Abstract
Nasopharyngeal carcinoma (NPC) is a prevalent malignancy in Southeast Asia among the Chinese population. Aberrant regulation of transcripts has been implicated in many types of cancers including NPC. Herein, we characterized mRNA and miRNA transcriptomes by RNA sequencing (RNASeq) of NPC model systems. Matched total mRNA and small RNA of undifferentiated Epstein–Barr virus (EBV)-positive NPC xenograft X666 and its derived cell line C666, well-differentiated NPC cell line HK1, and the immortalized nasopharyngeal epithelial cell line NP460 were sequenced by Solexa technology. We found 2812 genes and 149 miRNAs (human and EBV) to be differentially expressed in NP460, HK1, C666 and X666 with RNASeq; 533 miRNA–mRNA target pairs were inversely regulated in the three NPC cell lines compared to NP460. Integrated mRNA/miRNA expression profiling and pathway analysis show extracellular matrix organization, Beta-1 integrin cell surface interactions, and the PI3K/AKT, EGFR, ErbB, and Wnt pathways were potentially deregulated in NPC. Real-time quantitative PCR was performed on selected mRNA/miRNAs in order to validate their expression. Transcript sequence variants such as short insertions and deletions (INDEL), single nucleotide variant (SNV), and isomiRs were characterized in the NPC model systems. A novel TP53 transcript variant was identified in NP460, HK1, and C666. Detection of three previously reported novel EBV-encoded BART miRNAs and their isomiRs were also observed. Meta-analysis of a model system to a clinical system aids the choice of different cell lines in NPC studies. This comprehensive characterization of mRNA and miRNA transcriptomes in NPC cell lines and the xenograft provides insights on miRNA regulation of mRNA and valuable resources on transcript variation and regulation in NPC, which are potentially useful for mechanistic and preclinical studies.
Keywords: Nasopharyngeal carcinoma; RNA sequencing; Transcriptome analysis; Nasopharyngeal cell lines/xenograft (NP460, HK1, C666, X666); TP53
Abbreviations: NPC, nasopharyngeal carcinoma; EBV, Epstein–Barr virus; RNASeq, RNA sequencing; miRNA, microRNA; NGS, next-generation sequencing; SNP, single nucleotide polymorphism; INDEL, insertion and deletion; UTR, untranslated region; GO, gene ontology; ECM, extracellular matrix; EGFR, epidermal growth factor receptor; PI3K, phosphoinositide 3-kinase; EGR1, early growth response 1; GNG11, guanine nucleotide binding protein (G protein), Gamma 11; DKK1, Dickkopf-Like protein 1; MET, met proto-oncogene; CIITA, class II, major histocompatibility complex, transactivator; IL18, interleukin 18; TNFRSF9, tumour necrosis factor receptor superfamily, member 9; MMP19, matrix metallopeptidase 19; FBLN2, fibulin 2; LTBP2, latent transforming growth factor beta binding protein 2; PTEN, phosphatase and tensin homolog; LMP1, Epstein–Barr virus latent membrane protein 1; AIP, aryl hydrocarbon receptor interacting protein; BAX, BCL2-asscoiated X protein; GADD45, growth arrest and DNA-damage-inducible; MDM2, MDM2 oncogene, E3 ubiquitin protein ligase; GSTP1, glutathione S-transferase pi 1
Highlights
-
•
Using RNASeq we characterized the mRNA and miRNA transcriptomes in NPC and NP models.
-
•
2812 Genes and 149 miRNAs (human and EBV) were differentially expressed in NPC vs NP models.
-
•
533 miRNA–mRNA target pairs were inversely regulated in HK1, C666, and X666 vs NP460.
-
•
ECM, β1 integrin, PI3K/AKT, EGFR, ErbB, and Wnt pathways appeared to be deregulated in NPC.
-
•
A novel TP53 mutation was identified in NP460, HK1, and C666.
1. Introduction
Nasopharyngeal carcinoma (NPC) is a prevalent malignant disease in Southeast Asia among the Chinese population. According to the Hong Kong Cancer registry statistics, a high incidence rate of 15.1/100,000 in men has been observed in Hong Kong, while the incidence rate in Western countries is much lower (<1/100,000) [1]. NPC is classified by WHO into three types: type I keratinizing squamous carcinoma, type IIA non-keratinizing differentiated carcinoma, and type IIB non-keratinizing undifferentiated carcinoma [2]. The majority of NPC cases in Western countries are type I, such as in the United States, where it accounts for 75% of all NPC cases [3]. Most NPC cases from the Southeast Asia region are type II [4]. Type II NPC is constantly associated with Epstein–Barr virus (EBV) infection, but the association between type I NPC with EBV is still controversial [5].
In the past 30 years, several NPC cell lines and xenograft models have been established for the in vitro study of NPC. Examples such as the C666, CNE-1, CNE-2, HK1, HNE-1, and HONE-1 NPC cell lines were established from biopsies [6–9], while a series of nonmalignant nasopharyngeal epithelial cell lines were also established by immortalization from primary cultures [10]. Xenograft models such as X(eno)-666, X(eno)-2117, X(eno)-1915, C15, and C17 were established in a rodent xenograft system [11–14]. They are valuable models for research in NPC.
Aberrant transcript expression includes changes in expression levels, isoforms, and polymorphisms, which are commonly observed in cancer; these aberrations could alter biological pathways and disease phenotypes. Next-generation sequencing (NGS) of RNA (RNASeq) has become a popular tool for studying the comprehensive transcriptome in recent years. Despite the improving sensitivity and dynamic range of the gene expression array, RNASeq still plays a vital role in providing sequence information of the transcript that greatly enhances our knowledge of the transcriptome in cancer [15]. The microRNAs (miRNAs) are a class of small non-coding RNAs that regulate mRNA through sequence-specific binding to the UTR [16]. Studies on miRNA dysregulation in cancers have risen rapidly in recent years, including those in NPC. miRNAs such as hsa-mir-141, hsa-mir-138, hsa-mir-200a, and hsa-mir-26a are altered in NPC and regulate cell proliferation, cell cycle, extracellular matrix organization, migration, and invasion [17–20]. In addition to human miRNAs, NPC is also associated with EBV infection. The host-virus interaction has been thoroughly studied in B lymphocytes and it has been found that host transcriptional regulators play a role in EBV gene regulation, while EBV-encoded microRNAs induce cell transformation [21,22]. Regulation of both human and EBV gene expression by EBV-encoded miRNA has been observed in NPC. For example, expression of ebv-mir-BART-22 modulates expression of the EBV-encoded LMP2A protein, which is related to the host immune response [23]. The expression of ebv-mir-BART3 targets the DICE1 tumor suppressor that stimulates cell proliferation [24]. Several studies on expression profiling of miRNA from clinical biopsy samples have been reported on both EBV and human miRNAs [25–27]. However, studies on concurrent transcriptome characterization of both mRNA and miRNA are still lacking.
This study is to characterize the mRNA and miRNA transcriptome in NPC models, which provides a global view of transcript regulation in an in vitro system (Fig. 1A). NP460 is a well-established immortalized nasal epithelium cell line usually used as a control for mechanistic studies [10]. HK1 is one of the few well-differentiated NPC cell lines [8]. Xeno-666 (X666) and the subsequent cell line C666 are the only undifferentiated cell line and xenograft pairs that harbor EBV infection for EBV studies in NPC [6]. Biological pathways such as extracellular matrix organization, integrin signaling, angiogenesis, and hypoxia are commonly enriched in NPC cell lines. The miRNA-regulated pathways such as EGFR signaling are enriched in both HK1 and C666/X666. The miRNA-regulated nuclear beta catenin signaling is exclusively enriched in HK1, while the Wnt signaling pathway is enriched solely in C666/X666, respectively. Real-time quantitative PCR was performed on the selected miRNA-regulated biological networks like EGFR signaling and cytokine and interferon signaling in NPC and NP cell lines.
Fig. 1.
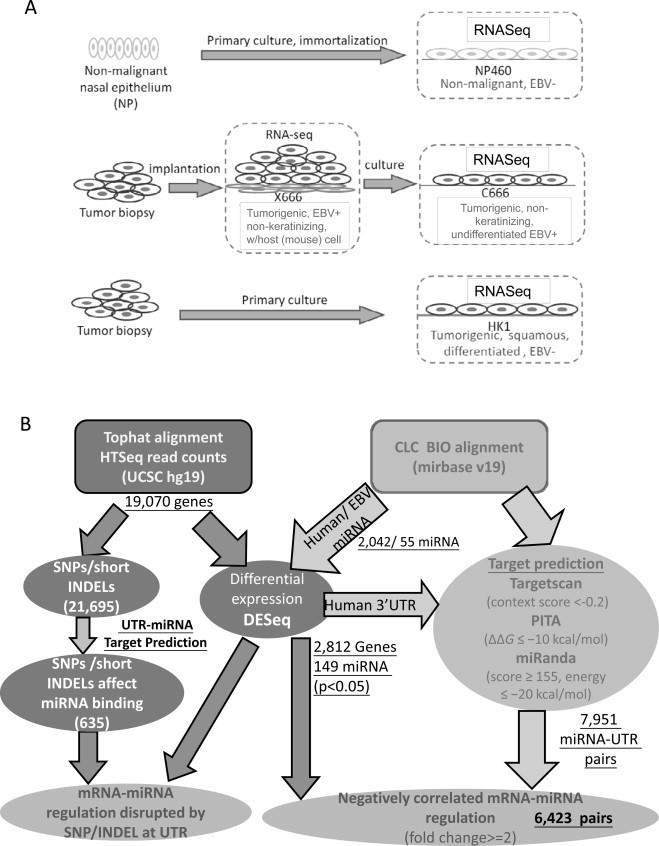
An overview of the samples used in this study and data analysis workflow. (A) Cell lines sequenced in this study. (B) Bioinformatics analysis workflow.
