Abstract
Background
Despite the high prevalence of parasitic infections, and their impact on global health and economy, the number of drugs available to treat them is extremely limited. As a result, the potential consequences of large-scale resistance to any existing drugs are a major concern. A number of recent investigations have focused on the effects of potential chemical inhibitors on bacterial and fungal carbonic anhydrases. Among the five classes of carbonic anhydrases (alpha, beta, gamma, delta and zeta), beta carbonic anhydrases have been reported in most species of bacteria, yeasts, algae, plants, and particular invertebrates (nematodes and insects). To date, there has been a lack of knowledge on the expression and molecular structure of beta carbonic anhydrases in metazoan (nematodes and arthropods) and protozoan species.
Methods
Here, the identification of novel beta carbonic anhydrases was based on the presence of the highly-conserved amino acid sequence patterns of the active site. A phylogenetic tree was constructed based on codon-aligned DNA sequences. Subcellular localization prediction for each identified invertebrate beta carbonic anhydrase was performed using the TargetP webserver.
Results
We verified a total of 75 beta carbonic anhydrase sequences in metazoan and protozoan species by proteome-wide searches and multiple sequence alignment. Of these, 52 were novel, and contained highly conserved amino acid residues, which are inferred to form the active site in beta carbonic anhydrases. Mitochondrial targeting peptide analysis revealed that 31 enzymes are predicted with mitochondrial localization; one was predicted to be a secretory enzyme, and the other 43 were predicted to have other undefined cellular localizations.
Conclusions
These investigations identified 75 beta carbonic anhydrases in metazoan and protozoan species, and among them there were 52 novel sequences that were not previously annotated as beta carbonic anhydrases. Our results will not only change the current information in proteomics and genomics databases, but will also suggest novel targets for drugs against parasites.
Keywords: Beta carbonic anhydrase, Inhibitor, Metazoa, Mitochondrial targeting peptide, Multiple sequence alignment, Protozoa
Background
Carbonic anhydrases (CAs) are ubiquitous metalloenzymes. They are encoded by five evolutionary divergent gene families and the corresponding enzymes are designated α, β, γ, δ and ζ-CAs. α-CAs are present in animals, some fungi, bacteria, algae, and cytoplasm of green plants. β-CAs are expressed mainly in fungi, bacteria, archaea, algae, and chloroplasts of monocotyledons and dicotyledons. γ-CAs are expressed in plants, archaea, and some bacteria. δ- and ζ-CAs are present in several classes of marine phytoplankton [1-6]. A total of 13 enzymatically active α-CAs have been reported in mammals: CA I, CA II, CA III, CA VII, and CA XIII are cytosolic enzymes; CA IV, CA IX, CA XII, CA XIV, and CA XV are membrane-bound; CA VA and CA VB are mitochondrial; CA VI is secreted and CA VIII, CA X, and CA XI are acatalytic CA-related proteins [3,7]. The active site of CA contains a zinc ion (Zn2+) which has a critical role in the catalytic activity of the enzyme. ζ-and γ-CAs represent exceptions to this rule since they can use cadmium (ζ), iron (γ), or cobalt (γ) as cofactors [8-10]. CAs are involved in many biological processes, such as respiration involving transport of CO2 and bicarbonate between metabolizing tissues, pH homeostasis, electrolyte transfer, bone resorption, calcification, and tumor progression. They also participate in some biosynthetic reactions, such as gluconeogenesis, lipogenesis, and ureagenesis [3,11-14].
The first β-CA was serendipitously discovered by Neish in 1939 [15]. In 1990, the cDNA sequence of spinach (Spinacea oleracea) chloroplast CA was determined, and found to be non-homologous to animal α-CA [16,17]. Thereafter, cDNA sequences of β-CA from pea (Pisium sativum) and Arabidopsis thaliana were determined [17-19]. It is believed that the plant β-CAs are distributed in the chloroplastic stroma, thylakoid space, and cytoplasm of plant cells [17]. Many putative β-CAs have been discovered since 1990, not only in photosynthetic organisms, but also in eubacteria, yeast, and archaea [17].
The first bacterial β-CA gene was named CynT and recognized in Escherichia coli[20,21]. Later, β-CA was identified in some other pathogenic bacteria, such as Helicobacter pylori, Mycobacterium tuberculosis, Salmonella typhimurium[17,22], Haemophilus influenzae[23,24], Brucella suis[24,25], Streptococcus pneumoniae[24,26], Salmonella enterica[24,27], and Vibrio cholerae[24,28,29]. β-CAs have also been identified in fungi, such as Candida albicans[1,30], Candida glabrata[1,31], Cryptococcus neoformans[1,32], and Sordaria macrospora[6,33]. This class of enzyme has also been discovered in a wide range of taxa, such as yeast (Saccharomyces cerevisiae) [34-36], cyanobacteria (Synechocystis sp. PCC6803) [37], carboxysomes of chemoautotrophic bacteria (Halothiobacillus neapolitanus) [38], green algae (Chlamydomonas reinhardtii) [39], red algae (Porphyridium purpureum) [40], nematodes (Caenorhabditis elegans)[41], and insects (Drosophila melanogaster)[4]. While β-CAs were initially thought to be expressed only in plants, this enzyme family is indeed present in a wide variety of species – from bacteria and archaea to invertebrate animals, missing only from vertebrates and most chordates, making it an attractive target for evolutionary studies [5].
β-CA is an important accessory enzyme for many CO2 or HCO3- utilizing enzymes (e.g. RuBisCO in chloroplasts, cyanase in E. coli[42], urease in H. pylori[43], and carboxylases in Corynebacterium glutamicum[44]). In cyanobacteria, β-CA is an essential component of the CO2-concentrating carboxysome organelle [17,45]. β-CA activity is required for growth of E. coli bacteria in air [46]; it is also indispensable if the atmospheric partial pressure of CO2 is high or during anaerobic growth in a closed vessel at low pH, where copious CO2 is generated endogenously. β-CA is also needed for growth of C. glutamicum[44,47] and some yeasts, such as S. cerevisiae[40]. In higher plants, the Flaveria bidentis genome contains at least three β-CA genes, named CA1, CA2, and CA3[48]. The functional roles of β-CAs in plants are not yet fully understood, even though a lot of new data has emerged in recent years. C3 and C4 plants have different mechanisms for carbon fixation and photosynthesis and, thus, β-CAs might possess different roles, depending on the location of the enzyme and the type of plant [49]. In plants, the highest CA activity has been found within the chloroplast stroma, but there is also some CA activity in the cytosol of mesophyll cells [50]. Carbon dioxide coming from the external environment must be rapidly hydrated by β-CA and converted into HCO3− for the phosphoenolpyruvate carboxylase enzyme [49]. Additionally, CAs play a role in photosynthesis by facilitating diffusion into and across the chloroplast, and by catalyzing HCO3- dehydration to supply CO2 for RuBisCO. Interestingly, both RuBisCO and β-CA expression levels increase together when P. sativum is transferred from an environment with high levels of CO2 to one with low levels [47].
Crystal structures of β-CAs reveal that a zinc ion (Zn2+) is ligated by two conserved cysteines and one conserved histidine [5]. Until now, the only X-ray crystallography structure defined for β-CAs in plants belongs to P. sativum[51]. E. coli was the first bacteria in which the β-CA crystal structure was determined [20]. β-CA can adopt a variety of oligomeric states with molecular masses ranging from 45 to 200 kDa [52].
