Background: The angiogenesis inhibitor, angiostatin, is an internal fragment of the blood zymogen, plasminogen.
Results: A fraction of plasminogen contains a reduced Cys462-Cys-541 disulfide bond.
Conclusion: Reduced plasminogen is the precursor for angiostatin.
Significance: The plasminogen disulfide bonds reduced during angiostatin formation are examples of allosteric disulfides, and their structures help define new allosteric disulfide bond configurations.
Keywords: Allosteric Regulation, Angiogenesis, Disulfide, Plasmin, Plasminogen
Abstract
Plasma plasminogen is the precursor of the tumor angiogenesis inhibitor, angiostatin. Generation of angiostatin in blood involves activation of plasminogen to the serine protease plasmin and facilitated cleavage of two disulfide bonds and up to three peptide bonds in the kringle 5 domain of the protein. The mechanism of reduction of the two allosteric disulfides has been explored in this study. Using thiol-alkylating agents, mass spectrometry, and an assay for angiostatin formation, we show that the Cys462-Cys541 disulfide bond is already cleaved in a fraction of plasma plasminogen and that this reduced plasminogen is the precursor for angiostatin formation. From the crystal structure of plasminogen, we propose that plasmin ligands such as phosphoglycerate kinase induce a conformational change in reduced kringle 5 that leads to attack by the Cys541 thiolate anion on the Cys536 sulfur atom of the Cys512-Cys536 disulfide bond, resulting in reduction of the bond by thiol/disulfide exchange. Cleavage of the Cys512-Cys536 allosteric disulfide allows further conformational change and exposure of the peptide backbone to proteolysis and angiostatin release. The Cys462-Cys541 and Cys512-Cys536 disulfides have −/+RHHook and −LHHook configurations, respectively, which are two of the 20 different measures of the geometry of a disulfide bond. Analysis of the structures of the known allosteric disulfide bonds identified six other bonds that have these configurations, and they share some functional similarities with the plasminogen disulfides. This suggests that the −/+RHHook and −LHHook disulfides, along with the −RHStaple bond, are potential allosteric configurations.
Introduction
Plasma plasminogen is the zymogen form of the serine protease, plasmin. Plasminogen is a ∼90-kDa protein containing three types of domain structures: an N-terminal Pan-apple domain, followed by five kringle domains, and a C-terminal serine protease domain. The zymogen is activated by urokinase or tissue plasminogen activator in the circulation through cleavage of the Arg561-Val562 peptide bond in the serine protease domain, and the Pan-apple domain is autoproteolytically released to produce mature plasmin. Plasmin is involved in thrombus dissolution by cleaving the fibrin meshwork but also activates other zymogens and cleaves components of the extracellular matrix (1).
Plasma plasmin is the precursor of the angiogenesis inhibitor, angiostatin (2). Angiostatin is an N-terminal fragment of plasmin consisting of kringle domains 1–4 and part of kringle 5. The generation of angiostatin involves cleavage of both disulfide and peptide bonds in plasmin. The kringle 5 Cys462-Cys541 and Cys512-Cys536 disulfide bonds are reduced which leads to cleavage of the Arg530-Lys531 peptide bond (3, 4). Smaller angiostatin molecules can also be generated by additional proteolysis at the Arg474-Val475 and/or Lys468-Gly469 peptide bonds. The peptide bond cleavage can be catalyzed by plasmin itself or by other serine- or metallo-proteases (3, 4).
The Cys462-Cys541 and Cys512-Cys536 plasmin disulfides are examples of allosteric disulfide bonds. By definition, allosteric disulfides are bonds that regulate the function of the mature protein in which they reside by triggering a change when they are cleaved (5–7). These bonds are cleaved by oxidoreductases or by thiol/disulfide exchange, and functional changes in ligand binding, substrate hydrolysis, proteolysis, or oligomer formation have been identified (for review, see 8, 9). The functional change upon cleavage of the plasmin allosteric disulfides is proteolysis of the protein.
Reduction of the kringle 5 allosteric disulfides involves interaction of plasmin with other proteins. A plasmin-binding protein that is secreted by tumor cells is the glycolytic enzyme, phosphoglycerate kinase (PGK)2 (10, 11). In glycolysis, PGK catalyzes phosphoryl transfer from 1,3-bisphosphoglycerate to ADP to form 3-phosphoglycerate and ATP. PGK secretion by cultured tumor cells positively correlates with oxygen tension (12), which is in accordance with hypoxia being a driver of tumor angiogenesis. In addition, plasma levels of PGK negatively correlate with tumor angiogenesis and tumor growth in mice (13–15).