We also explored sequence variants in transcripts such as SNVs and short INDELs in the NPC model system and integrated these transcriptomes to publicly available microarray data from clinical specimens. A novel TP53 variant in a transcript has been discovered from the SNV. The meta-analysis of these model systems to clinical specimens aids the choice of different cell lines in various NPC studies.
IsomiRs are heterogeneous variants of miRNAs in length and sequence. Recent studies suggested that isomiRs may interact with mRNA, affecting target selection, miRNA stability, and translational machinery [28]. Here we report a comprehensive collection of human and EBV-encoded isomiRs from the NPC model system and identified a number of substantially expressed EBV-encoded isomiRs, which may be considered as reference miRNAs in NPC. We discovered the existence of three previously reported novel EBV-miRNAs in C666 and X666 [26], which may play mechanistic roles in NPC. This report not only provides a fundamental picture of global gene and miRNA–mRNA regulation in NPC, but also provides potentially useful candidates for future mechanistic and preclinical studies.
2. Material and methods
2.1. Sample preparation, DNA and RNA extraction
Early passages of NPC cell lines, HK1, C666, and NP460, and the X666 xenograft were utilized for this study. DNAs were extracted from cell lines and the xenograft using the QIAGEN DNA extraction kit according to manufacturer instructions. DNA genotyping of cell lines authenticated their origins. Real-time quantitative EBV DNA PCR was performed on cell lines, as previously described [29]. EBV DNA was only detected in C666 and no EBV DNA was detected in HK1 and NP460 cell lines (data not shown). All the cell lines were re-grown in standard conditions [6,8,10]. Cells were collected from three subsequent culture passages upon reaching ∼80% confluence, as described. Total RNA was extracted with TRIZOL reagent (Invitrogen, NY, USA) according to standard procedures. Integrity of RNA was checked by Agilent 2100 bioanalyzer using the RNA 6000 Nano total RNA assay, with RIN value >8.9. RNA from the earliest passage of each cell line was selected for Solexa sequencing.
2.2. Solexa sequencing, read processing and sequence alignment
The sequencing library was prepared with the standard Illumina protocol. Briefly, total RNA was poly-A-selected to deplete the ribosomal RNA fraction. The cDNA was synthesized using random hexamers, end-repaired and ligated with appropriate adaptors for sequencing. The library then underwent size selection and PCR amplification, followed by PAGE purification before sequencing. Stranded small RNA libraries were prepared by ligating different 3′ and 5′ adaptors sequentially to the total RNA followed by reverse transcription and PCR amplification. Small RNAs with insert sizes of 20–70 bp were PAGE-purified for sequencing. Both mRNA and small RNA libraries were sequenced on the Illumina Solexa GAIIx sequencer with 58 bp single-end reads, according to the standard manufacturer's protocol. Raw RNASeq reads were filtered for adapters and ribosomal RNA, followed by alignment to the human genome (hg19) and mouse genome (mm9) using the Tophat [30,31] v2.0.3.1. Reads mapped to multiple locations were discarded using the −G option of Tophat. UCSC gene models were used for analysis with both software and downloaded from the Tophat website (http://tophat.cbcb.umd.edu/igenomes.html). CLC genomics workbench v5.5 (CLC bio, Denmark) was used for small RNA analysis. Adapters were trimmed under default parameter setting to retain only reads with lengths ≥15 bp. These reads were then mapped and annotated against the miRBase [32–35] (release 19). Read counts of the annotated miRNA were exported from CLC genomics workbench and RPM (Reads per million base pairs) were calculated using customs scripts. Non-annotated reads were further mapped to Ribosomal RNA (rRNA), Transfer RNA (tRNA), Small nucleolar RNA (snoRNA), Messenger RNA (mRNA), Small nuclear RNA (snRNA), and genomic repeats. IsomiRs were analyzed based on CLC results using custom scripts based on mature reference sequences from miRBase and novel EBV-miRNA from Chen et al. [26].
2.3. miRNA target prediction
The miRNA target prediction was done by scripts from Targetscan [36], PITA [37], and miRanda [38]. Human UTRs were downloaded from the Targetscan website sourced from UCSC genome informatics; mature miRNAs were downloaded from the miRBase (release 19). Human UTRs, which encode genes that were significantly expressed (p < 0.05) in at least two samples, were further selected for the algorithms to predict against those human and EBV miRNAs, which are significantly expressed in at least two samples. High efficacy targets were selected by the following cutoffs: (i) Targetscan: sum of the context score <−0.2; (ii) PITA: sites with ΔΔG ≤ −10 kcal/mol; (iii) miRanda: score ≥ 155, energy ≤ −20 kcal/mol. Predicted miRNA–UTR pairs were further selected according to the integrating mRNA and miRNA transcript expression data. A two-fold cutoff was set for defining up/down regulation and target pairs with inverse expression. For miRNA-binding UTR analysis, 25 base pairs upstream/downstream of the UTR sequence from the SNP/INDEL were downloaded from UCSC and predicted the miRNA binding site against all human and EBV miRNAs from the miRBase by the three prediction algorithms (Targetscan, PITA, and miRanda). For the predicted binding site by at least 2 of the 3 aforementioned algorithms, we replaced the reference sequence with the variant and carried out the prediction again with all the reference and variant sequences.
2.4. Biological network and function enrichment analysis
Network analysis, biological pathway, and gene ontology (GO) term enrichment were done using the Reactome FI [39] plug-in of Cytoscape [40]. Gene lists from each expression set with significant expression (p < 0.05) in the respective comparisons were loaded as input and the biological network was constructed by the gene set analysis of Reactome FI. Biological pathway and Gene Ontology Biological Function terms enrichment were performed using input genes from the biological network. Pathway and GO terms with adjusted p < 0.05 were considered to be enriched in the biological network.
miRNA–mRNA target expression pairs, which have significant expression in respective comparison from the previous section, were chosen. Then the expression networks of those selected pairs were constructed by defining the target pair interaction between miRNA and its target gene. These expression networks were further merged with the respective biological network, which was constructed from the Reactome FI containing all mRNA genes coming from the same expression set. Biological network of the miRNA–mRNA target plus the first-degree neighbor of the mRNA target were selected as the miRNA regulatory network. Pathway and function term enrichment was further performed as stated above.
2.5. SNP/INDEL detection and characterization
Variant calling was done using VarScan [41] v2.2.11 with filtering criteria of at least 10× coverage, variation frequency of more than 10%, and base quality of more than 15. Variants that passed the filtering criteria were annotated using ANNOVAR with COSMIC64 annotation [42]. DNA sequence logos were generated using WebLogo generator [43].
2.6. Statistical analysis
The following statistical analyses were performed on R (version 2.15.1) with respective packages and a p-value of <0.05 was considered as significant. Expression analysis of mRNA and miRNA was done using the DESeq package [44], which uses the read counts as input. Meta-analysis of mRNA expression and microarray expression data downloaded from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) were performed by the MetaDE R package [45]. Fisher exact test was used and a p-value of <0.05 was set as the cutoff.
2.7. Reverse transcription (RT) and real-time quantitative PCR (QPCR)
To quantitate mRNA expression, the total RNA from each sample was reverse transcribed into cDNA using MMLV (USB, Cleveland, OH). Specific primers were either from the literature or designed using Primer 3 software [46]; the sequences and references of the primers are listed in Supplement 1. Real-time QPCR was performed on Light-Cycler Roche 480 (Roche Molecular Systems). PCR was performed in a total volume of 10 μl containing 50 ng of total cDNA, 1× FastStart Universal SYBR Green master mix (Roche) and a final primer concentration as stated in Supplement 1. Detection of mature miRNA expression was performed by TaqMan miRNA assay (Applied Biosystems, Foster City, CA, USA), normalized using small nucleolar RNA RNU44 as a control. The relative expression level was analyzed by the ΔΔCt method [47] or using the Pfaffl model [48]. One-way ANOVA was followed by Tukey post hoc analysis, where a p < 0.05 was considered statistically significant.
3. Results
3.1. Overview of transcriptome sequencing results of NPC cell lines
To reveal the transcriptome of NPC model systems, we sequenced the mRNA and small RNA from the same total RNA samples from the early passages of model NPC cell lines and xenograft using Solexa GAIIx. From the four mRNA libraries, an average of 30,277,744 single-end 58 bp reads were generated (Supplement 2); four small RNA libraries were also sequenced and 25,613,996 single-end 58 bp reads were generated on average. After performing the quality filter, mRNA sequence reads were aligned to the human (hg19) and mouse (mm9) genomes, while small RNA sequence reads were aligned to miRBase (see Section 2). A total of 2812 mRNAs and 149 miRNAs (human and EBV) were differentially expressed (p < 0.05) with any combination of the four samples sequenced (Fig. 1B). Detailed sequencing data has been uploaded to the GEO database with accession number GSE54174.
We then analyzed the differences and similarity of the differentially-expressed transcripts that contribute to carcinogenesis. A Venn diagram of differentially-expressed mRNAs and miRNAs against the immortalized nasopharyngeal epithelial cell line NP460 shows that 368 protein-coding genes and 12 miRNAs were significantly expressed in common among the NPC cell lines (Fig. 2A). The 749 mRNAs and 37 miRNAs show significant expression changes only in HK1. On the other hand, 963 mRNAs and 62 miRNAs are differentially expressed in C666, but not HK1 (Fig. 2A). For the cell line samples, less than 0.03% of the transcripts were aligned to the mouse genome, while 5% of the transcripts aligned to mouse for the X666 xenograft. Considering the possible contamination of mouse homolog transcript by host cells from the xenograft, transcripts expressed significantly in the X666 xenograft were not included in the analysis (Supplement 2).
Fig. 2.
