Abstract
Members of the Importin-β family recognize nuclear localization signals (NLS) and nuclear export signals (NES). These proteins play important roles in various nucleocytoplasmic transport processes in cells. Here, we examined the expression patterns of 21 identified Importin-β genes in mouse embryonic stem cells (mESCs), mouse embryonic fibroblast (MEF) and mESCs differentiated into neural ectoderm (NE) or mesoendoderm (ME). We observed striking differences in the Importin-β mRNA expression levels within these cell types. We also found that knockdown of selected Importin-β genes led to suppression of Nanog, and altered the balance of Oct4/Sox2 expression ratio, which is important for NE/ME lineage choice. Furthermore, we demonstrated that knockdown of XPO4, RanBP17, RanBP16, or IPO7 differentially affected the lineage selection of differentiating mESCs. More specifically, knockdown of XPO4 selectively stimulated the mESC differentiation towards definitive endoderm, while concomitantly inhibiting NE differentiation. RanBP17 knockdown also promoted endodermal differentiation with no effect on NE differentiation. RanBP16 knockdown caused differentiation into ME, while IPO7 knockdown inhibited NE differentiation, without obvious effects on the other lineages. Collectively, our results suggest that Importin-βs play important roles in cell fate determination processes of mESCs, such as in the maintenance of pluripotency or selection of lineage during differentiation.
Keywords: Importin, Exportin, ESC, Germ layers, Differentiation
Abbreviations: ME, mesoendoderm; MEF, mouse embryonic fibroblast; mESCs, mouse embryonic stem cells; NE, neural ectoderm; NES, nuclear export signals; NLS, nuclear localization signals; NPC, nuclear pore complex; RA, retinoic acid
Highlights
-
•
Importin-β expression patterns are distinct in mESCs, MEFs, NE and ME cells.
-
•
Importin-β may modulate differentiation and lineage selection in mESCs.
-
•
Suppression of either XPO4 or RanBP17 induces endodermal differentiation in mESCs.
-
•
RanBP16 suppression induces a ME differentiation in mESCs.
-
•
XPO4 and IPO7 are essential for mESC differentiation into NE cells.
1. Introduction
The importin-β family, comprising importins and exportins, is a group of proteins of molecular weights ranging from 90 to150 kDa. Proteins belonging to this family have low sequence identity (10–20%) and all contain helical HEAT repeats [1–3]. These proteins recognize nuclear localization and export signals (NLS/NES, respectively), bind weakly to phenylalanine-glycine (FG)-repeats in the nuclear pore complex (NPC), and play roles in the nucleocytoplasmic transport processes of various proteins [4,5]. Importin-β–cargo interactions are regulated by the small GTPase, Ran [3]. Because the number of Importin-βs is limited, each member protein mediates the transport of multiple protein cargoes [6]; thus, Importin-βs are essential for diverse cellular processes such as gene expression, signal transduction, and oncogenesis [3]. Moreover, they are involved in non-transport processes such as mitosis, centrosomal duplication, and nuclear envelope assembly [7].
The Importin-β family comprises at least 20 proteins in humans and 14 in Saccharomyces cerevisiae [6,7]. Approximately 11 of these proteins in humans and 10 in S. cerevisiae are reported to mediate nuclear import through recognition of NLS [3]. However, limited data are available for mouse models, particularly with regard to embryonic stem cells (mESCs).
Thus, this study was conducted to understand the roles of Importin-βs in the different cellular events of mESCs. To our knowledge, this is the first study on the genetic expression patterns of the Importin-β family in mESCs and their differentiated germ layer cells. Our results reveal a possible association between the expression of some Importin-βs and the maintenance of pluripotency or lineage selection during the differentiation of mESCs.
2. Materials and methods
2.1. Culture of mouse embryonic stem cells
Feeder-free mouse embryonic stem cells (EB3) [8] were used for all experiments. EB3 cells were maintained on 0.1% gelatin-coated surfaces in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM of MEM nonessential amino acid (GIBCO), 100 mM of MEM sodium pyruvate (GIBCO), 0.1 mM β-mercaptoethanol (Sigma Chemical) and LIF at 37 °C in 5% CO2. Cells were passaged every 2 or 3 days.
2.2. Culture of mouse embryonic fibroblasts
Mouse embryonic fibroblasts (MEFs) isolated from C57BL/6NCrSlc (SLC) were cultured in DMEM supplemented with 10% FBS, at 37 °C in 5% CO2.
2.3. Mouse embryonic stem cell differentiation
For differentiation toward neural ectoderm (NE) or mesoendoderm (ME) lineages, 2 × 106 feeder-free EB3 cells were plated and incubated for 48 h on a 0.1% gelatin-coated surface of 100 mm culture dish with serum-free N2B27 media without LIF [9,10]. This was followed by the addition of 500 nM retinoic acid (RA) for NE differentiation [11] or 3 μM CHIR99021 for ME differentiation [12]. Treated cells were incubated for an additional 48 h before they were trypsinized and collected for quantitative PCR analysis.
2.4. RNA extraction and reverse transcription
For all cells, RNA was extracted with TRIZOL (Invitrogen), DNase treated (Zymo Research), and reverse transcribed using Transcriptor First Strand cDNA Synthesis Kit (Roche). All procedures were performed according to the manufacturer's recommendations. The reverse transcription was performed at 25 °C for 10 min, 50 °C for 60 min, and 85 °C for 5 min.
2.5. Reverse transcription PCR and quantitative PCR
Reverse transcription (RT) PCR was conducted using the initial step discussed in Section 2.4. Following cDNA synthesis, a 40 ng template for each of the test samples was amplified in GeneAmp™ PCR System 9700 (Applied Biosystems) using KOD Plus (Invitrogen), according to the manufacturer's recommendation. The PCR conditions were set at a pre-denaturation temperature of 94 °C for 2 min, 35 cycles of denaturation temperature at 94 °C for 15 s, annealing temperature at 55 °C for 30 s (for Brachyury and Actin) or 60 °C for 30 s (for Sox1), and extension at 68 °C for 30 s. This was followed by a final extension temperature of 72 °C for 5 min.