The mechanism of reduction of the Cys462-Cys541 and Cys512-Cys536 allosteric disulfides has been investigated in this study. We have observed that the Cys462-Cys541 disulfide bond is already cleaved in a fraction of plasma plasminogen, and we show that this reduced plasminogen is the precursor for PGK-mediated angiostatin formation. Moreover, the plasmin(ogen) allosteric disulfides have configurations that are shared by allosteric disulfides in other proteins.
EXPERIMENTAL PROCEDURES
Generation of Angiostatin from Plasmin
Plasminogen was purified from fresh frozen human plasma according to published procedures (16). Plasmin was generated by incubating plasminogen (20 μm) with urokinase plasminogen activator (20 nm; Medac) for 30 min in 20 mm Hepes, pH 7.4 buffer containing 0.14 m NaCl (Hepes-buffered saline) at 37 °C. Full-length recombinant human PGK was produced and purified as described (10). Plasmin was incubated with PGK in Hepes-buffered saline at 37 °C, and the kringle-containing fragments were either isolated using lysine-Sepharose and revealed by silver stain, or visualized by blotting unfractionated reactions with anti-plasminogen polyclonal antibodies (3, 4, 10, 17).
Quantification of Thiols in Plasminogen
Different plasminogen batches (15–27 μm) in Hepes-buffered saline containing 1 mm EDTA were incubated with 500 μm 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; Sigma) for 10 min, and the absorbance of TNB at 412 nm was measured using a Molecular Devices Thermomax Plus microplate reader. The extinction coefficient for the TNB dianion at pH 7.0 is 14,150 m−1cm−1 at 412 nm (18).
Labeling of the Free Thiols in Plasminogen and Fragments
Plasminogen fragments were generated by limited proteolysis with porcine elastase and purified by a combination of lysine-Sepharose affinity and Sephacryl S-100 gel filtration chromatography as described (19). Unpaired cysteine thiols in plasminogen or plasminogen fragments were labeled with either 3-(N-maleimidylpropionyl)biocytin (MPB; Invitrogen) or pegylated-maleimide (TMM(PEG)12, Thermo Scientific). Plasminogen or fragments in Hepes-buffered saline were incubated with MPB (100 μm) for 4–24 h or pegylated-maleimide (6.25 mm) for 1.5 h at 22 °C. The unreacted MPB was quenched with 200 μm reduced glutathione for 10 min at 22 °C. Samples of the reactions were resolved on a NuPAGE Novex 4–12% Bis-Tris gel (Invitrogen) with MOPS running buffer under nonreducing conditions and either stained with colloidal Coomassie or transferred to a polyvinylidene difluoride membrane (Millipore), blotted with a 1:5000 dilution of streptavidin-horseradish peroxidase (Invitrogen), then developed and visualized using chemiluminescence (PerkinElmer Life Sciences).
Mapping the Free Thiols in Plasminogen
2-Nitro-5-thiocyanobenzoic acid (NTCB; Sigma) specifically S-cyanylates cysteine thiols, and the peptide bond on the N-terminal side of the cyanylated cysteine is then cleaved under mildly alkaline conditions (20, 21). Plasminogen (5 μm) in 0.1 m Tris, pH 8.0 buffer was incubated with NTCB (20 mm) for 1 h at 37 °C to cyanylate the cysteine thiols. The pH of the reaction was adjusted to 9.0 using 3.0 m Tris and incubated at 37 °C for a further 24 h. The resulting fragments were reduced with 100 mm dithiothreitol and resolved on a NuPAGE Novex 4–12% Bis-Tris gel with MES running buffer. The bands were stained by SYPRO Ruby (Invitrogen), cut from the gel, and analyzed by mass spectrometry.
