Abstract
Phytoremediation is an attractive alternative to excavating and chemically treating contaminated soils. Certain plants can directly bioremediate by sequestering and/or transforming pollutants, but plants may also enhance bioremediation by promoting contaminant-degrading microorganisms in soils. In this study, we used high-throughput sequencing of bacterial 16S rRNA genes and the fungal internal transcribed spacer (ITS) region to compare the community composition of 66 soil samples from the rhizosphere of planted willows (Salix spp.) and six unplanted control samples at the site of a former petrochemical plant. The Bray–Curtis distance between bacterial communities across willow cultivars was significantly correlated with the distance between fungal communities in uncontaminated and moderately contaminated soils but not in highly contaminated (HC) soils (>2000 mg kg−1 hydrocarbons). The mean dissimilarity between fungal, but not bacterial, communities from the rhizosphere of different cultivars increased substantially in the HC blocks. This divergence was partly related to high fungal sensitivity to hydrocarbon contaminants, as demonstrated by reduced Shannon diversity, but also to a stronger influence of willows on fungal communities. Abundance of the fungal class Pezizomycetes in HC soils was directly related to willow phylogeny, with Pezizomycetes dominating the rhizosphere of a monophyletic cluster of cultivars, while remaining in low relative abundance in other soils. This has implications for plant selection in phytoremediation, as fungal associations may affect the health of introduced plants and the success of co-inoculated microbial strains. An integrated understanding of the relationships between fungi, bacteria and plants will enable the design of treatments that specifically promote effective bioremediating communities.
Keywords: plant–microbe interactions, Salix, phytoremediation, hydrocarbon contamination, 16S rRNA, fungal ITS
Introduction
Remediation of contaminated soils using in situ phytoremediation is an attractive alternative to soil excavation and ex situ chemical treatment. Phytoremediation is cost-effective, can be applied to large areas of contaminated land, and introduced plants may even act as pioneer species for the regeneration of disturbed ecosystems. Several plant species are known to directly decontaminate polluted soils by sequestering and/or breaking down toxic compounds within their tissues (Peuke and Rennenberg, 2005; Pilon-Smits, 2005; Doty et al., 2007), but plants may also promote soil remediation by stimulating rhizosphere-inhabiting microorganisms that can degrade pollutants such as petroleum hydrocarbons (Yergeau et al., In press). To date, however, the introduction of pollutant-tolerant plants has had mixed effects on the extent of hydrocarbon biodegradation performed by soil microbial populations (for example, Rentz et al., 2004; Sipilä et al., 2008; Phillips et al., 2009; Cébron et al., 2011), and this is likely related in part to which specific taxa are promoted. Plants may encourage the growth of hydrocarbon-degrading microorganisms in the rhizosphere in order to protect their roots from the toxic effects of petroleum contaminants (Siciliano et al., 2001), but other factors can also drive differences between rhizosphere communities. In uncontaminated soils, rhizosphere microbial community composition has been shown to vary based on plant identity (Haichar et al., 2008; Berg and Smalla, 2009), and specific rhizosphere microorganisms may benefit plant partners by increasing nutrient acquisition (Richardson et al., 2009), facilitating adaptation to environmental change (Lau and Lennon, 2012) and protecting plants against pathogens (St-Arnaud and Vujanovic, 2007; Sikes et al., 2009; Mendes et al., 2011) or drought (Marasco et al., 2012).
Mycorrhizae, the close and generally mutualistic relationships between plants and fungi, have now been described as ‘tripartite associations', as they are influenced by the interactive activity of plants, fungi and bacteria (Bonfante and Anca, 2009). The symbiotic association between mycorrhizal fungi and plants is well established, but bacteria may occupy a broader range of niches (de Boer et al., 2005), and their role in the rhizosphere is less clear. Soil-inhabiting fungal and bacterial taxa are often grouped together under the broad term ‘microbial community', but these groups may not respond similarly to either hydrocarbon contaminants or plant introduction because of differences in physiology and ecology. Factors such as trace-metal concentration and pH have had differing and sometimes opposing effects on the activity, growth and diversity of fungal and bacterial populations (Rajapaksha et al., 2004; Stefanowicz et al., 2008; Rousk et al., 2009, 2010). Fungi and bacteria are also frequently described as antagonists in the soil environment (de Boer et al., 2005; Rousk et al., 2008; Bonfante and Anca, 2009; Schrey et al., 2012), and the competition between these groups has been shown to reduce denitrification activity (Siciliano et al., 2009) and fungal growth (Mille-Lindblom et al., 2006; Meidute et al., 2008). Some bacteria even inhabit hyphae (Hoffman and Arnold, 2010; Ghignone et al., 2012), but it is unknown whether these relationships are mutually beneficial, as the role of these bacteria is yet to be demonstrated.
Despite substantial recent advances in our understanding of the environmental factors that shape soil bacterial and fungal communities, the link between these communities is not well characterized. A previous study showed that the composition of communities of fungi (specifically arbuscular mycorrhizal fungi (AMF)) and bacteria was closely correlated in the rhizosphere of grass species from uncontaminated soils (Singh et al., 2008), but community relationships may be affected by contaminant introduction, as increasing disturbance has been shown to augment interspecies competition (Violle et al., 2010). In a greenhouse experiment, the addition of AMF positively affected plant growth in soils that were mildly or highly contaminated (HC) with zinc (Glassman and Casper, 2012). The addition of non-AMF microorganisms further enhanced plant growth in the mildly contaminated soils but reduced the positive influence of AMF as well as plant zinc uptake at high contaminant concentrations, suggesting enhanced intermicrobial competition. It is also unclear whether plant identity has a similar role in shaping communities in uncontaminated and contaminated soils. Disturbance from pollutants will reduce the number of active microbial taxa to only those that are pollutant-tolerant; therefore, microorganisms that are generally associated with a particular plant species may no longer be a relevant component of the community at high contaminant concentrations. This may lead to a reduction in the specificity of plant–microbe interactions.
In this study, we planted 11 cultivars of willow (Salix spp. L. Salicaceae) at the site of a former petrochemical plant in both uncontaminated soils and soils that were primarily contaminated with petroleum. We examined the rhizosphere-associated bacterial and fungal communities after the first 2 months of growth using high-throughput sequencing of partial 16S rRNA gene and internal transcribed spacer region (ITS) amplicons, respectively. Our aims were to determine whether fungal and bacterial communities are influenced to a similar extent by contaminant disturbance and the introduction of willow cultivars. In addition, we examined whether willow identity relates to microbial community composition and whether this relationship is modified by contaminant disturbance of the soil environment.
Materials and methods
Design of field experiment
Sampling occurred within a pilot project that is aimed at determining the feasibility of willow phytoremediation in Quebec soils. The project is located at the site of a former petrochemical plant in Varennes, Quebec, Canada (45°43 N, 73°22 W), and the area allocated to the project is ∼5000 m2. Initial planting of fresh willow clippings occurred on 6–7 June 2011. Willow clippings produce clones of the progenitor plant, and roots and shoots developed on site. Soil sampling occurred on 6 August 2011 for uncontaminated blocks and on 8 August 2011 for contaminated blocks. Mean temperature at the site during the growth period (6 June 2011–8 August 2011) was 21.9 °C, and there was a total of 196.9 mm of precipitation (Environment Canada—Verchères Weather Station; http://www.climate.weatheroffice.ec.gc.ca/index.html).
