in Table 2. Approximately, 66% of southern Siberian mtDNAs were found to belong to the “Asian” macro-haplogroup M (defined by presence of the 10394 DdeI and 10397 AluI sites) and its derivatives. The remaining mtDNAs belonged to the macro-haplogroup N, lacking both the 10394 DdeI and 10397 AluI sites, or alternatively having the 10394 DdeI site but lacking the 10397 AluI site.
Table 1.
mtDNA diversity in nine Siberian populations
| Haplogroup | RFLPs | HVS-I (−16000) | Populations and number of subjectsa
|
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TB | TV | BR | TF | EV | NG | UL | NV | UD | ||||
| A1 | +663e | 192 223 290 319 362 | 8 | 8 | ||||||||
| +663e | 242 290 293 319 | 1 | 1 | |||||||||
| +663e | 086 223 290 293 319 362 | 1 | 1 | |||||||||
| +663e +15606a | 039 189 223 290 319 356 362 | 1 | 1 | |||||||||
| +663e +15606a | 039 189 218 223 290 319 356 362 | 1 | 1 | |||||||||
| +663e −9751l/+15606a | 039 189 223 290 319 356 362 | 1 | 1 | |||||||||
| +663e −9751l/+15606a −15883e | 039 189 223 290 319 356 362 | 1 | 1 | |||||||||
| B1 | 9bp del −6022a | 086 136 189 217 519 | 1 | 1 | ||||||||
| 9bp del −6022a −9052n/+9612k | 086 136 189 217 519 | 2 | 1 | 3 | ||||||||
| B3 | 9bp del +13940f | 189 217 240 519 | 1 | 1 | ||||||||
| B4 | 9bp del | 167 189 217 261 317T 519 | 4 | 2 | 6 | |||||||
| 9bp del −14766u | 129 189 217 261 356 | 1 | 1 | |||||||||
| B5 | 9bp del (+/−) | 140 189 243 519 | 1 | 1 | ||||||||
| 9bp del (+/−) +14465s | 140 148 189 218 266 243 519 | 1 | 1 | |||||||||
| 9bp del (+/−) +4895k (+/−) −10971g +15221j −15925l | 111 129 140 189 234 243 244 463 519 | 4 | 4 | |||||||||
| F | +4732k −12406h/−12629b | 189 232A 249 304 311 519 | 1 | 1 | 1 | 3 | ||||||
| +4732k −12406h/−12629b | 172 189 232A 249 286 304 311 | 3 | 3 | |||||||||
| +4732k −8391e −12406h/−12629b | 172 189 232A 249 286 304 311 | 1 | 1 | |||||||||
| −12406h/ | 189 304 327 519 | 3 | 3 | |||||||||
| +545g −9052n/−12406h/−13268g | 162 172 304 519 | 1 | 1 | |||||||||
| Y | +7933j −8391e (+/−) | 126 231 266 304 519 | 1 | 1 | 2 | |||||||
| +7933j −8391e (+/−) −13608l | 126 231 266 304 519 | 1 | 1 | |||||||||
| −7641a +7933j −8391e (+/−) | 126 231 266 304 380 519 | 1 | 1 | |||||||||
| +7933j −8391e (+/−) | 126 189 231 519 | 11 | 11 | |||||||||
| +7933j −8391e (+/−) | 126 189 231 266 519 | 5 | 18 | 10 | 2 | 35 | ||||||
| +7933j −8391e (+/−) | 126 189 266 519 | 1 | 1 | |||||||||
| +7933j −8391e (+/−) | 126 189 231 292 519 | 3 | 3 | |||||||||
| +7933j −8391e (+/−) | 126 189 266 292 519 | 1 | 1 | |||||||||
| +7933j −8391e (+/−) | 126 189 231 266 292 519 | 2 | 2 | |||||||||
| +7933j −8391e (+/−) | 126 189 231 266 294 519 | 1 | 1 | 2 | ||||||||
| +7933j −8391e (+/−) | 126 140 189 231 266 399 519 | 1 | 1 | |||||||||
| −322e +7933j −8391e (+/−) | 126 189 231 266 519 | 5 | 5 | |||||||||
| −322e +7933j −8391e (+/−) | 093h 126 189 231 266 519 | 1 | 1 | |||||||||
| −322e +7933j −8391e (+/−) | 126 189 231 266 294 519 | 1 | 1 | |||||||||
| −322e +7933j −8391e (+/−) | 126 189 266 519 | 1 | 1 | |||||||||
| +7933j −8391e (+/−) 4bp ins | 126 189 231 519 | 4 | 4 | |||||||||
| +7933j −8391e (+/−) +11900k | 126 189 266 292 519 | 1 | 1 | |||||||||
| +7933j −8391e (+/−) +11900k | 126 189 231 266 292 519 | 4 | 4 | |||||||||
| +7933j −8391e (+/−) +14168l | 126 189 231 266 519 | 2 | 1 | 3 | ||||||||
| −853c/+7933j −8391e (+/−) −15375g | 126 189 231 519 | 1 | 1 | |||||||||
| H | −7025a −14766u | 519 | 1 | 1 | ||||||||
| −7025a −14766u | 288 362 | 3 | 3 | |||||||||
| −7025a −14766u | 311 519 | 3 | 3 | |||||||||
| −7025a −9380f −14766u | 362 519 | 1 | 1 | |||||||||
| −7025a −7598f +14249f −14766u | 093 129 316 519 | 1 | 1 | |||||||||
| V | −4577q −14766u +15904u | 153 298 | 1 | 1 | ||||||||
| J | +4216q (+/−) −13704t | 069 126 519 | 1 | 1 | ||||||||
| +4216q (+/−) −13704t | 069 126 145 172 261 278 | 1 | 1 | |||||||||
| +4216q −4685a (+/−) +11001n/ | ||||||||||||
| +11414a −13704t−15254s | 069 126 241 | 1 | 1 | |||||||||
| U4 | +4643k +11329a +12308g | 356 519 | 10 | 1 | 11 | |||||||
| +4643k +11329a +12308g | 311 356 519 | 3 | 3 | |||||||||
| U5 | +12308g | 189 260 270 519 | 2 | 2 | ||||||||
| +12308g | 192 241 256 270 287 304 325 399 | 4 | 4 | |||||||||
| +1718a +12308g +13634s | 093 234 270 | 3 | 3 | |||||||||
| U* | +12308g | 129C 189 214 258 362 519 | 2 | 2 | ||||||||
| W | +8249b/−8994e (12705) | 104 223 284 519 | 2 | 2 | ||||||||
| X | (−/−) +14465s (12705) | 189 223 278 519 | 1 | 1 | ||||||||
| N9 | (5417) (12705) | 111 129 223 257A 261 | 3 | 3 | ||||||||
| (5417) (12705) | 223 248 257A 261 311 519 | 5 | 5 | |||||||||
| (5417) −5742i (12705) | 189 223 | 2 | 2 | |||||||||
| (5417) −5742i (12705) | 129 189 223 311 | 1 | 1 | |||||||||
| (5417) (12705) −14258m/ | 223 519 | 3 | 14 | 17 | ||||||||
| R6 | (−/−) −12282a | 129 184i 190i 362 390 519 | 1 | 1 | ||||||||
| R* | (−/−) | 145 192 243 309 390 519 527 | 1 | 1 | ||||||||
| C1a | −7598f (+/+) −13259o/ | 223 298 325 327 356 | 1 | 1 | ||||||||
| C2 | (+/+) −13259o/ | 223 298 327 519 | 8 | 1 | 9 | |||||||
| −5983g/(+/+) −13259o/ | 223 298 327 519 | 2 | 2 | |||||||||
| (+/+) −13259o/+15606a | 223 298 327 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 129 223 298 327 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 223 264 298 327 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 223 298 311 327 519 | 2 | 2 | 3 | 5 | 12 | ||||||
| (+/+) −13259o/ | 219 223 298 327 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 223 294 298 311 327 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 223 291 298 327 519 | 1 | 2 | 3 | ||||||||
| (+/+) −13259o/ | 223 261 298 519 | 1 | 1 | |||||||||
| (+/+)−12170g/−13259o/ | 223 249 291 295 298 327 519 | 2 | 2 | |||||||||
| +8249b/(+/+) −13259o/ | 223 298 327 519 | 10 | 5 | 5 | 20 | |||||||
| +8249b/(+/+) −13259o/ | 093 223 298 327 519 | 1 | 1 | |||||||||
| +8249b/(+/+) −13259o/ | 093 129 223 298 327 519 | 1 | 1 | |||||||||
| +8249b/(+/+) −13259o/ | 190 223 232 298 327 519 | 1 | 1 | |||||||||
| +8249b/(+/+) −13259o/ | 223 259+A 298 327 519 | 2 | 2 | |||||||||
| +8249b/(+/+) −13259o/ | 223 262+C 291 298 327 519 | 2 | 2 | |||||||||
| −1715c (+/+) −13259o/ | 150 223 298 327 519 | 1 | 1 | |||||||||
| −1715c (+/+) −13259o/ | 129 150 223 298 327 519 | 1 | 1 | 1 | 3 | |||||||
| −1715c (+/+) −13259o/ | 093 129 223 298 327 519 | 1 | 1 | 2 | ||||||||
| −1715c (+/+) −13259o/ | 129 223 298 327 519 | 1 | 2 | 1 | 1 | 1 | 6 | |||||
| −1715c (+/+) −13259o/+15606a | 093 129 223 298 327 519 | 8 | 3 | 7 | 1 | 19 | ||||||
| −1715c (+/+) −13259o/+15606a | 093 129 223 235 298 327 390 519 | 10 | 10 | |||||||||
| −1715c −6850i (+/+) −13259o/+15606a | 093 129 223 298 327 519 | 1 | 1 | |||||||||
| −1715c (+/+) −13259o/+15606a | 129 223 298 327 519 | 4 | 4 | |||||||||
| −1715c −6850i (+/+) −13259o/+15606a | 129 223 298 327 519 | 2 | 2 | |||||||||
| −1004o −1715c (+/+) −13259o/ | 129 223 298 327 519 | 2 | 4 | |||||||||