All Quantitative (Q) PCR analysis was performed on a 384-well plate with an ABI PRISM 7900HT system (Applied Biosystems) using FastStart Universal SYBR Green Master [Rox] (Roche). The qPCR reaction consisted of a holding temperature of 95 °C for 30 s, and 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and a standard dissociation stage. Standard curves were generated for all target genes with serial dilutions of total RNA from EB3 cells at 0.8, 4, 20, and 100 ng. Total RNA from experimental cells was diluted to 20 ng and used as a template. The relative target mRNA expression levels were determined using the Pfaffl method and all values were normalized using GAPDH mRNA levels.
2.6. siRNA-oligonucleotide treatment
For all transfections, 2 × 105 feeder-free EB3 cells were seeded onto 0.1% gelatin-coated surfaces of 6-well plates with 2 mL of Dulbecco's modified Eagles medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM of MEM nonessential amino acid (GIBCO), 100 mM of MEM sodium pyruvate (GIBCO), 0.1 mM β-mercaptoethanol (Sigma), and LIF. Immediately after plating, the cells were transfected with 2 different siRNA constructs for each of the target genes (i.e., RanBP17, XPO4, IPO7) using Lipofectamine™ RNAiMAX (Invitrogen), and incubated at 37 °C, in 5% CO2. After 48 h incubation, the medium was changed using 2 mL of fresh medium without LIF, and another transfection was performed according to the same procedure. The cells were incubated for an additional 48 h before they were trypsinized and collected.
2.7. Induced differentiation in siRNA-oligonucleotide treated EB3 cells
All transfections were carried out using the method described in Section 2.6. After the initial 48 h incubation in enriched DMEM with LIF, siRNA-oligonucleotide treated EB3 cells were induced to differentiate toward either NE or ME with the addition of 500 nM retinoic acid (RA) or 3 μM CHIR99021, respectively. The RA- or CHIR99021-treated cells were maintained in LIF-withdrawn enriched DMEM for another 48 h before they were trypsinized and collected for quantitative PCR analysis.
2.8. Western blot
Cells were lysed with RIPA buffer (50 mM Tris–HCl at pH 8.0, 0.15 M NaCl, 1 mM EDTA at pH 8.0, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 6 mM NaF, 1 mM Na3VO4, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 μg/mL pepstatin) and incubated for 15 min on ice. The samples were centrifuged at 1500 rpm for 10 min at 4 °C. Supernatants from samples were collected and total protein concentrations were determined using a BCA protein assay kit (Pierce). Then, 20 μg of each protein sample was separated by SDS–PAGE and transferred onto a nitrocellulose membrane. After blocking with 3% skim-milk in TBST buffer (50 mM Tris–HCl (pH 8.0), 100 nM NaCl, and 0.1% Tween 20) for 30 min at room temperature, the membrane was incubated overnight at 4 °C with primary antibodies anti-mouse Oct3/4 (BD Transduction Laboratory), anti-mouse Nanog (ReproCELL), or anti-mouse Sox2 (EMD Millipore) as suggested by the manufacturer. After incubation for 45 min with secondary antibodies conjugated to horseradish peroxidase, bands were visualized using Pierce western blotting substrate (Thermo Scientific). All protein levels were normalized to GAPDH levels (Ambion).
2.9. Nucleotide sequences
All oligonucleotide sequences used for this study are summarized in Supplementary Table 1.
2.10. Statistical analysis
Statistical analysis was carried out using unpaired Student's t test. The p values ≤0.05 indicated a statistically significant difference, while p values ≤0.01 indicated a highly significant difference (*p < 0.05; **p < 0.01).
3. Results
3.1. Importin-β expression levels in mESCs vs. MEFs
We identified 21 mouse Importin-β genes in the database, designed primers and optimized conditions for qPCR analysis (Primer sequences are available in Supplementary Information). The mESCs, particularly EB3 cells, were maintained in the pluripotent state by using LIF and enriched DMEM with methods previously described. We also cultured mouse embryonic fibroblasts (MEFs) using the methods described earlier. We then compared the relative expression levels of Importin-βs in mESCs and MEFs by qPCR from 3 independent experiments (Fig. 1). Interestingly, the RanBP17 mRNA expression level was found to be much higher in mESCs than in MEFs. We also found that other Importin-βs such as IPO7, IPO11, XPO1, XPO4, and Cse1L were also more highly expressed in mESCs compared to MEFs. This result is similar to previous reports of highly expressed Importin-β genes in different pluripotent cells such as hESCs, rat iPS cells, human iPS cells, and mESCs using microarray analysis [13–15]. On the other hand, we also observed that the IPO4 mRNA expression level was much lower in mESCs than in MEFs. Importin-βs IPO13, RanBP10, XPOt, TRN2, RanBP6, XPO5, and TRN1 were expressed at lower levels in mESCs than in MEFs.
Fig. 1.
