Abstract
While natural antimicrobial peptides are potential therapeutic agents, their physicochemical properties and bioactivity generally need to be enhanced for clinical and commercial development. We have previously developed a cationic, amphipathic α-helical, 11-residue peptide (named herein GA-W2: FLGWLFKWASK-NH2) with potent antimicrobial and hemolytic activity, which was derived from a 24-residue, natural antimicrobial peptide isolated from frog skin. Here, we attempted to optimize peptide bioactivity by a rational approach to sequence modification. Seven analogues were generated from GA-W2, and their activities were compared with that of a 12-residue peptide, omiganan, which is being developed for clinical and commercial applications. Most of the modifications reported here improved antimicrobial activity. Among them, the GA-K4AL (FAKWAFKWLKK-NH2) peptide displayed the most potent antimicrobial activity with negligible hemolytic activity, superior to that of omiganan. The therapeutic index of GAK4AL was improved more than 53- and more than 31-fold against Gram-negative and Gram-positive bacteria, respectively, compared to that of the starting peptide, GAW2. Given its relatively shorter length and simpler amino acid composition, our sequence-optimized GA-K4AL peptide may thus be a potentially useful antimicrobial peptide agent.
Keywords: activity optimization, amphipathic helix, antimicrobial activity, antimicrobial peptide, hemolytic activity, sequence modification
INTRODUCTION
Natural antimicrobial peptides (AMPs), also known as genetically encoded antibiotic peptides, are an important component of innate immunity in most living organisms (Diamond et al., 2009; Kim et al., 2010; Zaiou, 2007). Generally, their antimicrobial spectra are broad, acting against fungi, viruses, parasites, and both Gram-positive and Gram-negative bacteria, including, in some cases, multi-drug resistant bacteria (Ginsburg and Koren, 2008; Jenssen et al., 2006; Rivas et al., 2009; Ulvatne, 2003). In addition, tumoricidal activities and insulinotropic activities have been also observed for certain AMPs (Abdel-Wahab et al., 2007; 2008; Gubern et al., 2006; Mader and Hoskin, 2006; Marenah et al., 2006; Kim et al., 2009; Papo and Shai, 2005). In particular, some natural AMPs that have potent antimicrobial activity without toxicity against eukaryotic cells have emerged as potential therapeutic agents. After the discovery of the magainins in 1978 (Zasloff, 1987), therapeutic and commercial development of novel anti-infective drugs has been attempted using natural AMPs and their analogues (Giuliani et al., 2007; Gordon et al., 2005; Oyston et al., 2009; Zasloff, 2002). However, there remain several obstacles to their commercial and clinical applications, including high manufacturing costs and poor pharmaceutical and pharmacokinetic properties (Giuliani et al., 2007; Gordon et al., 2005; Zaiou, 2007; Zasloff, 2002). Thus, despite many successful approaches to therapeutic applications, no AMP agent has yet received FDA approval (Giuliani et al., 2007; Gordon et al., 2005; Oyston et al., 2009; Zaiou, 2007; Zasloff, 2002). At present, omiganan (MBI-226), a 12-residue, indolicidin-based peptide variant that we used in this study as a positive control for activity tests, is the most developed AMP (Gordon et al., 2005; Oyston et al., 2009; Rubinchik et al., 2009; Zasloff, 2002), currently undergoing confirmatory Phase III clinical trials. To reduce production costs and facilitate pharmaceutical optimization, two important considerations for commercial development, AMPs with a shorter size and a simpler amino acid composition than omiganan would be more favorable lead molecules for studies. Then, for clinical development, bioactivity of any such peptide molecule can first be optimized in vitro. Towards this goal, we have used a natural AMP, brevinin-1EMa to develop short AMP variants with favorable bioactivity (Won et al., 2004).
Brevinin-1EMa, formerly known as gaegurin 5, is a 24-residue AMP isolated from the skin of a species of Korean frog, Glandirana emeljanovi, formerly classified as Rana rugosa(Conlon, 2008; Park et al., 1994; Won et al., 2009). The peptide belongs to the group of cationic, amphipathic α-helical AMPs, which represent a particularly abundant, widespread, and most well-characterized class of naturally occurring AMPs (Oren and Shai, 1998; Shai, 1999; Tossi et al., 2000; Zelezetsky and Tossi, 2006). They are known to kill bacteria by selectively disintegrating bacterial membranes. Their positive charges are important to discriminate between the anionic surface of bacterial membranes and the zwitterionic membrane surface of eukaryotic cells, and their amphipathic helical structure is critical to promote membrane permeation by the peptides.