| (+/+) −13259o/ | 223 298 327 344 357 519 | 1 | 2 | 2 | 5 | |||||||
| (+/+) −13259o/ | 171 223 327 344 357 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 171 223 298 327 344 357 519 | 1 | 2 | 3 | 6 | |||||||
| (+/+) −13259o/ | 171 223 224 298 327 344 357 519 | 2 | 2 | 4 | ||||||||
| (+/+) −13259o/ | 171 209 223 298 327 344 357 519 | 1 | 1 | |||||||||
| −1715c (+/+) −13259o/ | 093 129 167 223 298 327 357 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 171 188 223 261 298 327 344 357 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 167 223 298 327 344 357 519 | 1 | 1 | |||||||||
| −5983g/(+/+) −13259o/ | 171 223 298 327 344 357 519 | 2 | 2 | 4 | ||||||||
| (+/+) −13259o/ | 093 171 223 298 327 344 357 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 129 140 171 223 298 327 344 357 519 | 1 | 1 | |||||||||
| C3 | −1413l +8249b/(+/+) −13259o/ | 093 223 288 298 327 390 519 | 1 | 1 | ||||||||
| −1413l (+/+) −13259o/ | 093 223 288 298 327 390 519 | 7 | 7 | |||||||||
| (+/+) −13259o/ | 223 261 288 298 519 | 3 | 5 | 8 | ||||||||
| (+/+) −13259o/ | 223 288 298 327 390 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 093 223 288 298 327 390 519 | 1 | 1 | |||||||||
| (+/+) −13259o/(3460) | 093 223 288 298 327 390 519 | 1 | 1 | |||||||||
| (+/+) −13259o/ | 093 223 288 291 298 327 518T 519 | 2 | 2 | |||||||||
| (+/+) −13259o/ | 093 223 288 298 327 390 500A 519 | 1 | 1 | |||||||||
| −1715c +3397k (+/+) −13259o/9bp ins | 148 223 288 298 327 519 | 1 | 3 | 4 | ||||||||
| −1715c (+/+) −13259o/9bp ins | 148 223 288 298 327 519 | 1 | 2 | 3 | ||||||||
| +1718e −7297e (+/+) −13259o/ | 148 164 223 288 298 327 519 | 2 | 2 | |||||||||
| D | −5176a (+/+) | 223 362 | 6 | 1 | 7 | |||||||
| −5176a (+/+) | 223 291 362 | 1 | 3 | 4 | ||||||||
| −5176a (+/+) | 223 362 368 | 1 | 1 | |||||||||
| −5176a (+/+) | 174 223 362 | 4 | 4 | |||||||||
| −5176a (+/+) | 223 362 519 | 2 | 2 | |||||||||
| −5176a (+/+) | 189 223 362 519 | 1 | 1 | |||||||||
| −5176a (+/+) | 223 263 362 519 | 4 | 4 | |||||||||
| −5176a (+/+) | 145 311 362 368 | 1 | 1 | |||||||||
| −5176a (+/+) | 223 274 362 368 | 2 | 1 | 3 | ||||||||
| −5176a (+/+) | 223 245 311 362 368 | 2 | 2 | |||||||||
| −5176a (+/+) | 093 223 232 290 362 | 1 | 1 | |||||||||
| −5176a (+/+) | 148 223 263 362 519 | 1 | 1 | 2 | ||||||||
| −5176a −5971f (+/+) | 223 362 | 1 | 1 | |||||||||
| +1973g −5176a (+/+) | 223 286 362 | 1 | 1 | |||||||||
| −5176a (+/+) +14583c | 223 245 362 | 1 | 1 | |||||||||
| −5176a −8074a (+/+) | 223 286 362 | 1 | 1 | |||||||||
| −5176a −9294e (+/+) | 223 362 519 | 1 | 1 | |||||||||
| −5176a (+/+) +14465s (14470) | 223 362 519 | 1 | 1 | |||||||||
| −5176a −7859j (+/+) | 223 263 362 519 | 2 | 2 | |||||||||
| −5176a (+/+) +10407k | 129 223 362 519 | 2 | 2 | |||||||||
| −5176a +7979e (+/+) | 184 223 311 362 | 1 | 1 | |||||||||
| −1715c −5176a (+/+) | 042 223 362 | 1 | 1 | |||||||||
| −1715c −5176a (+/+) | 042 214 223 362 | 1 | 10 | 11 | ||||||||
| −1715c −5176a (+/+) | 042 214 223 234 362 | 2 | 2 | |||||||||
| −1715c −5176a +8865j (+/+) | 042 172 223 362 | 1 | 1 | |||||||||
| D1a | −5176a (+/+) | 223 325 362 519 | 4 | 4 | ||||||||
| D3 | −5176a −10180l (+/+) | 223 319 362 | 1 | 1 | ||||||||
| −951j −5176a −10180l (+/+) +15437e | 223 319 362 | 2 | 2 | |||||||||
| −5176a −10180l (+/+) +13717a +14923c +15437e | 223 319 362 | 2 | 2 | |||||||||
| −951j +4133q −5176a −5823a −10180l (+/+) +15437e | 223 239 243 319 362 | 1 | 1 | |||||||||
| −5176a −10180l (+/+) +15437e | 093 172 173 215 223 319 362 519 | 1 | 1 | |||||||||
| D4 | −5176a (+/+) +10646k | 093 223 290 362 | 1 | 1 | ||||||||
| −5176a (+/+) +10646k −14015a | 093 223 232 290 362 | 3 | 3 | |||||||||
| −5176a (+/+) +10646k | 093 223 232 261 294 362 | 2 | 2 | |||||||||
| −5176a (+/+) +10646k | 093 223 232 290 362 | 2 | 2 | 4 | ||||||||
| −5176a +8249b/(+/+) +10646k | 093 223 232 261 290 362 | 2 | 2 | |||||||||
| −5176a (+/+) +10646k | 183 223 274 290 319 362 | 1 | 1 | |||||||||
| −5176a +6915k (+/+) +10646k | 183 223 274 290 319 362 519 | 1 | 1 | |||||||||
| −5176a (+/+) +10646k −13268g | 176 183 223 274 290 319 342 362 519 | 1 | 1 | |||||||||
| D5 | +4877a −5176a (−/−) +12026h/(12705) | 092 172 189 223 266 362 | 1 | 1 | ||||||||
| −5176a (−/−)+12026h/(12705) | 092 126 164 189 223 266 362 | 3 | 3 | |||||||||
| −5176a (−/−) +12026h/−13259o (12705) | 092 164 172 189 223 266 362 | 1 | 1 | |||||||||
| G1 | +4830n/+8198a (+/+) | 017 129 223 519 | 1 | 1 | ||||||||
| +4830n/+8198a (+/+) | 017 093 129 223 519 | 2 | 2 | 1 | 5 | |||||||
| +4830n/+8198a (+/+) | 017 093 129 223 303 519 | 1 | 1 | |||||||||
| +4830n/+8198a (+/+) | 017 051 093 207 223 399 519 | 1 | 1 | |||||||||
| +4830n/+8198a +9253k (+/+) | 017 093 129 223 311 519 | 1 | 1 | |||||||||
| +4830n/+8198a (+/+) +15494c | 017 129 223 519 | 1 | 1 | |||||||||
| +4830n/+8198a (+/+) +15494c | 017 093 129 207 223 519 | 2 | 2 | |||||||||
| +4830n/+8198a (+/+) +15494c | 017 093 207 223 244 399 519 | 1 | 1 | |||||||||
| +4830n/+8198a +8957l (+/+) +15494c | 017 051 093 207 223 399 519 | 1 | 1 | |||||||||
| +4830n/+6629a +8198a (+/+) | 017 129 223 519 | 1 | 1 | |||||||||
| +4830n/+6629a +8198a (+/+) | 017 093 129 223 519 | 5 | 5 | |||||||||
| +4830n/+6629a −7658j +8198a (+/+) | 017 093 129 223 313A 519 | 1 | 1 | |||||||||
| G2 | +4830n/−7598f (7600) (+/+) | 051 150 223 278 362 | 1 | 1 | ||||||||
| +4830n/−7598f (7600) (+/+) +12752a | 093 126 223 227 278 362 | 1 | 1 | |||||||||
| +4830n/−7598f (7600) (+/+) | 223 227 234 278 362 | 1 | 1 | |||||||||
| +4830n/−7598f (7600) (+/+) | 223 227 278 294 362 | 1 | 1 | |||||||||
| +4830n/−7598f (7600) +8249b/(+/+) | 223 227 278 362 519 | 1 | 1 | |||||||||
| +4830n/−7598f (7600) −9438e (+/+) | 220C 223 227 278 362 | 1 | 1 | |||||||||
| +4830n/−7598f (7600) (+/+) | 093 129 223 271 278 362 399 | 1 | 1 | 2 | ||||||||
| G3 | +4830n/(+/+) −12406h | 223 274 362 390 | 1 | 1 | ||||||||
| +4830n/(+/+) −12406h | 093 223 274 362 390 519 | 1 | 1 | |||||||||
| +4830n/(+/+) −12406h | 093 223 274 320 362 390 519 | 1 | 1 | |||||||||
| +4643k +4830n/(+/+) −12406h −15172e | 093 223 274 362 390 519 | 1 | 1 | |||||||||
| +4830n/−8858f (+/+) | 215 223 274 | 1 | 1 | |||||||||
| S | (+/+) +14465s (14470) | 184 223 293 298 319 519 | 1 | 7 | 8 | |||||||
| (+/+) +14465s (14470) | 184 189 223 293 298 319 | 1 | 1 | |||||||||
| (+/+) +14465s (14470) | 184 223 298 319 | 2 | 2 | |||||||||
| (+/+)−13608l +14465s (14470) | 184 223 298 319 | 1 | 1 | |||||||||
| +3744e (+/+) +14465s (14470) −15047e | 086 223 298 319 | 7 | 7 | |||||||||
| Z | (+/+) | 185 223 260 298 360 519 | 1 | 5 | 6 | |||||||
| (+/+) | 129 185 223 224 260 298 519 | 1 | 1 | |||||||||
| −9052n/(+/+) | 185 223 260 298 360 519 | 1 | 1 | |||||||||
| M7 | +9820g (+/+) | 223 245 311 362 368 | 1 | 1 | ||||||||
| +9820g (+/+) | 117 162delA 172 223 519 | 2 | 2 | |||||||||
| −4848e +9820g (+/+) | 173 223 362 519 | 1 | 1 | |||||||||
| +4092e +9820g (+/+) | 140 187 209 223 232 519 | 4 | 4 | |||||||||