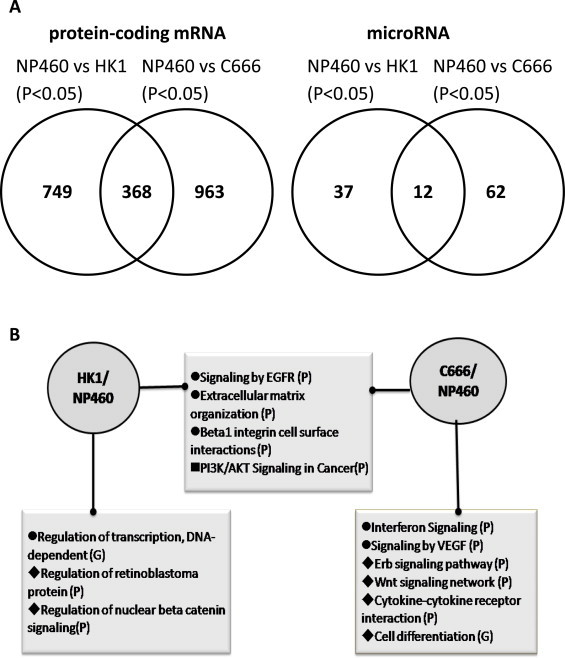
Differentially expressed genes/miRNAs in HK1 and C666 compared to NP460 and relevant biological pathways/functions in transcriptome data. (A) Venn diagram showing the number of protein coding gene/microRNA differentially expressed in HK1 and C666 compared to NP460. (B) List of Biological pathways and Gene Oncology Biological Functions from Reactome FI analysis. Terms begin with circle: enriched in mRNA only; terms begin with diamond: enriched in mRNA and miRNA; terms begin with rectangle: enriched in miRNA only; (P): term from Reactome pathway enrichment; (G): term from Gene Oncology Biological Process.
3.2. miRNA target prediction and integration of mRNA and miRNA expression profiles
To our knowledge, the resources available for the target prediction for EBV-encoded miRNAs to human 3′-UTR are limited; many of the public target prediction databases do not have the miRBase (release 19) for human miRNAs. Thus, we ran our in-house miRNA target prediction using scripts from Targetscan [36], PITA [37], and miRanda [38]. We ran the three prediction algorithms using the 149 miRNAs and UTR of the 2812 mRNAs, which are expressed significantly, as input. This resulted in 7951 miRNA–mRNA target pairs, which were predicted from at least two out of three algorithms with high efficacy. We further integrated transcript expression data into the predicted miRNA–UTR pairs to validate target pairs. We set the cutoff at two-fold for defining up/down regulation and focused on the pairs which are inversely expressed in terms of transcript expression. A total of 6423 target pairs were found inversely expressed in at least one combination of the expression comparisons. Of the 6423 target pairs, 4898 were inversely expressed in at least one NPC model system (HK1, C666, X666) against NP460, and 533 pairs are inversely regulated in all three NPC model systems against NP460 (Supplement 3).
3.3. Molecular pathway and functional enrichment from biological network analysis
Network analysis, biological pathway, and gene ontology (GO) enrichment were done for both the significantly expressed mRNA gene list and integrated panels of miRNA–mRNA regulation in common and uniquely expressed group of HK1 and C666 against NP460. In order to enrich the biological function arising from miRNA target pairs, integrated panels of miRNA regulation were generated by selecting the direct target of the miRNAs from the results of target prediction and the first neighbor of the genes that were significantly expressed from the same group from the mRNA network. Biological network analyses of these gene sets were done using the cytoscape plug-in of Reactome FI [39] and a summary of the significantly enriched terms (p < 0.05) in Reactome pathway and GO Biological Function is listed in Supplement 4. Fig. 2B shows the important pathway/GO terms enriched solely and coherently in NPC cell lines.
Biological pathways related to extracellular matrix (ECM) organization are explicitly enriched in the genes that are significantly expressed in both HK1 and C666 (Fig. 2B). Most of the commonly expressed genes from this pathway are down-regulated; two-thirds of the genes unique in HK1 are also decreased in expression and 40% of the unique genes in C666 are up-regulated (Supplement 4, Supplemental Fig. 1). Other ECM-related GO terms /pathways such as cell adhesion, beta-1 integrin cell surface interactions, and ECM-receptor interactions are also enriched in both HK1 and C666, implicating the importance of the extracellular matrix in NPC. EGFR and PI3K/AKT signaling are enriched only in the miRNA-regulated biological network in the two NPC cell lines (Supplement 4).
A total of 100 genes enriched in the GO term DNA-dependent transcription are significantly down-regulated in the HK1 only group (Supplement 4). The 73 genes from the ZNF family were down-regulated, of which 63 genes were located on chromosome 19 (Supplement 2). Several HK1 unique pathways are enriched in both mRNA and miRNA regulated biological networks. Pathways such as Regulation of retinoblastoma protein and nuclear beta catenin signaling were enriched in HK1 unique biological network regulated by hsa-mir-31–5p and hsa-mir-34b-5p (Supplemental Fig. 1).
Interferon and VEGF signaling pathways are enriched in the unique C666/X666 mRNA set. This involves up-regulation of MHC class II HLA genes (-DRA, -DQA1, -DRB1, -DMA and -DOA). Interferon signaling pathway that regulates the expression of this class of proteins is also up-regulated solely in C666/X666. Several biological networks enriched in C666/X666 are also regulated by EBV-encoded miRNA (Supplemental Fig. 1). Pathways such as Erb and Wnt signaling and cytokine-cytokine receptor interactions are enriched.
Fig. 3A shows miRNA regulated biological network enriched in EGFR, ErbB and Wnt signaling pathways. Expression trend of five protein-encoding genes (EGFR, EGR1, GNG11, DKK1, and MET) and five miRNAs (hsa-mir-141–5p, hsa-mir-200c-3p, EBV-mir-BART2–5p, EBV-mir-BART14–3p, EBV-mir-BART17–5p) from three subsequent passages were validated by real-time QPCR (Fig. 3B). EGFR, EGR1, and MET are direct targets of EBV-mir-BART14–3p, EBV-mir-BART17–5p, and EBV-mir-BART2–5p from our prediction algorithms validated by NGS expression, respectively. Three members (CIITA, IL18, and TNFRSF9) are in the immune-related biological network enriched in cytokine and interferon signaling (Fig. 4A). Their expression from subsequent passages was also validated by real-time QPCR (Fig. 4B).
Fig. 3.
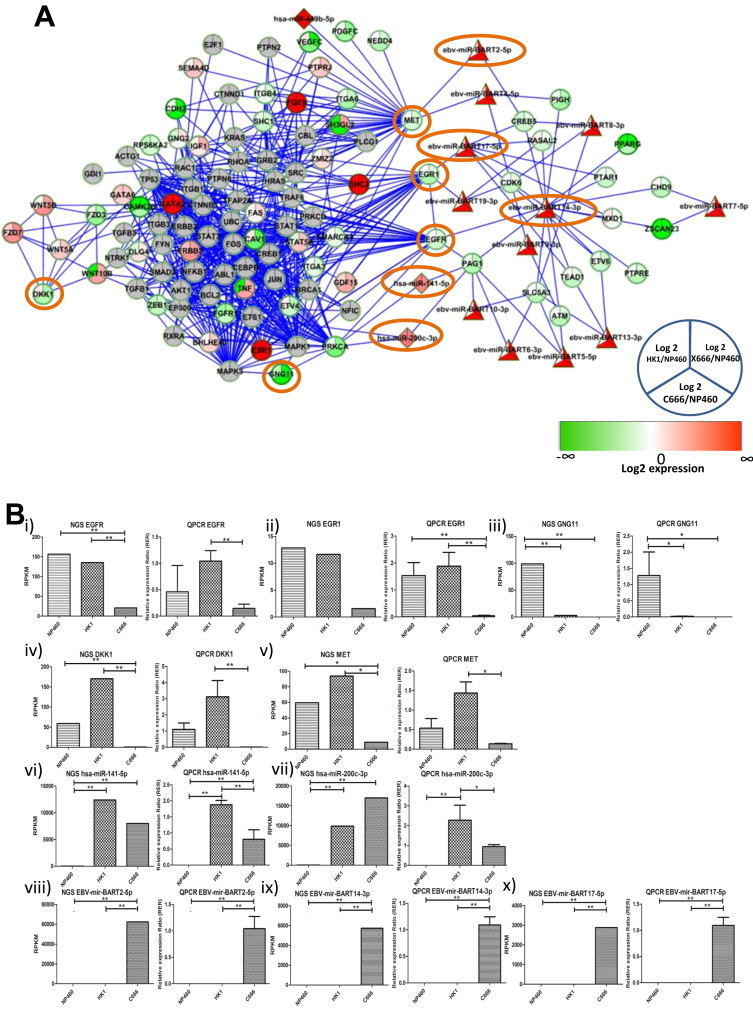
microRNA regulated biological network enriched in EGFR, ErbB and Wnt signaling pathways. (A) Network diagram showing Reactome biological network. The intensity of the node color indicates the degree of up-(red) or down (green)-regulation with respect to NP460. Round node: protein coding gene; diamond node: human encoded miRNA; triangle node: EBV encoded miRNA; orange circle: QPCR-validated genes from three subsequent passages. (B) NGS expression and QPCR expression on QPCR-validated genes. (i) EGFR: epidermal growth factor receptor; (ii) EGR1: early growth response protein 1; (iii) GNG11: guanine nucleotide binding protein (G Protein), Gamma 11; (iv) DKK1: Dickkopf-Like protein 1; (v) MET: Met Proto-Oncogene; (vi) hsa-miR-141-5p; (vii) has-miR-200c-3p; (viii) EBV-mir-BART2-5p; (ix) EBV-mir-BART14-3p; (x) EBV-mir-BART-17-5p. Error bar on QPCR plot: SD from three subsequent passages. *p < 0.05; **p < 0.01. p Value was estimated by DeSeq package in transcriptome data and by one-way ANOVA followed by Tukey post hoc analysis in QPCR, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