Gel slices containing the fragments were destained in 1:1 acetonitrile and 25 mm NH4HCO3 and dried before incubation with 10 mm dithiothreitol in 25 mm NH4HCO3 for 1 h at 56 °C and then with 50 mm iodoacetamide in 25 mm NH4HCO3 for a further 30 min at 25 °C in the dark. The gel slices were washed and dried before rehydration with 20 μl of 25 mm NH4HCO3 containing 20 ng/μl trypsin (Promega) and incubated at 30 °C overnight. Peptides were eluted from the slices with 5% formic acid, 50% acetonitrile for 30 min at 25 °C and separated by nano-LC on an Ultimate 3000 HPLC (Dionex) using a fritless nano column (75 μm × 10 cm) containing C18 medium (5 μm, 200 Å Magic; Michrom). Peptides were resolved using a linear gradient of H2O:CH3CN (98:2, 0.1% formic acid) to H2O:CH3CN (64:36, 0.1% formic acid) at 250 nl/min for 1 h. Positive ions were generated by electrospray and analyzed in a LTQ FT Ultra (Thermo Electron) mass spectrometer operated in data-dependent MS/MS acquisition mode. MS data were searched using Mascot (V2.2, Matrix Science) against the nonredundant database from the National Center for Biotechnology Information. Search parameters were: precursor tolerance 10 ppm and product ion tolerances ± 0.4 Da. Cys-carboxyamidomethyl and NTCB modifications were selected as variable modifications with full tryptic cleavage of up to nine missed cleavages.
Redox State of the Cys462-Cys541 Disulfide Bond
To characterize the redox state of the Cys462-Cys541 disulfide bond, we employed mass spectrometry with differential cysteine labeling as described previously (22). Purified plasma plasminogen (6 μg) was incubated with 50 mm iodoacetamide in the dark for 8 h at 22 °C. The band was resolved on NuPAGE Novex 4–12% Bis-Tris gel under reducing conditions and stained with colloidal Coomassie. The intact protein band was excised from the gel, destained, dried, and incubated with 10 mm dithiothreitol in 25 mm NH4HCO3 for 1 h at 56 °C. The gel slice was washed with NH4HCO3, dried, and incubated with 50 mm methyl methanethiolsulfonate (Sigma-Aldrich) in 25 mm NH4HCO3. The slices were washed and dried before digestion of the plasminogen with 50 ng/μl chymotrypsin (Roche Applied Science) in 25 mm NH4HCO3 for 16 h at 25 °C. Peptides were eluted from the slices, resolved by HPLC, and positive ions were generated and analyzed as described above. Cys-carboxyamidomethyl (the iodoacetamide adduct) and Cys-methyldisulfide (the methyl methanethiolsulfonate adduct) were selected as variable modifications with full chymotrypsin cleavage of up to five missed cleavages. To determine the extent of alkylated Cys462, the relative ion abundance of peptides containing Cys-carboxyamidomethyl and Cys-methyldisulfide was used. To calculate ion abundance of peptides, extracted ion chromatograms were generated using the XCalibur Qual Browser software (Version 2.0.7; Thermo Scientific). The area was calculated using the automated peak detection function built into the software. The ratio of carboxyamidomethyl to methyldisulfide labeling represents the fraction of the cysteine in the population that is in the reduced state.
Disulfide Bond Configurations
The Disulfide Bond Analysis tool (23) was used to determine the configurations of the allosteric disulfide bonds listed in Table 4.
TABLE 4.
Allosteric disulfide bonds with −/+RHHook or −LHHook configurations
| Protein | Disulfide cysteines | Configuration (PDB identifier) | References |
|---|---|---|---|
| Plasmin(ogen) | 462–541, 512–536 | −/+RHHook, −LHHook (4DUR) | (28) |
| β2-Glycoprotein I | 288–326 | −/+RHHook (1C1Z) | (39, 40, 52) |
| Angiotensinogen | 18–138 | −/+RHHook (2WXW) | (43) |
| Factor XI | 362–482 | −/+RHHook (2F83) | (44) |
| Interleukin receptor subunit γ | 183–232 | −LHHook (2ERJ, 2B5I) | (46, 54–56) |
| XRCC1 | 12–20 | −LHHook (3LQC, 3K75) | (47) |
| Vascular endothelial growth factors C and D | 156–165 | −LHHook (2X1W) | (49, 50, 52, 53) |
RESULTS
Only a Fraction of Plasmin Is a Substrate for PGK-mediated Angiostatin Formation
It has been consistently observed that not all plasmin is converted to angiostatin by PGK or other plasmin-binding proteins such as annexin A3-S100A10 heterotetramer (24), plasma-membrane associated β-actin (25), or truncated porcine plasminogen activator inhibitor 1 (residues 80–265) (26). This was further demonstrated in a PGK titration assay. Plasmin was incubated with PGK at PGK:plasmin molar ratios of 0.2–7.3, and angiostatin formation was measured (Fig. 1). There was little difference in the angiostatin generated at PGK:plasmin molar ratios from ∼0.5 to 7.3, and a significant fraction of the plasmin remained intact. This result further indicates that not all plasmin is a substrate for PGK. This prompted us to define the population of plasmin(ogen) that is converted to angiostatin. To this end we found that a fraction of plasma plasminogen contains a reduced disulfide bond, as reflected by the presence of one or more unpaired cysteine thiols.