The site is separated into two areas—one that has been contaminated with petroleum hydrocarbons (Blocks C3, C4 and C5) and an adjacent uncontaminated area (Blocks N1, N3 and N5; an initial survey in 2010 found that petroleum concentrations in this area are below the detection limit). A randomized complete block design with 3 × 300 m2 blocks was set up on each site. Within each block were 12–5 × 5 m plots: one for each of 11 Salix cultivars (Table 1) and an unplanted control plot. Each planted plot contained five rows with 15 plants per row and 30 cm between each plant. Rows were spaced evenly at 1-m intervals.
Table 1. A list of the Salix cultivars planted and their associated characteristics.
| Cultivar name | Authority | Genotype | Characteristics | Native area |
|---|---|---|---|---|
| Fish creek | SUNY-ESF, NY, USA | Salix purpurea | High biomass production. Whole-genome sequencing in progress Introduced species | Europe |
| SX67 | University of Toronto, ON, Canada | Salix miyabeana | High biomass production Cultivated species | Asia |
| SX61 | University of Toronto, ON, Canada | Salix sachalinensis | High biomass production Cultivated species | Asia |
| S05 | Unknown | Salix nigra | Indigenous species | North America |
| S25 | University of Toronto, ON, Canada | Salix eriocephala | Indigenous species | North America |
| S365 | University of Toronto, ON, Canada | Salix caprea | Introduced species | Europe |
| SV1 | University of Toronto, ON, Canada | Salix dasyclados | Cultivated species | Eurasia |
| S54 | Unknown | Salix acutifolia | High biomass production Cultivated species | Eurasia |
| S44 | Unknown | Salix alba | High biomass production Introduced species | Eurasia |
| S33 | Unknown | Salix viminalis | High biomass production Genetic maps available Introduced species | Europe |
| Millbrook | SUNY-ESF, NY, USA | Salix purpurea X Salix miyabeana | High biomass production | Hybrid |
Within each plot, five rhizosphere soil samples were collected from randomly selected trees that were destructively harvested and pooled. Approximately 100 g of root-adhering soil was collected from each tree, representing a depth of ∼0–15 cm. Within the unplanted control plots, five bulk soil samples were collected from the top 15 cm of soil and pooled in the same manner. Roughly 50 g of each pooled sample was flash-frozen on-site in dry ice and ethanol, brought to the laboratory within 12 h and stored in 50-ml Falcon tubes at −80 °C prior to DNA extraction.
Soil samples from each contaminated block (n=12) were sent to Maxxam Analytics (Montreal, Quebec, Canada) on 9 August 2011, where soil was analyzed for F1–F4 hydrocarbons (sum of all aromatic and aliphatic hydrocarbon compounds with chain lengths of C6–C50) according to the protocol set forth by The Canadian Council of Ministers of the Environment (http://www.maxxam.ca/solutions/sol_env_CCME_Petr_Hydroc_0805.pdf). Average total petroleum concentrations for each of the contaminated blocks were 709 mg kg−1 (±339 s.e.) in block C3, 2143 mg kg−1 (±551 s.e.) in block C4 and 3590 mg kg−1 (±760 s.e.) in block C5. Other soil characteristics were determined from a pooled sample for each block at Agridirect Inc (Longueuil, Quebec, Canada) according to their standard operating procedures, and the values are displayed in Table 2.
Table 2. Soil characteristics for each block used in this study.
| Soil | Contaminant level | pH | Organic matter (%) | CEC (meq per 100 g) | Ca (kg ha−1) | P (kg ha−1) | Al (ppm) | K (kg ha−1) | Mg (kg ha−1) |
|---|---|---|---|---|---|---|---|---|---|
| N1 | No contamination | 6.5 | 6.1 | 24.6 | 6637 | 182 | 850 | 734 | 1551 |
| N3 | No contamination | 5.4 | 4.9 | 17.7 | 4171 | 199 | 904 | 378 | 569 |
| N5 | No contamination | 5.6 | 7.2 | 22 | 5587 | 182 | 961 | 421 | 896 |
| C3 | Low contamination | 6.7 | 9.2 | 27.9 | 8423 | 209 | 627 | 665 | 1230 |
| C4 | High contamination | 7.4 | 5.5 | 37.5 | 14062 | 26 | 727 | 692 | 1434 |
| C5 | High contamination | 7.4 | 2.2 | 25.8 | 7659 | ⩽10 | 925 | 580 | 2166 |
Soil DNA extraction, gene amplification and high-throughput sequencing
Total soil DNA was extracted from all samples, partial 16S rRNA gene (bacteria) and ITS (fungi) amplicons were produced for each using barcoded primers, and high-throughput sequencing was performed using the 454 GS FLX Titanium platform (Roche, Branford, CT, USA). Details on these procedures are described in the Supplementary Material.
Sequence processing
Quality processing of 16S rRNA gene sequences was performed in Mothur (v.1.28.0) following mainly the 454 SOP that is outlined in Schloss et al. (2011). ITS sequences were also processed using Mothur but were clustered using CD-HIT (Li and Godzik, 2006). Details on these procedures are described in the Supplementary Material. The sequence data generated in this study were deposited in the NCBI Sequence Read Archive and are available under the project number SRP026572.
Determination of willow phylogeny
Willow phylogeny was determined using maximum likelihood analyses based on the ITS region. The CTAB method (Doyle and Doyle, 1987) was used to extract genomic DNA, as modified in Lauron-Moreau et al. (2013). PCR amplification, sequencing, alignment and maximum likelihood analyses were performed on 11 willow taxa (details in Supplementary Material). We added a specimen of Populus (P. deltoides) as the outgroup for maximum likelihood analyses. The resulting phylogeny is provided in Supplementary Figure 1.
Statistical analysis
All statistical analyses were conducted in Mothur, JMP 8.0 (SAS Institute, Cary, NC, USA) and R v.2.15.2 (R Foundation for Statistical Computing; available at http://www.R-project.org). We examined the effects of operational taxonomic unit (OTU) nucleotide similarity cutoffs on metrics such as diversity and community Bray–Curtis distance at 97, 95 and 90% however, as the observed patterns were very similar at each cutoff, we used only 97% OTUs (most analyses) and 90% OTUs (correlation network) for final analyses. Singleton sequences that appeared only once in the data set were removed, and each sample was subsampled with the Mothur command ‘sub.sample' to 1207 reads for 97% OTUs and 1260 for 90% OTUs, which was the minimum number of sequences remaining in a single sample. The minimum number of reads per sample is higher for 90% OTUs, as the number of singleton sequences is reduced because of inclusion within larger OTUs.
To look at the effect of contaminants on community composition, we used a detrended correspondence analysis. The detrended correspondence analysis transformation was performed in R using the ‘decorana' command in the ‘vegan' package with down-weighting of rare taxa. Significance of the effect of contaminants on community structure was confirmed with a PERMANOVA on community Bray–Curtis values using the ‘adonis' function in ‘vegan'. The number of OTUs that were shared between contaminant levels were visualized using the Mothur ‘venn' command.
To determine whether fungal and bacterial communities were similarly affected by the introduction of different willow cultivars, we used Mantel tests to compare fungal and bacterial community Bray–Curtis dissimilarity matrices from each block. This test determines whether the distance between all measured samples is significantly correlated for paired matrices (in our case, bacterial and fungal intercommunity similarity). In other words, for each block, we compared whether the community distances for each possible pairwise combination of samples (for example, Fish-SX67, Fish-Unplanted control, and so on) were related for fungal and bacterial communities. We also compared the mean Bray–Curtis dissimilarity value between all bacterial or fungal communities within each block and tested for significant differences using a one-way analysis of variance (ANOVA). Although 454 read abundance is not an exact reflection of the actual taxonomic abundance in situ (Amend et al., 2010), standardized processing allows the detection of relative shifts between microbial communities.