The first metazoan β-CAs were reported in 2010 [41]. In one of the studies [4,41], two genes encoding β-CAs (y116a8c.28 and bca-1) were identified in Caenorhabditis elegans. Another study reported a novel β-CA gene identified from FlyBase, which was named DmBCA (short for Drosophila melanogaster β-CA) [4]. Additionally, orthologs were retrieved from sequence databases, and reconstructed when necessary. The results confirmed the presence of β-CA sequences in 55 metazoan species, such as Aedes aegypti, Culex quinquefasciatus, Anopheles gambiae, Drosophila virilis, Tribolium castaneum, Nasonia vitripennis, Apis mellifera, Acyrthosiphon pisum, Daphnia pulex, Caenorhabditis elegans, Pristionchus pacificus, Trichoplax adhaerens, Caligus clemensi, Lepeophtheirus salmonis, Nematostella vectensis, Strongylocentrotus purpuratus, and Saccoglossus kowalevskii. The DmBCA enzyme was produced as a recombinant protein in Sf9 insect cells, and its kinetic and inhibition profiles were determined. The enzyme showed high CO2 hydratase activity, with a kcat of 9.5 × 105 s-1 and a kcat/KM of 1.1 × 108 M-1 s-1. DmBCA was inhibited by the clinically-used sulfonamide, acetazolamide, with an inhibition constant of 49 nM. Subcellular localization studies have indicated that DmBCA is probably a mitochondrial enzyme, as is also suggested by sequence analysis.
In this study, using bioinformatics tools, we discovered and verified the presence of β-CA in various other metazoan species, and, for the first time, in protozoa. Previously, most β-CA proteins have been identified in protein databases as ‘unknown’ proteins or ‘putative’ CAs, without a specific reference to β-CAs. Based on the present findings, new avenues will be opened to biochemically characterize β-CAs and their inhibitors in arthropods, nematodes and protozoans.
Methods
Identification of putative β-CA enzymes in protozoan and metazoan species and multiple sequence alignment
Identification of novel β-CAs was based on the presence of the highly-conserved amino acid sequence patterns of the active site, namely Cys-Xaa-Asp-Xaa-Arg and His-Xaa-Xaa-Cys also marked in Additional file 1: Figure S1. Alignment was visualized in Jalview [53]. In total, 75 invertebrate β-CA sequences were retrieved from Uniprot (http://www.uniprot.org/) for alignment analysis, and one bacterial sequence (Pelosinus fermentans) was included as an outgroup. All protein sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) [54]. The sequences were manually curated to remove residues associated with an incorrect starting methionine. A total of 90 residues were removed from the N-terminal end of Uniprot IDs D4NWE5_ADIVA, G0QPN9_ICHMG, D6WK56_TRICA, I7LWM1_TETTS and I7M0M0_TETTS. The modified protein sequences were then re-aligned. This protein alignment then served as the template for codon alignment of corresponding nucleotide sequences using the Pal2Nal program (http://www.bork.embl.de/pal2nal/) [55].
Phylogenetic analysis
The phylogenetic analysis was computed using Mr. Bayes v3.2 [56]. After 8 million generations using the GTR codon substitution model, with all other parameters as default, the standard deviation of split frequencies was 1.39 × 10-3. The final output tree was produced using 50% majority rule consensus. FigTree v1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) [56] was used to visualize the phylogenetic tree and the Pelosinus fermentans[57] sequence set as outgroup. Additional trees were constructed for comparison using maximum likelihood (PhyML)[58], UPGMA, and neighbor-joining methods within Geneious version 7.0.5 from Biomatters (Auckland, New Zealand) (http://www.geneious.com/).
Prediction of subcellular localization
Subcellular localization prediction of each identified invertebrate β-CA was performed using the TargetP webserver (http://www.cbs.dtu.dk/services/TargetP/). TargetP is built from two layers of neural networks, where the first layer contains one dedicated network for each type of pre-sequence [cTP (cytoplasmic targeting peptide), mTP (mitochondrial targeting peptide, or SP (secretory signal peptide)], and the second is an integrating network that outputs the actual prediction (cTP, mTP, SP, other). It is able to discriminate between cTPs, mTPs, and SPs with sensitivities and specificities higher than what has been obtained with other available subcellular localization predictors [59].
Results
Multiple sequence alignment