| +4092e +9820g (+/+) | 140 187 209 223 519 | 4 | 4 | |||||||||
| −1667c/+4092e +9820g (+/+) | 140 187 209 223 232 519 | 1 | 1 | |||||||||
| +5351f −7853o +9820g (+/+) | 129 152 179 189 223 362 | 1 | 1 | |||||||||
| M9 | +3391e (+/+) | 223 234 316 362 | 4 | 4 | ||||||||
| Total | 72 | 95 | 25 | 46 | 71 | 33 | 87 | 56 | 46 | 531 | ||
Note: RFLP sites are numbered from the first nucleotide of the enzyme recognition sequence. A “+” indicates the presence of the restriction site, a “−” the absence. The restriction enzymes are given using the following single–letter code: a = AluI; b = AvaII; c = DdeI; e = HaeIII; f = HhaI; g = Hinf I; h = HpaI; i = HpaII; j = MboI; k = RsaI; l = TaqI; m = BamHI; n = HaeII; o = HincII; q = NlaIII; r = BfaI; s = AccI; t = BstNI; u = MseI. A slash at the end of the restriction site implies the simultaneous presence/absence of a linked site due to the same nucleotide substitution. The presence/absence of the associated 10394 DdeI/10397 AluI sites is denoted through slash brackets (+/+), or (+/–). 9bp ins = 9bp COII/tRNALys triplication; 9bp del = 9bp COII/tRNALys deletion; 4bp ins = 4bp COII/tRNALys insertion; h = heteroplasmy. Additional mutations in the coding region verified through sequencing are shown in brackets. Founding RFLP/HVS-I haplotypes are shown in boldface. Only those nucleotide positions between 16013 and 16520 that differ from the revised Cambridge Reference Sequence (rCRS) (Andrews et al. 1999) are shown. Mutations are transitions, unless the base change is specified explicitly. The HVS-I mutations recognized also by RFLP analysis are shown only within the HVS–I sequence.
Population codes are as follows: TB = Tubalar; TV = Tuvan; BR = Buryat; TF = Tofalar; EV = Evenki; NG = Negidal; UL = Ulchi; NV = Nivkhi; UD = Udegey.
Table 2.
Frequencies of mtDNA haplogroups (%) in 16 Siberian populations
| Haplogroup | MN (98) | KT (38) | NS (24) | TB (72) | TV (95) | BR (25) | TF (46) | EV (71) | NG (33) | UL (87) | NV (56) | UD (46) | IT (47) | KR (155) | CH (66) | ES (79) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | 3.1 | 7.9 | - | 11.1 | 2.1 | - | - | 5.6 | - | - | - | - | 6.4 | 2.6 | - | - |
| A2 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.6 | 68.2 | 77.2 |
| B1 | - | - | - | 4.2 | 1.1 | - | - | - | - | - | - | - | - | - | - | - |
| B3 | - | - | - | - | - | 4.0 | - | - | - | - | - | - | - | - | - | - |
| B4 | - | - | - | - | 5.3 | - | 4.3 | - | - | - | - | - | - | - | - | - |
| B5 | - | - | - | - | 2.1 | - | - | - | 12.1 | - | - | - | - | - | - | - |
| F | 1.0 | 23.7 | - | 1.4 | 4.2 | - | 8.7 | 1.4 | - | 1.1 | - | - | - | - | - | - |
| Y | - | - | - | - | - | - | - | - | 21.2 | 37.9 | 66.1 | 8.7 | 4.1 | 9.7 | - | - |
| H | 14.3 | 10.5 | 8.4 | 5.6 | 4.2 | 4.0 | - | - | - | - | - | - | - | - | - | - |
| V | 1.0 | - | - | - | - | 4.0 | - | - | - | - | - | - | - | - | - | - |
| J | 12.2 | - | - | - | 2.1 | 4.0 | - | - | - | - | - | - | - | - | - | - |
| T | 7.2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| U | 25.4 | 34.2 | 25.0 | 26.4 | 3.2 | 12.0 | - | - | - | - | - | - | - | - | - | - |
| K | 3.1 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| W | - | 2.7 | - | - | 2.1 | - | - | - | - | - | - | - | - | - | - | - |
| X | - | - | - | 1.4 | - | - | - | - | - | - | - | - | - | - | - | - |
| N9 | - | - | - | 6.9 | 3.2 | - | - | - | - | 6.9 | - | 30.4 | - | - | - | - |
| R | - | - | - | 2.8 | - | - | - | - | - | - | - | - | - | - | - | - |
| C1a | - | - | - | - | - | - | - | - | - | 1.1 | - | - | - | - | - | - |
| C2 | 16.3 | 15.8 | 12.6 | 16.7 | 35.8 | 40.0 | 50.0 | 57.8 | 9.1 | 6.9 | - | 17.4 | 2.1 | 23.8 | 4.5 | 2.5 |
| C3 | 1.0 | - | 20.8 | 2.8 | 7.4 | - | 10.9 | 14.1 | 6.1 | 5.7 | - | - | 12.8 | 12.3 | 6.1 | - |
| D | 6.2 | 2.6 | 8.3 | 12.5 | 9.5 | 16.0 | - | 18.3 | 18.2 | 4.6 | 23.2 | - | - | - | - | - |
| D1a | - | - | - | - | - | - | - | - | - | 4.6 | - | - | - | - | - | - |
| D2 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 10.6 | 15.2 |
| D3 | 1.0 | - | 16.7 | 1.4 | 3.2 | - | - | 1.4 | - | 2.3 | - | - | - | 1.3 | 1.5 | 5.1 |
| D4 | - | - | 4.2 | 1.4 | - | - | - | - | 6.1 | 10.3 | 5.4 | - | - | - | - | - |
| D5 | 1.0 | - | - | 4.2 | 1.1 | - | - | 1.4 | - | - | - | - | - | - | - | - |
| G1 | - | - | - | - | - | - | - | - | 27.2 | 10.3 | 5.4 | - | 68.1 | 41.9 | 9.1 | - |
| G2 | 6.1 | - | - | - | 6.3 | 4.0 | - | - | - | 1.1 | - | - | - | - | - | - |
| G3 | - | - | - | - | 4.3 | 4.0 | - | - | - | - | - | - | - | - | - | - |
| S | - | - | - | - | - | 4.0 | 15.2 | - | - | 4.6 | - | 15.2 | - | - | - | - |
| Z | - | 2.6 | 4.2 | 1.4 | 1.6 | 4.0 | 10.9 | - | - | - | - | - | 6.4 | 5.8 | - | - |
| M | 1.0 | - | - | - | 1.6 | - | - | - | - | 2.3 | - | 28.3 | - | - | - | - |
Note: inferred from published data as follows: MN – Mansi (Derbeneva et al. 2002b); KT – Ket, NS - Nganasan (Derbeneva et al. 2002c); IT – Itelmen, KR – Koriak (Schurr et al. 1999); CH – Chukchi, ES – Eskimos (Starikovskaya et al. 1998). The double-letter code for populations in this study is given as in Table 1.
Macro-haplogroup N mtDNAs
Haplogroup A is defined by the 663 HaeIII site and the HVS-I motif 16223-16290-16319-16362. It is rare but widespread, having been found in some Western and Middle Siberian populations such as the Mansi, Ket, Tubalar, Tuvan and Evenki (Torroni et al. 1993a; Sukernik et al. 1996; Derbeneva et al. 2002b, c; this study). Haplogroup A mtDNAs in these populations lack the mutation at np 16111 and are classified within subhaplogroup A1 (Forster et al. 1996). In contrast, the Chukchi and Eskimo haplogroup A mtDNAs harbour the 16111 transition and belong to the same A subhaplogroup (A2) found in Native Americans (Starikovskaya et al. 1998). Thus, the homeland of A2 is most likely a Beringian refuge area, from where it subsequently expanded into the New World (Forster et al. 1996; Starikovskaya et al. 1998; Saillard et al. 2000). In the course of the present study, we completely sequenced two A1 mtDNAs (one Mansi from the Lower Ob and one Ket from the Lower Yenisei River) previously defined at the RFLP/HVS-I level (Derbeneva et al. 2002 b, c). While their control region harbored the CA deletion at nps 522–523, the coding region showed five transitions at nps 663, 1736, 4248, 4824 and 8794. These mutations apparently predated the split of A2 from A1, since they are also found in A2 mtDNAs (Ingman et al. 2000; Mishmar et al. 2003). Interestingly, two haplogroup A mtDNA samples from China, which were completely sequenced by Kong et al. (2003), harboured the same ancestral motif characterizing the A1 mtDNA subgroup in the Ket and Mansi. However, haplogroup A2 mtDNAs, as noted in both the Chukchi and Native Americans, can be easily distinguished by the additional transitions at nps 8027 and 12007 (Ingman et al. 2000; Herrnstadt et al. 2002; Mishmar et al. 2003; Silva et al. 2003).