As part of an effort to develop new, low molecular mass peptide antibiotics, we have extensively investigated peptide sequence modifications to search for the shortest bioactive analogue of brevinin-1EMa (Won et al., 2004). The N-terminal 11- residue fragment (sequence: FLGALFKVASK-NH2) of brevinin- 1EMa is completely inactive, but we found that certain amino acid substitutions could confer activity to the peptide. We previously found that the most potent such peptide, GA-W2 (sequence: FLGWLFKWASK-NH2), could be derived by substituting the amino acids at positions 4 and 8 with tryptophans. Despite the fact that GA-W2 is less than half the size of its parent molecule, it showed stronger bactericidal activity (Won et al., 2004). Unfortunately, the hemolytic activity of GA-W2 at high concentrations was also severe. Thus, in this study, further amino acid substitutions were attempted to suppress hemolytic activity while maintaining or enhancing antimicrobial activity. We directed these modifications to also simplify the amino acid composition of the peptide, thereby making it more amenable to commercial production. This paper reports peptides improved from GA-W2, which may serve as activity-optimized candidates superior to omiganan for the development of new peptide antibiotics.
MATERIALS AND METHODS
Structural parameters and peptide preparation
As structural parameters, the mean residue hydrophobicity (<H>) and the mean residue hydrophobic moment <μH>) were calculated from the amino acid sequences (Dathe et al., 2002; Nielsen et al., 2007), on the Eisenberg scale for hydrophobicity (Eisenberg, 1984), by using the HydroMCalc applet (http://www. bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html). After designing each peptide sequence, the chemically synthesized peptides, including the positive control omiganan (ILRWPWWPWRRK- NH2), were purchased as dry powders from the peptide manufacturer company AnyGen (Korea). All of the peptides were synthesized with C-terminal amidation to remove the Cterminal negative charge at neutral pH. The purity and the correct masses of the product peptides were determined by HPLC and mass spectrometry, respectively. For activity tests, a precise amount of each peptide powder was dissolved in PBS or in Luria-Bertani (LB) broth media to the designated concentration.
Antimicrobial assay
The antimicrobial activity of each peptide was determined against four Gram-positive bacterial strains (Bacillus subtilis ATCC 6633, Micrococcus luteus ATCC 10240, Staphylococcus aureus ATCC 6538p, and Staphylococcus epidermis ATCC 12228) and five Gram-negative strains (Escherichia coli ATCC 25922, Shigella dysentariae ATCC 9752, Salmonella typhomurium ATCC 14028, Klebsiella pneumoniae ATCC 10031, and Pseudomonas aeruginosa ATCC 27853). Antimicrobial susceptibility was assessed by the standard broth microdilution method to measure minimal inhibitory concentration (MIC) values. In brief, cell cultures (106-108 cells/ml) in Luria-Bertani (LB) broth media were incubated in the presence of various concentrations (1.6-200 μg/ml, two-fold serial dilutions) of peptides, and the MIC was defined as the lowest peptide concentration that completely inhibits cell growth. The tests were performed with triplicate samples, and MIC values that were reproduced twice or three times in the three independent measurements were recorded. When there was no detectable activity at the highest concentration tested (200 μg/ml), 400 μg/ml was used for calculation of geometric mean of MIC values (GM), since the test was carried out by two-fold serial dilutions (Chen et al., 2005).
Hemolytic assay and therapeutic index
In order to estimate hemolytic activities, suspensions of human red blood cells (10% v/v in PBS) were incubated for 30 min at 37℃ in the presence of various concentrations of peptides (1.6-100 μg/ml, two-fold serial dilutions). After centrifugation, the absorbance of the supernatant at 550 nm was measured. The relative attenuation, as compared with that of the blood suspension treated with 0.2% Triton X-100, was defined as the percentage of hemolysis. The tests were performed with triplicate samples, and the average values of the three independent measurements were recorded. The minimal hemolytic concentration (MHC) value was determined as the lowest peptide concentration that produces 5% or more hemolysis (Chen et al., 2002; Dathe et al., 2002; Zhu et al., 2007). When there was no significant hemolysis at the highest concentration tested (100 μg/ml), 200 μg/ml was used for calculation of the pseudotherapeutic index (TI′), since the test was carried out by twofold serial dilutions (Chen et al., 2005). TI′ was defined as the ratio of the MHC value to the GM (geometric mean of MICs) value (TI′ = MHC/GM).