The mean fungal and bacterial diversities at each contaminant level were compared using a one-way ANOVA. The mean change in diversity across willow cultivars relative to the unplanted block controls was also compared between bacteria and fungi at each contaminant level using two-tailed Student's t-tests. The composition of major fungal and bacterial classes was compared between willow cultivars at each contaminant level using UPGMA clustering. Taxonomic abundance data were first normalized using the ‘decostand' and ‘vegdist' commands in the ‘vegan' package of R. The resulting clustering trees were paired with a heatmap of abundance data created with ‘heatmap.2' from the ‘gplots' package and were compared with the willow phylogeny.
To calculate shifts in abundance correlations between organisms, we used a 90% OTU cutoff to simplify visualization and interpretation as in Barbéran et al. (2012). The aim was to observe shifts in abundance relationships between OTUs in response to willow introductions. For each willow cultivar, only those OTUs that were observed in at least three blocks for both the unplanted control plots and the cultivar were compared in order to reduce noise from rare OTUs. Spearman's rank correlations were calculated for each species pair in R using the ‘ade4' package and were kept only if the R2 value was higher than 0.531 (P=0.05 for one-tailed tests). From this data set, correlations were selected if they switched signs between unplanted plots and the willow cultivar (that is, positive to negative correlation or negative to positive correlation). The resulting correlation network was then visualized using Cytoscape (Smoot et al., 2011).
Results
Effect of contaminants on community divergence
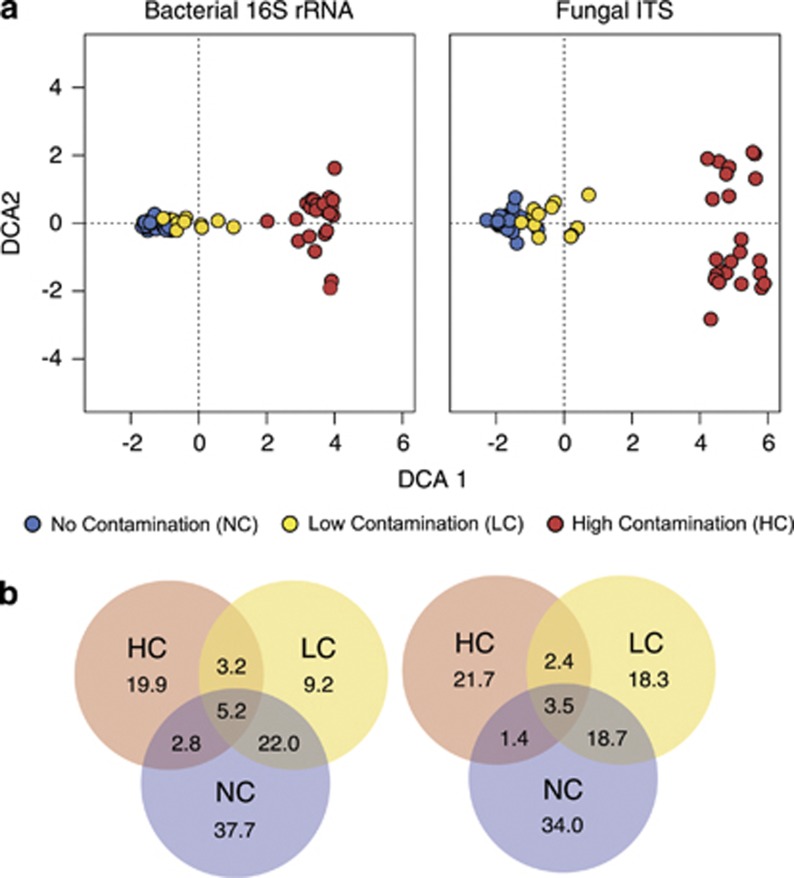
After sequence-quality filtering, we obtained a total of 232 021 usable 16S rRNA gene reads and 634 476 usable ITS reads. After standardizing the number of sequences per sample at a 97% OTU cutoff, detrended correspondence analyses of OTU matrices showed that the concentration of hydrocarbon contaminants in each block had a strong and significant effect on both bacterial and fungal community structures (Figure 1a; P<0.01 for both bacteria and fungi using a PERMANOVA). Communities from the HC blocks (>2000 mg kg−1 soil) were clearly distinct from all others, whereas communities from the low-contaminant (LC) block (709 mg kg−1) were distinct but with slight overlap with the non-contaminated (NC) communities. The percent of OTUs shared between contaminant types (for example, HC and LC) was similar for fungal and bacterial communities (Figure 1b). The percent of OTUs unique to each contaminant type was also similar, with the exception of LC, in which the percent of unique OTUs in fungal communities was nearly double of that observed for bacteria.
Figure 1.
Comparison of bacterial 16S rRNA and fungal ITS communities by contaminant level. Community relatedness shown using (a) detrended correspondence analyses, and (b) Venn diagrams showing OTUs shared between contaminant levels at 97% genetic similarity. Data derived from rhizosphere samples of each of the 11 willow cultivars and unplanted controls from each block (no contamination=blocks N1, N3, N5; low contamination=block C3; high contamination=blocks C4, C5).
Interactive effects of contaminants and willow rhizosphere
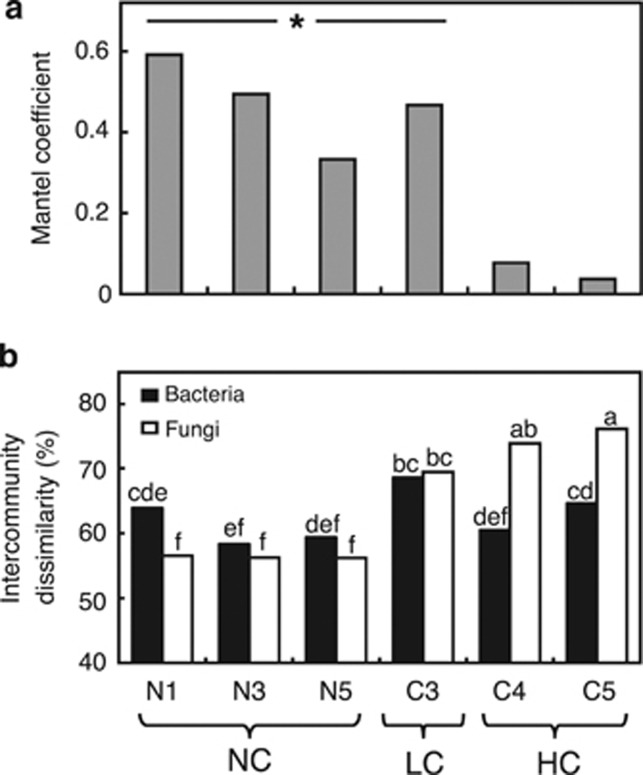
In the NC and LC blocks, Mantel correlation coefficients showed that significant relationships (P<0.05) existed between the fungal and bacterial communities, but there was no significant relationship in the HC blocks (Figure 2a). The mean Bray–Curtis dissimilarity within each block varied significantly (one-way ANOVA, F=29.02, P<0.001) both across blocks and between bacterial and fungal communities (Figure 2b). The highest divergence of fungal communities occurred in the HC blocks, although the mean Bray–Curtis dissimilarity in block C4 could not be significantly differentiated from either bacterial or fungal communities in the LC block (Tukey's HSD test, P<0.05).