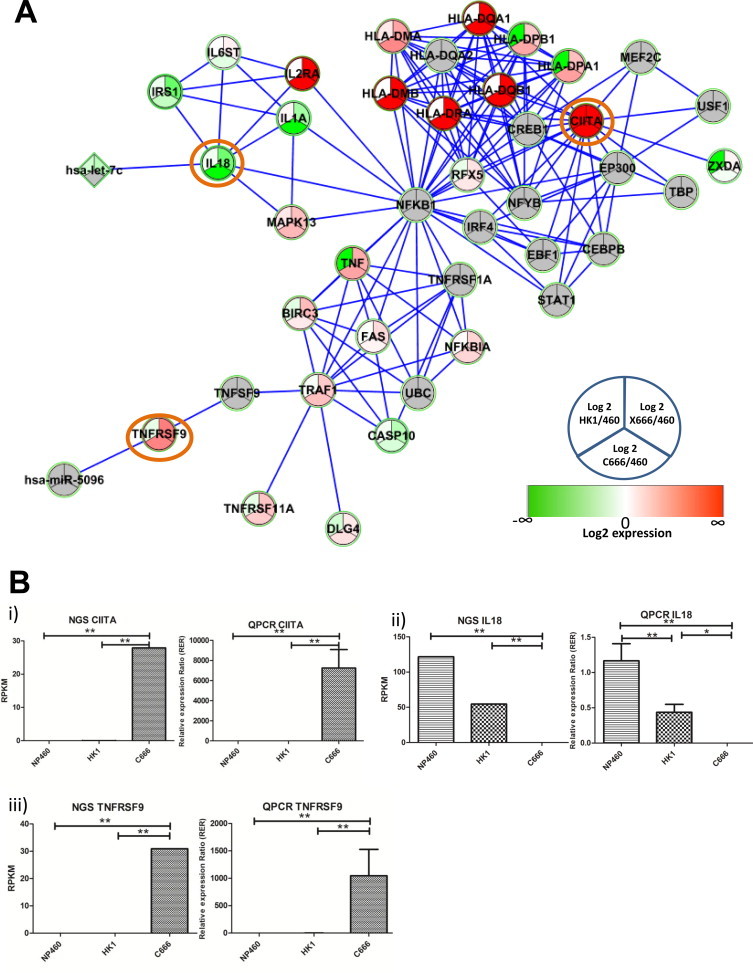
Biological network enriched in cytokine and interferon signaling. (A) Network diagram showing Reactome biological network relevant to the genes differentially expressed and regulated by miRNA in C666/X666. The intensity of the node color indicates the degree of up-(red) or down (green)-regulation with respect to NP460. Round node: protein coding gene; diamond node: human encoded miRNA; triangle node: EBV encoded miRNA; orange circle: QPCR-validated genes from three subsequent passages. (B) NGS expression and QPCR expression on QPCR-validated genes. (i) CIITA: MHC class II transactivator type III; ii) IL18: interleukin 18; iii) TNFRSF9: tumor necrosis factor receptor superfamily, member 9. Error bar on QPCR plot: SD from three subsequent passages. *p < 0.05; **p < 0.01. p Value was estimated by DeSeq package in transcriptome data and by one-way ANOVA followed by Tukey post hoc analysis in QPCR, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Sequence variants in NPC transcriptome
Sequence variants analysis detecting SNP and short INDELS was carried out using VarScan [41]. The 17,389 SNP/INDELS were detected from NP460, HK1, and C666 and the results are summarized in Fig. 5A and Supplement 5. Sequence variants, which are not solely from X666, are also listed in Supplement 5, while variants solely from X666 were removed due to possible contamination of mouse sequences. A total of 62% (10,929/17,389) of variants are from non-protein coding regions, while around 37% (6460/17,389) are from the exonic regions of the genome (Fig. 5A). Of the exonic variants, around one-third (2743/6460) are non-synonymous or frameshift or stopgain/loss, which affect the protein products of the gene. Moreover, 98 out of 2743 protein-affected variants are genes from the catalogue of somatic mutations in the cancer (COSMIC) panel [49]. Of note, we discovered a novel TP53 mutation at Chr17:7,579,335T>G in NP460, HK1, and C666 cell lines. Differences in polymorphism of GSTP1 (Chr11:67,352,689A>G) across NP460, HK1, and C666-1 were also found from the NGS data (Fig. 5B).
Fig. 5.
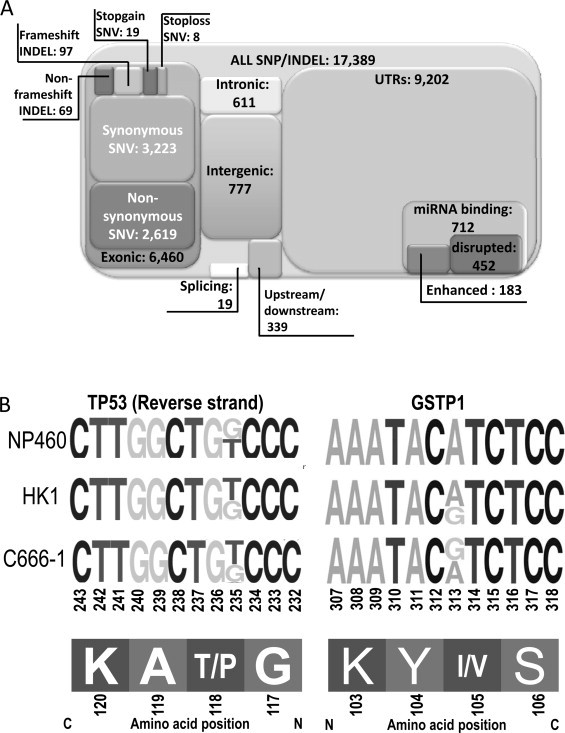
Sequence variants identified from transcriptome data. (A) An overview of sequence variants identified from transcriptome NGS. (B) Single nucleotide variants in TP53 and GSTP1 detected in NPC and NP cell lines. Height of sequence logos represents the frequency of nucleotides detected at each position.
Sequence variation in the UTR may affect the binding of its regulatory miRNA and influence the level of mRNA expression [50]. We sought to analyze the UTR variant that disrupts the miRNA binding and affects mRNA expression in cell lines. The 712 UTRs from the reference sequence were predicted to be miRNA binding sites. Of these miRNA binding sequences, 452 miRNA binding sites were disrupted by the variation and 184 target sites were predicted to be enhanced (Supplement 6). We further examined the disrupted/ enhanced miRNA-binding pairs by referring to the expression of their corresponding mRNAs and miRNAs. Compared to NP460, almost 100% of the genes with enhanced variants at the UTR in HK1 and C666 responded to the changes of the paired miRNAs, while only 41% and 50% of the genes with disrupted variants at the UTR in HK1 and C666 did so. Three examples of the genes with and without disrupted/enhanced variants are illustrated in Supplemental Fig. 2.
From the small library NGS data, we have analyzed human and EBV-encoded isomiRs based on miRBase and these are listed in Supplements 7, 8 and 9, respectively. Supplement 9 shows the selected top three EBV-encoded isomiRs from C666 and X666 with substantial expression of isomiRs discovered in RNASeq. The most abundant sequences in 18 C666 and 19 X666 EBV miRNAs are not the reference ones. Four miRNAs differed from their most abundant isomiRs in C666 and X666. No reference reads of BART-19–5p were detected in X666, while only one read detected in C666, mature reference sequence was out of top three of all detected sequences in mir-BART18-3p in both C666 and X666 (Supplement 9). Mature BHRF-1-1-5p was detected in both C666 and X666, while BHRF1-2-3p and BHFR1-2-5p were detected in X666 only (Table 1a). Four novel EBV-encoded miRNAs have been reported by Chen et al. [26] from clinical samples, which are not included in the miRBase and further reports are lacking. We have detected three (BART16-3p, BART22-5p, BART12-5p) in both C666 and X666 samples; the mature sequences as described by Chen et al. [26], are the most abundant in BART16-3p and BART22-5p (Table 1b).
Table 1a.
Top three isomirs of EBV-BHRF miRNA from mature miRbase annotated sequence.
| EBV-miRNA | Match typea | Read sequence | C666 readb,d | X666 readb,d | C666 rankc,d | X666 rankc,d |
|---|---|---|---|---|---|---|
| mir-BHRF1-1-5p | **Mature 5′ | TAACCTGATCAGCCCCGGAGTT | 3 | 8 | 1st | 1st |
| Mature 5′ sub | AACCTGATCAGCCCCGGAGTT | 2 | N/A | 2nd | N/A | |
| mir-BHRF1-2-3p | Mature 3′ sub | TATCTTTTGCGGCAGAAATTG | N/A | 2 | N/A | 1st |
| **Mature 3′ | TATCTTTTGCGGCAGAAATTGA | N/A | 1 | N/A | 2nd | |
| mir-BHRF1-2-5p | Mature 5′ sub | AAATTCTGTTGCAGCAGATAG | N/A | 1 | N/A | 1st |
Mature 3′/5′: **Mature miRbase annotated sequence; Mature 3′/5′ sub: observed tag is shorter than reference sequence; Mature 3′/5′ super: observed tag is longer than the annotated mature sequence; Mature 3′/5′ sub/super variant: observed tag with mismatches to annotated sequence.
Number of reads detected in NGS. N/A: no read detected in this sample. Total read counts of EBV miRNA of C666: 1761771; X666: 469388.
Rank in N/A: ranking of the read in that sample is out of top3. d No mature sequence nor isomirs of mir-BART15-5p found in C666-1 and X666 NGS read.
Table 1b.