RESULTS AND DISCUSSION
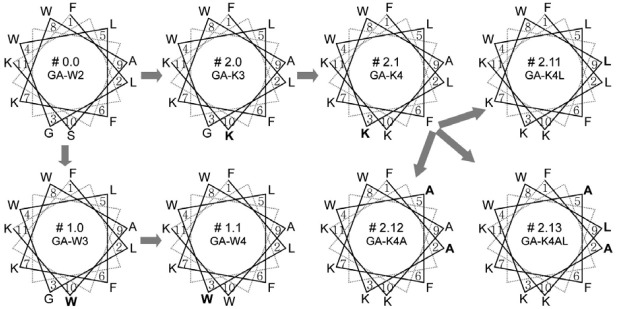
Amino acid sequences of the peptide analogues generated in this study are summarized in Fig. 1. To design each peptide sequence, we used the helical wheel diagrams shown in Fig. 2. The starting molecule, GA-W2 (peptide no. 0.0), has been previously revealed to adopt an amphipathic α-helical structure by converging the hydrophobic residues to one side and the hydrophilic residues to the other side of the helical axis (Won et al., 2004; 2006). The two tryptophan residues were located at the critical amphipathic interface between the end of the hydrophilic side and the start of the hydrophobic side, as seen in the helical wheel projection (Fig. 2). This specific location of tryptophan is critical for peptide activity, as it stabilizes the helical structure and anchors it into the bacterial membrane (Won et al., 2002; 2004; 2006). Thus, in all of the present variants, the two tryptophans (W4 and W8) were conserved. Also, two hydrophobic phenylalanines (F1 and F6) and two positive-charged lysines (K7 and K11) were fixed to maintain the fundamentally amphipathic structure and positive charges, which are known to be important for activity (Won et al., 2002; 2004; 2006). In contrast, the non-positive, hydrophilic S10 and the neutral G3 were targeted for initial substitutions.
Fig. 1. Amino acid sequences of the undecapeptides generated in this study. Each peptide sequence is labeled with the serial number on the left and the peptide name on the right. The regions conserved in all peptides are boxed, and amino acid variations from the starting peptide (no. 0.0) are indicated in bold letters.
Fig. 2. Helical wheel diagrams for the undecapeptides generated in this study. The serial number is presented with the peptide name, in the middle of each diagram. The order of design is depicted by the flow of the gray arrows. The amino acid variations from the preceding number of peptide are indicated in bold letters.
The first set of changes tested additional tryptophanyl substitutions. S10 alone or both the G3 and S10 residues were substituted by tryptophans, thereby generating the GA-W3 (peptide no. 1.0) and GA-W4 (peptide no. 1.1) variants, respectively. Judging from GM value of GA-W3 (Table 1), the S10W substitution of GA-W2 increased antimicrobial activity more than 2- fold. In contrast, the hemolytic activity of GA-W3 was significantly lower than that of GA-W2 (Fig. 3), although it was still severe at high concentrations (MHC value of 50 μg/ml; Table 2). The antimicrobial activity enhancing effect by the S10W substitution was more remarkable against Gram-positive than Gramnegative bacteria and was further increased by the G3W/S10W double substitution (Table 1; GA-W4, peptide no. 1.2). However, the double tryptophanyl substitution almost abolished antibacterial activity against Gram-negative bacteria (Table 1), and hardly decreased hemolytic activity (Fig. 3).
Table 1.