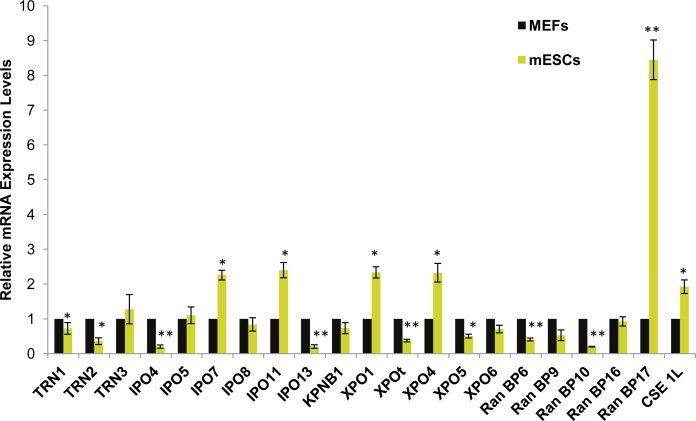
Expression of Importin-βs in mouse embryonic stem cells (mESCs) was assessed by QPCR and the changes are presented as a fold change relative to their expression levels in mouse embryonic fibroblasts (MEFs) used as controls. Importin-βs mRNA levels were normalized to GAPDH levels. Significance was assessed and compared with the levels in the control using unpaired Student's t test (*p < 0.05; **p < 0.01). Error bars represent SEM from 3 independent experiments.
Interestingly, the closely-related RanBP16 (also designated XPO7) and RanBP17, which reportedly share 67% amino acid sequence identity [16,17], exhibited different expression levels; that is, the RanBP16 mRNA expression levels were similar in both mESCs and MEFs, while RanBP17 was highly expressed in mESCs, but not in MEFs. This suggests that RanBP16 and RanBP17 may vary in function in mESCs, which is consistent with results from a previous study reporting different activities of these proteins in other cells [18].
3.2. Importin-β expression levels in cells differentiated into germ layer progenitors from mESCs in vitro
We also determined the relative expression levels of Importin-β genes in cells differentiated from mESCs to germ layer progenitors, namely, cells of the neural ectoderm (NE) and mesoendoderm (ME). We propagated EB3 cells maintained in a pluripotent state by using LIF in enriched DMEM; transferred them to N2B27, a defined medium without differentiation signals [9,10]; and incubated them for 48 h as previously described. The 48 h “temporal window” is needed for cells removed from pluripotency-promoting conditions to respond to either NE- or ME-inducing signals as reported by Jackson et al. [19]. We then induced the cells to transform into either NE or ME cells using retinoic acid (RA) or CHIR99021, respectively.
After 48 h in N2B27 supplemented with RA, the cells exhibited signs of NE differentiation and subsequently triggered the activation of the NE marker, Sox1 (Fig. 2A and B). This result was consistent with those of published studies reporting Sox1 activation following RA addition [11,20,21]. Likewise, cells responded to CHIR99021 and differentiated into ME with the activation of the core mesodermal regulator Brachyury (Fig. 2A and B), as reported previously [21].
Fig. 2.
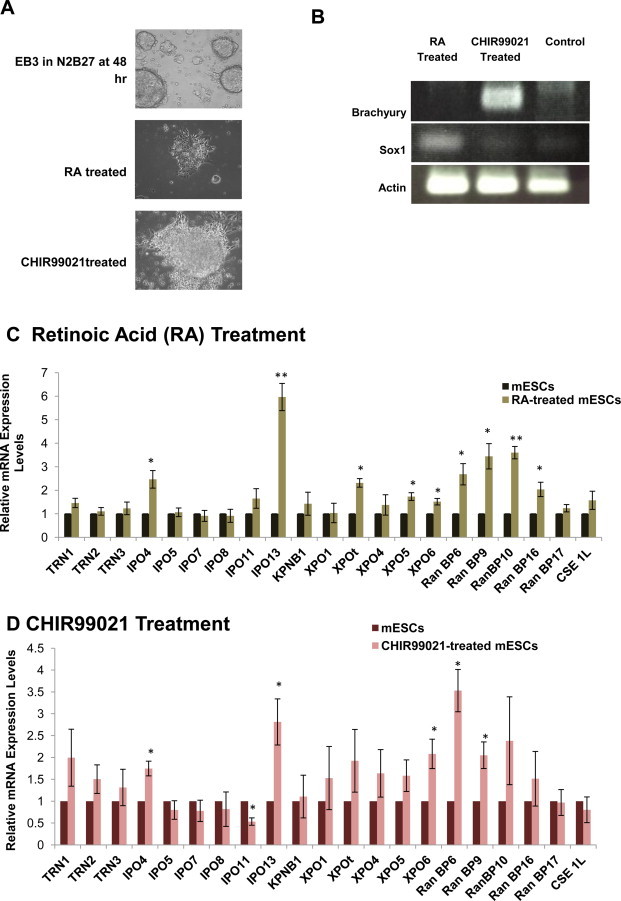
Importin-β mRNA expression levels of mESCs differentiate into germ layer progenitors in vitro. (A) Phase-contrast images of EB3 cells exhibiting signs of differentiation following retinoic acid (RA) or CHIR99021 treatment. (B) RT-PCR expression analysis of early lineage markers Brachyury (for mesoderm) and Sox1 (for neural ectoderm). (C,D) Expression of Importin-βs in RA- or CHIR99021-treated mESCs was assessed by QPCR and is presented as a fold change relative to their expression levels in non-treated mESCs used as controls. Importin-βs mRNA levels were normalized to GAPDH levels. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05; **p < 0.01). Error bars represent SEM from 3 experiments.
A comparison of the mRNA expression levels of Importin-βs in progenitor germ layer cells and mESCs (Fig. 2C and D) revealed a striking difference between levels in NE and ME, and levels in mESCs (Fig. 1). In NE cells, we observed that IPO13 was the most highly expressed gene, although RanBP10, RanBP9, RanBP6, IPO4, XPOt, RanBP16, XPO5, and XPO6 were also highly expressed. On the other hand, the other Importin-β members in this study were expressed in NE at levels comparable to the levels in mESCs. In ME cells, RanBP6 was the most abundantly expressed Importin-β gene, while IPO11 was found to be expressed at the lowest level. IPO13, IPO4, XPO6, and RanBP9 were also readily detected, whereas the remaining Importin-βs were expressed at levels similar to those in mESCs.