The Uniprot search of potential β-CA sequences, and the subsequent multiple sequence alignment, identified 75 β-CAs in metazoan and protozoan species, of which 23 sequences were reported as β-CAs previously [4]. Thus, 52 metazoan and protozoan β-CA sequences were novel and reported here for the first time. All 75 β-CAs in metazoan and protozoan species are shown in Table 1. The multiple sequence alignment results of these 75 β-CAs, plus a bacterial β-CA sequence from Pelosinus fermentans, are shown as Additional file 1: Figure S1. Multiple sequence alignment of all animal β-CAs confirmed conservation of the known active site motifs CxDxR and HxxC in all identified enzymes. Several other key residues were also highly conserved. Notably, all β-CA sequences from Leishmania species (Leishmania donovani, Leishmania infantum, Leishmania major, and Leishmania mexicana) contained a 71 residue N-terminal extension not present in any other sequences.
Table 1.
Identified β-CAs in protozoan and metazoan species
| Species | β- CA ID | Entry ID | Gene name | Protein name |
|---|---|---|---|---|
|
Acromyrmex echinatior
|
BCA |
F4WAG3 |
G5I_02499 |
Beta carbonic anhydrase 1 |
|
Acyrthosiphon pisum
|
BCA1 |
J9K706 |
Uncharacterized |
Uncharacterized |
| |
BCA2 |
C4WVD8 |
ACYPI006033 |
ACYPI006033 |
| |
BCA3 |
J9JZY3 |
XM_001950078.1 |
Uncharacterized |
|
Adineta vaga
|
BCA |
D4NWE5 |
Uncharacterized |
Putative uncharacterized protein |
|
Aedes aegypti
|
BCA |
Q17N64 |
AAEL000816 |
AAEL000816-PA |
|
Ancylostoma caninum
|
BCA |
FC551456 |
Uncharacterized |
Uncharacterized protein |
|
Anopheles darlingi
|
BCA |
E3X5Q8 |
AND_14274 |
Uncharacterized protein |
|
Anopheles gambiae
|
BCA |
Q5TU56 |
AGAP002992 AgaP_AGAP002992 |
AGAP002992-PA |
|
Apis mellifera
|
BCA |
H9KS29 |
Uncharacterized |
Uncharacterized protein |
|
Ascaris suum
|
BCA |
F1LE18 |
Uncharacterized |
Beta carbonic anhydrase 1 |
|
Caenorhabditis brenneri
|
BCA1 |
G0MSW4 |
Cbn-bca-1 CAEBREN_17105 |
CBN-BCA-1 protein |
| |
BCA2 |
G0MRG1 |
Cbn-bca-2 CAEBREN_06024 |
CBN-BCA-2 protein |
|
Caenorhabditis briggsae
|
BCA1 |
A8XKV0 |
bca-1 CBG14861 |
Beta carbonic anhydrase 1 |
| |
BCA2 |
A8WN21 |
bca-2 Cbr-bca-2 cbr-bca-2 CBG00424 CBG_00424 |
Protein CBR-BCA-2 |
|
Caenorhabditis elegans
|
BCA1 |
Q22460 |
bca-1 T13C5.5 |
Beta carbonic anhydrase 1 |
| |
BCA2 |
Q2YS41 |
bca-2 Y116A8C.28 |
Protein BCA-2 |
|
Caenorhabditis remanei
|
BCA1 |
E3LDN3 |
Cre-bca-1 CRE_00190 |
CRE-BCA-1 protein |
| |
BCA2 |
E3MK96 |
Cre-bca-2 CRE_28742 |
CRE-BCA-2 protein |
|
Caligus clemensi
|
BCA |
C1C2M7 |
CYNT |
Carbonic anhydrase |
|
Camponotus floridanus
|
BCA |
E2ANQ9 |
EAG_05651 |
Carbonic anhydrase |
|
Culex quinquefasciatus
|
BCA |
B0WKV7 |
CpipJ_CPIJ007527 |
Carbonic anhydrase |
|
Danaus plexippus
|
BCA |
G6D7Z4 |
Uncharacterized |
Putative carbonic anhydrase |
|
Daphnia pulex
|
BCA |
E9GLB5 |
CAB |
Beta-carbonic anhydrase |
|
Dendroctonus ponderosae
|
BCA |
J3JTM9 |
Uncharacterized |
Uncharacterized protein |
|
Drosophila ananassae
|
BCA |
B3LZ10 |
GF17694 Dana\GF17694 Dana_GF17694 |
GF17694 |
|
Drosophila erecta
|
BCA |
B3P1V8 |
GG13874 Dere\GG13874 Dere_GG13874 |
GG13874 |
|
Drosophila grimshawi
|
BCA |
B4JHY1 |
GH19010 Dgri\GH19010 Dgri_GH19010 |
GH19010 |
|
Drosophila melanogaster
|
BCA |
Q9VHJ5 |
CAHbeta CG11967 Dmel_CG11967 |
CG11967 |
|
Drosophila mojavensis
|
BCA |
B4KDC1 |
GI23065 Dmoj\GI23065 Dmoj_GI23065 |
GI23065 |
|
Drosophila persimilis
|
BCA |
B4GFA1 |
GL22171 Dper\GL22171 Dper_GL22171 |
GL22171 |
|
Drosophila pseudoobscura
|
BCA |
Q296E4 |
GA11301 Dpse\GA11301 Dpse_GA11301 |
GA11301 |
|
Drosophila sechellia
|
BCA |
B4HKY7 |
GM23772 Dsec\GM23772 Dsec_GM23772 |
GM23772 |
|
Drosophila simulans
|
BCA |
B4QXC5 |
GD18582 Dsim\GD18582 Dsim_GD18582 |
GD18582 |
|
Drosophila virilis
|
BCA |
B4LZE7 |
CAHbeta Dvir\GJ24578 GJ24578 Dvir_GJ24578 |
GJ24578 |
|
Drosophila willistoni
|
BCA |
B4NBB9 |
GK11865 Dwil\GK11865 Dwil_GK11865 |
GK11865 |
|
Drosophila yakuba
|
BCA |
B4PTY0 |
GE25916 Dyak\GE25916 Dyak_GE25916 |
GE25916 |
|
Entamoeba dispar
|
BCA |
B0E7M0 |
EDI_275880 |
Carbonic anhydrase |
|
Entamoeba histolytica
|
BCA |
C4LXK3 |
EHI_073380 |
Carbonic anhydrase |
|
Entamoeba nuttalli
|
BCA |
K2GQM0 |
ENU1_204230 |
Carbonate dehydratase domain containing protein |
|
Harpegnathos saltator
|
BCA |
E2B2Q1 |
EAI_05019 |
Carbonic anhydrase |
|
Heliconius melpomene
|
BCA |
HMEL015257 |
Uncharacterized |
Uncharacterized protein |
|
Hirudo medicinalis
|
BCA |
EY481200 |
Uncharacterized |
Uncharacterized protein |
|
Ichthyophthirius multifiliis
|
BCA |
G0QPN9 |
IMG5_069900 |
Carbonic anhydrase |
|
Leishmania donovani
|
BCA |
E9B8S3 |
LDBPK_060630 |
Carbonic anhydrase |
|
Leishmania infantum
|
BCA |
A4HSV2 |
LINJ_06_0630 |
Carbonic anhydrase |
|
Leishmania major
|
BCA |
Q4QJ17 |
LMJF_06_0610 |
Carbonic anhydrase |
|
Leishmania mexicana
|
BCA |
E9AKU0 |
LMXM_06_0610 |
Carbonic anhydrase |
|
Lepeophtheirus salmonis
|
BCA |
D3PI48 |
BCA1 |