Antimicrobial activities of the undecapeptides generated in this study
| Minimal inhibitory | omiganan | GA-W2 | GA-W3 | GA-W4 | GA-K3 | GA-K4 | GA-K4L | GA-K4A | GA-K4AL |
|---|---|---|---|---|---|---|---|---|---|
| concentration (μg/ml) | (MBI-226) | (#0.0)a | (#1.0) | (#1.1) | (#2.0) | (#2.1) | (#2.11) | (#2.12) | (#2.13) |
| Gram-positive bacteria | |||||||||
| B. subtilis | 6.3 | 25 | 6.3 | 3.1 | 6.3 | 3.1 | 6.3 | 25 | 6.3 |
| M. luteus | 6.3 | 12.5 | 3.1 | 3.1 | 3.1 | 3.1 | 3.1 | 25 | 6.3 |
| S. aureus | 6.3 | 25 | 6.3 | 6.3 | 12.5 | 6.3 | 12.5 | 100 | 6.3 |
| S. epidermis | 12.5 | 50 | 6.3 | 3.1 | 6.3 | 6.3 | 12.5 | 100 | 6.3 |
| Gram-negative bacteria | |||||||||
| E. coli | 25 | 50 | 25 | > 200 | 12.5 | 12.5 | 50 | 100 | 12.5 |
| S. dysentariae | 50 | 25 | 12.5 | 200 | 25 | 25 | 25 | 50 | 12.5 |
| S. typhimorium | 100 | 200 | 100 | > 200 | 50 | 50 | 200 | 200 | 25 |
| K. pneumoniae | 50 | 50 | 12.5 | 200 | 12.5 | 12.5 | 50 | 200 | 6.3 |
| P. aeruginosa | 200 | 200 | > 200 | > 200 | 25 | 12.5 | 25 | 200 | 12.5 |
| GM (geometric mean of MICs) | |||||||||
| Gram (+)b | 7.5 | 25.0 | 5.3 | 3.7 | 6.3 | 4.4 | 7.4 | 50.0 | 6.3 |
| Gram (-)c | 66.0 | 75.8 | ≥ 43.5 | ≥ 303.1 | 21.8 | 18.9 | 50.0 | 132.0 | 12.5 |
| Gram (+,-)d | 25.1 | 46.3 | ≥ 17.0 | ≥ 42.8 | 12.5 | 9.9 | 21.4 | 85.7 | 9.2 |
aAmino acid sequences are represented by the corresponding model numbers (refer to Fig. 1)
bagainst 4 Gram-positive strains tested
cagainst 5 Gram-negative strains tested
dagainst all of the 9 strains tested
Fig. 3. Hemolytic activities of the undecapeptides tested in this study. Hemolysis (%) by the peptides, defined as the relative value to 100% hemolysis of human red blood cells treated with 0.2% Triton X-100, are plotted along the peptide concentration.
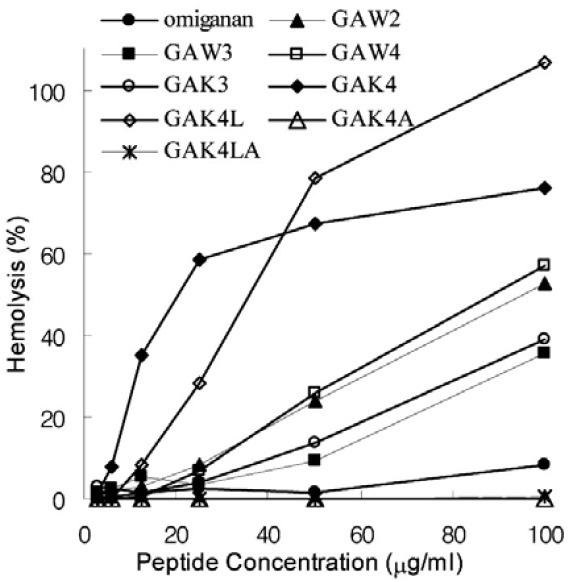
Table 2.