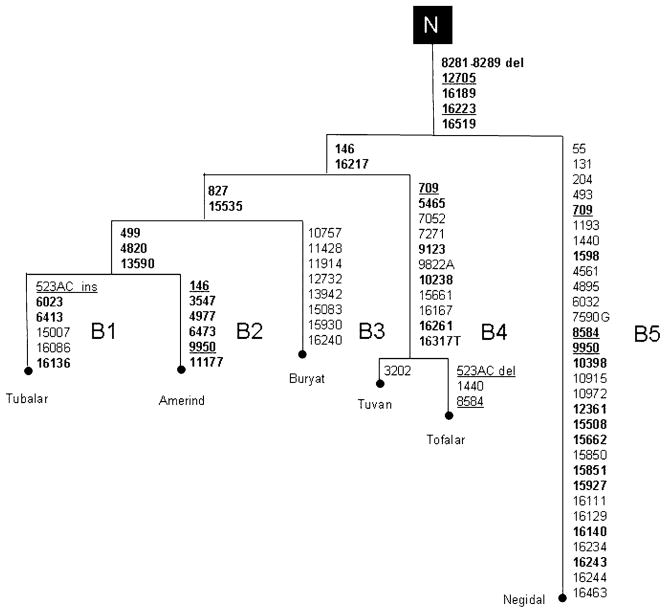
Haplogroup B mtDNAs are defined by the 9-bp deletion at nps 8281–8289 and the control-region motif 16189–16519. Several subhaplogroups of B have been delineated in Siberians (Table 1). B1 mtDNAs were found in a Tubalar and a Tuvan. They are distinguished by the 6022 AluI site loss and the control region motif 16086-16136-16189-16217-16519 (Table 1). A similar (at the HVS-I level) haplogroup B mtDNA has been detected in the skeletal remains exhumed from a 2000 year-old cemetery in northern Mongolia (Keyser-Tracqui et al. 2003). The complete sequence of the Tubalar mtDNA revealed five mutations (Figure 2) (499, 827, 4820, 13590, and 15535) which are shared with the Native American haplogroup B mtDNAs sequenced by Ingman et al. (2000) while two mutations (6023 and 6413) are shared with some, but not all, Asian haplogroup B mtDNAs (Ingman et al. 2000; this study). At the coding region level, the Native American branch (B2) of haplogroup B (Herrnstadt et al. 2002) differs from B1 for the additional transitions at nps 3547, 4977, 6473, 9950, 11177 and the lack of the mutations at nps 6023 and 6413. We also sequenced the haplogroup B mtDNA found in the Buryat (Table 1). It lacked three (499, 4820, 13590) of the transitions shared by the Siberian B1 and the Native American B2. In addition, its coding region harboured seven transitions, and we classified this mtDNA within the novel subhaplogroup B3. An additional subset of B mtDNAs, which we designated B4, is defined by the HVS-I motif 16189-16217-16261 (Table 1) and was observed in five Tuvans and two Tofalars. This mtDNA subset seems to be widely dispersed in adjacent Mongolia and China (Kolman et al. 1996; Yao et al. 2000, 2002). We sequenced two B4 mtDNAs (one Tuvan and one Tofalar) and compared them with those obtained from Korean and Samoan haplogroup B mtDNAs (Ingman et al. 2000). The comparison showed that they all share the coding region variants 709, 5465, 9123, and 10238, suggesting that subhaplogroup B4 originated in either central or southeastern Asia. Finally, we observed two Tuvan and four Negidal mtDNAs belonging to subhaplogroup B5 (Kolman et al. 1996; Yao et al. 2002). This lineage, in contrast to the other subsets of B, harbours the 10394 DdeI site. The complete sequence of one Negidal B5 mtDNA revealed numerous mutations (Figure 2). A similar complete mitochondrial genome was reported in a Japanese cardiomyopathy patient (Shin et al. 2000). Lineage B5 is most likely widespread in central/eastern Asia since it has been previously described at the RFLP/HVS-I level in a few Mongolians (Kolman et al. 1996), Chinese (Yao et al. 2000; 2002), and one Tuvan (Derenko et al. 2000). Overall, our analysis of haplogroup B mtDNAs from southern Siberia has revealed four distinct subhaplogroups (B1, B3, B4 and B5), but only one (B1) is closely related to B2, the branch of B found in Native Americans.
Figure 2.
Schematic phylogeny of haplogroup B mtDNAs found in Native Siberians and Native Americans. The phylogeny is rooted from macrohaplogroup N as inferred from the complete sequences of Siberian/Asian haplogroup A, B, and Y (Ingman et al. 2000; Herrnstadt et al. 2002; Mishmar et al. 2003; this study). Mutation positions, relative to the revised Cambridge Reference Sequence (Andrews et al. 1999), are transitions unless the base change is specified. Underlining indicates recurrent mutations. Founding motifs are shown in bold type.
Haplogroup F mtDNAs characterized by the 4732 RsaI site, the lack of the 12406 HpaI/HincII and 12629 AvaII sites, and the HVS-I motif 16189-16232A-16249-16304-16311, were observed in the Tubalar, Tofalar, and Tuvan of the Altai-Sayan, the Mansi of the Lower Ob River, and the Ket and Evenki of the Middle Yenisei River (Derbeneva et al. 2002a,c, this study). A related mtDNA type was found in three Tuvan and one Ulchi but it lacked the 4732 RsaI site and was very different in HVS-I. Thus, this mtDNA type appears to be more closely related to the one which is common in Tibet (Torroni et al. 1994).
Haplogroup Y is characterized by the 7933 MboI and 10394 DdeI sites, the lack of the 8391 HaeIII site, the control-region motif 16126–16519, and was first localized in the Russian Far East (Torroni et al. 1993a; Schurr et al. 1999). The complete sequence of one Ulchi haplogroup Y mtDNA revealed five coding region transitions (3834, 7933, 8392, 14178, 14693) that distinguish haplogroup Y from other Asian haplogroups of the macro-cluster N (Mishmar et al. 2003). Haplogroup Y is restricted to the Lower Amur/Sea of Okhotsk region and adjacent Kamchatka. It reaches its maximum frequency in Sakhalin Island, where it represents 66% of the Nivkhi mtDNAs (Tables 1 and 2). Since Y has a limited geographic range and shows a relatively low level of intra-group diversity, it must be of recent origin, arising long after the migration of the progenitors of Native Americans to the New World.
A new unique “east” Eurasian haplogroup within macrohaplogroup N was found in three Tuvan, six Ulchi and 14 Udegey. It harbours transitions at nps 12705 and 16223 and is defined by the transition at np 5417. Several “west” Eurasian haplogroups, including H, V, J, U4, U5, W, and X, were also detected. Their confinement to the south-west part of Siberia might be indicative of an Upper Paleolithic dispersal from the Middle East/southeastern Europe, the traces of which have not been erased by subsequent migrations and gene flow. Alternatively, a relatively recent gene flow mediated by women of European/West Asian ancestry could have occurred at the time of the expanding Mongolian Empire.