Top three isomirs of novel EBV-BART miRNA from Chen et al. [26].
| EBV-miRNA | Match typea | Read sequence | C666 readb,d | X666 readb,d | C666 rankc,d | X666 rankc,d |
|---|---|---|---|---|---|---|
| mir-BART16-3p | ##Mature 3′ | ATCACCACCCTCTATCCATAT | 41 | 5 | 1st | N/A |
| Mature 3′ sub/super | AGATCACCACCCTCTATCCATA | 40 | 11 | 2nd | 1st | |
| Mature 3′ sub/super | AGATCACCACCCTCTATCCAT | 39 | 7 | 3rd | 3rd | |
| Mature 3′ super | AGATCACCACCCTCTATCCATAT | 9 | 8 | N/A | 2nd | |
| Mature 3′ super | GATCACCACCCTCTATCCATAT | 11 | 8 | N/A | 2nd | |
| mir-BART22-5p | ##Mature 5′ | TGCTAGACCCTGGAGTTGAACC | 121 | 23 | 1st | 1st |
| Mature 5′ sub | TGCTAGACCCTGGAGTTGAAC | 55 | 13 | 2nd | 2nd | |
| Mature 5′ sub | TGCTAGACCCTGGAGTTG | 3 | N/A | 3rd | N/A | |
| Mature 5′ super variant | TGCTAGACCCTGGAGTTGAACCt | 3 | 1 | 3rd | N/A | |
| Mature 5′ sub | TGCTAGACCCTGGAGTT | N/A | 3 | N/A | 3rd | |
| Mature 5′ variant | TGCTAGACCCTGGAGTTGAACa | 1 | 3 | N/A | 3rd | |
| mir-BART12-5p | Mature 5′ super | ACCCGCCCATCACCACCGGACA | 50 | 2 | 1st | 1st |
| Mature 5′ super variant | gACCCGCCCATCACCACCGGA | 10 | N/A | 2nd | N/A | |
| ##Mature 5′ | ACCCGCCCATCACCACCGGAC | 7 | 1 | 3rd | 2nd | |
| Mature 5′ sub | ACCCGCCCATCACCACCGGA | 7 | N/A | 3rd | N/A | |
| Mature 5′ super variant | ACCCGCCCATCACCACCGGACt | 5 | 1 | N/A | 2nd |
Mature 3′/5′: ## Mature sequence from Chen et al. [26]; Mature 3′/5′ sub: observed tag is shorter than reference sequence; Mature 3′/5′ super: observed tag is longer than the annotated mature sequence; Mature 3′/5′ sub/super variant: observed tag with mismatches to annonated sequence.
Number of reads detected in NGS. N/A: no read detected in this sample. Total read counts of EBV miRNA of C666: 1761771; X666: 469388.
Rank in N/A: ranking of the read in that sample is out of top3. dNo mature sequence nor isomirs of mir-BART15–5p found in C666–1 and X666 NGS read.
3.5. Integrated analysis of transcriptome from model system and clinical specimen
In order to evaluate the importance of different NPC model systems for translational application, integrated analysis to compare transcriptomes from cell line systems and clinical specimens was carried out. Meta-analysis of the mRNA gene expression used MetaDE algorithms [45] to compare our transcriptome expression against the microarray datasets from biopsies available from GEO. We combined the dataset from GSE12452 [18] and GSE13597 [51], according to their stages (Fig. 6A). The dataset from GSE34573-GPL570 [52] has no stage information available and we combined the dataset with the transcriptome according to normal and cancer groups (Fig. 6B). C666 and X666, which were alike in origin, are on the same cluster as their transcript expression patterns are similar. The sample from the C666 microarray GSE34573 (C666.1) and NGS in this study are clustered close together, validating both studies (Fig. 6B). We integrated the transcript variants to the DNA SNP array data (GPL3718/GPL3720) from clinical specimens in GEO [52]. Five SNPs from the coding region and 7 from the miRNA-binding UTR region were found to have SNP information from these public datasets (Table 2). C666 RNA and DNA have been used in NGS and DNA SNP array as control samples, respectively. Four SNPs differ in genotype between C666 RNA NGS and DNA SNP array, while five are in common.
Fig. 6.
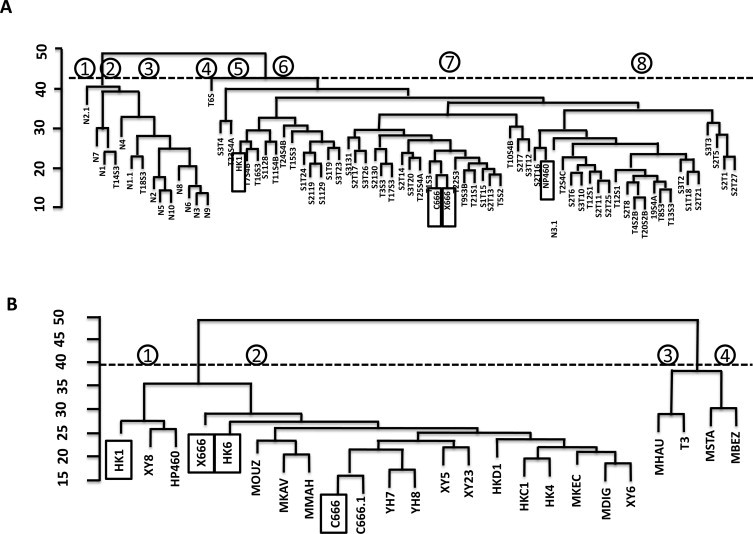
Meta-analysis of NGS data with clinical data from public database. (A) Meta-analysis on GSE12452, GSE13597 and NGS data; (B)meta-analysis on GSE34573 and NGS data; Dotted line: threshold for cluster analysis; circled number: cluster number according to threshold. Both dendrograms in (A) and (B) were generated by MetaDE using complete linkage method. (C) Common SNPs detected in NGS and GPL3718/ GPL3720 SNP array. ND: SNP not detected.
Table 2.
| Transcriptome NGS result |
GPL3718/ GPL3720 SNP array result |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VarFreq: ratio B/A |
C666 | NPC biopsies (15) |
Control blood (5) |
|||||||||
| Gene | Function | dbSNP135 | NP460 VarFreq | HK1 VarFreq | C666 VarFreq | AA | AB | BB | AA | AB | BB | |
| CCDC99 | Exonic | rs3797713 | 1 | 0.6034 | 1 | AA | 7 | 6 | 0 | 2 | 3 | 0 |
| CEP55 | Exonic | rs2293277 | 0.4123 | 1 | 0.4487 | AB | 7 | 7 | 1 | 2 | 3 | 0 |
| PIGC | Exonic | rs1063412 | 1 | 0.5946 | 1 | AB | 0 | 13 | 1 | 0 | 2 | 1 |
| GNL3 | Exonic | rs2289247 | 0.4375 | 1 | 1 | AB | 11 | 4 | 0 | 2 | 2 | 0 |
| PPA2 | Exonic | rs13787 | 0.4058 | 1 | 0.3667 | AB | 6 | 4 | 1 | 0 | 1 | 3 |
| PDLIM5 | UTR3 | rs13141591 | 1 | 1 | ND | BB | 0 | 0 | 17 | 0 | 0 | 5 |
| SQSTM1 | UTR3 | rs1065154 | ND | ND | 0.6486 | AB | 3 | 7 | 3 | 3 | 0 | 1 |
| DUSP7 | UTR3 | rs3821841 | 0.4731 | 1 | ND | AB | 4 | 7 | 0 | 5 | 0 | 0 |
| DLX4 | UTR3 | rs1058564 | ND | ND | 0.6538 | AB | 10 | 4 | 0 | 3 | 2 | 0 |
| RAB31 | UTR3 | rs557706 | 1 | 0.4528 | ND | ND | 2 | 8 | 1 | 1 | 0 | 4 |
| CERS2 | UTR3 | rs267739 | 1 | 1 | 1 | AB | 13 | 1 | 0 | 5 | 0 | 0 |
| CANT1 | UTR5 | rs3744171 | ND | ND | 0.5 | AB | 6 | 5 | 1 | 4 | 1 | 0 |
ND: SNP not detected.
4. Discussion
This study provides a global overview of the NPC transcriptome, explores different variants, and reveals the miRNA regulation of transcript expression by integrated analysis of mRNA and miRNA. We sequenced the mRNAs and small RNAs from the same early passage of immortalized NP cell line (NP460), two NPC cell lines (HK1 and C666), and the NPC xenograft (X666). In consideration of genomic and epigenetic changes arising from longterm in vitro cultivation [53], we chose the earliest passage available for sequencing in order to minimize variation introduced from passaging. HK1 and C666 are two NPC cell lines with a great difference in terms of phenotype and genetic context; the results of this study provide further comparative insight in molecular pathways deregulated in NPC.
In this study we used RNASeq as a platform of analysis. It has a better sensitivity and dynamic range over traditional microarrays and enables identification of novel as well as more detailed studies of biological pathways in NPC. From the biological network analysis, biological pathways such as extracellular matrix (ECM) organization, PI3K/AKT signaling, and EGFR signaling are commonly enriched in HK1 and C666 NPC cell line over the immortalized nasopharyngeal epithelial cell line NP460 (Fig. 2B, Supplemental Fig. 1). A number of studies reported dysregulation of ECM pathways in NPC. Members of ECM pathway such as MMP19, FBLN2, and LTBP-2 had been studied as tumor suppressors in NPC [26,54–56], while down-regulation of hsa-miR-29c was associated with up-regulation of several ECM proteins such as collagens and fibronectins in NPC tumor tissues [57]. The PI3K/AKT pathways are important in NPC development and are reactivated by several factors such as PTEN, EGFR, and LMP1 [58,59]. The commonly enriched pathways are important in both NPC cell lines as well as NPC development. However, we observed differences in enrichment and expression levels between HK1 and C666 cell lines in the same pathway, such as the difference in the number of genes enriched and expressed in the EGFR pathway (Supplement 4). This may account for the different phenotypes observed between different NPC cell types.