Figure 2.
Comparison of bacterial 16S rRNA and fungal ITS Bray–Curtis community dissimilarity matrices across willow cultivars and unplanted control by block. Similarity in Bray–Curtis distance matrices for bacteria and fungi was (a) tested for significance using Mantel correlation tests. Columns indicated by the star and horizontal line represent significant Mantel coefficients (P<0.05). The mean Bray–Curtis dissimilarity was also compared between bacteria and fungi by (b) averaging Bray–Curtis distances between all plots within each block. Different letters over columns indicate significantly different means according to Tukey's HSD test (P<0.05). Contaminant levels are indicated by NC (not contaminated), LC (low contamination) and HC (high contamination).
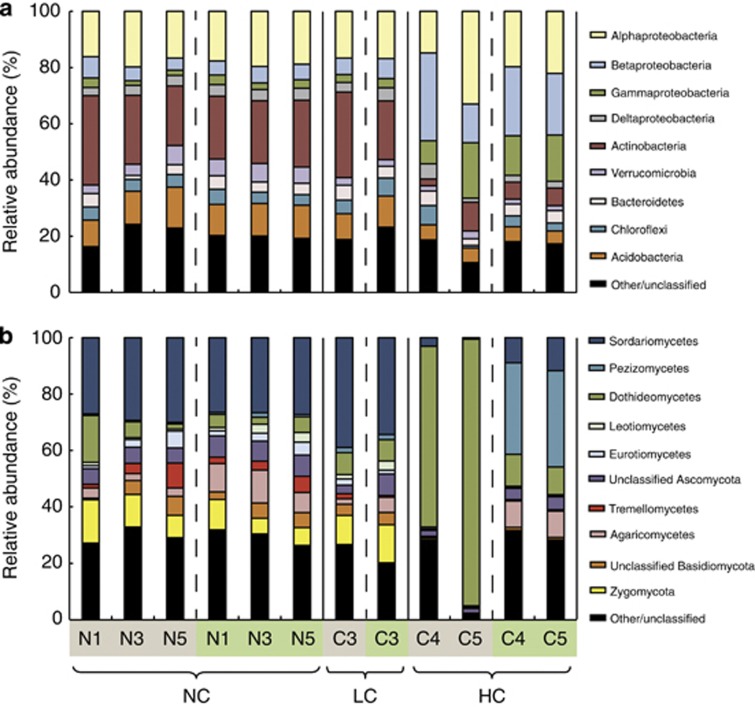
Relative to the NC and LC blocks, there was a reduction in the HC blocks in the abundance of Actinobacteria as well as several groups that are not known to contain important hydrocarbon-biodegrading organisms (for example, Acidobacteria, Verrucomicrobia), while the Beta- and Gammaproteobacteria generally increased, as well as the Alphaproteobacteria in the unplanted control from block C5 (Figure 3a). These classes of Proteobacteria were represented by several taxa in which hydrocarbon degradation is commonly known to occur, including the Sphingomonadaceae, the Burkholderiales and the Xanthomonadaceae (Supplementary Figure 2). Similar abundances of the major groups were observed with and without the presence of willows, although the mean effect of willow introduction on the bacterial community structure appears to be a reduction in the block-to-block variation observed in the unplanted controls (Figure 3a).
Figure 3.
Community composition of major (a) bacterial phyla (and classes of Proteobacteria), and (b) classes of fungi by block for both unplanted controls and averaged across willow cultivars. Block labels shaded in brown indicate unplanted controls, whereas labels shaded in green indicate community composition averaged across the 11 willow cultivars of that block. Contaminant levels are indicated by NC (not contaminated), LC (low contamination) and HC (high contamination).
Larger shifts were observed in the affiliation of classified fungal sequences (Figure 3b). Communities from the NC and LC blocks were represented by a variety of fungal classes, with Sordariomycetes appearing as the most abundant group; however, HC-unplanted control plots were dominated by Dothideomycetes (Figure 3b), primarily by Pleosporales inc. sed. and Sporormiaceae (Supplementary Figures 3 and 4). The addition of willows drastically reduced the abundance of Dothideomycetes and allowed several other groups to increase in abundance, especially the Pezizomycetes group (Figure 3b), which were represented almost exclusively by the genus Sphaerosporella.
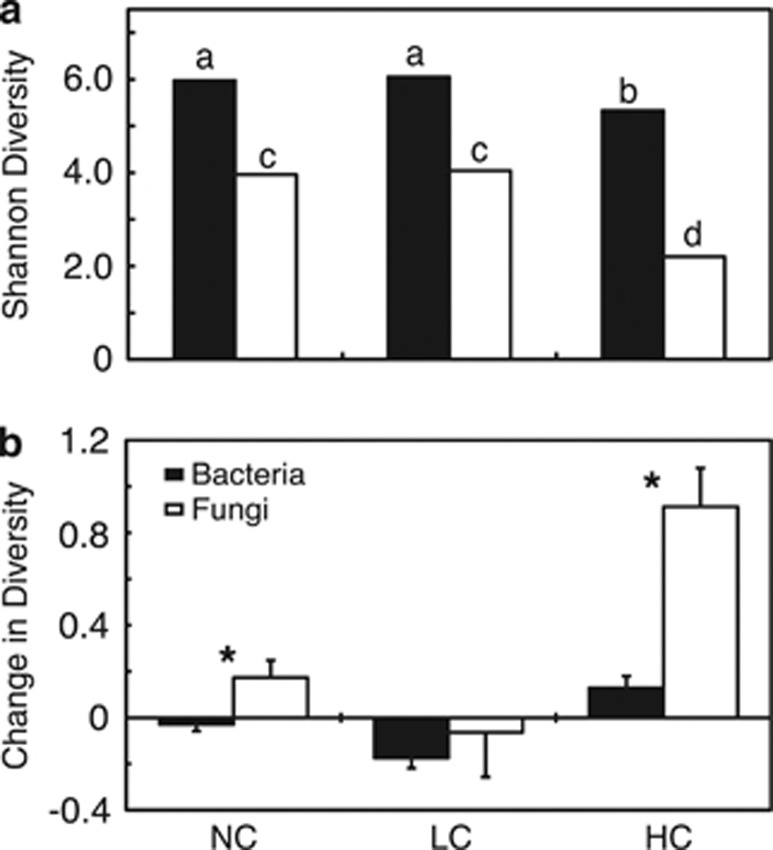
The qualitative shifts in community composition were corroborated by differences in OTU Shannon diversity. Diversity differed significantly between fungal and bacterial populations (one-way ANOVA, F=310.84, P<0.001) (Figure 4a), with lower overall diversity in fungal populations across all contaminant levels and significant reductions in both fungal and bacterial diversity in the HC blocks (n=24) relative to the NC (n=36) and LC (n=12) blocks (Tukey's HSD test, P<0.05). While comparing shifts in diversity following willow introduction for fungal and bacterial populations at each contaminant level, fungal diversity increased significantly more than did bacterial diversity in both NC (n=33) and HC (n=22) blocks (Student's t-test, P<0.05) (Figure 4b).
Figure 4.
Comparison of Shannon diversity between bacteria and fungi by contaminant level using (a) the mean of all rhizosphere communities and unplanted controls, and (b) change in diversity relative to unplanted control for each block. Different letters over columns indicate significantly different means according to Tukey's HSD test (P<0.05). Stars over paired columns indicate that the means are significantly different according to the Student's t-test (P<0.05).