FIGURE 1.
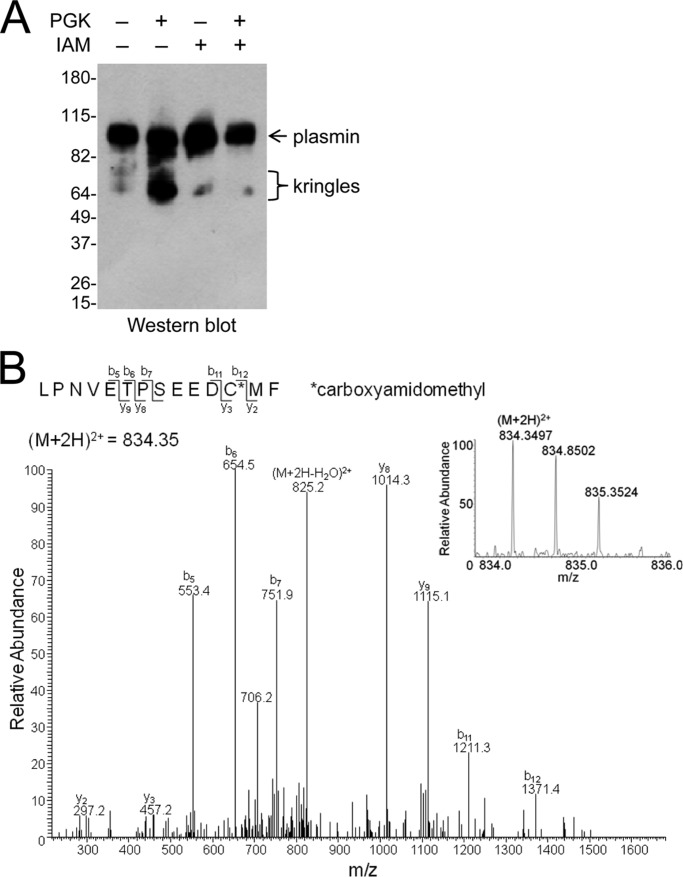
Only a fraction of plasmin is a substrate for PGK-mediated angiostatin formation. Plasmin (20 μg) was incubated with PGK (0–80 μg) in 0.5 ml of Hepes-buffered saline for 30 min at 37 °C. Kringle-containing products were collected on lysine-Sepharose, resolved on SDS-PAGE, and silver stained. The three angiostatin fragments are indicated. The positions of molecular mass markers are shown at left.
Plasminogen Contains Unpaired Cysteine Thiols
Purified plasma plasminogen incorporated conjugates of maleimide with biotin (MPB) and polyethylene glycol. Maleimide is a thiol-specific alkylator at neutral pH. Alkylation of plasminogen with either MPB (Fig. 2A) or pegylated maleimide (Fig. 2B) resulted in a broadening of the plasminogen band on SDS-PAGE, which is indicative of labeling. Incorporation of MPB was also detected by blotting with streptavidin-peroxidase (Fig. 2A). Preincubation of plasminogen with the small thiol-specific alkylator, iodoacetamide, significantly reduced labeling with MPB (Fig. 2A), confirming the thiol specificity of labeling of the maleimides.
FIGURE 2.