Beta carbonic anhydrase 1 |
|
Nasonia vitripennis
|
BCA |
K7IWK8 |
Uncharacterized |
Uncharacterized protein |
|
Nematostella vectensis
|
BCA |
A7S717 |
v1g186479 |
Predicted protein |
|
Paramecium tetraurelia
|
BCA1 |
A0BD61 |
GSPATT00004572001 |
Carbonic anhydrase |
| |
BCA2 |
A0E8J0 |
GSPATT00024336001 |
Carbonic anhydrase |
| |
BCA3 |
A0CEX6 |
GSPATT00037782001 |
Carbonic anhydrase |
| |
BCA4 |
A0BDB1 |
GSPATT00004622001 |
Carbonic anhydrase |
| |
BCA5 |
A0C922 |
GSPATT00006595001 |
Carbonic anhydrase |
|
Saccoglossus kowalevskii
|
BCA |
187043763 |
Uncharacterized |
Uncharacterized protein |
|
Schistosoma mansoni
|
BCA |
G4V6B2 |
Smp_004070 |
Putative carbonic anhydrase |
|
Solenopsis invicta
|
BCA |
E9IP13 |
SINV_09652 |
Putative carbonic anhydrase |
|
Strigamia maritima
|
BCA |
SMAR006741 |
Uncharacterized |
Uncharacterized protein |
|
Strongylocentrotus purpuratus
|
BCA |
H3I177 |
Uncharacterized |
Uncharacterized protein |
|
Tetrahymena thermophila
|
BCA1 |
Q22U21 |
TTHERM_00263620 |
Carbonic anhydrase |
| |
BCA2 |
Q22U16 |
TTHERM_00263670 |
Carbonic anhydrase |
| |
BCA3 |
I7MDL7 |
TTHERM_00373840 |
Carbonic anhydrase |
| |
BCA4 |
I7LWM1 |
TTHERM_00558270 |
Carbonic anhydrase |
| |
BCA5 |
I7M0M0 |
TTHERM_00374880 |
Carbonic anhydrase |
| |
BCA6 |
I7MD92 |
TTHERM_00541480 |
Carbonic anhydrase |
| |
BCA7 |
I7M748 |
TTHERM_00374870 |
Carbonic anhydrase |
| |
BCA8 |
Q23AV1 |
TTHERM_00654260 |
Carbonic anhydrase |
|
Tribolium castaneum
|
BCA |
D6WK56 |
TcasGA2_TC014816 |
Putative uncharacterized protein |
|
Trichinella spiralis
|
BCA |
E5SH53 |
Uncharacterized |
Carbonic anhydrase |
|
Trichomonas vaginalis
|
BCA1 |
A2ENQ8 |
TVAG_005270 |
Carbonic anhydrase |
| |
BCA2 |
A2DLG4 |
TVAG_268150 |
Carbonic anhydrase |
|
Trichoplax adhaerens
|
BCA |
B3S5Y1 |
TRIADDRAFT_29634 |
Putative uncharacterized protein |
| Xenoturbella bocki | BCA | 117195962 | Uncharacterized | Uncharacterized protein |
Phylogenetic analysis
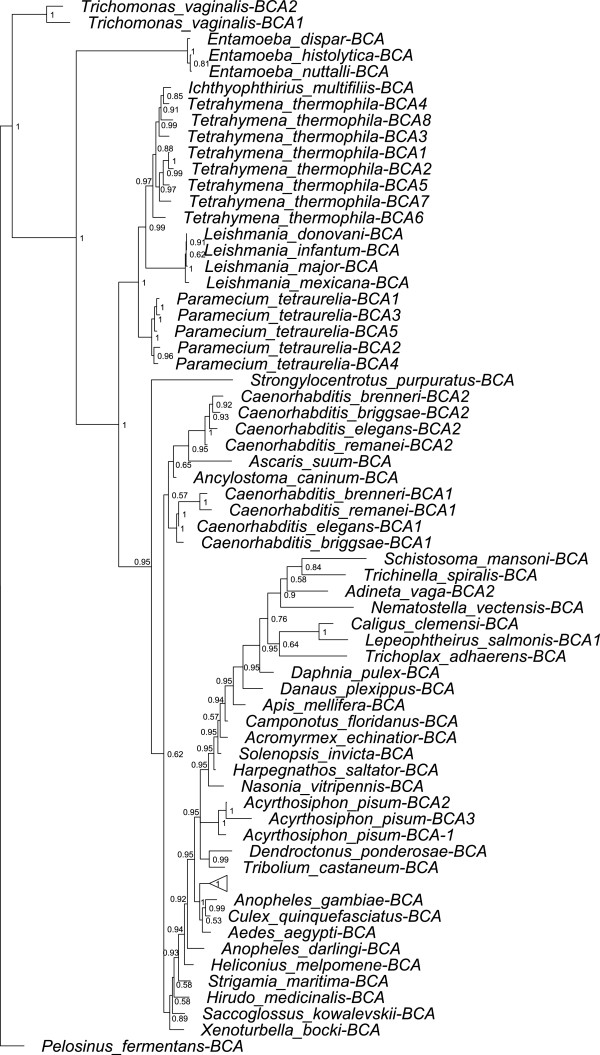
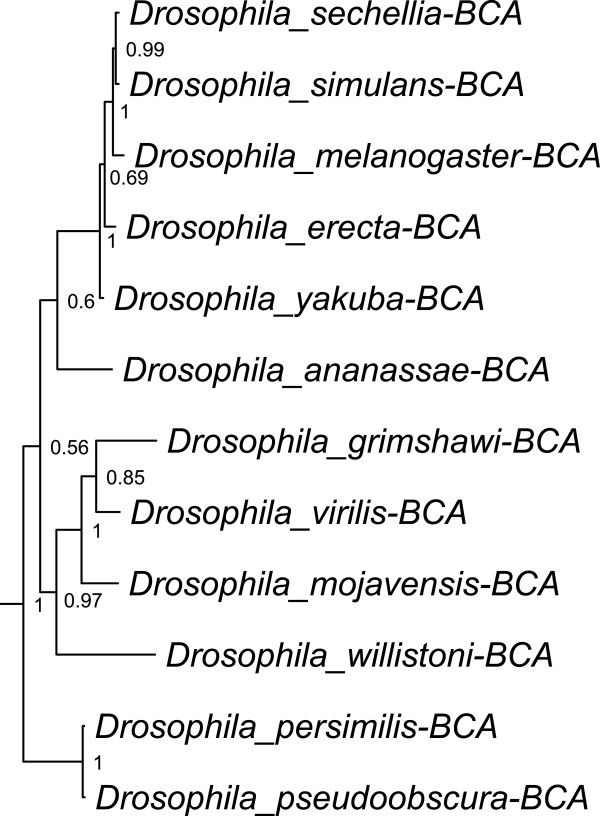
The results of the phylogenetic analysis of 75 β-CAs in metazoan and protozoan species are shown in Figure 1. A β-CA sequence from the Pelosinus fermentans bacterium was used as an outgroup [60]. The phylogenetic results represent the evolutionary root of β-CAs in metazoan and protozoan species, the similarity between them, and duplications that have occurred. The branching pattern and branch lengths reveal interesting evolutionary relationships of β-CAs in various invertebrate species. There is a close relationship between our bacterial outgroup and Trichomonas vaginalis β-CAs, both having originated well before the other species within the tree. β-CAs of nematodes and arthropods are located in the lower evolutionary branches. In the protozoan Tetrahymena thermophilia and Paramecium tetraurelia clades significant duplications of β-CA have occurred, with 8 and 5 distinct proteins respectively. Meanwhile, metazoan and nematode species tend to have just one or two β-CAs. Surprisingly, β-CAs of the nematode Trichinella spiralis and trematode Schistosoma mansoni appear more closely related to arthropod than to nematode enzymes. The triangle located near the bottom of Figure 1 represents the clade of β-CAs in different Drosophila species. The details of the phylogenetic tree of β-CAs in Drosophila species are shown in Figure 2. The likely presence of inaccuracies in some of the database sequences, and inherent limitations of Bayesian inference, prompted use of additional phylogenetic methods. These analyses generally supported the major features of the final tree achieved via Bayesian inference.
Figure 1.
Phylogenetic analysis of 75 metazoan and protozoan β-CAs. The position of β-CAs of Drosophila species has been represented at the bottom of the phylogenetic tree by a triangle shape. The details of β-CAs of Drosophila species in the phylogenetic tree are shown in Figure 2.
Figure 2.
Phylogenetic analysis of β-CAs of Drosophila species. This tree represents the expanded view of the triangle located near the bottom of the main phylogenetic tree of β-CAs in Figure 1.
Subcellular localization of β-CAs
The predictions for subcellular localization of the 75 β-CAs are shown in Table 2. The results reveal that 31 are predicted to have a mitochondrial localization, one (Anopheles darlingi, Uniprot ID: E3X5Q8) was predicted to be secreted, and the remaining 43 were predicted to have other cellular localizations. The predictions were based on the analysis of 175 N-terminal amino acids of each sequence. In the Name column, there are both IDs of the β-CAs in Uniprot database and scientific name of the metazoan and protozoan species.
Table 2.