From the miRNA and human UTR sequences, a stringent prediction of miRNA targets has been made using three different target prediction programs. The target pairs were further validated by comparing the expression value of miRNA and target pairs, which can serve as references for further studies on miRNA and its target (Supplement 3). We have further validated the expression level of miRNA-regulated biological networks enriched in EGFR, Erb, and Wnt signaling (Fig. 3). Both hsa-mir-141–5p and hsa-mir-200c-3p are up-regulated in all NPC cell lines and belong to the miR-200 family, which controls the epithelial-to-mesenchymal transition (EMT) process and regulates EGFR activity in bladder cancer [60]. Hsa-mir-141 is up-regulated in NPC biopsies and targets UBAP1, BRD3, and PTEN in the 5-8F NPC cell line [20]. C666/X666 are the only samples carrying EBV in this study and encoding EBV-BART miRNA as described in previous studies [26,61]. The MET receptor tyrosine kinase and FAT1 protocadherin related to the Wnt signaling pathway [62,63] are the predicted targets of ebv-mir-BART2-5p and ebv-mir-BART11-5p, respectively. EGR1, a transcription factor predicted as a target of ebv-mir-BART17-5p, regulates transcription of multiple genes such as EGFR, IL1A, and the ErbB ligand AREG [64,65]. Lowered expression of EGFR was also observed in C666 compared with NP460 and HK1 cell lines, consistent with the previous finding on the EGFR inhibitor trial in vitro [66]. Dickkopf-Like protein 1 (DKK1) is a secreted protein that regulates Wnt signaling, which binds to the frizzled receptor and LRP5/6, blocks interaction with Wnt1 and degrades beta-catenin [67]. DKK1 is over-expressed in a number of cancers such as lung, esophageal, and hepatoblastoma [68], while this over-expression declines, when prostate cancer develops from primary tumor to metastasis [69]. The over-expression of DKK1 in well-differentiated HK1 and down-regulation in the undifferentiated C666 may imply a similar model for DKK1 during NPC progression.
The MHC class II transactivator (CIITA) is regarded as the “master control factor” of MHC class II (MHCII) genes including HLA class II genes such as HLA-DQ, -DR and -DP molecules [70]. MHCII molecules are an important class of molecules that control and regulate adaptive immune responses, which are expressed at high levels in the majority of NPC tumors, which are EBV positive [71]. No transcript of CIITA has been detected in NP460, whilst there is a significant, but low expression in HK1, and its highest expression is in C666/X666 (Fig. 4B). This suggested that CIITA expression has been initiated, even at a low level, in the differentiated NPC cell stage. Detection of CIITA in EBV-positive clinical samples [71] also suggests that CIITA expression is related to EBV infection. TNFRSF9/CD137 (Tumor necrosis factor receptor superfamily, member 9) is a receptor protein that is essential for M (microfold) cell maturation in nasopharyngeal associated lymphoid cells [72]. M cells are specialized for transepithelial transport of foreign antigens and microorganisms to lymphoid tissue, and most of the studies are limited to the intestinal cells only [73]. The up-regulation of TNFRSF9 in C666 may relate to the immune response against foreign antigens such as EBV. IL18 is a pro-inflammatory cytokine that is a major inducer of IFN-gamma, which may serve as a protective cytokine against cancer; however, blocking of IL18 in the melanoma model suppresses tumor burden and metastasis [74]. From our RNASeq and QPCR results, lower transcript expression of IL18 in HK1 and C666 compared to NP460 has been observed, However, IL18 protein has been reported to be expressed in NPC biopsies, but not normal NP biopsies and mRNA expression has been detected in HK1 [75]. In the previous literature, only mRNA expression of HK1 and CNE2 has been reported and no comparison is reported for HK1 against the NP cell line NP460 as well as C666. The lower mRNA expression of IL18 in NPC cells, especially in C666, may suggest the pro-inflammatory function of IL18 lowering immunological defense against cancer. The mechanism for IL18 being down-regulated in HK1 and C666, but up-regulated in NPC biopsies remains unclear and requires further investigation. Epigenetic change is a common mechanism in gene regulation. The altered epigenetic signature at this gene in NPC cell lines and biopsies might be one explanation for the discrepancy. Nevertheless, caution must be taken to explain the disparity observed for IL18 in different NPC model systems.
Apart from expression analysis, we have analyzed the sequence variants and their relationship to expression. Besides proteins encoding SNV/INDEL, a large portion of sequence variants are located in UTRs of the mRNA transcript whose function still remains unknown (Fig. 5A). The analysis of SNV, short INDEL, and UTR sequence may provide a possible opportunity for further mechanistic studies (Supplements 5 and 6). From the TP53 mutant database [76], we discovered a novel TP53 mutation at the HCDII binding domain. From the RNASeq data of NP460, HK1, and C666 cell lines (Fig. 5B), this T-to-G variation occurs in the range of 30% to 50% of the total transcripts (Supplement 5). The mutation causes an amino acid change (Thr118>Pro) at the DNA binding domain, reducing transactivation activity in various promoters (14-3-3-σ, AIP, BAX, GADD45, MDM2, NOXA, p53R2, WAF1) from 50% to 90% in a TP53 yeast mutant library [77,78]. TP53 is a tumor suppressor protein that highly expressed in NPC [79]; several TP53 polymorphisms have been reported in NPC. However, the results are controversial and the function of TP53 in NPC is still inconclusive [80]. This novel TP53 variant may suggest a new mechanism for the TP53 pathway in the nasopharyngeal cell and NPC development. We also identified a differential variation of GSTP1 SNV in NP and NPC cell lines. All of the transcripts from RNASeq were in T-to-C variant (rs1695/rs947894) form in C666, whilst only wild-type GSTP1 was found in NP460, and around 65% of the variants are found in HK1 (Fig. 5B). This polymorphism is reported to lower enzyme activity of GSTP1 [81] and has been reported in NPC and the Han Chinese population [82]. Since GSTP1 is an enzyme detoxification enzyme, as well as the regulator of MAP kinase pathway in cancer [83], caution must be taken when using NP and/or NPC cell line models with different degrees of GSTP1 functional polymorphism.
IsomiRs are heterogeneous variants of miRNAs in length and sequences, which may affect target selection, miRNA stability, and translational machinery. Recently, some isomiRs are reported to have biological significance in plants and animals [28]. We have listed the isomiRs from the NGS data for both human and EBV-encoded miRNAs for future reference (Supplements 8 and 9). Furthermore, we analyzed the EBV-encoded isomiRs from the C666 and X666 samples, to compare against a previous report of the EBV miRNAome from clinical tissues [26]. Most isomiRs are length variants; some of the isomiRs are expressed in even higher levels than the reference mature sequence (Supplement 9). IsomiRs of EBV-miR-BART19-5p have been found highly expressed (top three) in both C666 and X666 samples, consistent with the genomic variant discovered in C666 from a previous report [26]. Four novel EBV-encoded miRNAs have been identified from clinical samples in Ref. [26]. However, they are not in the miRBase (release 19) collection and further reports of these miRNAs were limited. Here we have identified expression of three out of four novel miRNAs and their isomiRs (EBV-miR-BART16-3p, EBV-miR-BART22-5p, EBV-miR-BART12-5p) in both C666/X666 samples (Table 1b). We have detected EBV-miR-BART12-5p isomiRs as the most abundant reads in C666/X666, which is expressed in undetectable levels in previous reports, respectively. The function of these isomiRs in NPC development remains elusive. It has been reported that the isomiRs might functionally cooperate with canonical miRNAs to target pathways of functionally related genes [84]. Only precursors, but no mature reads of EBV-miR-BART22-5p, have been detected from the RNASeq results (data not shown), whilst detection of this miRNA has been described in C666 in previous reports [23,26]. We also detected EBV-encoded BHRF mature miRNA in a low number of reads. In contrast to a previous report on detection of BHRF1-1-5p,1-2-3p and BHRF-1-3 in C666 [85], mature BHRF-1-1-5p was detected in both C666 and X666, while BHRF1-2-3p and BHFR1-2-5p were detected in X666 only (Table 1a), and only the precursor of BHRF1-3 has been detected (data not shown).
To explore the translational relevance of the transcriptome in model systems, we have integrated our NGS data from model systems to the expression/SNP array data from clinical specimens. Fig. 6a and b shows the clustering result of meta-analysis of NGS data to the public data according to the stage information and cancer/non-cancer information, respectively. The NPC cell lines were clustered with different groups of clinical samples, suggesting that the two NPC cell lines can be used as models of different types of NPC. C666 and X666 were in the same cluster as they are from the same cell origin. To our surprise, the expression pattern of NP460 does not cluster with the normal control samples for all three microarray studies from biopsies. Furthermore, we have analyzed the known SNV in the SNP database with the DNA SNP array result (Table 2). Due to the few SNPs available from array data in the literature, the number of SNPs consistent with the NGS data is limited; C666 has not been analyzed specifically as a control. Four SNPs have a different genotype between C666 RNASeq and DNA SNP array, while five are common. The difference may imply technical difference and/or post-transcriptional editing in RNA. Post-transcriptional editing of RNA in cancer has been demonstrated in hepatocellular carcinoma (HCC) [86], but examples in NPC have yet to be discovered. rs557706 (3′UTR of RAB31) has the same genotype (BB) in NP460 and 4/5 of the controls, and a similar genotype (AB) in HK1 8/15 of the NPC biopsies, suggesting that NP460 and HK1 can be used as the model system for mechanistic studies in this SNP. However, the numbers of publicly available high-throughput data sets are limited in NPC and are conducted in a different platform without proper internal control such as using C666 as a calibrating sample, increasing difficulties in analyzing data across studies. More high-throughput studies on clinical samples with detailed information and proper controls would be needed in the future.
Here we report a comprehensive characterization of mRNA and miRNA transcriptome in NPC cell lines and xenograft from expression level and sequence variation, miRNA/mRNA regulation on biological network, integrated analysis on expression-to-sequence characteristics and model system-to-clinical specimen. This will provide a valuable reference for future studies in transcript variation and regulation of NPC in in vitro model cell lines and xenograft and will potentially be useful for mechanistic and preclinical studies.