Macro-haplogroup M mtDNAs
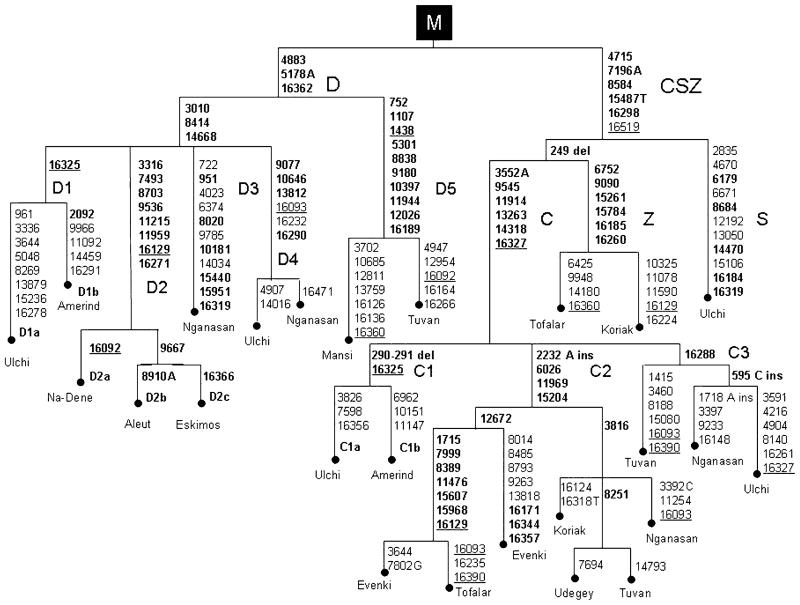
Haplogroup C is defined by the 13262 AluI site and the HVS-I motif 16223-16298-16327. It is the most common and widespread throughout Siberia and can be divided into several subhaplogroups. One of these, which we termed C1 (Figure 3), is defined by the HVS-I transition at np 16325. It was found in one Ulchi of the Lower Amur, and has previously been reported in two Mongols and one Kirgiz (Kolman et al. 1996; Comas et al. 1998). The Ulchi C mtDNA with the HVS-I transition at np 16325 was completely sequenced and compared with Native American and Asian haplogroup C mtDNAs (this study). All were found to share the motif 3552A-9545-11914-13263-14318-16327, but the Ulchi mtDNA, in addition to the transition at np 16325, also harboured the deletion in HVS-II of nps 290–291 ubiquitously present in Amerindian haplogroup C mtDNAs (Ingman et al. 2000; Moraga et al. 2000; Malhi et al. 2001). Network analysis confirmed that the Ulchi mtDNA (C1a) and Native American mtDNA (C1b) are the most similar, while many of the Tuvan, Tofalar, Evenki, Nganasan, Udegey and Koriak complete sequences are distinguished by the HVS-I/coding region motif 2233 ins-6026-11969-15204 (Mishmar et al. 2003; this study), and are members of a different cluster that we termed C2. This lineage appears to be the most common and widespread in Siberia (Table 2). An additional lineage of haplogoup C has been discerned through complete sequencing of one of the Nganasan and Ulchi mtDNA samples (this study), and one Tuvan mtDNA with 3460A primary LHON mutation (unpublished data of Volodko et al.). We termed this lineage C3 as it lacked the coding region motif 2233 ins-6026-11969-15204 but has the control region variant marked by 16288. At the HVS-I level it has been observed not only in the populations of southern Siberia but also among the circumpolar groups, the Nganasan, Chukchi, Koriak and Itelmen (Starikovskaya et al. 1998; Schurr et al. 1999; Derbeneva et al. 2002b, c). The network analysis of haplogroup C2 and C3 complete sequences has revealed several distinct sub-lineages. Thus, the sublineage marked by 3816 (C2) is particularly widespread, being shared by the Buryat, Kirgiz, Koriak, Udegey, Tuvan and Nganasan haplogroup C mtDNA samples that were subjected to complete sequencing (Ingman et al. 2000; Mishmar et al. 2003; this study). It is worth noting the reversion in HVS-I of the 16327 mutation, which was found to characterize a C haplotype shared by three Tuvan and six Ulchi mtDNA samples (Table 1).
Figure 3.
Schematic phylogeny of haplogroup C, D, S and Z mtDNAs found in Native Siberians in comparison with their Native American counterparts. The phylogeny is rooted from macrohaplogroup M. Note that a transition at np 13879 in the Amerindian D1b mtDNA was erroneously shown in the legend of Figure 4 by Derbeneva et al. (2002a).
Haplogroup D is defined by the lack of the 5176 AluI site and the HVS-I motif 16223–16362. The majority of the haplogroup D haplotypes from southern Siberia did not exhibit sub-haplogroup-specific mutations, and thus they were attributed to a general haplogroup D category. However, we were able to identify at least five distinct sub-clusters (D1–D5) branching off from the root of D. Lineage D1 is defined by the HVS-I transition at np 16325 and was found exclusively in four Ulchi of the Lower Amur. This transition is typically observed in Native American D mtDNAs (Torroni et al. 1993b; Moraga et al. 2000; Mahli et al. 2001, 2003) but has never been observed in central-eastern Asian populations (Kolman et al. 1996; Comas et al. 1998; Yao et al. 2002; Keyser-Tracqui et al. 2003). Thus, the sharing of the mutation at np 16325 in the Lower Amur and the Americas suggests a possible association between the Lower Amur and Native Americans.
Subhaplogroup D2 harbours the 8700 AluI site and lacks the 3315 HaeIII site in the presence of the HVS-I transition at np 16271. Complete sequence analyses show that overall it is defined by the motif 3316-7493-8703-9536-11215-11959-16129-16271. Lineage D2 was most likely restricted to a relatively small area of Greater Beringia, since today it is concentrated in the Chukchi and Eskimo-Aleuts (Starikovskaya et al. 1998; Derbeneva et al. 2002a).
Subhaplogroup D3 at the RFLP/HVS-I level is distinguished by the presence of the 10180 TaqI and 15437 HaeIII sites, and the transition at np 16319. This lineage is uncommon but widespread in Siberia being represented in the Tubular, Mansi, Nganasan, Tuvan, Evenki, Ulchi, Chukchi, and Siberian Eskimos. One Nganasan D3 mtDNA, which was completely sequenced, exhibited the motif 951-8020-10181-15440-15951-16319 (Derbeneva et al. 2002a).
Subhaplogroup D4 is distinguished by the presence of the 10646 RsaI site linked with the transition at np 16290. It is common among the Negidal, Ulchi and Nivkhi of the Lower Amur, and has also been observed at low frequencies in the Tubular and Naganasan. The complete sequencing of two D4 mtDNA samples, one Ulchi of the Lower Amur and one Nganasan of the Taimir Peninsula, isolated by a distance of some 5,000 km, has revealed that they share the three coding region transitions, 9077, 10646, and 13812, thus delineating the D4 founding motif (Debeneva et al. 2002a; this study).
Finally, there is subhaplogroup D5 which, in contrast to the other subsets of D, has back mutated at the 10394 DdeI and 10397 AluI sites. D5 mtDNAs have been observed in three Tubular, one Tuvan and one Evenki. The lack of the diagnostic 10394 DdeI and 10397 AluI sites has been observed also in one Mansi mtDNA, and the sequencing of the segment encompassing nps 10394–10397 revealed the expected macrohaplogroup M transitions at nps 10398 and 10400. However, a transition at np 10397 was also present that accounted for the simultaneous elimination of the adjacent restriction sites (Derbeneva et al. 2002b). The complete sequencing of both Mansi and Tuvan D5 mtDNAs, aside from the transition at np 10397, revealed in their coding regions seven additional transitions, namely at nps 752, 1107, 5301, 8838, 9180, 11944 and 12026 (this study). Furthermore, they differed from other derivatives of the M cluster by lacking the transition at np 1438 (Figure 3). Interestingly, a similar kind of variation is present in one of two Chinese haplogroup D5 mtDNAs that was subjected to complete sequencing by Kong et al. (2003). The lack of the 10394 DdeI and 10397 AluI sites in haplogroup D mtDNAs was previously reported in the Taiwanese Han, Sabah aborigines (Ballinger et al. 1992), Tibetans (Torroni et al. 1994), and Mongolians (Kolman et al. 1996). A similar haplotype defined by the 10397, 10398 and 10400 variants has been reported among the Chinese Han (Yao et al. 2002), thus indicating a northward expansion of D5 from East Asia.
Haplogroup G is defined by the 4830/4831 HaeII/HhaI sites and is widespread in Siberian and adjacent Asian populations (Ballinger et al. 1992; Torroni et al. 1994; Yao et al. 2002; present study). It can be subdivided into three distinct subhaplogroups (Tables 1 and 2). Subhaplogroup G1 is distinguished by the 8198 AluI site and the transition at np 16017 in HVS-I. Geographically, G1 is restricted to the Sea of Okhotsk region, being the most prevalent in the Koriak and Itelmen of Kamchatka (Schurr et al. 1999). A variant, observed solely in four Chukchi of adjacent Chukotka, lacks the 8198 AluI site (Starikovskaya et al. 1998). Lineage G2 is characterized by the lack of the 7598 HhaI site, caused by a G-A transition at np 7600, and the HVS-I motif 16223-16278-16362. A Koriak G1 mtDNA and a Tuvan G2 mtDNA studied through complete sequencing were found to share the coding region motif 709-4833-5108-14569, thus forming the founding haplotype for haplogroup G (Mishmar et al. 2003; this study). In addition, the Koriak G1 showed the distinguishing motif 8200-12361-12972-15323-15497, while the Tuvan G2 harboured 1189-5601-7600-9377-9575-12372-13563-14198-15355. Subhaplogroup G2 appears to be widespread in central/eastern Asia as Korean haplogroup G sequences also harbour the motif 5601-7600-9377-9575-13563 (Snall et al. 2002). It has also been observed in the Mansi of Siberia’s northwest (Derbeneva et al. 2002b), though it is rare or absent in the Russian Far East. Subhaplogroup G3 is distinguished from the root of G by transitions at nps 12408, 16274 and 16390 and has been seen in several Tuvan and one Buryat. The identity of this lineage was confirmed by sequencing the 12406 HpaI-HincII region which revealed that the distinguishing mutation is a transition at np 12408. However, one Tuvan G3 mtDNA was found to lack this transition, together with the 16390 variant (Table 1).
One haplogroup M lineage was characterized by the HVS-I motif 16223-16298-16319 and the 14465 AccI site. This novel lineage, which we termed haplogroup S, was observed in one Buryat, seven Tofalar, four Ulchi and seven Udegey. A similar mtDNA type, designated M8a, has been recently described in China by Yao et al. (2002). One of the Ulchi haplogroup S mtDNA was completely sequenced. It shared with haplogroup C the coding region motif 4715-71976A-8584-15487T, revealing that these mutations predate the radiation of haplogroups C, S and Z. In addition, the Ulchi mtDNA also harboured the coding region variants 2835, 4670, 6179, 6671, 7196A, 8684, 12192, 13050, 14470 and 15106. Three of these (6179, 8684, and 14470) are shared with an Asian mtDNA described by Herrnstadt et al. (2002). The confinement of haplogroup S to southern and southeastern Siberia, inhabited by Mongolic-, Turkic-, and Tungusic-speaking populations which are members of the Altaic linguistic family, implies that it probably arose in one of the hunting groups ancestral to the Altaic-speakers. We presume that this lineage originated in the putative homeland (Mongolia/Manchuria) of the Altaic-speakers and was then dispersed by their geographical expansion.