Cultivar-specific influences
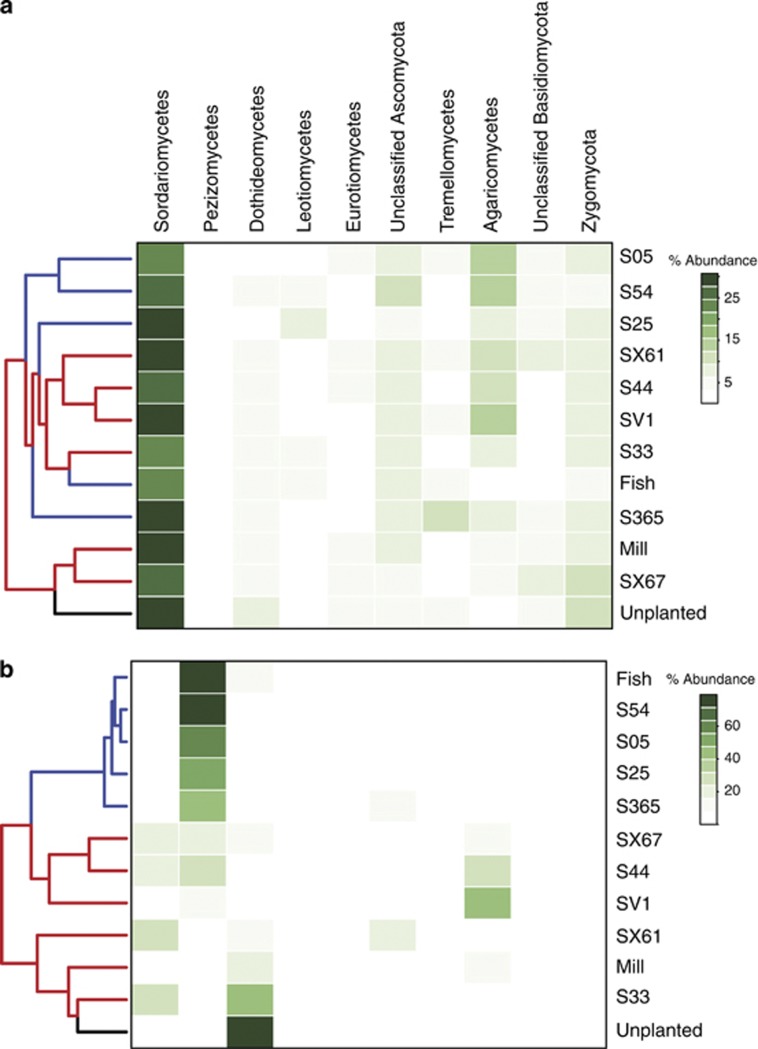
The clustering of cultivar-specific bacterial communities at the class level did not reveal any close associations between willow types at any contaminant level (data not shown); however, the same analysis indicated close similarities of certain fungal communities. Although the abundance of the dominant Sordariomycetes was similar across cultivars in the NC blocks (Figure 5a), fungal communities occurring in the rhizosphere of the Fish creek, S54, S05, S365 and S25 cultivars in the HC blocks were tightly associated following the UPGMA clustering, driven mainly by a high abundance of Pezizomycetes (Figure 5b). This cluster was not clearly associated with any of the early measurements of plant growth (that is, tree height, diameter, number of shoots and seedling survival) from the site (data not shown), but was related to willow phylogeny (phylogenetic tree of willow cultivars provided as Supplementary Figure 1).
Figure 5.
Heatmap showing differences in composition of major classified fungal classes by cultivar for communities from (a) non-contaminated soils and (b) highly contaminated soils. Note the change in scale between panels. The tree to the left indicates similarity in fungal community composition by cultivar based on the UPGMA clustering. Similarly colored tree branches (blue/red) indicate cultivars that formed monophyletic clades according to ITS phylogeny mapping of the willows used in this study (see Supplementary Figure 1). The unplanted control communities are indicated in black.
Aside from changes in the abundance of specific taxa, the biotic environment of many groups was influenced by the introduction of willows across contaminant levels and was variable between cultivars (Figure 6; Supplementary Table 1). We targeted OTUs with at least four changed associations to indicate ‘hubs' of change within the biotic environment. Only the portion of the network connected to hubs is depicted, although this represents the vast majority of each network. The number of hubs varied widely between cultivars, ranging from 2 (S25) to 12 (S54), and was generally not associated with large changes in abundance in the OTU itself. When OTUs were ranked by average change in abundance by block in the presence of each cultivar, only 12/70 of these hubs were among the top 10 OTUs in changed abundance, and only 14/70 were among the top 20. Hubs represented both bacteria (47 nodes) and fungi (27 nodes), showing that large changes were not limited to the groups that were most directly affected by willow introduction.
Figure 6.
Graphical representation of correlations between taxa (P<0.05) that change sign (positive to negative or vice versa) between unplanted controls and the rhizosphere of each willow cultivar. Nodes represent OTUs (90% nucleotide similarity), and nodes with four or more changed correlations are enlarged. The consensus taxonomy for each major node is provided in the legend. Node shapes indicate whether the OTU belongs to the Bacteria (circle) or Fungi (triangle). Node coloring indicates whether the OTU is more abundant in the unplanted control plots (gray) or the rhizosphere of the specified cultivar (black). Connecting lines indicate whether the correlation has switched to a positive correlation (solid blue) or a negative correlation (dashed red) in the presence of the cultivar. The legend for node labels is provided as Supplementary Table 1.
Discussion
Plants influence and are influenced by many aspects of the soil environment, including soil structure, nutrient availability and the biotic landscape (Van Breemen and Finzi, 1998; Reynolds et al., 2003). In this study, we examined how the introduction of the 11 Salix cultivars influenced the composition and differentiation of soil bacterial and fungal communities, and whether this effect varied in the presence of hydrocarbon contaminants. Bacterial and fungal intercommunity distance between rhizosphere samples (that is, Bray–Curtis dissimilarity between Fish-SX67, Fish-Unplanted control, and so on) was significantly correlated for bacterial and fungal communities in soils that were uncontaminated or contained low concentrations of hydrocarbons, but diverged in soils that were highly hydrocarbon-contaminated. This shows that there was a differential effect of willows on bacteria and fungi in HC soils. In addition, both fungal and bacterial diversity declined significantly in HC soils, but fungal diversity was increased by willow introduction significantly more than was bacterial diversity, suggesting that phytoremediation may have a disproportionate direct effect on fungi in highly disturbed environments. Nevertheless, the shift in the biotic environment of both bacteria and fungi caused by willow introduction potentially affects interactions between bacteria, fungi and plants, which may lead to other indirect effects on community functioning.