Acknowledgements
We thank Dr. Wei Dai for her detailed editorial and statistical review for preparation of this manuscript. This work was supported by Research Grants Council, HKSAR Area of Excellence grant AoE/M-06/08 to M.L.L.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2014.01.004. Data has been uploaded to the GEO database with accession number GSE54174.
Appendix. Supplementary materials
Supplementary Table 1 Primer list for SYBR green real time QPCR.
Supplementary Table 2 Summary of reads and expression.
Supplementary Table 3 miRegbase1.1: Database of miRNA target prediction.
Supplementary Table 4 Summary of Reactome and Gene Oncology terms.
Supplementary Table 5 Summary of SNPs and INDELs discovered from transcriptome data.
Supplementary Table 6 Summary of mutations in miRNA binding UTRs.
Supplementary Table 7 miRNA reads from CLC alignment.
Supplementary Table 8 Summary of human miRNA isomirs.
Supplementary Table 9 Summary of EBV miRNA isomirs.
Supplementary Fig. 1. Biological networks enriched in NPC model systems.
Supplementary Fig. 2. Integrated analysis on the selected candidates with sequence variation in miRNA binding UTRs.
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Boyle, P. and Levin, B. (2008) World cancer report 2008.
- 3.Marks J.E., Phillips J.L., Menck H.R. The National Cancer Data Base report on the relationship of race and national origin to the histology of nasopharyngeal carcinoma. Cancer. 1998;83:582–588. doi: 10.1002/(sici)1097-0142(19980801)83:3<582::aid-cncr29>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 4.Smerq J., Sharma M. The risk factor: nasopharyngeal carcinoma. Int. J. Pharm. Clin. Res. 2011;3:48–51. [Google Scholar]
- 5.Spano J.P., Busson P., Atlan D., Bourhis J., Pignon J.P., Esteban C., Armand J.P. Nasopharyngeal carcinomas: an update. Eur. J. Cancer. 2003;39:2121–2135. doi: 10.1016/s0959-8049(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 6.Cheung S.T., Huang D.P., Hui A.B., Lo K.W., Ko C.W., Tsang Y.S., Wong N., Whitney B.M., Lee J.C. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer. 1999;83:121–126. doi: 10.1002/(sici)1097-0215(19990924)83:1<121::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 7.Glaser R., Zhang H.Y., Yao K.T., Zhu H.C., Wang F.X., Li G.Y., Wen D.S., Li Y.P. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl. Acad. Sci. USA. 1989;86:9524–9528. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang D.P., Ho J.H., Poon Y.F., Chew E.C., Saw D., Lui M., Li C.L., Mak L.S., Lai S.H., Lau W.H. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. Int. J. Cancer. 1980;26:127–132. doi: 10.1002/ijc.2910260202. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S., Wu Y., Zeng Y., Zech L., Klein G. Cytogenetic studies on an epithelioid cell line derived from nasopharyngeal carcinoma. Hereditas. 1982;97:23–28. doi: 10.1111/j.1601-5223.1982.tb00706.x. [DOI] [PubMed] [Google Scholar]
- 10.Li H.M., Man C., Jin Y., Deng W., Yip Y.L., Feng H.C., Cheung Y.C., Lo K.W., Meltzer P.S., Wu Z.G., Kwong Y.L., Yuen A.P., Tsao S.W. Molecular and cytogenetic changes involved in the immortalization of nasopharyngeal epithelial cells by telomerase. Int. J. Cancer. 2006;119:1567–1576. doi: 10.1002/ijc.22032. [DOI] [PubMed] [Google Scholar]
- 11.Busson P., Ganem G., Flores P., Mugneret F., Clausse B., Caillou B., Braham K., Wakasugi H., Lipinski M., Tursz T. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int. J. Cancer. 1988;42:599–606. doi: 10.1002/ijc.2910420422. [DOI] [PubMed] [Google Scholar]
- 12.Huang D.P., Ho J.H., Chan W.K., Lau W.H., Lui M. Cytogenetics of undifferentiated nasopharyngeal carcinoma xenografts from southern Chin. Int. J. Cancer. 1989;43:936–939. doi: 10.1002/ijc.2910430535. [DOI] [PubMed] [Google Scholar]
- 13.Huang D.P., Ng H.K., Ho Y.H., Chan K.M. Epstein-Barr virus (EBV)-associated undifferentiated carcinoma of the parotid gland. Histopathology. 1988;13:509–517. doi: 10.1111/j.1365-2559.1988.tb02075.x. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez S., Khabir A., Keryer C., Perrot C., Drira M., Ghorbel A., Jlidi R., Bernheim A., Valent A., Busson P. Conventional and array-based comparative genomic hybridization analysis of nasopharyngeal carcinomas from the Mediterranean area. Cancer Genet. Cytogenet. 2005;157:140–147. doi: 10.1016/j.cancergencyto.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Lv X.B., Wang X.P., Sang Y., Xu S., Hu K., Wu M., Liang Y., Liu P., Tang J., Lu W.H., Feng Q.S., Chen L.Z., Qian C.N., Bei J.X., Kang T., Zeng Y.X. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- 18.Sengupta S., den Boon J.A., Chen I.H., Newton M.A., Dahl D.B., Chen M., Cheng Y.J., Westra W.H., Chen C.J., Hildesheim A., Sugden B., Ahlquist P. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 19.Xia H., Ng S.S., Jiang S., Cheung W.K., Sze J., Bian X.W., Kung H.F., Lin M.C. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem. Biophys. Res. Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Deng T., Li X., Liu H., Zhou H., Ma J., Wu M., Zhou M., Shen S., Niu Z., Zhang W., Shi L., Xiang B., Lu J., Wang L., Li D., Tang H., Li G. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis. 2010;31:559–566. doi: 10.1093/carcin/bgp335. [DOI] [PubMed] [Google Scholar]
- 21.Arvey A., Tempera I., Tsai K., Chen H.S., Tikhmyanova N., Klichinsky M., Leslie C., Lieberman P.M. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vereide D.T., Seto E., Chiu Y.F., Hayes M., Tagawa T., Grundhoff A., Hammerschmidt W., Sugden B. Epstein-Barr virus maintains lymphomas via its miRNAs. Oncogene. 2013 doi: 10.1038/onc.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lung R.W., Tong J.H., Sung Y.M., Leung P.S., Ng D.C., Chau S.L., Chan A.W., Ng E.K., Lo K.W., To K.F. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–1184. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei T., Yuen K.S., Xu R., Tsao S.W., Chen H., Li M., Kok K.H., Jin D.Y. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int. J. Cancer. 2012 doi: 10.1002/ijc.28007. [DOI] [PubMed] [Google Scholar]
- 25.Chen H.C., Chen G.H., Chen Y.H., Liao W.L., Liu C.Y., Chang K.P., Chang Y.S., Chen S.J. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br. J. Cancer. 2009;100:1002–1011. doi: 10.1038/sj.bjc.6604948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S.J., Chen G.H., Chen Y.H., Liu C.Y., Chang K.P., Chang Y.S., Chen H.C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong A.M., Kong K.L., Tsang J.W., Kwong D.L., Guan X.Y. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 28.Neilsen C.T., Goodall G.J., Bracken C.P. IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012;28:544–549. doi: 10.1016/j.tig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Lo Y.M., Chan L.Y., Lo K.W., Leung S.F., Zhang J., Chan A.T., Lee J.C., Hjelm N.M., Johnson P.J., Huang D.P. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 30.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozomara A., Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 38.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croft D., O’Kelly G., Wu G., Haw R., Gillespie M., Matthews L., Caudy M., Garapati P., Gopinath G., Jassal B., Jupe S., Kalatskaya I., Mahajan S., May B., Ndegwa N., Schmidt E., Shamovsky V., Yung C., Birney E., Hermjakob H., D’Eustachio P., Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A.R., Vailaya A., Wang P.L., Adler A., Conklin B.R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G.J., Ideker T., Bader G.D. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Kang D.D., Shen K., Song C., Lu S., Chang L.C., Liao S.G., Huo Z., Tang S., Ding Y., Kaminski N., Sibille E., Lin Y., Li J., Tseng G.C. An R package suite for microarray meta-analysis in quality control, differentially expressed gene analysis and pathway enrichment detection. Bioinformatics. 2012;28:2534–2536. doi: 10.1093/bioinformatics/bts485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., Leunissen J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winer J., Jung C.K., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 48.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J.W., Campbell P.J., Stratton M.R., Futreal P.A. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen K., Song F., Calin G.A., Wei Q., Hao X., Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 51.Bose S., Yap L.F., Fung M., Starzcynski J., Saleh A., Morgan S., Dawson C., Chukwuma M.B., Maina E., Buettner M., Wei W., Arrand J., Lim P.V., Young L.S., Teo S.H., Stankovic T., Woodman C.B., Murray P.G. The ATM tumour suppressor gene is down-regulated in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2009;217:345–352. doi: 10.1002/path.2487. [DOI] [PubMed] [Google Scholar]
- 52.Hu C., Wei W., Chen X., Woodman C.B., Yao Y., Nicholls J.M., Joab I., Sihota S.K., Shao J.Y., Derkaoui K.D., Amari A., Maloney S.L., Bell A.I., Murray P.G., Dawson C.W., Young L.S., Arrand J.R. A global view of the oncogenic landscape in nasopharyngeal carcinoma: an integrated analysis at the genetic and expression levels. PLoS One. 2012;7:e41055. doi: 10.1371/journal.pone.0041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Chan K.C., Ko J.M., Lung H.L., Sedlacek R., Zhang Z.F., Luo D.Z., Feng Z.B., Chen S., Chen H., Chan K.W., Tsao S.W., Chua D.T., Zabarovsky E.R., Stanbridge E.J., Lung M.L. Catalytic activity of Matrix metalloproteinase-19 is essential for tumor suppressor and anti-angiogenic activities in nasopharyngeal carcinoma. Int. J. Cancer. 2011;129:1826–1837. doi: 10.1002/ijc.25855. [DOI] [PubMed] [Google Scholar]