Prediction of the subcellular localization of 75 β-CAs of metazoan and protozoan species
| Species | β- CA ID | Entry ID | mTP | SP | Other | Loc | RC |
|---|---|---|---|---|---|---|---|
|
Acromyrmex echinatior
|
BCA |
F4WAG3 |
0.199 |
0.054 |
0.86 |
- |
2 |
|
Acyrthosiphon pisum
|
BCA1 |
J9K706 |
0.473 |
0.05 |
0.631 |
- |
5 |
| |
BCA2 |
C4WVD8 |
0.579 |
0.043 |
0.536 |
M |
5 |
| |
BCA3 |
J9JZY3 |
0.579 |
0.043 |
0.534 |
M |
5 |
|
Adineta vaga
|
BCA |
D4NWE5 |
0.509 |
0.102 |
0.375 |
M |
5 |
|
Aedes aegypti
|
BCA |
Q17N64 |
0.589 |
0.029 |
0.491 |
M |
5 |
|
Ancylostoma caninum
|
BCA |
FC551456 |
0.466 |
0.046 |
0.514 |
- |
5 |
|
Anopheles darlingi
|
BCA |
E3X5Q8 |
0.044 |
0.836 |
0.144 |
S |
2 |
|
Anopheles gambiae
|
BCA |
Q5TU56 |
0.713 |
0.03 |
0.34 |
M |
4 |
|
Apis mellifera
|
BCA |
H9KS29 |
0.126 |
0.08 |
0.875 |
- |
2 |
|
Ascaris suum
|
BCA |
F1LE18 |
0.388 |
0.079 |
0.406 |
- |
5 |
|
Caenorhabditis brenneri
|
BCA1 |
G0MSW4 |
0.522 |
0.036 |
0.518 |
M |
5 |
| |
BCA2 |
G0MRG1 |
0.52 |
0.051 |
0.473 |
M |
5 |
|
Caenorhabditis briggsae
|
BCA1 |
A8XKV0 |
0.392 |
0.047 |
0.615 |
- |
4 |
| |
BCA2 |
A8WN21 |
0.546 |
0.048 |
0.466 |
M |
5 |
|
Caenorhabditis elegans
|
BCA1 |
Q22460 |
0.475 |
0.039 |
0.549 |
- |
5 |
| |
BCA2 |
Q2YS41 |
0.465 |
0.05 |
0.529 |
- |
5 |
|
Caenorhabditis remanei
|
BCA1 |
E3LDN3 |
0.327 |
0.045 |
0.69 |
- |
4 |
| |
BCA2 |
E3MK96 |
0.51 |
0.051 |
0.48 |
M |
5 |
|
Caligus clemensi
|
BCA |
C1C2M7 |
0.21 |
0.04 |
0.873 |
- |
2 |
|
Camponotus floridanus
|
BCA |
E2ANQ9 |
0.325 |
0.051 |
0.735 |
- |
3 |
|
Culex quinquefasciatus
|
BCA |
B0WKV7 |
0.573 |
0.032 |
0.507 |
M |
5 |
|
Danaus plexippus
|
BCA |
G6D7Z4 |
0.793 |
0.032 |
0.273 |
M |
3 |
|
Daphnia pulex
|
BCA |
E9GLB5 |
0.157 |
0.055 |
0.843 |
- |
2 |
|
Dendroctonus ponderosae
|
BCA |
J3JTM9 |
0.27 |
0.064 |
0.742 |
- |
3 |
|
Drosophila ananassae
|
BCA |
B3LZ10 |
0.537 |
0.041 |
0.518 |
M |
5 |
|
Drosophila erecta
|
BCA |
B3P1V8 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila grimshawi
|
BCA |
B4JHY1 |
0.605 |
0.037 |
0.454 |
M |
5 |
|
Drosophila melanogaster
|
BCA |
Q9VHJ5 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila mojavensis
|
BCA |
B4KDC1 |
0.556 |
0.039 |
0.511 |
M |
5 |
|
Drosophila persimilis
|
BCA |
B4GFA1 |
0.595 |
0.037 |
0.466 |
M |
5 |
|
Drosophila pseudoobscura
|
BCA |
Q296E4 |
0.595 |
0.037 |
0.466 |
M |
5 |
|
Drosophila sechellia
|
BCA |
B4HKY7 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila simulans
|
BCA |
B4QXC5 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila virilis
|
BCA |
B4LZE7 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila willistoni
|
BCA |
B4NBB9 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Drosophila yakuba
|
BCA |
B4PTY0 |
0.531 |
0.04 |
0.53 |
M |
5 |
|
Entamoeba dispar
|
BCA |
B0E7M0 |
0.114 |
0.158 |
0.766 |
- |
2 |
|
Entamoeba histolytica
|
BCA |
C4LXK3 |
0.113 |
0.151 |
0.779 |
- |
2 |
|
Entamoeba nuttalli
|
BCA |
K2GQM0 |
0.132 |
0.142 |
0.763 |
- |
2 |
|
Harpegnathos saltator
|
BCA |
E2B2Q1 |
0.248 |
0.055 |
0.801 |
- |
3 |
|
Heliconius melpomene
|
BCA |
HMEL015257 |
0.77 |
0.032 |
0.302 |
M |
3 |
|
Hirudo medicinalis
|
BCA |
EY481200 |
0.121 |
0.098 |
0.778 |
- |
2 |
|
Ichthyophthirius multifiliis
|
BCA |
G0QPN9 |
0.181 |
0.04 |
0.872 |
- |
2 |
|
Leishmania donovani
|
BCA |
E9B8S3 |
0.106 |
0.13 |
0.826 |
- |
2 |
|
Leishmania infantum
|
BCA |
A4HSV2 |
0.106 |
0.13 |
0.826 |
- |
2 |
|
Leishmania major
|
BCA |
Q4QJ17 |
0.108 |
0.124 |
0.822 |
- |
2 |
|
Leishmania mexicana
|
BCA |
E9AKU0 |
0.109 |
0.135 |
0.82 |
- |
2 |
|
Lepeophtheirus salmonis
|
BCA |
D3PI48 |
0.126 |
0.068 |
0.889 |
- |
2 |
|
Nasonia vitripennis
|
BCA |
K7IWK8 |
0.388 |
0.046 |
0.713 |
- |
4 |
|
Nematostella vectensis
|
BCA |
A7S717 |
0.775 |
0.052 |
0.211 |
M |
3 |
|
Paramecium tetraurelia
|
BCA1 |
A0BD61 |
0.196 |
0.045 |
0.843 |
- |
2 |
| |
BCA2 |
A0E8J0 |
0.107 |
0.056 |
0.909 |
- |
1 |
| |
BCA3 |
A0CEX6 |
0.28 |
0.045 |
0.725 |
- |
3 |
| |
BCA4 |
A0BDB1 |
0.073 |
0.065 |
0.938 |
- |
1 |
| |
BCA5 |
A0C922 |
0.178 |
0.056 |
0.826 |
- |
2 |
|
Saccoglossus kowalevskii
|
BCA |
187043763 |
0.565 |
0.049 |
0.463 |
M |
5 |
|
Schistosoma mansoni
|
BCA |
G4V6B2 |
0.388 |
0.064 |
0.605 |
- |
4 |
|
Solenopsis invicta
|
BCA |
E9IP13 |
0.326 |
0.052 |
0.756 |
- |
3 |
|
Strigamia maritima
|
BCA |
SMAR006741 |
0.683 |
0.046 |
0.28 |
M |
3 |
|
Strongylocentrotus purpuratus
|
BCA |
H3I177 |
0.804 |
0.047 |
0.16 |
M |
2 |
|
Tetrahymena thermophila
|
BCA1 |
Q22U21 |
0.092 |
0.064 |
0.92 |
- |
1 |
| |
BCA2 |
Q22U16 |
0.087 |
0.075 |
0.918 |
- |
1 |
| |
BCA3 |
I7MDL7 |
0.659 |
0.067 |
0.203 |
M |
3 |
| |
BCA4 |
I7LWM1 |
0.115 |
0.058 |
0.871 |
- |
2 |
| |
BCA5 |
I7M0M0 |
0.087 |
0.034 |
0.947 |
- |
1 |
| |
BCA6 |
I7MD92 |
0.058 |
0.069 |
0.941 |
- |
1 |
| |
BCA7 |
I7M748 |
0.09 |
0.047 |
0.933 |
- |
1 |
| |
BCA8 |
Q23AV1 |
0.187 |
0.123 |
0.758 |
- |
3 |
|
Tribolium castaneum
|
BCA |
D6WK56 |
0.054 |
0.097 |
0.938 |
- |
1 |
|
Trichinella spiralis
|
BCA |
E5SH53 |
0.876 |
0.028 |
0.177 |
M |
2 |
|
Trichomonas vaginalis
|
BCA1 |
A2ENQ8 |
0.043 |
0.137 |
0.933 |
- |
2 |
| |
BCA2 |
A2DLG4 |
0.073 |
0.061 |
0.937 |
- |
1 |
|
Trichoplax adhaerens
|
BCA |
B3S5Y1 |
0.582 |
0.038 |
0.459 |
M |
5 |
| Xenoturbella bocki | BCA | 117195962 | 0.222 | 0.056 | 0.78 | - | 3 |
Discussion
This study shows that the β-CA enzyme is present in a range of protozoans and metazoans. A total of 75 sequences were identified and a phylogenetic tree constructed. The multiple sequence alignment results revealed that all 75 sequences have the highly conserved residues (Cysteine, Aspartic acid, Arginine, and Histidine) consistent with a β-CA enzyme (Additional file 1: Figure S1). Most of the metazoan and protozoan β-CAs, and corresponding coding sequences, were designated as uncharacterized sequences or CAs with no class specification. These can be now assigned to β-CAs in proteomics and genomics databases.