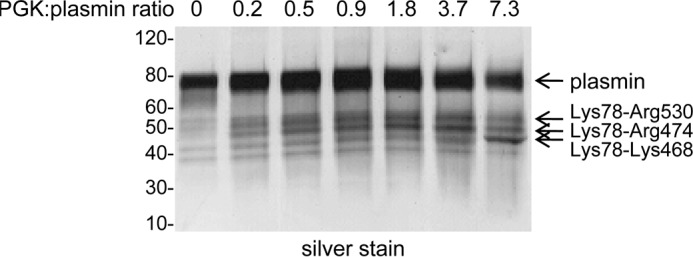
Plasminogen contains unpaired cysteine thiols. A, purified plasma plasminogen was incubated without or with 20 mm iodoacetamide (IAM) for 24 h prior to incubation with the biotin-linked maleimide (MPB) for a further 24 h. Unreacted MPB was quenched with glutathione, and the plasminogen was resolved on SDS-PAGE and either stained with colloidal Coomassie or transferred to PVDF membrane and blotted with streptavidin-peroxidase to detect the MPB label. B, purified plasma plasminogen was incubated without or with 10 mm pegylated maleimide for 1 h, resolved on SDS-PAGE, and stained with colloidal Coomassie. C, K1–3 or K5-serine protease plasminogen fragments were generated by limited proteolysis with porcine elastase. The fragments (5 μg) were resolved on SDS-PAGE and stained with colloidal Coomassie. D, K1–3 or K5-serine protease fragments were incubated with MPB for 4 h, the unreacted MPB was quenched with glutathione, and the fragments (1 μg) were resolved on SDS-PAGE and blotted with streptavidin-peroxidase to detect the MPB label. E, K5-serine protease was incubated with MPB for 1, 4, or 24 h, the unreacted MPB was quenched with glutathione, and the fragment was resolved on SDS-PAGE and blotted with streptavidin-peroxidase. The positions of molecular mass markers are shown at left.
The thiol content of four different plasminogen batches was determined using DTNB. DTNB reacts with cysteine thiols, and the absorbance of the liberated TNB dianion is used to quantitate the number of thiols in the reaction. A range of 0.03–0.36 mol of thiols/mol of plasminogen was determined for the four preparations (Table 1). Two mol of thiols/mol of plasminogen is expected if one disulfide bond is cleaved in a homogeneous preparation, and the interaction of the unpaired cysteine thiols with DTNB is not sterically hindered.
TABLE 1.
Number of thiols in different plasma plasminogen batches determined using DTNB
| Plasminogen batch | mol thiols/mol plasminogen (± S.D., n = 3) |
|---|---|
| 1 | 0.359 ± 0.160 |
| 2 | 0.032 ± 0.004 |
| 3 | 0.192 ± 0.039 |
| 4 | 0.129 ± 0.024 |
To determine whether the unpaired cysteine thiols were in the N- or C-terminal half of plasminogen, kringle 1–3 (K1–3) and kringle 5-serine protease (K5-serine protease) fragments of plasminogen were produced by limited proteolysis with elastase (Fig. 2C) and labeled with MPB (Fig. 2D). MPB labeled the K5-serine protease fragment but not the K1–3 fragment. To assess the accessibility of the unpaired thiols, the time dependence of labeling of K5-serine protease with MPB was performed. MPB incorporation increased over 24 h (Fig. 2E), which implies a restricted access to the thiols. Maleimides react rapidly with exposed thiols such as reduced glutathione with a second order rate constant of ∼10,000 m−1s−1 at pH 8 (27).
The Cys462-Cys541 Disulfide Bond Is Reduced in a Fraction of Plasminogen
Cysteine-specific chemical cleavage and mass spectrometry was used to establish the position of the unpaired cysteine residues in plasminogen (20, 21). NTCB S-cyanylates unpaired cysteine residues at pH 8, and cleavage of the peptide bond N-terminal to the cyanylated cysteine is achieved by transfer/migration of the cysteine residue nitrogen to the cyano group at pH 9, forming a 2-iminothiazolidine-4-carboxylyl (ITC) peptide. This cleavage will occur at every cyanylated cysteine, and therefore the number of ITC peptides corresponds to the number of unpaired cysteines in the protein.