- 55.Chen H., Ko J.M., Wong V.C., Hyytiainen M., Keski-Oja J., Chua D., Nicholls J.M., Cheung F.M., Lee A.W., Kwong D.L., Chiu P.M., Zabarovsky E.R., Tsao S.W., Tao Q., Kan R., Chan S.H., Stanbridge E.J., Lung M.L. LTBP-2 confers pleiotropic suppression and promotes dormancy in a growth factor permissive microenvironment in nasopharyngeal carcinoma. Cancer Lett. 2012;325:89–98. doi: 10.1016/j.canlet.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 56.Law E.W., Cheung A.K., Kashuba V.I., Pavlova T.V., Zabarovsky E.R., Lung H.L., Cheng Y., Chua D., Lai-Wan Kwong D., Tsao S.W., Sasaki T., Stanbridge E.J., Lung M.L. Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma. Oncogene. 2012;31:728–738. doi: 10.1038/onc.2011.272. [DOI] [PubMed] [Google Scholar]
- 57.Sengupta S., den Boon J.A., Chen I.H., Newton M.A., Stanhope S.A., Cheng Y.J., Chen C.J., Hildesheim A., Sugden B., Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl. Acad. Sci. USA. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedrero J.M., Carracedo D.G., Pinto C.M., Zapatero A.H., Rodrigo J.P., Nieto C.S., Gonzalez M.V. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int. J. Cancer. 2005;114:242–248. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 59.Yip W.K., Leong V.C., Abdullah M.A., Yusoff S., Seow H.F. Overexpression of phospho-Akt correlates with phosphorylation of EGF receptor, FKHR and BAD in nasopharyngeal carcinoma. Oncol. Rep. 2008;19:319–328. [PubMed] [Google Scholar]
- 60.Adam L., Zhong M., Choi W., Qi W., Nicoloso M., Arora A., Calin G., Wang H., Siefker-Radtke A., McConkey D., Bar-Eli M., Dinney C. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lo A.K., Dawson C.W., Jin D.Y., Lo K.W. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J. Pathol. 2012;227:392–403. doi: 10.1002/path.4025. [DOI] [PubMed] [Google Scholar]
- 62.Kim K.H., Seol H.J., Kim E.H., Rheey J., Jin H.J., Lee Y., Joo K.M., Lee J., Nam D.H. Wnt/beta-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol. 2013;15:161–171. doi: 10.1093/neuonc/nos299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris L.G., Kaufman A.M., Gong Y., Ramaswami D., Walsh L.A., Turcan S., Eng S., Kannan K., Zou Y., Peng L., Banuchi V.E., Paty P., Zeng Z., Vakiani E., Solit D., Singh B., Ganly I., Liau L., Cloughesy T.C., Mischel P.S., Mellinghoff I.K., Chan T.A. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat. Genet. 2013 doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olayioye M.A., Neve R.M., Lane H.A., Hynes N.E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao F., Xuan Z., Liu L., Zhang M.Q. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Res. 2005;33:D103–107. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sung F.L., Poon T.C., Hui E.P., Ma B.B., Liong E., To K.F., Huang D.P., Chan A.T. Antitumor effect and enhancement of cytotoxic drug activity by cetuximab in nasopharyngeal carcinoma cells. In Vivo. 2005;19:237–245. [PubMed] [Google Scholar]
- 67.Forget M.A., Turcotte S., Beauseigle D., Godin-Ethier J., Pelletier S., Martin J., Tanguay S., Lapointe R. The Wnt pathway regulator DKK1 is preferentially expressed in hormone-resistant breast tumours and in some common cancer types. Br. J. Cancer. 2007;96:646–653. doi: 10.1038/sj.bjc.6603579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamabuki T., Takano A., Hayama S., Ishikawa N., Kato T., Miyamoto M., Ito T., Ito H., Miyagi Y., Nakayama H., Fujita M., Hosokawa M., Tsuchiya E., Kohno N., Kondo S., Nakamura Y., Daigo Y. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas. Cancer Res. 2007;67:2517–2525. doi: 10.1158/0008-5472.CAN-06-3369. [DOI] [PubMed] [Google Scholar]
- 69.Hall C.L., Daignault S.D., Shah R.B., Pienta K.J., Keller E.T. Dickkopf-1 expression increases early in prostate cancer development and decreases during progression from primary tumor to metastasis. Prostate. 2008;68:1396–1404. doi: 10.1002/pros.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.LeibundGut-Landmann S., Waldburger J.M., Krawczyk M., Otten L.A., Suter T., Fontana A., Acha-Orbea H., Reith W. Mini-review: specificity and expression of CIITA, the master regulator of MHC class II genes. Eur. J. Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 71.Yao Y., Minter H.A., Chen X., Reynolds G.M., Bromley M., Arrand J.R. Heterogeneity of HLA and EBER expression in Epstein-Barr virus-associated nasopharyngeal carcinoma. Int. J. Cancer. 2000;88:949–955. doi: 10.1002/1097-0215(20001215)88:6<949::aid-ijc18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 72.Hsieh E.H., Fernandez X., Wang J., Hamer M., Calvillo S., Croft M., Kwon B.S., Lo D.D. CD137 is required for M cell functional maturation but not lineage commitment. Am. J. Pathol. 2010;177:666–676. doi: 10.2353/ajpath.2010.090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kraehenbuhl J.P., Neutra M.R. Epithelial M cells: differentiation and function. Annu. Rev. Cell Dev. Biol. 2000;16:301–332. doi: 10.1146/annurev.cellbio.16.1.301. [DOI] [PubMed] [Google Scholar]
- 74.Dinarello C.A. The paradox of pro-inflammatory cytokines in cancer. Cancer Metastasis Rev. 2006;25:307–313. doi: 10.1007/s10555-006-9000-8. [DOI] [PubMed] [Google Scholar]
- 75.Hu H., Tang K.F., Chua Y.N., Lu J., Feng P., Chew C.T., Chan S.H. Expression of interleukin-18 by nasopharyngeal carcinoma cells: a factor that possibly initiates the massive leukocyte infiltration. Hum. Pathol. 2004;35:722–728. doi: 10.1016/j.humpath.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 76.Leroy B., Fournier J.L., Ishioka C., Monti P., Inga A., Fronza G., Soussi T. The TP53 website: an integrative resource centre for the TP53 mutation database and TP53 mutant analysis. Nucleic Acids Res. 2013;41:D962–969. doi: 10.1093/nar/gks1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kato S., Han S.Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shiraishi K., Kato S., Han S.Y., Liu W., Otsuka K., Sakayori M., Ishida T., Takeda M., Kanamaru R., Ohuchi N., Ishioka C. Isolation of temperature-sensitive p53 mutations from a comprehensive missense mutation library. J. Biol. Chem. 2004;279:348–355. doi: 10.1074/jbc.M310815200. [DOI] [PubMed] [Google Scholar]
- 79.Sheu L.F., Chen A., Tseng H.H., Leu F.J., Lin J.K., Ho K.C., Meng C.L. Assessment of p53 expression in nasopharyngeal carcinoma. Hum. Pathol. 1995;26:380–386. doi: 10.1016/0046-8177(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 80.Zhuo X.L., Cai L., Xiang Z.L., Zhuo W.L., Wang Y., Zhang X.Y. TP53 codon 72 polymorphism contributes to nasopharyngeal cancer susceptibility: a meta-analysis. Arch. Med. Res. 2009;40:299–305. doi: 10.1016/j.arcmed.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 81.Watson M.A., Stewart R.K., Smith G.B., Massey T.E., Bell D.A. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. doi: 10.1093/carcin/19.2.275. [DOI] [PubMed] [Google Scholar]
- 82.Guo X., Zeng Y., Deng H., Liao J., Zheng Y., Li J., Kessing B., O’Brien S.J. Genetic polymorphisms of CYP2E1, GSTP1, NQO1 and MPO and the risk of nasopharyngeal carcinoma in a Han Chinese population of Southern China. BMC Res. Notes. 2010;3:212. doi: 10.1186/1756-0500-3-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Townsend D.M., Tew K.D. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene. 2003;22:7369–7375. doi: 10.1038/sj.onc.1206940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cloonan N., Wani S., Xu Q., Gu J., Lea K., Heater S., Barbacioru C., Steptoe A.L., Martin H.C., Nourbakhsh E., Krishnan K., Gardiner B., Wang X., Nones K., Steen J.A., Matigian N.A., Wood D.L., Kassahn K.S., Waddell N., Shepherd J., Lee C., Ichikawa J., McKernan K., Bramlett K., Kuersten S., Grimmond S.M. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011;12:R126. doi: 10.1186/gb-2011-12-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cosmopoulos K., Pegtel M., Hawkins J., Moffett H., Novina C., Middeldorp J., Thorley-Lawson D.A. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen L., Li Y., Lin C.H., Chan T.H., Chow R.K., Song Y., Liu M., Yuan Y.F., Fu L., Kong K.L., Qi L., Li Y., Zhang N., Tong A.H., Kwong D.L., Man K., Lo C.M., Lok S., Tenen D.G., Guan X.Y. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Primer list for SYBR green real time QPCR.
Supplementary Table 2 Summary of reads and expression.
Supplementary Table 3 miRegbase1.1: Database of miRNA target prediction.
Supplementary Table 4 Summary of Reactome and Gene Oncology terms.
Supplementary Table 5 Summary of SNPs and INDELs discovered from transcriptome data.
Supplementary Table 6 Summary of mutations in miRNA binding UTRs.
Supplementary Table 7 miRNA reads from CLC alignment.
Supplementary Table 8 Summary of human miRNA isomirs.
Supplementary Table 9 Summary of EBV miRNA isomirs.
Supplementary Fig. 1. Biological networks enriched in NPC model systems.
Supplementary Fig. 2. Integrated analysis on the selected candidates with sequence variation in miRNA binding UTRs.