Haplogroup Z is distinguished by the HVS-I motif 16185–16260 and was first identified in the Itelmen and Koriak of Kamchatka (Schurr et al. 1999). This haplogroup was also noted amongst the Entsi/Nganasan and Ket of the Lower Yenisei (Derbeneva et al. 2002c). In the southern part of Siberia, we found this haplogroup in one Northern Altaian (1.4%), one Tuvan (1.7%), one Buryat (4.0%), and five Tofalar (10.9%) mtDNAs. The sequence of one Tofalar and one Koriak haplogroup Z mtDNA revealed that they differ from the common root of C–Z for the 6752, 9090, 15261, 15784, 16185, 16260 variants (Mishmar et al. 2003; this study), mutations that they also share with the Finnish/Saami haplogroup Z mtDNAs (Finnila et al. 2001; Majamaa, personal communication). Thus, these six transitions are characteristic of the ancestral haplogroup Z haplotype. Based on the distribution of haplogroups C, S, Z and the number of variants encompasses, we postulate that south-central Siberia was the place of origin for the ancestral haplotype, and the populations bearing haplogroup Z must have spread into northern Eurasia after the ancestors of the Amerindians left the region.
The lineages of macro-haplogroup M were also found mainly in the populations of extreme south and southeastern Siberia (Table 1). One lineage marked by the 9820 HinfI site was initially detected in the Vietnamese and Taiwanese Han (Ballinger et al. 1992). Another lineage of M harbouring the 3391 HaeIII site (T3394C) in conjunction with the HVS-I motif 16223-16234-16316-16362 was found in four Udegey mtDNAs. A similar motif was recently observed in Chinese M mtD-NAs, suggesting an East Asian origin for this lineage (Yao et al. 2002).
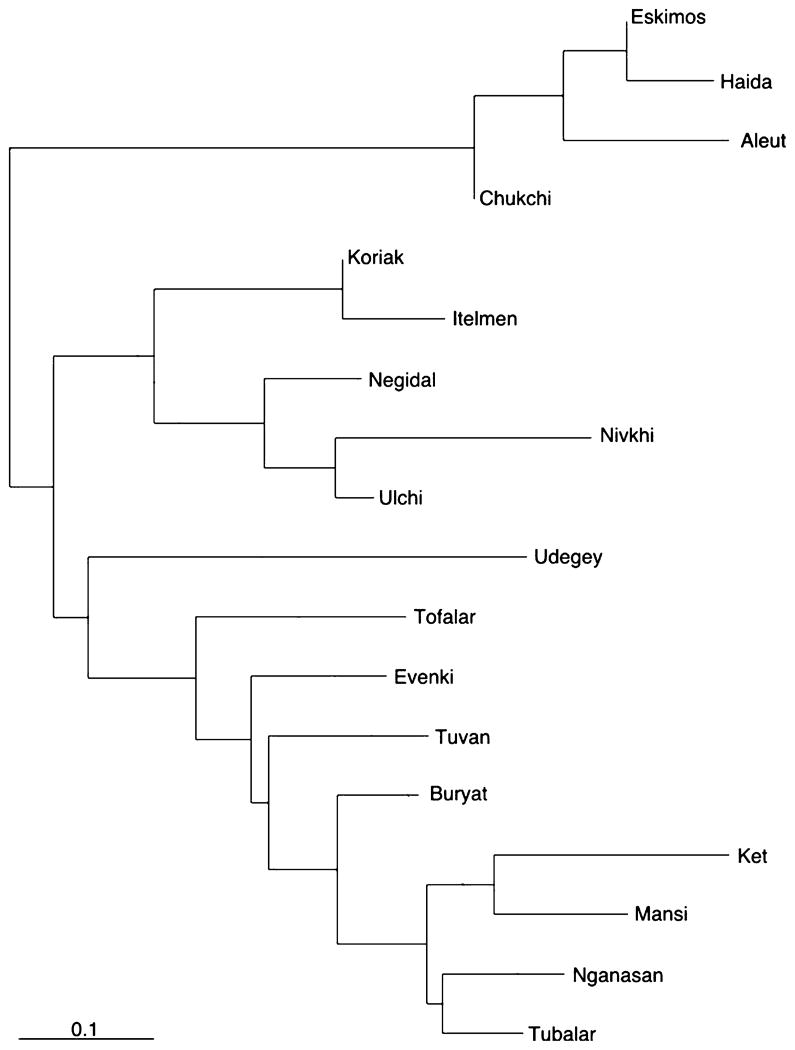
NJ analysis
The genetic distances between interior Siberian and north Pacific populations were computed from their subhaplogroup frequencies (Table 2), and analysed with the help of a dendrogram (Figure 4). The tree is drawn by mid-point rooting. The dendrogram supports the hypothesis that the Chukchi, Eskimos, Aleut and Haida, though quite divergent from one another, represent a detached population cluster isolated by sufficient time to be relatively distinct over the average genetic background of Siberia. In addition, the branching patterns of the Lower Amur and adjacent Okhotsk Sea coast/Kamchatkan populations, relative to those located in Western and Middle Siberia, may reflect relatively recent demic expansions which have brought haplogroups G, S, Y and Z into the Sea of Okhotsk/Kamchatka region. The observed geographical distribution of these haplogroups (Tables 1 and 2) would be expected from anthropological, archaeological and linguistic evidence indicating the expansion of the Altaic-speakers from the Lower Amur/Manchuria region during the Neolithic (Krauss, 1988; Turner, 1994; Janhunen, 1996; Kozintsev et al. 1999).
Figure 4.
Phylogenetic relationships between 16 Siberian and two Native American populations of the North Pacific Rim (Aleut and Haida), determined by neighbor-joining using the frequencies of the 31 observed subhaplogroups. Haplogroup composition and frequencies in Haida and Aleut mtDNAs were adopted from Ward et al. (1993), and Derbeneva et al. (2002) and Rubicz et al. (2003) respectively.
Discussion
A large variety of mtDNA haplogroups and subhaplogroups have been found in Siberia. However, only one or at most two subhaplogroups of A, B, C, and D appear to have contributed to the human colonization of the Americas, confirming that only a limited number of founder mtDNAs gave rise to the ancestral Native American populations (Wallace et al. 1985; Neel et al. 1994; Starikovskaya et al. 1998). Indeed, the evidence provided by the analysis of variation in the Y chromosome types, using almost the same set of Siberian DNAs, is also consistent with Native Americans being founded by a limited number of individuals (Lell et al. 2002). Furthermore, many HLA class II genes (i.e., DRB1 allelic lineages) observed in the Siberian interior are absent in the New World (Uinuk-ool et al. 2002, 2004; Volodko et al. 2003).
Many of the mtDNA haplotypes found in Siberia are shared between culturally and linguistically distinct populations, indicating that extensive gene flow has occurred from the Ural Mountains to the Pacific Ocean, and from Mongolia/Manchuria/Southeastern Siberia (former Greater Manchuria) to the upper Arctic, since the early Holocene. However, still there exists a remarkable geographical and even tribal specificity for some of the Siberian subhaplogroups, and this provides major clues about the origin of Native Americans.
The trans-Beringian Relationships of Haplogroup A and B mtDNAs
Almost all Native American haplogroup A mtDNAs are characterized by the three distinguishing mutations, at nps 8027, 12007 and 16111, thus belonging to subhaplogroup A2 (Ingman et al. 2000; Silva et al. 2003; Mishmar et al. 2003). The phylogenetic relationship of A1 and A2 mtDNAs and their pattern of spatial distribution in Siberia/Asia (Starikovskaya et al. 1998; Schurr et al. 1999; Yao et al. 2002; Kong et al. 2003; this study) suggests that the A1 mtDNAs observed in the Mansi and Ket are part of an A1 northward dispersal that presumably originated in the Altai-Sayan region, and gave rise to A2 after expanding into eastern Beringia.
Haplogroup B is extremely diverse; several subsets of this haplogroup (B1, B3, B4, B5) that share a common root, but diverged early with very different subsequent evolutionary histories, have been found in southern Siberia. Subhaplogroup B1 is rare, but it is the only one that shares the coding region motif 827-4820-13590-15535 with B2, the Native American branch of haplogroup B. In our Siberian sample, B1 mtDNAs were found only in one Tuvan and three Tubalar, suggesting that the common ancestor of Siberian B1 and Amerind B2 could be traced back to the Altai-Sayan Upland. The failure to identify B1 mtDNAs north and northeast of the Altai-Sayan may be explained by their extinction due to genetic drift and/or selection in small populations inhabiting Siberian arctic and sub-arctic prior to the last glacial maximum (>24,000 YBP).
Siberian Affinities of Native American Haplogroup C and D mtDNAs
A combined phylogenetic analysis of mtDNAs belonging to haplogroups C, D, S and Z revealed that, relative to the CRS, they all harbour the distinguishing M motif: 8701-9540-10398-10400-10873-12705-14783C-15043-15301-16223. However, while haplogroup D forms a cluster by itself within the macrogroup M, haplogroups C, S, and Z are sister groups sharing the motif 4715-7196A-8584-15487T-16298 (Figure 3). The origin of the Asian CSZ cluster appears to be rooted in central/eastern Asia during the Upper Paleolithic period, and to have later emerged as the progenitor of the Native American haplogroup C. However, since neither S nor Z mtDNAs have been reported in Native Americans mtDNAs, these haplogroups could have appeared in the Altai-Sayan-Baikal Upland and the Amur region after the immediate ancestors of Paleoindians had already migrated from the southern Siberia.