3.3. Effect of knockdown of importin-βs on the expression of Nanog
Given the variation in Importin-β gene expression patterns in mESCs, MEF, NE cells, and ME cells, we considered whether this might have a functional impact on either the maintenance of pluripotency or lineage selection during differentiation. To address this, we selected and knocked down three Importin-β genes that were highly expressed in mESCs, namely, RanBP17, XPO4, and IPO7. We also targeted RanBP16, despite its moderate expression in mESCs, because of its high sequence identity with RanBP17. We speculated that LIF withdrawal after 48 h is necessary to enhance the effects of transfection on EB3 cells. Furthermore, we suggested that the 48 h window following the second transfection was critical, because it falls within the “temporal window” [19] where ES cells removed from pluripotency-promoting factors are still nonresponsive to differentiation-inducing agents. Therefore, at 48 h, we re-transfected the cells, changed the medium to LIF-withdrawn enriched DMEM, and maintained them for another 48 h before collection.
We successfully knocked down all target genes by using 2 different siRNAs in EB3 cells as analyzed by qPCR, with a non-targeting siRNA used as a control (Fig. 3A). We evaluated the effect of Importin-β knockdown on the ability of mESCs to maintain pluripotency by analyzing the changes in expression level of Nanog, Oct4 and Sox2; these transcriptional factors cooperatively maintain the regulatory network responsible for self-renewal and pluripotency in mESCs by coregulating large sets of genes and co-occupying many regulatory loci [22–24]. Interestingly, all siRNA treatments resulted in lower Nanog expression levels compared to that in the control (Fig. 3B and E). Thomson et al. [21] demonstrated the necessity of Nanog downregulation for differentiation and lineage selection. Thus, our results indicated that knockdown of select Importin-β genes may predispose mESCs to cellular differentiation, suggesting that these genes play important roles in the maintenance of mESC pluripotency.
Fig. 3.
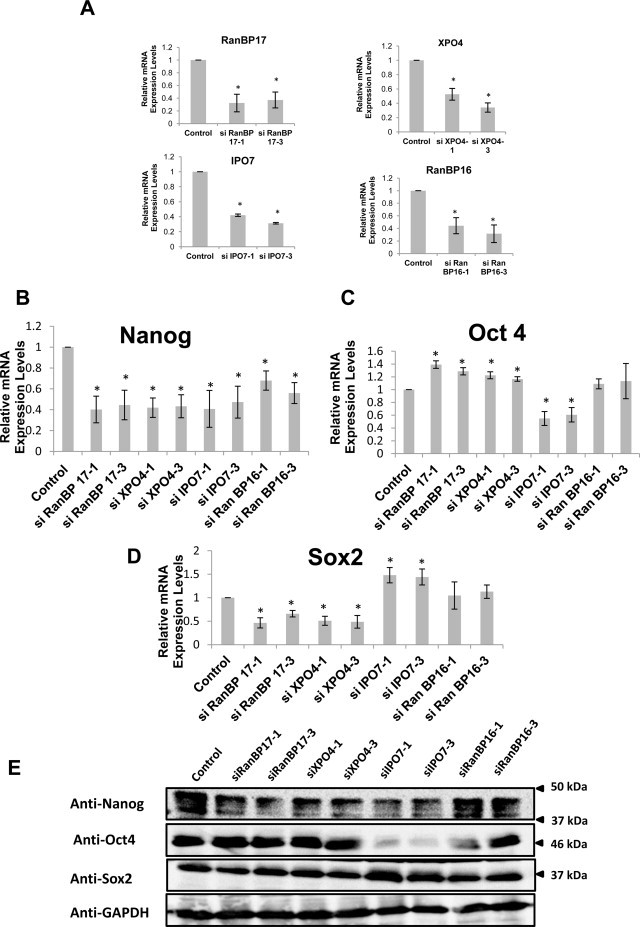
Effects of Importin-β knockdown on the expression of pluripotency markers. (A) Analysis of knockdown efficiency determined by qPCR analysis for RanBP17, XPO4, IPO7, and RanBP16 in siRNA-treated EB3 cells, using 2 variants of siRNA for each gene at 96 h incubation. All samples were normalized against GAPDH levels, and expression level of each gene is presented as a fold change relative to the expression level of the same gene in non-targeting siRNA treated EB3 cells used as control. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05). Error bars represent SEM from 3 independent experiments. (B) qPCR analysis of Nanog in siRNA-treated EB3 cells incubated for 96 h in an enriched DMEM with LIF for the first 48 h and in LIF-withdrawn enriched DMEM for the next 48 h. All samples were normalized against GAPDH levels, and the expression of Nanog in siRNA-treated EB3 cells is shown as a fold change relative to its expression level in non-targeting siRNA treated EB3 cells used as control. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05). Error bars represent SEM from 4 independent experiments. (C) qPCR analysis of Oct4 performed as in (B). (D) qPCR analysis of Sox2 performed as in (B). (E) Protein expression levels of Nanog, Oct4 and Sox2 in Importin-β siRNA-treated mESCs and in non-targeting siRNA-treated mESCs used as controls. Cell lysates (20 μg) were used for western blotting, and GAPDH was used as the loading control.
3.4. Effect of Importin-β knockdown on the expression of Oct4 and Sox2
Likewise, we observed Oct4 and Sox2 expression level changes in siRNA treated mESCs. As indicated in Fig. 3C–E, an opposing expression pattern was observed for Oct4 and Sox2 from the different siRNA treatments. Knockdown of RanBP17 and XPO4 induced slightly higher Oct4 and lower Sox2 expression levels, while knockdown of IPO7 resulted in lower Oct4 and higher Sox2 expression levels. Reduction of RanBP16, however, revealed comparable levels with the control for both genes. The observed variation in the Oct4/Sox2 expression levels precede cell fate selection as previously reported in mESCs [21].
