Abstract
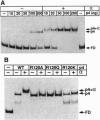
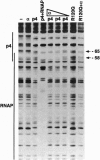
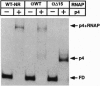
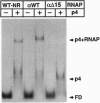
Regulatory protein p4 from Bacillus subtilis phage phi29 activates transcription from the viral late A3 promoter by stabilizing sigmaA-RNA polymerase at the promoter as a closed complex. Activation requires an interaction between protein p4 and RNA polymerase mediated by the protein p4 carboxyl-end, mainly through residue Arg-120. We have obtained derivatives of B. subtilis RNA polymerase alpha subunit with serial deletions at the carboxyl-end and reconstituted RNA polymerase holoenzymes harboring the mutant alpha subunits. Protein p4 promoted the binding of purified B. subtilis RNA polymerase alpha subunit to the A3 promoter in a cooperative way. Binding was abolished by deletion of the last 15 amino acids of the alpha subunit. Reconstituted RNA polymerases with deletions of 15 to 59 residues at the alpha subunit carboxyl-end could recognize and transcribe viral promoters not activated by protein p4, but they had lost their ability to recognize the A3 promoter in the presence of protein p4. In addition, these mutant reconstituted RNA polymerases could not interact with protein p4. We conclude that protein p4 activation of the viral A3 promoter requires an interaction between the carboxyl-end of protein p4 and the carboxyl-end of the alpha subunit of B. subtilis RNA polymerase that stabilizes the RNA polymerase at the promoter.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldus J. M., Buckner C. M., Moran C. P., Jr Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995 Jul;17(2):281–290. doi: 10.1111/j.1365-2958.1995.mmi_17020281.x. [DOI] [PubMed] [Google Scholar]
- Barthelemy I., Lázaro J. M., Méndez E., Mellado R. P., Salas M. Purification in an active form of the phage phi 29 protein p4 that controls the viral late transcription. Nucleic Acids Res. 1987 Oct 12;15(19):7781–7793. doi: 10.1093/nar/15.19.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy I., Salas M. Characterization of a new prokaryotic transcriptional activator and its DNA recognition site. J Mol Biol. 1989 Jul 20;208(2):225–232. doi: 10.1016/0022-2836(89)90384-7. [DOI] [PubMed] [Google Scholar]
- Blatter E. E., Ross W., Tang H., Gourse R. L., Ebright R. H. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell. 1994 Sep 9;78(5):889–896. doi: 10.1016/s0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- Chang B. Y., Doi R. H. Overproduction, purification, and characterization of Bacillus subtilis RNA polymerase sigma A factor. J Bacteriol. 1990 Jun;172(6):3257–3263. doi: 10.1128/jb.172.6.3257-3263.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Busby S. The Escherichia coli RNA polymerase alpha subunit: structure and function. Curr Opin Genet Dev. 1995 Apr;5(2):197–203. doi: 10.1016/0959-437x(95)80008-5. [DOI] [PubMed] [Google Scholar]
- Fredrick K., Caramori T., Chen Y. F., Galizzi A., Helmann J. D. Promoter architecture in the flagellar regulon of Bacillus subtilis: high-level expression of flagellin by the sigma D RNA polymerase requires an upstream promoter element. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2582–2586. doi: 10.1073/pnas.92.7.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward R. S., Igarashi K., Ishihama A. Functional specialization within the alpha-subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991 Sep 5;221(1):23–29. doi: 10.1016/0022-2836(91)80197-3. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Fujita N., Ishihama A. Identification of a subunit assembly domain in the alpha subunit of Escherichia coli RNA polymerase. J Mol Biol. 1991 Mar 5;218(1):1–6. [PubMed] [Google Scholar]
- Igarashi K., Ishihama A. Bipartite functional map of the E. coli RNA polymerase alpha subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991 Jun 14;65(6):1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993 May;175(9):2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R., Sharif K. A., Krakow J. S. Evidence for contact between the cyclic AMP receptor protein and the delta 70 subunit of Escherichia coli RNA polymerase. J Biol Chem. 1995 Aug 18;270(33):19213–19216. doi: 10.1074/jbc.270.33.19213. [DOI] [PubMed] [Google Scholar]
- Kimura M., Fujita N., Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino-terminal assembly domain. J Mol Biol. 1994 Sep 16;242(2):107–115. doi: 10.1006/jmbi.1994.1562. [DOI] [PubMed] [Google Scholar]
- Kuldell N., Hochschild A. Amino acid substitutions in the -35 recognition motif of sigma 70 that result in defects in phage lambda repressor-stimulated transcription. J Bacteriol. 1994 May;176(10):2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Grimes B., Fujita N., Makino K., Malloch R. A., Hayward R. S., Ishihama A. Role of the sigma 70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol. 1994 Jan 14;235(2):405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Li M., Moyle H., Susskind M. M. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994 Jan 7;263(5143):75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- Makino K., Amemura M., Kim S. K., Nakata A., Shinagawa H. Role of the sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993 Jan;7(1):149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- Mencía M., Monsalve M., Salas M., Rojo F. Transcriptional activator of phage phi 29 late promoter: mapping of residues involved in interaction with RNA polymerase and in DNA bending. Mol Microbiol. 1996 Apr;20(2):273–282. doi: 10.1111/j.1365-2958.1996.tb02616.x. [DOI] [PubMed] [Google Scholar]
- Mencía M., Salas M., Rojo F. Residues of the Bacillus subtilis phage phi 29 transcriptional activator required both to interact with RNA polymerase and to activate transcription. J Mol Biol. 1993 Oct 20;233(4):695–704. doi: 10.1006/jmbi.1993.1546. [DOI] [PubMed] [Google Scholar]
- Monsalve M., Mencía M., Rojo F., Salas M. Transcription regulation in Bacillus subtilis phage phi 29: expression of the viral promoters throughout the infection cycle. Virology. 1995 Feb 20;207(1):23–31. doi: 10.1006/viro.1995.1048. [DOI] [PubMed] [Google Scholar]
- Negishi T., Fujita N., Ishihama A. Structural map of the alpha subunit of Escherichia coli RNA polymerase: structural domains identified by proteolytic cleavage. J Mol Biol. 1995 May 12;248(4):723–728. doi: 10.1006/jmbi.1995.0254. [DOI] [PubMed] [Google Scholar]
- Nuez B., Rojo F., Salas M. Phage phi 29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein-protein contacts. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11401–11405. doi: 10.1073/pnas.89.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L., Ross W., Appleman J. A., Gaal T., Leirmo S., Schlax P. J., Record M. T., Jr, Gourse R. L. Factor independent activation of rrnB P1. An "extended" promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994 Feb 4;235(5):1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- Rojo F., Salas M. A DNA curvature can substitute phage phi 29 regulatory protein p4 when acting as a transcriptional repressor. EMBO J. 1991 Nov;10(11):3429–3438. doi: 10.1002/j.1460-2075.1991.tb04907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F., Zaballos A., Salas M. Bend induced by the phage phi 29 transcriptional activator in the viral late promoter is required for activation. J Mol Biol. 1990 Feb 20;211(4):713–725. doi: 10.1016/0022-2836(90)90072-t. [DOI] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Inciarte M. R., Corral J., Viñuela E., Salas M. RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage phi29. J Mol Biol. 1979 Feb 5;127(4):411–436. doi: 10.1016/0022-2836(79)90230-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yura T., Ishihama A. Genetics of bacterial RNA polymerases. Annu Rev Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]