Structural and activity parameters of the undecapeptides generated in this study
| Peptides | <H>a | <μH> b | MHCc (μg/ml) | TI′ (MHC/GM)d | ||
|---|---|---|---|---|---|---|
| (+)e | (-)f | (+ , -) g | ||||
| Omiganan | -0.31 | 0.28 | 100 | 13.4 | 1.5 | 4.0 |
| GA-W2 (#0.0) | 0.08 | 0.30 | 25 | 1.0 | 0.3 | 0.5 |
| GA-W3 (#1.0) | 0.14 | 0.28 | 50 | 9.5 | ≤ 1.1 | ≤ 2.9 |
| GA-W4 (#1.1) | 0.16 | 0.27 | 25 | 6.8 | ≤ 0.1 | ≤ 0.6 |
| GA-K3 (#2.0) | 0.01 | 0.34 | 50 | 8.0 | 2.3 | 4.0 |
| GA-K4 (#2.1) | -0.10 | 0.44 | 6.3 | 1.4 | 0.3 | 0.6 |
| GA-K4L (#2.11) | -0.07 | 0.47 | 12.5 | 1.7 | 0.3 | 0.6 |
| GA-K4A (#2.12) | -0.15 | 0.41 | ≥ 200 | ≥ 4.0 | ≥ 1.5 | ≥ 2.3 |
| GA-K4AL (#2.13) | -0.12 | 0.43 | ≥ 200 | ≥ 31.7 | ≥ 16.0 | ≥ 21.7 |
amean residue hydrophobicity
bmean residue hydrophobic moment
cminimal hemolytic concentration
dpseudo-therapeutic index
eagainst 4 Gram-positive strains tested
fagainst 5 Gram-negative strains tested
gagainst all of the 9 strains tested
Other substitutions were directed to increase positive charges by introducing more lysines. The GA-K3 (peptide no. 2.0) and GA-K4 (peptide no. 2.1) were generated by a S10K and a G3K/S10K double substitution, respectively (Fig. 2). The S10K substitution increased antibacterial activity (approximately 3.7- fold, based on GM values of GA-W2 and GA-K3 in Table 1) and significantly decreased hemolytic activity (compare between GA-W2 and GA-K3 in Fig. 3), although it was still severe at high concentrations (MHC value of 50 μg/ml; Table 2). Then, the antimicrobial activity of GA-K4 with the G3K/S10K double substitution was somewhat stronger than that of GA-K3, with just the S10K substitution (Table 1). However, hemolytic activity was significantly increased in the GA-K4 variant (Fig. 3). It seemed that the tightly organized amphipathicity by the G3K/ S10K double substitution stabilizes membrane interactions at both bacterial cells and human erythrocytes. This explanation is consistent with the result that GA-W4 was also more hemolytic than GA-W3 (Fig. 3).
Because it exhibited the most potent antimicrobial activity, despite strong hemolytic activity, the GA-K4 peptide was employed as a template for further tests of amino acid substitutions, to alleviate its severe hemolytic activity. Five structural parameters, including conformation (χ), charge (Q), hydrophobicity (H), hydrophobic moment (μH) or amphipathicity (A), and polar angle (θ) are generally known as structural determinants of antimicrobial peptide activity (Chou et al., 2008; Dathe et al., 2002; Nielsen et al., 2007; Yeaman and Yount, 2003). In particular, it has been suggested that high hydrophobicity and hydrophobic moment are correlated with increased hemolytic activity, whilst antimicrobial activity is less dependent on these factors (Chou et al., 2008; Yeaman and Yount, 2003). Therefore, we anticipated that the hemolytic activity of GA-K4 could be suppressed by modulating the mean residue hydrophobicity (<H>) and hydrophobic moment (<μH>) values (Table 2). To test this possibility, three analogues of GA-K4 were generated by leucine to alanine or alanine to leucine substitutions. As expected from the increased values of <H> and <μH> (Table 2), the A9L substitution of GA-K4 resulted in significant increase of hemolytic activity (Fig. 3; GA-K4L, peptide no. 2.11) and moderate decrease of antimicrobial activity (Table 1). In particular, the GA-K4L peptide that has the largest <μH> value among all of the present peptides showed the strongest hemolytic activity at high concentrations (Fig. 3), evidencing the fact that hydrophobic moment is one of the critical factors for hemolytic activity. In contrast, reduced hemolytic activity could be observed from the GA-K4A (peptide no. 2.12), where both leucines of GA-K4 were substituted with less hydrophobic alanines, thereby lowering both the <H> and <μH> values (Table 2). Particularly, the GA-K4A peptide that has the lowest <H> value among all of the present peptides showed negligible hemolytic activity even at high concentrations, in support of the fact that hydrophobicity is also one of the critical factors for hemolytic activity. Unfortunately, however, antimicrobial activity was also significantly impaired in the GA-K4AL peptide (Table 1). Thus, we generated GA-K4AL (peptide no. 2.13), in expectation of reduced hemolytic activity without decrease of antimicrobial activity. By substituting the two leucines of GA-K4 with alanines and the alanine with a leucine (Figs. 1 and 2), the <H> and <μH> values of GA-K4AL was adjusted to be lower than those of GA-K4 but higher than those of GA-K4A. Finally, the most favorable activity could be obtained from the GA-K4AL peptide. Its hemolytic activity was almost negligible as observed for GA-K4A (Fig. 3), while the potent antimicrobial activity was maintained or a little bit more improved; i.e. the GM value of GA-K4AL is slightly lower than that of GA-K4 (Table 1).