3.5. Effect of Importin-β knockdown on the expression of ME- or NE-specific markers
Considering the relationship between Importin-β expression levels and the changes in the expression levels of pluripotency markers (i.e., Nanog, Oct4 and Sox2), our results suggest that modulation of Importin-β expression may induce differentiation in mESCs. In order to see if the knockdown of Importin-βs in mESCs can also induce lineage specific differentiation, we examined the activation and expression patterns of several differentiation markers including FGF5, Brachyury, FoxA2, Sox1 and Nestin. Except for FGF5, which is an early differentiation marker for primitive ectodermal differentiation, the other markers (Brachyury, FoxA2, Sox1 and Nestin) are known to be germ layer specific, and their high expression levels in populations of differentiating cells indicate a definitive lineage fate. Brachyury and FoxA2 are main mesoendodermal (ME) regulators. Brachyury is specific for mesodermal differentiation, whereas FoxA2 is a regulator of endodermal differentiation [21,25]. On the other hand, Sox1 and Nestin are readily detectable in early developing neuroectodermal (NE) cells [9,20,21]. However, our results showed either comparable or lower expression levels of FGF5, Brachyury, FoxA2, Sox1 and Nestin from the different Importin-β siRNA-treated cells in comparison with the control siRNA-treated cells (Fig. 4A–E). Interestingly, the knockdown of XPO4 or RanBP16 in mESCs resulted in lower basal expression levels of Sox1compared to that in control siRNA-treated cells, while reduction of IPO7 resulted in reduced basal expression levels of both Sox1 and Nestin. Thus, knockdown of Importin-βs does not induce a lineage specific differentiation within a 48 h incubation period following LIF withdrawal. However, knockdown of Importin-βs within this “temporal window” predisposes the early differentiating mESCs toward a specific lineage.
Fig. 4.
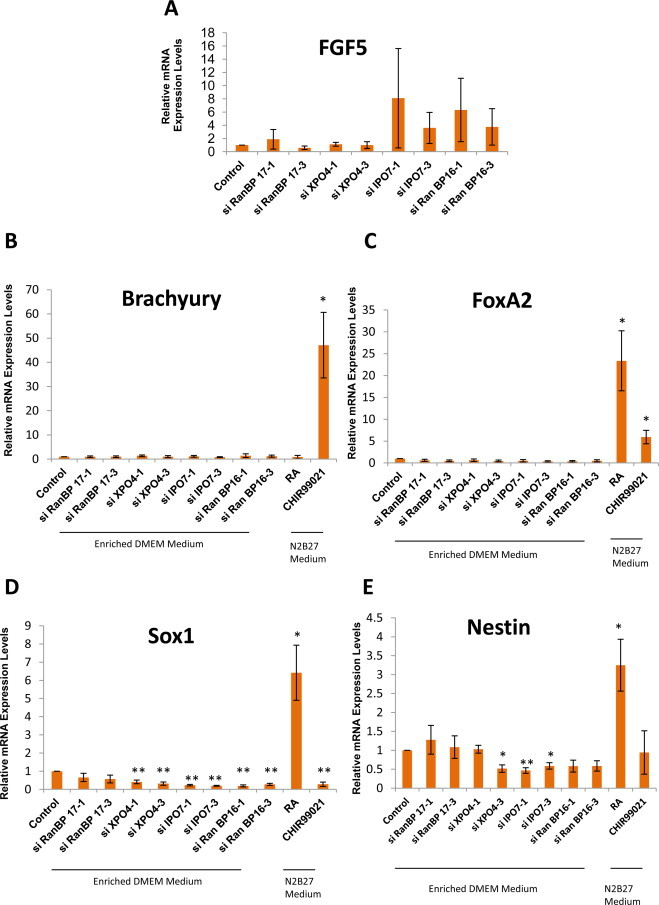
Effects of Importin-β knockdown on the expression of early differentiation markers and lineage-specific markers. (A) qPCR analysis of FGF5 in siRNA-treated EB3 cells incubated for 96 h in enriched DMEM with LIF for the first 48 h and in LIF-withdrawn enriched DMEM for the next 48 h. All samples were normalized against GAPDH levels, and the expression of FGF5 in siRNA-treated EB3 cells is shown as a fold change relative to its expression in non-targeting siRNA treated EB3 cells used as control. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05; **p < 0.01). Error bars represent SEM from 4 independent experiments. (B) qPCR analysis of Brachyury performed as in (A) and from EB3 cells induced to differentiate using either CHIR99021 or RA maintained in N2B27 medium. (C) qPCR analysis of FoxA2 performed as in (B). (D) qPCR analysis of Sox1 performed as in (B). (D) qPCR analysis of Nestin performed as in (B).
3.6. ME-specific marker expression in CHIR99021-treated Importin-β knockdown cells
Next, we examined the effects of Importin-β knockdown on the cell differentiation process, which was promoted either by CHIR99021 or by RA treatment. As expected, we observed an upregulation of Brachyury in CHIR99021-treated cells, while the RA-treated cells showed very high expression levels of Sox1 and Nestin (Fig. 4B, D and E). In both treatments, relatively higher expression levels of FoxA2 were also recorded (Fig. 4C), which suggests that in either RA or CHIR99021 supplementation, there is a certain population of cells undergoing an endoderm differentiation, as described previously [25,26].
Next, we induced ME differentiation using CHIR99021 in siRNA-treated EB3 cells, and analyzed the resulting expressions of Brachyury and FoxA2. Knockdown of RanBP16 resulted in a higher expression of Brachyury, which indicates the induction of mesodermal differentiation, while knockdown of other Importin-βs showed no obvious effects (Fig. 5A). High expression of FoxA2 was also observed from RanBP16 knockdown cells (Fig. 5B), which further indicated enhanced endoderm differentiation. Comparatively similar to higher FoxA2 expression levels were also observed in RanBP17, XPO4 and IPO7 knockdown cells. Therefore, these results suggest that RanBP16 impedes ME differentiation, while XPO4 and RanBP17 are associated with endodermal differentiation.