While lineages C2 and C3 are present at significant frequencies in Uralic, Kettic, Altaic, Chukchi/Koriak speakers, they are absent in Native Americans. The geographic distribution and phylogenetic relationships of C2 and C3 suggest that they arose in the Altai-Sayan-Baikal area. The subhaplogroup C1a mtDNA found in one Ulchi, similarly to Amerindian C1b mtDNAs, harbours the HVS-I marker 16325 and lack C2 and C3 markers. Moreover, C1a and C1b share the deletion of nps 290–291. This finding may imply that C1 and its distinguishing mutations (16325 and the deletion of nps 290–291) originated in the Amur River region and that from that region, the Native American branch C1b arose.
As was found in the course of this and our previous studies (Starikovskaya et al. 1998; Derbeneva et al. 2002a, b, c), Siberian haplogroup D mtDNAs belong to a wide range of distinct subhaplogroups. Subhaplogroup D1a, which was observed in four Ulchi, is the only one which is more closely related to D1b, the Amerindian-specific branch of haplogroup D, than the other sublineages of D in Siberia/Asia. However, Amerindian D1b mtDNAs are commonly associated with a transition at np 2092 (Ingman et al. 2000; Herrnstadt et al. 2002), which was not found in the Siberian D1a mtD-NAs. The finding of both C1a and D1a only on the southwestern edge of former Beringia is intriguing, and indicates that the Amur River region could have been the main Siberian source for Amerindian haplogroups C and D. In contrast, subhaplogroup D2 is confined to Chukotka, Alaska and the Aleutian archipelago with a great geographical/tribal specificity; D2a in the Na-Dene, D2b in the Aleut and D2c in the Eskimos (Figure 3). The geographic specificity and phylogeny of haplogroup D complete sequences (Derbeneva et al. 2002a; this study) support the refugial hypothesis (Rogers et al. 1991), which proposes that the founding population of Eskimo-Aleut and Na-Dene Indians originated in the eastern Beringian/Alaskan refuge area during the early postglacial period. However, the genealogical relationships among the lineages of the exceptionally diverse haplogroup D cluster are not explicitly clear. Some intermediate haplotypes may be missing, suggesting a relatively long evolutionary history of Native American D.
Many factors must have influenced the success of human biological adaptation to the harsh glacial environment of Beringia and further movements into the New World. Late glacial and early postglacial climatic fluctuations have influenced population size and structure and, hence, the number of founder haplogroups. Natural selection might have either impoverished or enriched certain mtDNA haplogroups and subhaplogroups as people migrated into sub-arctic and then arctic regions (Mishmar et al. 2003; Ruiz-Pesini et al. 2004).
Conclusion
This study revealed that the remnants of ancient east Eurasian populations are still evident in Siberia due to the occurrence of lineages A1 and B1 in the Altai-Sayan area, and C1 and D1 in the Lower Amur. Assuming low effective population size, reduced diversity and a uniform distribution of mtDNA lineages prior to the last glacial maximum, the lineages A1, B1, C1 and D1 constituted the core genetic makeup of the Siberian progenitors of Paleoindians that spread northwards as a uniform entity. However, it is more likely that the current mtDNA differences between Altai-Sayan Upland, where A1 and B1 occur, and the Amur region, where the traces of C1 and D1 are found, are due to genetic differentiation of the Upper Paleolithic inhabitants of Siberia. Hence, while subhaplogroups A1 and B1 could be a major part of the population dispersal from the Altai-Sayan area downstream via the Yenisei and Lena rivers and then toward eastern Beringia and Alaska, the migration originated in the Amur river basin brought the founders of C1 and D1 to the Americas along the Siberian Pacific. Indeed, a mosaic-like picture of different cultural variants (mammoth, bison and reindeer hunters, fishing and sea-mammal hunting populations) is evidenced by the diverse lithic industries that persisted in the river valleys and along the Siberian Arctic and Pacific regions (Dikov, 1994; Khlobystin, 1998). Moreover, the earliest traces of modern human occupation of northeastern Siberia (at 71°N) indicate that the ancestors of Paleoindians may have colonized the Lower Yenisei and Lena rivers and the now submerged Laptev, Eastern Siberian and Chukchi Sea coastal shelves during a relatively warm interstadial episode around 30,000-28,000 YBP (Vasil’ev, 2002; Pitulko et al. 2004). Since the glaciation of mountaineous Chukotka hampered faunal and human migrations from the Lena river basin, this was the only practical route from the Middle Siberian Plateau to Alaska. Consequently, the bearers of the Amur microlithic tradition (Derev’anko & Volkov, 1997), lacking subhaplogroup B1, might have reached and occupied the maritime region of southwestern Beringia generally later. The phylogeny of haplogroup A, B, C and D lineages revealed through this study is consistent with the hypothesis that the direct ancestors of Native Americans were a hybrid product of different Siberian groups that had migrated to eastern Beringia at different times and following different routes (Kunz & Reanier, 1994).
Acknowledgments
We dedicate this paper to the memory of Dr. James V. Neel who, more than anybody else, encouraged collaborative molecular and evolutionary studies on the gene pool of indigenous Siberian and Native American populations. We are indebted to the native people of Siberia for their participation, and the personnel of the village hospitals and medical posts for their invaluable help and hospitality. Thanks to Tatiana V. Karamysheva and Theodor G. Schurr for their technical assistance at the earliest phase of this research. We also thank Dr. Naoko Takezaki for her advice pertinent to the NJ analysis, and Dr. Kari Majamaa for an access to unpublished haplogroup Z. This work received support from the NIH Institutes of Health grants TW1175 (to M.D.B. and R.I.S.) and NS21328, AG13154, NS41850, HL64017 (to D.C.W.), the U.S. Civilian Research and Development Foundation for the Independent States of the Former Soviet Union (CRDF, #RB1-2501-NO-03 to R.I.S and D.C.W), the Italian Ministry of the University (Progetti Ricerca Interesse Nazionale 2002 and 2003) (to A.T.), Progetti CNR-MIUR Genomica Funzionale-Legge 449/97 (to A.T.), the Wenner-Gren Foundation for Anthropological Research (grant GR 6383 to R.I.S) and the Russian Academy of Science Program on Physical Chemistry and Biology (to R.I.S.).
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, Wallace DC. Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient mongoloid migrations. Genetics. 1992;130:139–152. doi: 10.1093/genetics/130.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC. mtDNA haplogroup X: an ancient link between Europe/Western Asia and North America? Am J Hum Genet. 1998;63:1852–1861. doi: 10.1086/302155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black LT. Peoples of the Amur and maritime regions. In: Fitzhugh WW, Crowell A, editors. Crossroads of continents: cultures of Siberia and Alaska. Smithsonian Institution Press; Washington, DC: 1988. pp. 24–31. [Google Scholar]
- Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bosch E, Martinez-Arias R, Clarimon J, Facchini F, Fiori G, Luiselli D, Pettener D, Bertranpetit J. Trading genes along the silk road: mtDNA sequences and the origin of central Asian populations. Am J Hum Genet. 1998;63:1824–38. doi: 10.1086/302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton R. The coast road. Nature. 2003;422:10–12. doi: 10.1038/422010a. [DOI] [PubMed] [Google Scholar]
- Derbeneva OA, Sukernik RI, Volodko NV, Hosseini SH, Lott MT, Wallace DC. Analysis of mitochondrial DNA diversity in the Aleuts of the Commander Islands and its implications for the genetic history of Beringia. Am J Hum Genet. 2002a;71:415–421. doi: 10.1086/341720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI. Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am J Hum Genet. 2002b;70:1009–1014. doi: 10.1086/339524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Volodko NV, Wallace DC, Sukernik RI. Mitochondrial DNA variation in Kets and Nganasans and the early peopling of Northern Eurasia. Genetika (Russian J Genetics) 2002c;38:1554–1560. [PubMed] [Google Scholar]
- Derenko MV, Malyarchuk BA, Dambueva IK, Shaikhaev GO, Dorzhu CM, Nimaev DD, Zakharov IA. Mitochondrial DNA variation in two south Siberian aboriginal populations: implications for the genetic history of North Asia. Hum Biol. 2000;72:945–973. [PubMed] [Google Scholar]
- Derenko MV, Grzybowsky T, Malyarchuk BA, Czarny J, Miscicka-Sliwka D, Zakharov IA. The pre-sense of mitochondrial haplogroup X in Altayans from south Siberia. Am J Hum Genet. 2001;69:237–241. doi: 10.1086/321266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko MV, Grzybowsky T, Malyarchuk BA, Dambueva IK, Denisova GA, Czarny J, Dorzhu CM, Kakpakov VT, Miscicka-Sliwka D, Wozniak M, Zakharov IA. Diversity of mitochondrial DNA lineages in South Siberia. Ann Hum Genet. 2003;67:391–411. doi: 10.1046/j.1469-1809.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- Derev’anko AP, Volkov PV. Evolution of the paleoeconomy of the ancient population of the Amur region (from the Upper Palaeolithic to Neolithic)//Suyanggae and her neighbors – Chungju. 1997. pp. 35–44. [Google Scholar]