Activities of the present peptide analogues were compared with that of the known peptide agent, omiganan. Since our peptides are one residue shorter than omiganan, comparable activities can indicate that may be superior to omiganan. As shown in Table 1, some peptides, including GA-W3, GA-K3, GA-K4, GA-K4L, and GA-K4AL, were observed to even be more potent than omiganan. To evaluate the peptides for their potential as therapeutic candidates, their pseudo-therapeutic index (TI′) values were estimated and compared with the value of omiganan (Table 2). Although it is distinct from the typical therapeutic index (TI = LD50/ED50), the TI′ value is useful for relative comparison of selectivity or safety between peptides, since it represents the balance between antimicrobial and hemolytic activities (Chen et al., 2005; Chou et al., 2008; Zhu et al., 2007). As shown in Table 2, the TI′ value was more than 43-folds improved from 0.5 for the initial template, GA-W2, to 21.7 for the final analogue, GA-K4AL. It corresponds to 32- and 53-fold increase against Gram-positive and Gram-negative bacteria, respectively. Finally, GA-K4AL could possess more than 5-fold higher TI′ even than omiganan.
The mode of action of the present peptides remains to be investigated in detail. However, it has generally been accepted that the membrane permeation of amphipathic α-helical AMPs is mainly accomplished via either a ‘pore-forming’ mechanism or a ‘carpet-like’ mechanism (Chia et al., 2002; Shai and Oren, 2001; Tossi et al., 2000; Zelezetsky and Tossi, 2006). In particular, when acting via the pore-forming mechanism, in which the peptide oligomers form helical bundles upon membrane binding, the peptides tend to be selectively active against Gram-positive bacteria, because they do not readily cross the outer membrane of Gram-negative bacteria (Chia et al., 2002; Shai and Oren, 2001). In contrast, peptides acting via the carpet- like mechanism are expected to possess activity against both Gram-positive and Gram-negative bacteria. Thus, the GA-W4 peptide in this study, which showed a marked Gram-positive selectivity, may act via the pore-forming mechanism, while the others with rather higher activity against Gram-positive than against Gram-negative strains act via the carpetlike or via both mechanisms.
In summary, this study aimed to generate useful candidates for developing novel peptide antibiotics. Thus, using an antimicrobial and hemolytic peptide, GA-W2, as a template, we used a rational approach to sequence modification to optimize peptide activity. The properties of the GA-W2 peptide could be improved, thereby generating better candidates with increased antimicrobial activity and/or decreased hemolytic activity. Among the GA-W2 variants, the GA-K4AL peptide appears to be the best candidate, as it was the most potent molecule with no significant hemolytic activity. Compared to omiganan, which has already reached clinical trials, the GA-K4A has better properties: shorter length (11 residues versus 12 in omiganan), simpler composition (four kinds of amino acids versus six in omiganan), stronger antimicrobial activity (Table 1), and weaker hemolytic activity (Table 2 and Fig. 3).
Taken together, we expect that our undecapeptides can serve as useful lead molecules for novel antibiotic development, and some of them, including GA-K4AL, are currently undergoing preclinical tests to explore the potential for further clinical and commercial development.
Acknowledgments
This work was supported by the Innovative Drug Research Center for Metabolic and Inflammatory Disease of the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean Government (MOST); Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea [A092006 to H.W. and B.L.]; and Regional Research Universities Program [Chungbuk BIT Research- Oriented University Consortium to H.W. and W.C.] of the Korea Research Foundation (KRF) grant funded by the Korean Government (MOEHRD).