Fig. 5.
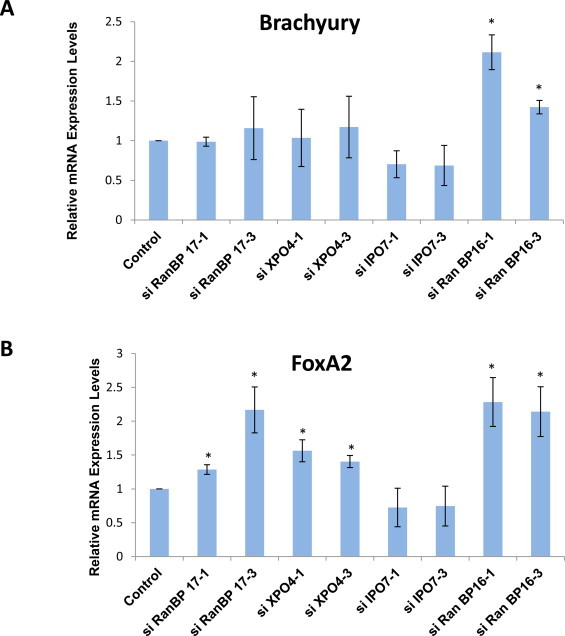
Expression of ME-specific markers from CHIR99021-treated Importin-β knockdown cells. (A) qPCR analysis of Brachyury in siRNA-treated EB3 cells incubated for 96 h in an enriched DMEM with LIF for the first 48 h and followed by incubation in CHIR99021- supplemented, LIF-withdrawn enriched DMEM for the next 48 h. All samples were normalized against GAPDH levels, and expression of Brachyury in siRNA-treated EB3 cells is shown as a fold change relative to its expression in non-targeting siRNA treated EB3 cells used as control. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05; **p < 0.01). Error bars represent SEM from 4 independent experiments. (B) qPCR analysis of FoxA2 as performed in (A).
3.7. NE- and endoderm-specific marker expression in RA-treated Importin-β knockdown cells
We induced NE differentiation using RA in siRNA-treated EB3 cells. The collected cells were then analyzed for Sox1, Nestin and FoxA2 expression levels. Knockdown of XPO4 or IPO7 significantly decreased the expression levels of Sox1, while the rest of the siRNA treatments showed no obvious effects (Fig. 6A). Analogous with their Sox1 expressions, XPO4 and IPO7 knockdown also resulted in very low expression levels of the NE marker Nestin (Fig. 6B). This indicates that reducing the levels of XPO4 or IPO7 may inhibit NE differentiation in RA-treated mESCs.
Fig. 6.
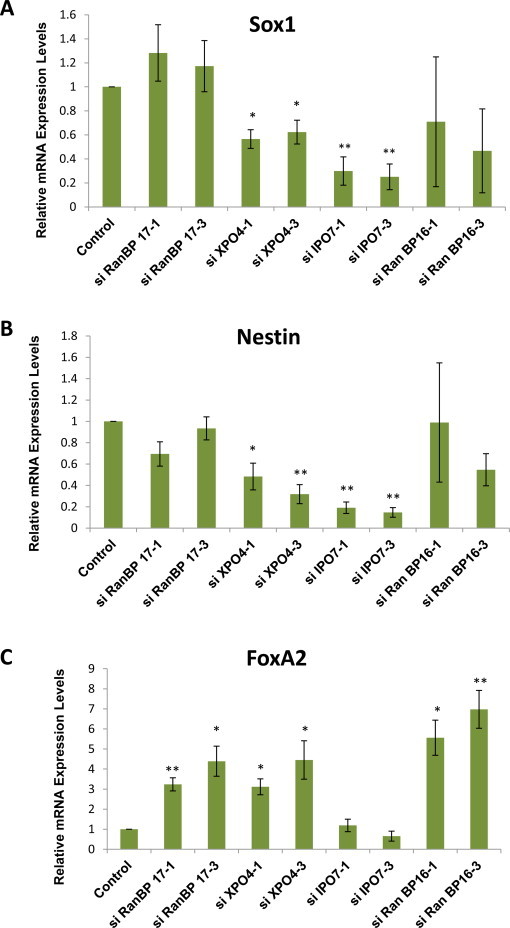
Expression of NE-and endoderm-specific markers from RA-treated Importin-β knockdown cells. (A) qPCR analysis of Sox1 in siRNA-treated EB3 cells incubated for 96 h in an enriched DMEM with LIF for the first 48 h, followed by incubation in retinoic acid (RA)-supplemented, LIF-withdrawn enriched DMEM for the next 48 h. All samples were normalized against GAPDH levels, and expression of Sox1 in siRNA-treated EB3 cells is shown as a fold change relative to its expression in non-targeting siRNA treated EB3 cells used as control. Significance was assessed and compared with the control using unpaired Student's t test (*p < 0.05; **p < 0.01). Error bars represent SEM from 4 independent experiments. (B) qPCR analysis of Nestin as performed in (A). (C) qPCR analysis of FoxA2 as performed in (A).
We also analyzed the FoxA2 expression level of RA treated Importin-β knockdown cells, since we observed a significant induction of FoxA2 expression in RA-treated EB3 cells grown in N2B27 medium (Fig. 4C). Interestingly, as seen in Fig. 6C, we noticed that the reduction in RanBP16 caused a very high expression level of FoxA2, similar to what was observed in CHIR99021-treated cells (Fig. 5B). In addition, higher expression levels were also seen in RanBP17 and XPO4 knockdown cells. These findings further emphasized the involvement of RanBP16, RanBP17 and XPO4 in endodermal differentiation of mESCs.