Hydrocarbon contamination had a large effect on the structuring of both bacterial and fungal communities, as has been shown previously (Törneman et al., 2008; Bell et al., 2011; Liang et al., 2011; Bell et al., 2013a, 2013b). The observed reduction in bacterial diversity in the HC soils is in line with what was observed by Bell et al. (2013b). The decline in Actinobacteria and promotion of Proteobacteria in these low organic-matter soils, however, are in contrast with those results, in which Actinobacteria were generally dominant in low organic-matter soils following the addition of diesel and nutrient amendments. This difference may be related to the fact that willows can provide a steady source of organic material through root exudation, thus increasing the organic-matter supply to the rhizosphere environment. Less is known about hydrocarbon-tolerant fungal populations, although most identified hydrocarbon-degrading fungi are restricted to the Ascomycota and Basidiomycota (Harms et al., 2011). Cultivable fungal diversity has previously been shown to decrease at high hydrocarbon concentrations, which is similar to what we observed (Ferrari et al., 2011). Although the same study showed that the abundance of cultured Dothideomycetes increased at high hydrocarbon concentrations (Ferrari et al., 2011), the complete dominance of this group in the unplanted controls was unexpected. Interestingly, several genera of Dothideomycetes, including Phoma and Preussia, which dominated the unplanted HC plots, have been shown to harbor endohyphal bacteria from groups that are capable of hydrocarbon biodegradation, such as the Xanthomonadales, Pseudomonadales, Burkholderiales and Sphingomonadales (Hoffman and Arnold, 2010), which are bacterial groups that were also abundant in our HC plots (Supplementary Figures 2 and 4). Fungi have also been suspected to facilitate hydrocarbon bioremediation in soils by translocating hydrocarbon-degrading bacteria through their hyphae, thereby increasing bacterial access to hydrophobic substrates (Wick et al., 2010). Phoma may be more tolerant than other fungal groups to the conditions in the HC blocks, as this genus is known to both produce and degrade hydrocarbon compounds (Oikawa et al., 2001; Cerniglia and Sutherland, 2010; Strobel et al., 2011), whereas Preussia possesses alkane monoxygenase genes (Shahsavari et al., 2013). Differences between the HC and uncontaminated blocks may also be partly related to a notable pH shift, although an increase to a neutral soil pH (7.4) should not be expected to lead to drastic reductions in microbial diversity (Fierer and Jackson, 2006; Rousk et al., 2010).
Interestingly, the effect of willows on bacterial and fungal communities changed depending on the contaminant concentration. Although the similarity of bacterial and fungal communities was significantly correlated in the NC and LC blocks, this was not the case in the HC blocks. This was partly because of a large decline in the fungal OTU diversity from the uncontaminated to HC soils (∼50%), whereas the positive influence of willow introduction on diversity was significantly greater in both NC and HC plots. This points to a closer association between plants and fungi than between plants and bacteria, and especially that phytoremediation may have a more direct influence on the fungal than the bacterial population in HC soils. Bacteria are generally considered to be the primary hydrocarbon degraders in contaminated soils (Song et al., 1986; Harrison and Betts, 1996), and the extent of hydrocarbon degradation in soils has been correlated with the abundance of specific bacterial groups (Bell et al., 2013b). As a result, one of the main roles of effective plants in phytoremediation of hydrocarbons may be in selecting fungi that are synergistic with, or at least not antagonistic to, target hydrocarbon-degrading bacteria. Reduced abundance of specific microbial groups has previously been shown to enhance bioremediation in soil (Fournier and Fournier, 1993; Bell et al., 2013a), and microbial competition also apparently affects the success of plant growth and pollutant uptake (Glassman and Casper, 2012).
De Deyn et al. (2011) showed that whereas AMF abundance was related to plant identity, the abundance of saprobic fungi and bacteria was related to plant biomass. In our study, we found no obvious effect of plant biomass measures on bacterial or fungal community structure, and interestingly, plant–fungi specificity was more apparent in HC soils. Whether coevolution has an important ongoing role in most plant–mycorrhizal relationships is unknown (Hoeksema, 2010); however, our data suggest that the importance of some relationships may be cryptic and only evident in disturbed environments when selection pressures are enhanced. Although the history of many willow cultivars is complex and incompletely cataloged, willows that promoted high Pezizomycete abundance were closely related phylogenetically, and were primarily North American varieties, either indigenous (S05, S25) or naturalized in the case of Fish Creek and S365 (Argus, 2007; Lin et al., 2009). The exception was S. acutifolia, a cultivar native to Russia (Zapesochnaya et al., 2002), for which little information is available. This suggests that these varieties may have evolved natural relationships with indigenous microbial species, whereas more distantly related varieties formed nonspecific associations with the fungi that were best able to capitalize on the particular rhizosphere environment that was created. The benefits of this association to willows remain to be shown, however, and it may be that the promoted Pezizomycetes have adapted to exploit plant species that are similar to those that they have previously encountered. The majority of Pezizomycete sequences were identified as Sphaerosporella brunnea, which is an ectomycorrhizal fungus with a rapid growth rate (Danielson, 1984) that has been shown to be a successful pioneer symbiont following fire disturbance (Egger and Paden, 1986).
The implications of this finding on plant selection for phytoremediation are two-fold. Firstly, the health of locally adapted and foreign cultivars may be differentially affected by the strength of their associations with indigenous fungi. For instance, fungi grown in their soil of origin with native plant species have been shown to be more mutualistic (more arbuscules) than combinations with foreign plant species (Johnson et al., 2010). Sphaerosporella was previously shown to have no significant effect on willow growth in an uncontaminated soil (Fernando and Currah, 1996) but may affect plant health in other respects (for example, increased pathogen protection or susceptibility, adaptation to high levels of stress and so on). Modeling suggests that a limited number of specific fungal symbionts may actually benefit plants more than a diverse community, as the number of defectors that do not contribute to plant health is limited (Veresoglou and Halley, 2012). Secondly, the interactions between Sphaerosporella and other microorganisms may affect bioremediation performance. Bioaugmentation, the addition of cultured microbial strains to contaminated soils, has frequently been ineffective with bacteria, as added strains tend to be outcompeted by the existing microbial population (Thomassin-Lacroix et al., 2002; Thompson et al., 2005). As the fungal community associated with some willow cultivars is specific and predictable in HC soils, interactions between putative bioaugmentation strains and the existing community can be tested thoroughly in vitro to determine antagonistic or synergistic relationships. Alternatively, the less specific associations between fungi and foreign willow cultivars may actually facilitate the introduction and persistence of target species. Strain selection for bioaugmentation must consider factors that allow introduced strains to survive in the target environment (Thompson et al., 2005), and this necessarily includes interactions with the indigenous community.
Phylogenetic markers such as the 16S and 18S rRNA subunits and the fungal ITS region do not carry explicit functional information. Nevertheless, community assembly patterns will likely influence the functioning of microbial communities, as many, if not most, functional traits appear to be linked to specific phylogenetic lineages among fungi (Maherali and Klironomos, 2007) and even prokaryotes that engage in horizontal gene transfer (Martiny et al., 2013). In the context of hydrocarbon bioremediation, community composition has previously been correlated to function (Bell et al., 2013b). We also show here that changes within the rhizosphere community are not limited to the microorganisms that are most directly affected by plant introduction, as willows shift the potential interaction network of many taxa. Extraction of total soil DNA only allows us to examine microbial communities at a coarse scale, and we cannot determine whether taxa occupy similar microenvironments and actually interact. Nevertheless, it is interesting to consider the fact that plants may be able to indirectly influence microorganisms that do not directly associate with roots or root-produced compounds. The distinct communities selected by the 11 willow cultivars should vary in their potential to biodegrade hydrocarbons, although actual activity will also be influenced by environmental factors and the abundance of alternative carbon substrates.
Conclusions
Close interactions between plants, bacteria and fungi occur within the rhizosphere; however, the link between these groups is altered in soils that have been disturbed by high concentrations of hydrocarbon contaminants. Fungal communities were disproportionately sensitive to hydrocarbons relative to bacteria. The introduction of willows promoted more diverse fungal communities on average; however, fungal communities diverged based on Salix identity, moreso than bacterial communities. The promotion of a very specific Pezizomycete-dominated community by phylogenetically similar willow cultivars suggests that the evolutionary history of plants should be considered when selecting varieties for phytoremediation. This study highlights the need to closely examine interactions between rhizosphere-inhabiting organisms in the context of phytoremediation. An integrated understanding of community dynamics will further clarify why plant species promote specific microorganisms, especially those that are proficient in bioremediation.