β-CAs have been identified in the mitochondria of a variety of different organisms, such as plants [61], green algae [62], fungi [1,63], and Drosophila melanogaster[4]. Our results of subcellular localization prediction (Table 2) suggested that 31 of the β-CAs are targeted to mitochondria. In mitochondrial targeting peptides (mTPs), Arginine, Alanine and Serine are over-represented, while negatively charged amino acid residues (Aspartic acid and Glutamic acid) are rare. Furthermore, mTPs are believed to form an amphiphilic α-helix, which is important for the import of the nascent protein into the mitochondrion [59]. The successful construction of the TargetP predictor demonstrates that protein sorting signals can be recognized with reasonable reliability from amino acid sequence data alone, thus, to some extent, mimicking the cellular recognition processes [59]. The prediction of the mitochondrial localization for many of the proteins studied is also supported by the previous experimental data, showing that recombinant DmBCA protein is indeed located in mitochondria of insect cells [4]. As mitochondrial proteins the β-CAs may contribute to key metabolic functions. Among the mammalian α-CAs, CA VA and CA VB are the only enzymes that have been exclusively located to mitochondria. Functional studies, summarized in [64], have indicated them in several metabolic processes, such as gluconeogenesis, urea synthesis, and fatty acid synthesis. It has been shown previously that the gluconeogenic enzyme, pyruvate carboxylase, is expressed in protozoan (Toxoplasma gondii) mitochondria [65]. This enzyme utilizes bicarbonate to convert pyruvate to oxaloacetate. Mitochondrial CA V is also involved in lipid synthesis through pyruvate carboxylation reaction [66]. Importantly, lipid metabolism is of crucial importance for parasites. Lipids serve as cellular building blocks, signaling molecules, energy stores, posttranslational modifiers, and pathogenesis factors [67]. Parasites rely on complex metabolic systems to satisfy their lipid needs. The present findings open a new avenue to investigate whether mitochondrial β-CAs are functionally involved in these processes.
The single β-CA of Anopheles darlingi is the first predicted secretory β-CA. Among the various α-CAs, the first secreted form (CA VI) was identified in human saliva in 1987 [68], and in 2011 another α-CA was identified in the salivary gland of Aedes aegypti[69]. Complementary research, such as morphological, biochemical, and spatial mapping of gene expression in Anopheles darlingi will clarify the exact expression pattern of β-CA in this mosquito [69,70].
The TargetP predictor defined 43 β-CAs with ‘other’ cellular localizations. Although it is possible that β-CAs are truly located in different subcellular compartments depending on the species, these results should be interpreted with caution. Both the common errors in full genomic DNA, cDNA, or protein sequences in databases, and the potential inaccuracy of TargetP predictor could contribute to the observed deviations of the results. The highest prediction accuracy, with appropriate selection of specificity and sensitivity, is 90% [59].
Among the species mentioned in Table 1, some have important medical relevance, such as Aedes aegypti, Anopheles darlingi, Anopheles gambiae, Ascaris suum (Ascaris lumbricoides), Culex quinquefasciatus, Entamoeba histolytica, Hirudo medicinalis, Leishmania species, Schistosoma mansoni, Trichinella spiralis, and Trichomonas vaginalis. In the past decade, inhibition profiles of β-CAs of bacteria [24,31,71] and fungi [72-75] have been investigated with various inhibitors. Our results suggest that various protozoans and metazoans express β-CAs and that these molecules represent protein targets appropriate for inhibitor development. These proteins are not restricted to nematodes, insects, or protozoa causing human diseases, but are also present in many species with relevance to agriculture or veterinary medicine. These species include: Acyrthosiphon pisum, Ancylostoma caninum, Ascaris suum, Caligus clemensi, Camponotus floridanus, Culex quinquefasciatus, Dendroctonus ponderosae, Entamoeba species, Ichthyophthirius multifiliis, Solenopsis invicta, Tribolium castaneum, Trichinella spiralis, and Trichoplax adhaerens. Therefore, our findings also suggest that it might be possible to develop specific β-CA inhibitors as pesticides for the protection of crops and other natural resources against pathogens and pests.
Conclusions
The present data identifies β-CA enzymes that are expressed in a number of protozoans and metazoans. Metazoan and protozoan β-CAs represent promising diagnostic and therapeutic targets for parasitic infections, because this CA family is absent from mammalian proteomes. Many of these enzymes are predicted to be present in mitochondria where they might contribute to cell metabolism by providing bicarbonate for biosynthetic reactions and regulating intra-mitochondrial pH.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RZE, HB, MEET, CO carried out the bioinformatics searches on metazoan and protozoan species. RZE and HB participated in the sequence alignment and made the phylogenetic analysis. RZE performed the mitochondrial targeting peptide prediction. All authors participated in the design of the study. RZE and HB drafted the first version of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Multiple sequence alignment of all 75 β-CAs in metazoan and protozoan species with β-CA of Pelosinus fermentans (a bacterial out group). β-CAs contain two highly conserved active site motifs, CxDxR as well as HxxC (C=Cysteine, D=Aspartic acid, R=Arginine, H=Histidine, C=Cysteine) which are indicated by arrows. Alignment was visualized in Jalview [53].
Contributor Information
Reza Zolfaghari Emameh, Email: reza.zolfaghari.emameh@uta.fi.
Harlan Barker, Email: harlan.barker@uta.fi.
Martti E E Tolvanen, Email: martti.tolvanen@uta.fi.
Csaba Ortutay, Email: csaba.ortutay@uta.fi.
Seppo Parkkila, Email: seppo.parkkila@uta.fi.
Acknowledgement
To perform these studies RZE received a scholarship support from the Ministry of Science, Research and Technology, and National Institute of Genetic Engineering and Biotechnology of Islamic Republic of Iran. This study was also funded by Finnish Cultural Foundation (HB), Academy of Finland (SP), Sigrid Juselius Foundation (SP), Jane and Aatos Erkko Foundation (SP), Tampere Tuberculosis Foundation (SP), and Competitive Research Funding of the Tampere University Hospital (SP; 9 N054).