Together, these results demonstrate that Importin-β family members, such as RanBP16, RanBP17, XPO4 and IPO7, are differentially involved in the lineage commitment of mESCs.
4. Discussion
This is the first report on the mRNA expression patterns of Importin-β genes in mESCs and their differentiated germ layer cells (Figs. 1 and 2C, D). Knockdown of highly expressed Importin-β genes in mESCs was performed to further understand their relationships to cell fate determination processes such as maintenance of pluripotency or lineage selection during differentiation.
4.1. Importin-β suppression promotes differentiation in mESCs
Our data revealed a decrease in Nanog expression in Importin-β-knockdown cells (Fig. 3B and E). Nanog affects both pluripotency and differentiation propensity in mESCs [27,28]. Nanog undergoes autorepressive regulation that is Oct4/Sox2-independent [29]. Moreover, Thomson et al. [21] suggested that Nanog downregulation could be an early and causal event for moving embryonic stem cells into the responsive state as an initial step towards differentiation. This suggests possible direct or indirect association of RanBP17, XPO4, IPO7, and RanBP16 with the maintenance of pluripotency in mESCs and the suppression of at least one of these genes creates a condition that promotes differentiation. Protein-binding analysis of these Importin-β proteins will be required to further elucidate whether they associate with Nanog and other cargoes in mESCs, and how this is related to pluripotency.
4.2. Importin-β suppression in mESCs modulates a differential Oct4 and Sox2 level that leads to lineage-specific differentiation
Our data also demonstrated variation in terms of Oct4 and Sox2 expression in response to Importin-β knockdown in mESCs (Fig. 3C–E). Oct4 and Sox2 are transcription factors that, aside from their functions in the maintenance of pluripotency, have been reported to integrate external signals and control lineage selection. Specifically, Oct4 suppresses NE differentiation and promotes ME differentiation, while Sox2 hinders ME differentiation and promotes NE differentiation in mESCs and hESCs [21,30]. According to our results, XPO4 knockdown caused a concomitant reduction in Sox2 expression. This observation is consistent with an earlier study that indicated that XPO4 mediates Sox2 import [31]. The inability to import Sox2 after XPO4 knockdown may in turn affect Sox2 expression, since Sox2 itself undergoes transcriptional autoregulation [32]. Consistent with this, a significant reduction in the expression of Importin-βs may change the nucleocytoplasmic traffic efficiency of Sox2 or other transcription factors involved in their transcriptional regulation. On the other hand, it has been shown that Importin-βs, aside from their transport roles, may interact directly with transcriptional factors and regulate their actions [17,33]. Depending on the Importin-β targeted, this would lead to high Oct4 and low Sox2 levels, or vice versa, thereby, predisposing mESCs to differentiate into ME or NE cells. However, we observed that the lineage-specific differentiation in knockdown cells is not yet readily detectable at this time. Nevertheless, following supplementation of inducing agents (i.e., CHIR99021 or RA) within the same “48 h window,” we were able to observe the effects of Importin-β knockdown on the lineage-specific differentiation of mESCs.
4.3. XPO4 and RanBP17 suppression induce an endodermal differentiation in mESCs
From this study, we found that reduction of XPO4 and RanBP17 in mESCs resulted in slightly higher Oct4 and lower Sox2 levels after 48 h incubation in a LIF withdrawn medium (Fig. 3C–E), which is a condition favorable for ME differentiation. The specificity of XPO4 knockdown cells to differentiate into endodermal cells was demonstrated by their selective up-regulation of FoxA2 compared with Brachyury, Sox1 and Nestin from the respective treatments (Figs. 5A, B and 6A–C). Similarly, a significant induction of FoxA2 expression was also observed in RA-treated RanBP17 knockdown cells, however, only a comparable Brachyury level was observed following CHIR99021 treatment (Fig. 5A and B). These results indicated that reductions in both XPO4 and RanBP17 augment endodermal differentiation in mESCs.
4.4. RanBP16 suppression induces a ME differentiation in mESCs
Although, RanBP16 knockdown cells showed no definite lineage specificity based on their Oct4 and Sox2 expression patterns (Fig. 3C–E), the reduction of RanBP16 in mESCs was shown to strongly promote ME differentiation as it led to very high expressions of Brachyury and FoxA2 following treatment with CHIR99021 and RA, respectively (Figs. 5A, B and 6C). Collectively these findings suggest that RanBP16 in mESCs may inhibit ME differentiation. Considering the different responses observed with RanBP16 and RanBP17 knockdown cells (Figs. 3C, D, 5A, B and 6A–C), this study demonstrates the functional differences between these two homologous proteins.
4.5. XPO4 and IPO7 are essential for mESC differentiation into NE cells
As previously discussed, the knockdown of XPO4 in mESCs concomitantly resulted in lower expressions of NE markers following RA treatment (Fig. 6A and B). Conversely, knockdown of IPO7 resulted in lower Oct4 and higher Sox2 levels after a 48 h incubation period in a LIF withdrawn medium, which is a condition favorable for NE differentiation (Fig. 3C–E). Surprisingly, IPO7 reduction after RA treatment followed very low expressions of NE markers, Sox1 and Nestin (Fig. 6A–C), suggesting that XPO4 is important for the earlier stage of lineage commitment to NE, while IPO7 is involved at the later stage of NE differentiation, although, the exact mechanism of their association in these cellular events is still unknown.
Taken together, our findings indicate that the expression patterns of Importin-β proteins in mESCs are distinct from their differentiated progenitor cells. Moreover, the appropriate expression patterns of these proteins in mESCs are important in the maintenance of pluripotency and lineage choice during differentiation.