Acknowledgments
We thank Werther Guidi for providing descriptive plant data from with our site to compare with our microbial community analyses. We also thank Werther, Marie-Claude Turmel, Sébastien Halary and Yves Terrat for their assistance with field sampling and advice on data analysis. We also thank ConocoPhillips for providing us with access to the Varennes field site. This work was supported by the Genome Quebec and Genome Canada funded GenoRem Project. TH Bell was also supported in part by a grant from the Fonds québécois de la recherche sur la nature et les technologies.
The GenoRem project contains several industrial partners, but these partners have in no way influenced or modified this manuscript or the analysis of the results presented.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amend AS, Seifert KA, Bruns TD. Quantifying microbial communities with 454 pyrosequencing: does read abundance count. Mol Ecol. 2010;19:5555–5565. doi: 10.1111/j.1365-294X.2010.04898.x. [DOI] [PubMed] [Google Scholar]
- Argus GW. Salix (Salicaceae) distribution maps and a synopsis of their classification in North America, North of Mexico. Harv Paper Bot. 2007;12:335–368. [Google Scholar]
- Barbéran A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TH, Yergeau E, Martineau C, Juck D, Whyte LG, Greer CW. Identification of nitrogen-incorporating bacteria in petroleum-contaminated Arctic soils by using [15N]DNA-based stable isotope probing and pyrosequencing. Appl Environ Microbiol. 2011;77:4163–4171. doi: 10.1128/AEM.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TH, Yergeau E, Juck D, Whyte LG, Greer CW. Alteration of microbial community structure affects diesel degradation in an Arctic soil. FEMS Microb Ecol. 2013a;85:51–61. doi: 10.1111/1574-6941.12102. [DOI] [PubMed] [Google Scholar]
- Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW. Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J. 2013b;7:1200–1210. doi: 10.1038/ismej.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009;68:1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Anca IA. Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol. 2009;63:363–383. doi: 10.1146/annurev.micro.091208.073504. [DOI] [PubMed] [Google Scholar]
- Cébron A, Louvel B, Faure P, France-Lanord C, Chen Y, Murrell JC, et al. Root exudates modify bacterial diversity of phenanthrene degraders in PAH-polluted soil but not phenanthrene degradation rates. Environ Microbiol. 2011;13:722–736. doi: 10.1111/j.1462-2920.2010.02376.x. [DOI] [PubMed] [Google Scholar]
- Cerniglia CE, Sutherland JB.2010Degradation of polycyclic aromatic hydrocarbons by fungiIn: Timmis KN (ed)Handbook of Hydrocarbon and Lipid Microbiology Springer: Berlin, Heidelberg; 2079–2110. [Google Scholar]
- Danielson RM. Ectomycorrhiza formation by the operculate discomycete Sphaerosporella brunnea (Pezizales) Mycologia. 1984;76:454–461. [Google Scholar]
- de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- De Deyn GB, Quirk H, Bardgett RD. Plant species richness, identity and productivity differentially influence key groups of microbes in grassland soils of contrasting fertility. Biol Lett. 2011;7:75–78. doi: 10.1098/rsbl.2010.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty SL, James CA, Moore AL, Vajzovic A, Singleton GL, Ma C, et al. Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. P Natl Acad Sci USA. 2007;104:16816–16821. doi: 10.1073/pnas.0703276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Egger KN, Paden JW. Biotrophic associations between lodgepole pine seedlings and post-fire ascomycetes (Pezizales) in monoxenic culture. Can J Bot. 1986;64:2719–2725. [Google Scholar]
- Fernando AA, Currah RS. A comparative study of the effects of the root endophytes Leptodontidium orchidicola and Phialocephala fortinii (Fungi Imperfecti) on the growth of some subalpine plants in culture. Can J Bot. 1996;74:1071–1078. [Google Scholar]
- Ferrari BC, Zhang C, van Dorst J. Recovering greater fungal diversity from pristine and diesel fuel contaminated sub-Antarctic soil through cultivation using both a high and a low nutrient media approach. Front Microbiol. 2011;2:217. doi: 10.3389/fmicb.2011.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. P Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier G, Fournier JC. Effect of microbial competition on the survival and activity of 2,4-D-degrading Alcaligenes xylosoxidans subsp. denitrificans added to soil. Lett Appl Microbiol. 1993;16:178–181. [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L, et al. The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 2012;6:136–145. doi: 10.1038/ismej.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman SI, Casper BB. Biotic contexts alter metal sequestration and AMF effects on plant growth in soils polluted with heavy metals. Ecology. 2012;93:1550–1559. doi: 10.1890/10-2135.1. [DOI] [PubMed] [Google Scholar]
- Haichar FE, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- Harms H, Schlosser D, Wick LY. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol. 2011;9:177–192. doi: 10.1038/nrmicro2519. [DOI] [PubMed] [Google Scholar]
- Harrison AB, Betts WB. Assay of bacterial and fungal activity in diesel contaminated soil using a 14C-glucose utilization method. Lett Appl Microbiol. 1996;23:43–46. [Google Scholar]
- Hoeksema JD. Ongoing coevolution in mycorrhizal interactions. New Phytol. 2010;187:286–300. doi: 10.1111/j.1469-8137.2010.03305.x. [DOI] [PubMed] [Google Scholar]
- Hoffman MT, Arnold AE. Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microb. 2010;76:4063–4075. doi: 10.1128/AEM.02928-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NC, Wilson GWT, Bowker MA, Wilson JA, Miller RM. Resource limitation is a driver of local adaptation in mycorrhizal symbioses. P Natl Acad Sci USA. 2010;107:2093–2098. doi: 10.1073/pnas.0906710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JA, Lennon JT. Rapid responses of soil microorganisms improve plant fitness in novel environments. P Natl Acad Sci USA. 2012;109:14058–14062. doi: 10.1073/pnas.1202319109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauron-Moreau A, Pitre FE, Brouillet L, Labrecque M. Microsatellite markers of willow species and characterization of 11 polymorphic microsatellites for Salix eriocephala (Salicaceae), a potential native species for biomass production in Canada. Plants. 2013;2:203–210. doi: 10.3390/plants2020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WZ, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liang Y, Van Nostrand JD, Deng Y, He Z, Wu L, Zhang X, et al. Functional gene diversity of soil microbial communities from five oil-contaminated fields in China. ISME J. 2011;5:403–413. doi: 10.1038/ismej.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Gibbs JP, Smart LB. Population genetic structure of native versus naturalized sympatric shrub willows (Salix; Salicaceae) Am J Bot. 2009;96:771–785. doi: 10.3732/ajb.0800321. [DOI] [PubMed] [Google Scholar]
- Maherali H, Klironomos JN. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science. 2007;316:1746–1748. doi: 10.1126/science.1143082. [DOI] [PubMed] [Google Scholar]