References
- 1.Abdel-Wahab Y.H., Marenah L., Flatt P.R., Conlon J.M. Insulin releasing properties of the temporin family of antimicrobial peptides. Protein Pept. Lett. (2007);14:702–707. doi: 10.2174/092986607781483822. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Wahab Y.H., Power G.J., Ng M.T., Flatt P.R., Conlon J.M. Insulin-releasing properties of the frog skin peptide pseudin-2 and its [Lys18]-substituted analogue. Biol. Chem. (2008);389:143–148. doi: 10.1515/BC.2008.018. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Mant C.T., Farmer S.W., Hancock R.E.W., Vasil M.L., Hodges R.S. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/thera-peutic index. J. Biol. Chem. (2005);280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia C.S., Torres J., Cooper M.A., Arkin I.T., Bowie J.H. The orientation of the antibiotic peptide maculatin 1.1 in DMPG and DMPC lipid bilayers. Support for a pore-forming mechanism. FEBS Lett. (2002);512:47–51. doi: 10.1016/s0014-5793(01)03313-0. [DOI] [PubMed] [Google Scholar]
- 5.Chou H.T., Kuo T.Y., Chiang J.C., Pei M.J., Yang W.T., Yu H.C., Lin S.B., Chen W.J. Design and synthesis of cationic antimicrobial peptides with improved activity and selectivity against Vibrio spp. Int. J. Antimicrob. Agents. (2008);32:130–138. doi: 10.1016/j.ijantimicag.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Conlon J.M. Reflections on a systematic nomenclature for antimicrobial peptides from the skins of frogs of the family Ranidae. Peptides. (2008);29:1815–1819. doi: 10.1016/j.peptides.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 7.Dathe M., Meyer J., Beyermann M., Maul B., Hoischen C., Bienert M. General aspects of peptide selectivity towards lipid bilayers and cell membranes studied by variation of the structural parameters of amphipathic helical model peptides. Biochim. Biophys. Acta. (2002);1558:171–186. doi: 10.1016/s0005-2736(01)00429-1. [DOI] [PubMed] [Google Scholar]
- 8.Diamond G., Beckloff N., Weinberg A., Kisich K.O. The roles of antimicrobial peptides in innate host defense. Curr. Pharm. Des. (2009);15:2377–2392. doi: 10.2174/138161209788682325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu. Rev. Biochem. (1984);53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- 10.Ginsburg I., Koren E. Are cationic antimicrobial peptides also ‘double-edged swords? Expert. Rev. Anti. Infect. Ther. (2008);6:453–462. doi: 10.1586/14787210.6.4.453. [DOI] [PubMed] [Google Scholar]
- 11.Giuliani A., Pirri G., Nicoletto S.F. Antimicrobial peptides: an overview of a promising class of therapeutics. Cent. Eur. J. Biol. (2007);2:1–33. [Google Scholar]
- 12.Gordon Y.J., Romanowski E.G., McDermott A.M. A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs. Curr. Eye Res. (2005);30:505–515. doi: 10.1080/02713680590968637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gubern C., López-Bermejo A., Biarnés J., Vendrell J., Ricart W., Fernández-Real J.M. Natural antibiotics and insulin sensitivity: the role of bactericidal/permeability-increasing protein. Diabetes. (2006);55:216–224. [PubMed] [Google Scholar]
- 14.Jenssen H., Hamill P., Hancock R.E.W. Peptide antimicrobial agents. Clin. Microbiol. Rev. (2006);19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J.H., Lee J.O., Jung J.H., Lee S.K., You G.Y., Park S.J., and Kim H.S. Gaegurin-6 stimulates insulin secretion through calcium influx in pancreatic β Rin5mf cells. Regul. Pept. (2009);159:123–128. doi: 10.1016/j.regpep.2009.07.014. [DOI] [PubMed] [Google Scholar]
- 16.Kim S.R., Hong M.Y., Park S.W., Choi K.H., Yun E.Y., Goo T.W., Kang S.W., Suh H.J., Kim I., Hwang J.S. Characterization and cDNA cloning of a cecropin-like antimicrobial peptide, papiliocin, from the swallowtail butterfly, Papilio xuthus. Mol. Cells. (2010);29:419–423. doi: 10.1007/s10059-010-0050-y. [DOI] [PubMed] [Google Scholar]
- 17.Mader J.S., Hoskin D.W. Cationic antimicrobial peptides as novel cytotoxic agents for cancer treatment. Expert. Opin. Investig. Drugs. (2006);15:933–946. doi: 10.1517/13543784.15.8.933. [DOI] [PubMed] [Google Scholar]