Acknowledgements
We are indebted to all the members of the Biomolecular Dynamic Group for valuable discussions during the writing of this paper. Special thanks to Dr. H. Niwa (Riken, Japan) for the EB3 cells and Dr. Y. Kamikawa for his technical advice on mESC differentiation. We would also like to extend our deepest gratitude to M. Okamura, R. Tanaka, and M. Harayama for their administrative assistance. This work was supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology, the Japan Society for the Promotion of Science, and by the Core Research for Evolutional Science and Technology (CREST) program of the Japan Science and Technology Agency (JST).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2014.01.001.
Appendix. Supplementary materials
Supplementary Table 1. Oligonucleotide sequences used in the study.
References
- 1.Strom A.-C., Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2(6) doi: 10.1186/gb-2001-2-6-reviews3008. reviews3008.1–reviews3008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chook Y.M., Blobel G. Karyopherin and nuclear import. Curr. Opin. Struct. Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 3.Yuh M.C., Suel K.E. Nuclear import by karyopherin-βs: recognition and inhibition. Biochem. Biophys. Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 5.Gorlich D., Kutay U. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 6.Pemberton L.F., Paschal B.M. Mechanisms of receptor mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 7.Mosammaparast N., Pemberton L.F. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H., Masui S., Chambers I., Smith A.G., Miyazaki J. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol. Cell Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying Q.L., Smith A.G. Defined conditions for neural commitment and differentiation. Methods Enzymol. 2003;365:327–341. doi: 10.1016/s0076-6879(03)65023-8. [DOI] [PubMed] [Google Scholar]
- 10.Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying Q.L., Stavridis M., Griffiths D., Li M., Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 12.ten Berge D., Koole W., Fuerer C., Fish M., Eroglu E., Nusse R. Wnt signaling mediates self-organization and axis formation in embryoid bodies. Cell Stem Cell. 2008;3:508–518. doi: 10.1016/j.stem.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R., Plath K., Hochedlinge K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. (supplementary table) [DOI] [PubMed] [Google Scholar]
- 14.Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y., Li H., Jia N., Cheng L., Xiao H., Xiao L. Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell. 2009;4:11–15. doi: 10.1016/j.stem.2008.11.013. (supplementary table) [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. (supplementary table) [DOI] [PubMed] [Google Scholar]
- 16.Kutay U., Hartmann E., Treichel N., Calado A., Carmo-Fonseca M., Prehn S., Kraft R., Gorlich D., Bischoff F.R. Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J. Biol. Chem. 2000;275:40163–40168. doi: 10.1074/jbc.M006242200. [DOI] [PubMed] [Google Scholar]
- 17.Koch P., Bohlmann I., Schafer M., Hansen-Hagge T.E., Kiyoi H., Wilda M., Hameister H., Bartram C.R., Janssen J.W.G. Identification of a novel putative Ran-binding protein and its close homologue. Biochem. Biophys. Res. Commun. 2000;278:241–249. doi: 10.1006/bbrc.2000.3788. [DOI] [PubMed] [Google Scholar]
- 18.Dorfman J., Macara I.G. STRADalpha regulates LKB1 localization by blocking access to import-alpha, and by association with Crm1 and exportin-7. Mol. Biol. Cell. 2008;19:1614–1626. doi: 10.1091/mbc.E07-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson S.A., Schiesser J., Stanley E.G., Elefanty A.G. Differentiating embryonic stem cells pass through ‘temporal window’ that mark responsiveness to exogenous and paracrine mesoendoderm inducing signals. PLoS ONE. 2010;5:e10706. doi: 10.1371/journal.pone.0010706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abranches E., Silva M., Pradier L., Schulz H., Hummel O., Henrique D., Bekman E. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomson M., Liu S.J., Zou L.N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova I., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 24.Loh Y.H., Wuh Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 25.Kim P.T.W., Hoffman B.G, Plesner A., Helgason C.D., Verchere C.B., Chung S.W., Warnock G.L., Mui A.L.F., Ong C.J. Differentiation of mouse embryonic stem cells into endoderm without embryoid body formation. PLoS ONE. 2010;5:e14146. doi: 10.1371/journal.pone.0014146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illing A., Stockmann M., Telugu N.S., Linta L., Russell R., Muller M., Seufferlein T., Liebau S., Kleger A. Definitive endoderm formation from plucked human hair-derived induced pluripotent stem cells and SK channel regulation. Stem Cell Int. 2013;2013::1–13. doi: 10.1155/2013/360573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 28.Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 29.Navarro P., Festuccia N., Colby D., gagliardi A., Mullin N.P., Zhang W., Karwacki V.N., Osorno R., Kelly D., Robertson M., Chambers I. Oct/Sox2-independent Nanog autorepression modulates heterogeneous Nanog gene expression in mouse ES cells. Embo J. 2012;31:4547–4562. doi: 10.1038/emboj.2012.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for Nanog, Oct4 and Sox2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 31.Gontan C., Guttler T., Engelen E., Demmers J., Fornerod M., Grosveld F., Tibboel D., Gorlich D., Poot R., Rottier R. Exportin 4 mediates a novel nuclear import pathway for Sox family transcription factors. J. Cell Biol. 2008;185:27–34. doi: 10.1083/jcb.200810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chew J.-L., Loh Y.-H., Zhang W., Chen X., Tam W.-L., Yeap L.-S., Li P., Ang Y.-S., Lim B., Robson P., Ng H.-H. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuchiya M., Ogawa H., Suzuki T., Sugiyama M., Haraguchi T., Hiraoka Y. Exportin 4 interacts with Sox9 through the HMG box and inhibits the DNA binding of Sox9. PLoS ONE. 2011;6(10):e10706. doi: 10.1371/journal.pone.0025694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Oligonucleotide sequences used in the study.