References
- Elleuche S, Poggeler S. Carbonic anhydrases in fungi. Microbiology. 2010;156(Pt 1):23–29. doi: 10.1099/mic.0.032581-0. [DOI] [PubMed] [Google Scholar]
- Huang S, Hainzl T, Grundstrom C, Forsman C, Samuelsson G, Sauer-Eriksson AE. Structural studies of beta-carbonic anhydrase from the green alga Coccomyxa: inhibitor complexes with anions and acetazolamide. PLoS One. 2011;6(12):e28458. doi: 10.1371/journal.pone.0028458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7(2):168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- Syrjanen L, Tolvanen M, Hilvo M, Olatubosun A, Innocenti A, Scozzafava A, Leppiniemi J, Niederhauser B, Hytonen VP, Gorr TA. et al. Characterization of the first beta-class carbonic anhydrase from an arthropod (Drosophila melanogaster) and phylogenetic analysis of beta-class carbonic anhydrases in invertebrates. BMC Biochem. 2010;11:28. doi: 10.1186/1471-2091-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp BC, Smith K, Ferry JG. Carbonic anhydrase: new insights for an ancient enzyme. J Biol Chem. 2001;276(52):48615–48618. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Poggeler S. Evolution of carbonic anhydrases in fungi. Curr Genet. 2009;55(2):211–222. doi: 10.1007/s00294-009-0238-x. [DOI] [PubMed] [Google Scholar]
- Esbaugh AJ, Tufts BL. The structure and function of carbonic anhydrase isozymes in the respiratory system of vertebrates. Respir Physiol Neurobiol. 2006;154(1–2):185–198. doi: 10.1016/j.resp.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lane TW, Saito MA, George GN, Pickering IJ, Prince RC, Morel FM. Biochemistry: a cadmium enzyme from a marine diatom. Nature. 2005;435(7038):42. doi: 10.1038/435042a. [DOI] [PubMed] [Google Scholar]
- Xu Y, Feng L, Jeffrey PD, Shi Y, Morel FM. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature. 2008;452(7183):56–61. doi: 10.1038/nature06636. [DOI] [PubMed] [Google Scholar]
- Ferry JG. The gamma class of carbonic anhydrases. Biochim Biophys Acta. 2010;1804(2):374–381. doi: 10.1016/j.bbapap.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterio V, Vitale RM, Monti SM, Pedone C, Scozzafava A, Cecchi A, De Simone G, Supuran CT. Carbonic anhydrase inhibitors: X-ray and molecular modeling study for the interaction of a fluorescent antitumor sulfonamide with isozyme II and IX. J Am Chem Soc. 2006;128(25):8329–8335. doi: 10.1021/ja061574s. [DOI] [PubMed] [Google Scholar]
- Nishimori I, Minakuchi T, Onishi S, Vullo D, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. DNA cloning, characterization, and inhibition studies of the human secretory isoform VI, a new target for sulfonamide and sulfamate inhibitors. J Med Chem. 2007;50(2):381–388. doi: 10.1021/jm0612057. [DOI] [PubMed] [Google Scholar]
- Vullo D, Franchi M, Gallori E, Antel J, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Inhibition of mitochondrial isozyme V with aromatic and heterocyclic sulfonamides. J Med Chem. 2004;47(5):1272–1279. doi: 10.1021/jm031057+. [DOI] [PubMed] [Google Scholar]
- Vullo D, Innocenti A, Nishimori I, Pastorek J, Scozzafava A, Pastorekova S, Supuran CT. Carbonic anhydrase inhibitors. Inhibition of the transmembrane isozyme XII with sulfonamides-a new target for the design of antitumor and antiglaucoma drugs? Bioorg Med Chem Lett. 2005;15(4):963–969. doi: 10.1016/j.bmcl.2004.12.053. [DOI] [PubMed] [Google Scholar]
- Neish AC. Studies on chloroplasts: Their chemical composition and the distribution of certain metabolites between the chloroplasts and the remainder of the leaf. Biochem J. 1939;33(3):300–308. doi: 10.1042/bj0330300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell JN, Gibbs MJ, Mason JG. Spinach chloroplastic carbonic anhydrase: nucleotide sequence analysis of cDNA. Plant Physiol. 1990;92(1):37–40. doi: 10.1104/pp.92.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett RS. Structure and catalytic mechanism of the beta-carbonic anhydrases. Biochim Biophys Acta. 2010;1804(2):362–373. doi: 10.1016/j.bbapap.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Fett JP, Coleman JR. Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol. 1994;105(2):707–713. doi: 10.1104/pp.105.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau N, Coleman JR. Nucleotide sequence of a complementary DNA encoding tobacco chloroplastic carbonic anhydrase. Plant Physiol. 1992;100(2):1077–1078. doi: 10.1104/pp.100.2.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronk JD, Endrizzi JA, Cronk MR, O’Neill JW, Zhang KY. Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 2001;10(5):911–922. doi: 10.1110/ps.46301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloton MB, Korte JJ, Lamblin AF, Fuchs JA, Anderson PM. Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J Biol Chem. 1992;267(6):3731–3734. [PubMed] [Google Scholar]
- Smith KS, Ferry JG. A plant-type (beta-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J Bacteriol. 1999;181(20):6247–6253. doi: 10.1128/jb.181.20.6247-6253.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett RS, Tu C, Lee J, Herman AG, Chapnick DA, Shah SH, Gareiss PC. Allosteric site variants of Haemophilus influenzae beta-carbonic anhydrase. Biochemistry. 2009;48(26):6146–6156. doi: 10.1021/bi900663h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran CT. Bacterial carbonic anhydrases as drug targets: toward novel antibiotics? Front Pharmacol. 2011;2:34. doi: 10.3389/fphar.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P, Turtaut F, Ouahrani-Bettache S, Montero JL, Nishimori I, Minakuchi T, Vullo D, Scozzafava A, Kohler S, Winum JY. et al. Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Brucella suis. J Med Chem. 2010;53(5):2277–2285. doi: 10.1021/jm901855h. [DOI] [PubMed] [Google Scholar]
- Burghout P, Vullo D, Scozzafava A, Hermans PW, Supuran CT. Inhibition of the beta-carbonic anhydrase from Streptococcus pneumoniae by inorganic anions and small molecules: Toward innovative drug design of antiinfectives? Bioorg Med Chem. 2011;19(1):243–248. doi: 10.1016/j.bmc.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Vullo D, Nishimori I, Minakuchi T, Scozzafava A, Supuran CT. Inhibition studies with anions and small molecules of two novel beta-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium. Bioorg Med Chem Lett. 2011;21(12):3591–3595. doi: 10.1016/j.bmcl.2011.04.105. [DOI] [PubMed] [Google Scholar]
- Abuaita BH, Withey JH. Bicarbonate Induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun. 2009;77(9):4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G, Lin W, Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol. 2010;192(16):4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Liang WJ, Chaloupka J, Ruoff C, Schroppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF. et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15(22):2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti A, Leewattanapasuk W, Muhlschlegel FA, Mastrolorenzo A, Supuran CT. Carbonic anhydrase inhibitors. Inhibition of the beta-class enzyme from the pathogenic yeast Candida glabrata with anions. Bioorg Med Chem Lett. 2009;19(16):4802–4805. doi: 10.1016/j.bmcl.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol. 2005;15(22):2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Elleuche S, Poggeler S. A cyanase is transcriptionally regulated by arginine and involved in cyanate decomposition in Sordaria macrospora. Fungal Genet Biol. 2008;45(11):1458–1469. doi: 10.1016/j.fgb.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Amoroso G, Morell-Avrahov L, Muller D, Klug K, Sultemeyer D. The gene NCE103 (YNL036w) from Saccharomyces cerevisiae encodes a functional carbonic anhydrase and its transcription is regulated by the concentration of inorganic carbon in the medium. Mol Microbiol. 2005;56(2):549–558. doi: 10.1111/j.1365-2958.2005.04560.x. [DOI] [PubMed] [Google Scholar]