- Dikov NN. The Palaeolithic of Kamchatka and Chukotka and the problem of the peopling of America. In: Fitzhugh WW, Crowell A, editors. Crossroads of continents: cultures of Siberia and Alaska. Smithsonian Institution Press; Washington, DC: 1994. pp. 87–95. [Google Scholar]
- Finnila S, Lehtonen MS, Majamaa K. Phylogenetic network for European mtDNA. Am J Hum Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Goebel T, Waters MR, Dikova M. The archeology of Ushki Lake, Kamchatka, and the Pleistocene peopling of the Americas. Science. 2003;301:501–505. doi: 10.1126/science.1086555. [DOI] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Janhunen J. An Ethnic History. Printed by the Finno-Ugrian Society at Vammalan Kirjapaino Oy; Helsinki: 1996. Manchuria. [Google Scholar]
- Keyser-Tracqui C, Crubezi E, Ludes B. Nuclear and mitochondrial analysis of a 2,000-year-old necropolis in the Egyn Go Valley of Mongolia. Am J Hum Genet. 2003;73:247–260. doi: 10.1086/377005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khlobystin LP. In: Ancient history of Taimyr and the formation of the North Eurasian cultures. Pitulko VV, Shumkin VYa, editors. Publisher - Dmitriy Bulanin; St. Petersburg: 1998. (in Russian) [Google Scholar]
- Kolman CJ, Sambuughin N, Bermingham E. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics. 1996;142:1321–1334. doi: 10.1093/genetics/142.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP. Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozintsev AG, Gromov AV, Moiseyev VG. Collateral relatives of American Indians among the Bronze Age populations of Siberia? Am J Phys Anthropol. 1999;108:193–204. doi: 10.1002/(SICI)1096-8644(199902)108:2<193::AID-AJPA5>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Krauss ME. Many tongues-ancient tales. Peoples of the Amur and maritime regions. In: Fitzhugh WW, Crowell A, editors. Crossroads of continents: cultures of Siberia and Alaska. Smithsonian Institution Press; Washington, DC: 1988. pp. 145–150. [Google Scholar]
- Kuntz ML, Reanier RE. Paleoindians in Beringia: evidence from arctic Alaska. Science. 1994;263:660–662. doi: 10.1126/science.263.5147.660. [DOI] [PubMed] [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet. 2002;70:192–206. doi: 10.1086/338457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MG, Potapov LP. The Peoples of Siberia. The University of Chicago Press; Chicago and London: 1964. [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A. The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi RS, Schultz BA, Smith DG. Distribution of mitochondrial DNA lineages among Native American tribes of northeastern North America. Hum Biol. 2001;73:17–55. doi: 10.1353/hub.2001.0008. [DOI] [PubMed] [Google Scholar]
- Malhi RS, Mortensen HM, Eshleman JA, Kemp BM, Lorenz JG, Kaestle FA, Johnson JR, Gorodezky C, Smith DG. Native American mtDNA prehistory in the American Southwest. Am J Phys Anthropol. 2003;120:108–124. doi: 10.1002/ajpa.10138. [DOI] [PubMed] [Google Scholar]
- Middendorf A. Travelling to the north and east of Siberia (Puteshestvi’e na Sever i vostok Sibiri) S-Petersburg; 1869. pp. 707–711. Part 2. (In Russian) [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraga ML, Rocco P, Miquel JF, Nervi F, Llop E, Chakraborty R, Rothhammer F, Carvallo P. Mitochondrial DNA polymorphisms in Chilean aboriginal populations: implications for the peopling of the southern cone of the continent. Am J Phys Anthropol. 2000;113:19–29. doi: 10.1002/1096-8644(200009)113:1<19::AID-AJPA3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Neel JV, Biggar RJ, Sukernik RI. Virologic and genetic studies relate Amerind origins to the indigenous people of the Mongolia/Manchuria/southeastern Siberia region. Proc Natl Acad Sci USA. 1994;91:10737–10741. doi: 10.1073/pnas.91.22.10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulko VV, Nikolsky PA, Girya E Yu, Basilyan AE, Tumskoy VE, Koulakov SA, Astakhov SN, Pavlova E Yu, Anisimov MA. The Yana RHS site: humans in the Artic before the Last Glacial Maximum. Science. 2004;303:52–56. doi: 10.1126/science.1085219. [DOI] [PubMed] [Google Scholar]
- Rogers RA, Rogers L, Hoffmann R, Martin LD. Native American biological diversity and the bio-geographic influence of Ice Age refugia. J Biogeogr. 1991;18:623–630. [Google Scholar]
- Rubicz R, Schurr TG, Babb PL, Crawford MH. Mitochondrial DNA variation and the origins of the Aleuts. Hum Biol. 2003;75:809–835. doi: 10.1353/hub.2004.0009. [DOI] [PubMed] [Google Scholar]
- Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC. Mitochondrial DNA variation in Koriaks and Itel’men: Population replacement in the Okhotsk Sea - Bering Sea region during the Neolithic. Am J Phys Anthropol. 1999;108:1–42. doi: 10.1002/(SICI)1096-8644(199901)108:1<1::AID-AJPA1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Shin WS, Tanaka M, Suzuki J, Hemmi C, Toyo-Oka T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am J Hum Genet. 2000;67:1617–1620. doi: 10.1086/316896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snall N, Savontaus ML, Kares S, Lee MS, Cho EK, Rinne JO, Huoponen K. A rare mitochondrial DNA haplotype observed in Koreans. Hum Biol. 2002;74:253–262. doi: 10.1353/hub.2002.0024. [DOI] [PubMed] [Google Scholar]
- Silva WA, Jr, Bonatto SL, Holanda A, Ribeiro-Dos-Santos AK, Paixao BM, Goldman GH, Abe-Sandes K, Rodriguez-Delfin LA, Barbosa M, Pako-Larson ML, Petzl-Erler ML, Valente V, Santos SEB, Zago M. Correction: mitochondrial DNA variation in Amerindians. Am J Hum Genet. 2003;72:1346–1348. [Google Scholar]
- Starikovskaya YB, Sukernik RI, Schurr TG, Kogelnik A, Wallace DC. Mitochondrial DNA diversity in Chukchi and Siberian Eskimos: Implications for the genetic history of ancient Beringia and the peopling of the New World. Am J Hum Genet. 1998;63:1473–1491. doi: 10.1086/302087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukernik RI, Shurr TG, Starikovskaya YB, Wallace DC. Mitochondrial DNA variation in native inhabitants of Siberia, with reconstructions of the evolutionary history of American Indians. Restriction polymorphism. Genetika (Russian J Genetics) 1996;32:432–439. [PubMed] [Google Scholar]
- Takezaki N, Nei M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics. 1996;144:389–399. doi: 10.1093/genetics/144.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N. NJBAFD: neighbor-joining tree constructions from allele frequency data. [Google Scholar]
- Torroni A, Sukernik RI, Shurr TG, Starikovskaya YB, Cabell MF, Crawford MH, Comuzzie AG, Wallace DC. a) Analysis of mitochondrial DNA variation of indigenous Siberians confirms their genetic diversity and reveals distinctive affinity with Native Americans. Am J Hum Genet. 1993;53:591–608. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet. 1993b;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Miller JA, Moore LG, Zamudio S, Zhuang JG, Droma T, Wallace DC. Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol. 1994;93:189–199. doi: 10.1002/ajpa.1330930204. [DOI] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalocci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus ML, Bonne-Tamir B, Scozzari R. mtDNA analysis reveals a major late Palaeolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CG., II . New dental anthropological observations relevant to the human population system of the Greater Beringian Realm. In: Fitzhugh WW, Chassonet V, editors. Anthropology of the North Pacific Rim. Smithsonian Institution Press; Washington and London: 1994. pp. 97–106. [Google Scholar]
- Uinuk-ool TS, Takezaki N, Sukernik RI, Nagl S, Klein J. Origin and affinities of indigenous Siberian populations as revealed by HLA class II gene frequencies. Hum Genet. 2002;110:209–226. doi: 10.1007/s00439-001-0668-0. [DOI] [PubMed] [Google Scholar]
- Uinuk-ool TS, Takezaki N, Derbeneva OA, Volodko NV, Sukernik Variation of HLA class II genes in the Nganasan and Ket, two aboriginal Siberian populations. Eur J Immunogenet. 2004;31:43–51. doi: 10.1111/j.1365-2370.2004.00443.x. [DOI] [PubMed] [Google Scholar]
- Vasil’ev SA, Kuzmin YV, Orlova LA, Dementiev VN. Radiocarbon-based chronology of the Palaeolithic in Siberia and its relevance to the peopling of the New World. Radiocarbon. 2002;44:503–530. [Google Scholar]
- Volodko NV, Derbeneva OA, Uinuk-ool TS, Sukernik RI. Genetic history of Aleuts of the Commander Islands as revealed by HLA Class II gene frequencies. Genetika (Russian J Genetics) 2003;39:1710–1718. (in Russian) [PubMed] [Google Scholar]
- Wallace DC, Garrison K, Knowler WC. Dramatic founder effects in Amerindian mitochondrial DNAs. Am J Phys Anthropol. 1985;68:149–155. doi: 10.1002/ajpa.1330680202. [DOI] [PubMed] [Google Scholar]
- Ward RH, Redd A, Valencia D, Frazier B, Paabo S. Genetic and linguistic differentiation in the Americas. Proc Natl Acad Sci USA. 1993;90:10663–10667. doi: 10.1073/pnas.90.22.10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West FH. American beginnings, the prehistory and paleoecology of Beringia. University of Chicago Press; Chicago: 1996. [Google Scholar]
- Wright HE., Jr . Environmental condition for Paleoindian immigrations. In: Dillihay TD, Meltzer DJ, editors. The first Americans: search and research. CRS; Boca Raton, FL: 1991. pp. 113–135. [Google Scholar]
- Yao YG, Watkins WS, Zhang YP. Evolutionary history of the mtDNA 9-bp deletion in Chinese populations and its relevance to the peopling of East and Southeast Asia. Hum Genet. 2000;107:504–512. doi: 10.1007/s004390000403. [DOI] [PubMed] [Google Scholar]
- Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP. Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet. 2002;70:635–651. doi: 10.1086/338999. [DOI] [PMC free article] [PubMed] [Google Scholar]