- Marasco R, Rolli E, Ettoumi B, Vigani G, Mapelli F, Borin S, et al. A drought resistance-promoting microbiome is selected by root system under desert farming. PLoS One. 2012;7:e48479. doi: 10.1371/journal.pone.0048479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7:830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meidute S, Demoling F, Bååth E. Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol Biochem. 2008;40:2334–2343. [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- Mille-Lindblom C, Fischer H, Tranvik LJ. Antagonism between bacteria and fungi: substrate competition and a possible tradeoff between fungal growth and tolerance towards bacteria. Oikos. 2006;113:233–242. [Google Scholar]
- Oikawa H, Toshima H, Ohashi S, Konig H, Kenmoku H, Sassa T. Diversity of diterpene hydrocarbons in fungus Phoma betae. Tetrahedron Lett. 2001;42:2329–2332. [Google Scholar]
- Peuke AD, Rennenberg H. Phytoremediation: molecular biology, requirements for application, environmental protection, public attention and feasibility. EMBO Rep. 2005;6:497–501. doi: 10.1038/sj.embor.7400445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips LA, Greer CW, Farrell RE, Germida JJ. Field-scale assessment of weathered hydrocarbon degradation by mixed and single plant treatments. Appl Soil Ecol. 2009;42:9–17. [Google Scholar]
- Pilon-Smits E. Phytoremediation. Annu Rev Plant Biol. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- Rajapaksha RMCP, Tobor-Kaplon MA, Bååth E. Metal toxicity affects fungal and bacterial activities in soil differently. Appl Environ Microb. 2004;70:2966–2973. doi: 10.1128/AEM.70.5.2966-2973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentz JA, Alvarez PJJ, Schnoor JL. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ Microbiol. 2004;6:574–583. doi: 10.1111/j.1462-2920.2004.00589.x. [DOI] [PubMed] [Google Scholar]
- Reynolds HL, Packer A, Bever JD, Clay K. Grassroots ecology: plant-microbe-soil interactions as drivers of plant community structure and dynamics. Ecology. 2003;84:2281–2291. [Google Scholar]
- Richardson AE, Barea JM, McNeill AM, Prigent-Combaret C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009;321:305–339. [Google Scholar]
- Rousk J, Demoling LA, Bahr A, Bååth E. Examining the fungal and bacterial niche overlap using selective inhibitors in soil. FEMS Microbiol Ecol. 2008;63:350–358. doi: 10.1111/j.1574-6941.2008.00440.x. [DOI] [PubMed] [Google Scholar]
- Rousk J, Brookes PC, Bååth E. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl Environ Microb. 2009;75:1589–1596. doi: 10.1128/AEM.02775-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010;4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16 S rRNA-based studies. PLoS One. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrey SD, Erkenbrack E, Fruh E, Fengler S, Hommel K, Horlacher N, et al. Production of fungal and bacterial growth modulating secondary metabolites is widespread among mycorrhiza-associated streptomycetes. BMC Microbiol. 2012;12:164. doi: 10.1186/1471-2180-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahsavari E, Adetutu EM, Anderson PA, Ball AS. Plant residues: a low cost, effective bioremediation treatment for petrogenic hydrocarbon-contaminated soil. Sci Total Environ. 2013;443:766–774. doi: 10.1016/j.scitotenv.2012.11.029. [DOI] [PubMed] [Google Scholar]
- Siciliano SD, Fortin N, Mihoc A, Wisse G, Labelle S, Beaumier D, et al. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl Environ Microbiol. 2001;67:2469–2475. doi: 10.1128/AEM.67.6.2469-2475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano SD, Ma WK, Ferguson S, Farrell RE. Nitrifier dominance of Arctic soil nitrous oxide emissions arises due to fungal competition with denitrifiers for nitrate. Soil Biol Biochem. 2009;41:1104–1110. [Google Scholar]
- Sikes BA, Cottenie K, Klironomos JN. Plant and fungal identity determines pathogen protection of plant roots by arbuscular mycorrhizas. J Ecol. 2009;97:1274–1280. [Google Scholar]
- Singh BK, Nunan N, Ridgway KP, McNicol J, Young JPW, Daniell TJ, et al. Relationship between assemblages of mycorrhizal fungi and bacteria on grass roots. Environ Microbiol. 2008;10:534–541. doi: 10.1111/j.1462-2920.2007.01474.x. [DOI] [PubMed] [Google Scholar]
- Sipilä TP, Keskinen AK, Åkerman ML, Fortelius C, Haahtela K, Yrjälä K. High aromatic ring-cleavage diversity in birch rhizosphere: PAH treatment-specific changes of IE3 group extradiol dioxygenases and 16 S rRNA bacterial communities in soil. ISME J. 2008;2:968–981. doi: 10.1038/ismej.2008.50. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HG, Pedersen TA, Bartha R. Hydrocarbon mineralization in soil: relative bacterial and fungal contribution. Soil Biol Biochem. 1986;18:109–111. [Google Scholar]
- St-Arnaud M, Vujanovic V.2007Effect of arbuscular mycorrhizal symbiosis on plant diseases and pestsIn: Hamel C, Plenchette C (eds)Mycorrhizae in crop production Haworth Food & Agricultural Products Press: Binghampton: New York, NY, USA; 67–122. [Google Scholar]
- Stefanowicz AM, Niklinska M, Laskowski R. Metals affect soil bacterial and fungal functional diversity differently. Environ Toxicol Chem. 2008;27:591–598. doi: 10.1897/07-288.1. [DOI] [PubMed] [Google Scholar]
- Strobel G, Singh SK, Riyaz-Ul-Hassan S, Mitchell AM, Geary B, Sears J. An endophytic/pathogenic Phoma sp. from creosote bush producing biologically active volatile compounds having fuel potential. FEMS Microbiol Lett. 2011;320:87–94. doi: 10.1111/j.1574-6968.2011.02297.x. [DOI] [PubMed] [Google Scholar]
- Thomassin-Lacroix EJM, Eriksson M, Reimer KJ, Mohn WW. Biostimulation and bioaugmentation for on-site treatment of weathered diesel fuel in Arctic soil. Appl Microbiol Biot. 2002;59:551–556. doi: 10.1007/s00253-002-1038-0. [DOI] [PubMed] [Google Scholar]
- Thompson IP, van der Gast CJ, Ciric L, Singer AC. Bioaugmentation for bioremediation: the challenge of strain selection. Environ Microbiol. 2005;7:909–915. doi: 10.1111/j.1462-2920.2005.00804.x. [DOI] [PubMed] [Google Scholar]
- Törneman N, Yang XH, Bååth E, Bengtsson G. Spatial covariation of microbial community composition and polycyclic aromatic hydrocarbon concentration in a creosote-polluted soil. Environ Toxicol Chem. 2008;27:1039–1046. doi: 10.1897/07-440.1. [DOI] [PubMed] [Google Scholar]
- Van Breemen N, Finzi AC. Plant-soil interactions: ecological aspects and evolutionary implications. Biogeochemistry. 1998;42:1–19. [Google Scholar]
- Veresoglou SD, Halley JM. A model that explains diversity patterns of arbuscular mycorrhizas. Ecol Model. 2012;231:146–152. [Google Scholar]
- Violle C, Pu ZC, Jiang L. Experimental demonstration of the importance of competition under disturbance. Proc Natl Acad Sci USA. 2010;107:12925–12929. doi: 10.1073/pnas.1000699107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LY, Furuno S, Harms H.2010Fungi as transport vectors for contaminants and contaminant-degrading bacteriaIn: Timmis KN (ed).Handbook of Hydrocarbon and Lipid Microbiology Springer: Berlin, Heidelberg; 1555–1561. [Google Scholar]
- Zapesochnaya GG, Kurkin VA, Braslavskii VB, Filatova NV. Phenolic compounds of Salix acutifolia bark. Chem Nat Compd. 2002;38:314–318. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.