- 18.Marenah L., Flatt P.R., Orr D.F., Shaw C., Abdel-Wahab Y.H.A. Skin secretions of Rana saharica frogs reveal antimicrobial peptides esculentins-1 and -1B and brevinins-1E and -2EC with novel insulin releasing activity. J. Endocrinol. (2006);188:1–9. doi: 10.1677/joe.1.06293. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen S.L., Frimodt-Møller N., Kragelund B.B., Hansen P.R. Structure-activity study of the antibacterial peptide fallaxin. Protein Sci. (2007);16:1969–1976. doi: 10.1110/ps.072966007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oren Z., Shai Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers. (1998);47:451–463. doi: 10.1002/(SICI)1097-0282(1998)47:6<451::AID-BIP4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 21.Oyston P.C.F., Fox M.A., Richards S.J., Clark G.C. Novel peptides therapeutics for treatment of infections. J. Med. Microbiol. (2009);58:977–987. doi: 10.1099/jmm.0.011122-0. [DOI] [PubMed] [Google Scholar]
- 22.Papo N., Shai Y. Host defense peptides as new weapons in cancer treatment. Cell. Mol. Life Sci. (2005);62:784–790. doi: 10.1007/s00018-005-4560-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J.M., Jung J.E., Lee B.J. Antimicrobial peptides from the skin of a Korean frog, Rana rugosa. Biochem. Biophys. Res. Commun. (1994);209:948–954. doi: 10.1006/bbrc.1994.2757. [DOI] [PubMed] [Google Scholar]
- 24.Rivas L., Luque-Ortega J.R., Andreu D. Amphibian antimicrobial peptides and Protozoa: lessons from parasites. Biochim. Biophys. Acta. (2009);1788:1570–1581. doi: 10.1016/j.bbamem.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Rubinchik E., Dugourd D., Algara T., Pasetka C., Friedland H.D. Antimicrobial and antifungal activities of novel cationic antimicrobial peptide, omigana, in experimental skin colonization models. Int. J. Antimicrob. Agents. (2009);34:457–461. doi: 10.1016/j.ijantimicag.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta. (1999);1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 27.Shai Y., Oren Z. From “carpet” mechanism to de-novo designed diastereomeric cell-selective antimicrobial peptides. Peptides. (2001);22:1629–1641. doi: 10.1016/s0196-9781(01)00498-3. [DOI] [PubMed] [Google Scholar]
- 28.Tossi A., Sandri L., Giangaspero A. Amphipathic, α-helical antimicrobial peptides. Biopolymers. (2000);55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 29.Ulvatne H. Antimicrobial peptides: potential use in skin infections. Am. J. Clin. Dermatol. (2003);4:591–595. doi: 10.2165/00128071-200304090-00001. [DOI] [PubMed] [Google Scholar]
- 30.Won H.S., Park S.H., Kim H.E., Hyun B., Kim M., Lee B., Lee B.J. Effects of a tryptophanyl substitution on the structure and antimicrobial activity of C-terminally truncated gaegurin 4. Eur. J. Biochem. (2002);269:4367–4374. doi: 10.1046/j.1432-1033.2002.03139.x. [DOI] [PubMed] [Google Scholar]
- 31.Won H.S., Jung S.J., Kim H.E., Seo M.D., Lee B.J. Systematic peptide engineering and structural characterization to search for the shortest antimicrobial peptide analogue of gaegurin 5. J. Biol. Chem. (2004);279:14784–14791. doi: 10.1074/jbc.M309822200. [DOI] [PubMed] [Google Scholar]
- 32.Won H.S., Seo M.D., Jung S.J., Lee S.J., Kang S.J., Son W.S., Kim H.J., Park T.K., Park S.J., Lee B.J. Structural determinants for the membrane interaction of novel bioactive undecapeptides derived from gaegurin 5. J. Med. Chem. (2006);49:4886–4895. doi: 10.1021/jm050996u. [DOI] [PubMed] [Google Scholar]
- 33.Won H.S., Kang S.J., Lee B.J. Action mechanism and structural requirements of the antimicrobial peptides, gaegurins. Biochim. Biophys. Acta. (2009);1788:1620–1629. doi: 10.1016/j.bbamem.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Yeaman M.R., Yount N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. (2003);55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 35.Zaiou M. Multifunctional antimicrobial peptides: therapeutic targets in several human diseases. J. Mol. Med. (2007);85:317–329. doi: 10.1007/s00109-006-0143-4. [DOI] [PubMed] [Google Scholar]
- 36.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA. (1987);84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. (2002);415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 38.Zelezetsky I., Tossi A. Alpha-helical antimicrobial peptides-Using a sequence template to guide structure-activity relationships studies. Biochim. Biophys. Acta. (2006);1758:1436–1449. doi: 10.1016/j.bbamem.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 39.Zhu W.L., Nan Y.H., Hahm K.S., Shin S.Y. Cell selectivity of an antimicrobial peptide melittin diastereomer with Damino acid in the leucine zipper sequence. J. Biochem. Mol. Biol. (2007);40:1090–1094. doi: 10.5483/bmbrep.2007.40.6.1090. [DOI] [PubMed] [Google Scholar]