- Cleves AE, Cooper DN, Barondes SH, Kelly RB. A new pathway for protein export in Saccharomyces cerevisiae. J Cell Biol. 1996;133(5):1017–1026. doi: 10.1083/jcb.133.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz R, Gnann A, Zimmermann FK. Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast. 1999;15(10A):855–864. doi: 10.1002/(SICI)1097-0061(199907)15:10A<855::AID-YEA425>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- So AK, Espie GS. Cloning, characterization and expression of carbonic anhydrase from the cyanobacterium Synechocystis PCC6803. Plant Mol Biol. 1998;37(2):205–215. doi: 10.1023/A:1005959200390. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Cannon GC, Heinhorst S, Tanaka S, Williams EB, Yeates TO, Kerfeld CA. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J Biol Chem. 2006;281(11):7546–7555. doi: 10.1074/jbc.M510464200. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardestrom P, Samuelsson G. Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1996;93(21):12031–12034. doi: 10.1073/pnas.93.21.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Mizushima T, Yamashita E, Yamamoto M, Kumasaka T, Moriyama H, Ueki T, Miyachi S, Tsukihara T. X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO(2) hydration. J Biol Chem. 2000;275(8):5521–5526. doi: 10.1074/jbc.275.8.5521. [DOI] [PubMed] [Google Scholar]
- Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Molecular and biochemical analysis of the beta class carbonic anhydrases in Caenorhabditis elegans. Mol Biol Rep. 2010;37(6):2941–2950. doi: 10.1007/s11033-009-9857-z. [DOI] [PubMed] [Google Scholar]
- Guilloton MB, Lamblin AF, Kozliak EI, Gerami-Nejad M, Tu C, Silverman D, Anderson PM, Fuchs JA. A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol. 1993;175(5):1443–1451. doi: 10.1128/jb.175.5.1443-1451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimori I, Onishi S, Takeuchi H, Supuran CT. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des. 2008;14(7):622–630. doi: 10.2174/138161208783877875. [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S, Ohnishi J, Hayashi M, Ikeda M. A gene homologous to beta-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl Microbiol Biotechnol. 2004;63(5):592–601. doi: 10.1007/s00253-003-1402-8. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci U S A. 1992;89(10):4437–4441. doi: 10.1073/pnas.89.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C, Masters M, McAteer S, Coulson A. Why is carbonic anhydrase essential to Escherichia coli? J Bacteriol. 2003;185(21):6415–6424. doi: 10.1128/JB.185.21.6415-6424.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau N, Coleman JR. Effect of CO2 Concentration on Carbonic Anhydrase and Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase Expression in Pea. Plant Physiol. 1996;112(2):569–574. doi: 10.1104/pp.112.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetu SG, Tanz SK, Vella N, Burnell JN, Ludwig M. The Flaveria bidentis beta-carbonic anhydrase gene family encodes cytosolic and chloroplastic isoforms demonstrating distinct organ-specific expression patterns. Plant Physiol. 2007;144(3):1316–1327. doi: 10.1104/pp.107.098152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M. The molecular evolution of beta-carbonic anhydrase in Flaveria. J Exp Bot. 2011;62(9):3071–3081. doi: 10.1093/jxb/err071. [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Martin MV, Braun HP. A basal carbon concentrating mechanism in plants? Plant Sci: Int J Exp Plant Biol. 2012;187:97–104. doi: 10.1016/j.plantsci.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Kimber MS, Pai EF. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000;19(7):1407–1418. doi: 10.1093/emboj/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Mizushima T, Yamashita E, Miyachi S, Tsukihara T. Crystallization and preliminary X-ray diffraction studies of a beta-carbonic anhydrase from the red alga Porphyridium purpureum. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 2):210–211. doi: 10.1107/s0907444999015954. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. eb Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SD, Podar M, Klingeman DM, Johnson CM, Yang ZK, Utturkar SM, Land ML, Mosher JJ, Hurt RA Jr, Phelps TJ. et al. Draft genome sequences for two metal-reducing Pelosinus fermentans strains isolated from a Cr(VI)-contaminated site and for type strain R7. J Bacteriol. 2012;194(18):5147–5148. doi: 10.1128/JB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic biology. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Woese CR. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci U S A. 2000;97(15):8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre N, Reiter IM, Becuwe-Linka N, Genty B, Rumeau D. Characterization and expression analysis of genes encoding alpha and beta carbonic anhydrases in Arabidopsis. Plant Cell Environ. 2007;30(5):617–629. doi: 10.1111/j.1365-3040.2007.01651.x. [DOI] [PubMed] [Google Scholar]
- Mitra M, Lato SM, Ynalvez RA, Xiao Y, Moroney JV. Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 2004;135(1):173–182. doi: 10.1104/pp.103.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Muhlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9(6):572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Shah GN, Rubbelke TS, Hendin J, Nguyen H, Waheed A, Shoemaker JD, Sly WS. Targeted mutagenesis of mitochondrial carbonic anhydrases VA and VB implicates both enzymes in ammonia detoxification and glucose metabolism. Proc Natl Acad Sci U S A. 2013;110(18):7423–7428. doi: 10.1073/pnas.1305805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleige T, Pfaff N, Gross U, Bohne W. Localisation of gluconeogenesis and tricarboxylic acid (TCA)-cycle enzymes and first functional analysis of the TCA cycle in Toxoplasma gondii. Int J Parasit. 2008;38(10):1121–1132. doi: 10.1016/j.ijpara.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Hazen SA, Waheed A, Sly WS, LaNoue KF, Lynch CJ. Differentiation-dependent expression of CA V and the role of carbonic anhydrase isozymes in pyruvate carboxylation in adipocytes. FASEB J. 1996;10(4):481–490. doi: 10.1096/fasebj.10.4.8647347. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan S, Serricchio M, Striepen B, Butikofer P. Lipid synthesis in protozoan parasites: A comparison between kinetoplastids and apicomplexans. Prog Lipid Res. 2013;52(4):488–512. doi: 10.1016/j.plipres.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Sly WS. Purification and characterization of human salivary carbonic anhydrase. J Biol Chem. 1987;262(3):1382–1388. [PubMed] [Google Scholar]
- Juhn J, Naeem-Ullah U, Maciel Guedes BA, Majid A, Coleman J, Paolucci Pimenta PF, Akram W, James AA, Marinotti O. Spatial mapping of gene expression in the salivary glands of the dengue vector mosquito, Aedes aegypti. Parasit Vectors. 2011;4:1. doi: 10.1186/1756-3305-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Ferro CK, Marinotti O, Bijovsky AT. Morphological and biochemical analyses of the salivary glands of the malaria vector, Anopheles darlingi. Tissue Cell. 1999;31(3):264–273. doi: 10.1054/tice.1999.0057. [DOI] [PubMed] [Google Scholar]
- Maresca A, Scozzafava A, Kohler S, Winum JY, Supuran CT. Inhibition of beta-carbonic anhydrases from the bacterial pathogen Brucella suis with inorganic anions. J Inorg Biochem. 2012;110:36–39. doi: 10.1016/j.jinorgbio.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Isik S, Guler OO, Kockar F, Aydin M, Arslan O, Supuran CT. Saccharomyces cerevisiae beta-carbonic anhydrase: inhibition and activation studies. Curr Pharm Des. 2010;16(29):3327–3336. doi: 10.2174/138161210793429878. [DOI] [PubMed] [Google Scholar]
- Isik S, Kockar F, Aydin M, Arslan O, Guler OO, Innocenti A, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors: inhibition of the beta-class enzyme from the yeast Saccharomyces cerevisiae with sulfonamides and sulfamates. Bioorg Med Chem. 2009;17(3):1158–1163. doi: 10.1016/j.bmc.2008.12.035. [DOI] [PubMed] [Google Scholar]
- Monti SM, Maresca A, Viparelli F, Carta F, De Simone G, Muhlschlegel FA, Scozzafava A, Supuran CT. Dithiocarbamates are strong inhibitors of the beta-class fungal carbonic anhydrases from Cryptococcus neoformans, Candida albicans and Candida glabrata. Bioorg Med Chem Lett. 2012;22(2):859–862. doi: 10.1016/j.bmcl.2011.12.033. [DOI] [PubMed] [Google Scholar]
- Guzel O, Maresca A, Hall RA, Scozzafava A, Mastrolorenzo A, Muhlschlegel FA, Supuran CT. Carbonic anhydrase inhibitors. The beta-carbonic anhydrases from the fungal pathogens Cryptococcus neoformans and Candida albicans are strongly inhibited by substituted-phenyl-1H-indole-5-sulfonamides. Bioorg Med Chem Lett. 2010;20(8):2508–2511. doi: 10.1016/j.bmcl.2010.02.103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple sequence alignment of all 75 β-CAs in metazoan and protozoan species with β-CA of Pelosinus fermentans (a bacterial out group). β-CAs contain two highly conserved active site motifs, CxDxR as well as HxxC (C=Cysteine, D=Aspartic acid, R=Arginine, H=Histidine, C=Cysteine) which are indicated by arrows. Alignment was visualized in Jalview [53].