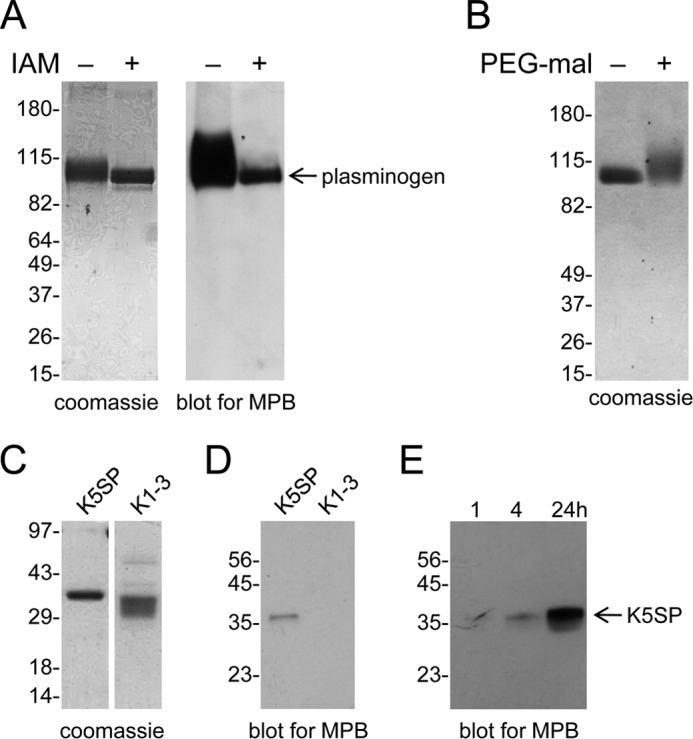
Plasminogen in pH 8.0 buffer was reacted with NTCB and the peptide bond N-terminal of the cyanylated cysteines was then cleaved at pH 9. The protein was reduced and alkylated and resolved by SDS-PAGE. Five distinct fragments were generated from a fraction of the plasminogen (Fig. 3A), and their pattern and molecular masses (Fig. 3B) corresponded to cyanylation of cysteines 462 and 541 in kringle 5. This is in accordance with MPB labeling of the K5-serine protease fragment. The identity of the plasminogen fragments were confirmed by mass spectrometry analysis of trypsin digests (Table 2).
FIGURE 3.
The Cys462-Cys541 disulfide bond is reduced in a fraction of plasminogen. A, plasminogen in pH 8.0 buffer was untreated or reacted with NTCB. The peptide bond N-terminal of the cyanylated cysteine was then cleaved at pH 9. The fragments were reduced and alkylated, resolved by SDS-PAGE, and stained with SYPRO Ruby. Five distinct fragments were generated (bands 1–5). Two nonspecific fragments that were present in the untreated plasminogen are indicated by dotted arrows. The positions of molecular mass markers are shown at left. B, the molecular masses of the five fragments are consistent with NTCB cleavage of a fraction of plasminogen containing unpaired cysteines at residues 462 and 541. Comparison of the measured and predicted fragment sizes based on NTCB cleavage at cysteines 462 and 541 is shown in the bar graph. The identity of the fragments was confirmed by mass spectrometry (Table 2). C, structure of plasminogen kringle 5 containing a reduced Cys462-Cys541 disulfide bond. Ribbon structure of kringle 5 (residues 460–543) of full-length human plasminogen shows the Cys462-Cys541 and Cys512-Cys536 disulfide bonds in yellow. The Cys462-Cys541 disulfide bond is reduced in the structure at right. The asterisk represents regions of the polypeptide chain of the reduced structure that were not resolved by x-ray crystallography. The structure is that of PDB identifier 4DUR (28).
TABLE 2.
Mass spectrometry characterization of the NTCB cleavage products
The five fragments shown in Fig. 3A were cut from the gel and digested with trypsin, and the resulting peptides were identified by mass spectrometry. The molecular masses and peptide fingerprints of the five NTCB fragments are consistent with cleavage of the Cys462-Cys541 disulfide bond in a fraction of plasminogen.
| Band | Predicted residues | Representative peptide |
|---|---|---|
| 2 | Glu1-Asp461 | Residues 118–134; FSPATHPSEGLEENYCR |
| 3 | Cys462-Asn791 | Residues 494–504; HSIFTPETNPR |
| 4 | Cys541-Asn791 | Residues 662–677; VIPACLPSPNYVVADR |
| 5 | Cys462-Gln540 | Residues 494–504; HSIFTPETNPR |
The recent crystal structure of plasminogen supports the labile nature of the Cys462-Cys541 disulfide (28). Plasma plasminogen crystallized as a dimer and the structure of the kringle 5 domain differed between the two molecules. The Cys462-Cys541 disulfide bond is oxidized in one molecule (chain A) and reduced in the other (chain B) (Fig. 3C). Cleavage of the Cys462-Cys541 bond changed the structure of kringle 5 as a number of regions of the reduced kringle were not resolved.
Alkylation of the Cys462 and Cys541 Plasmin Thiols Ablates PGK-mediated Angiostatin Formation
The effect of alkylating the unpaired Cys462 and Cys541 sulfur atoms in plasminogen on PGK-mediated angiostatin formation was examined. Alkylation of plasminogen with iodoacetamide blocked angiostatin formation from plasmin (Fig. 4A). Alkylation of Cys462 was confirmed by mass spectrometry (Fig. 4B).
FIGURE 4.
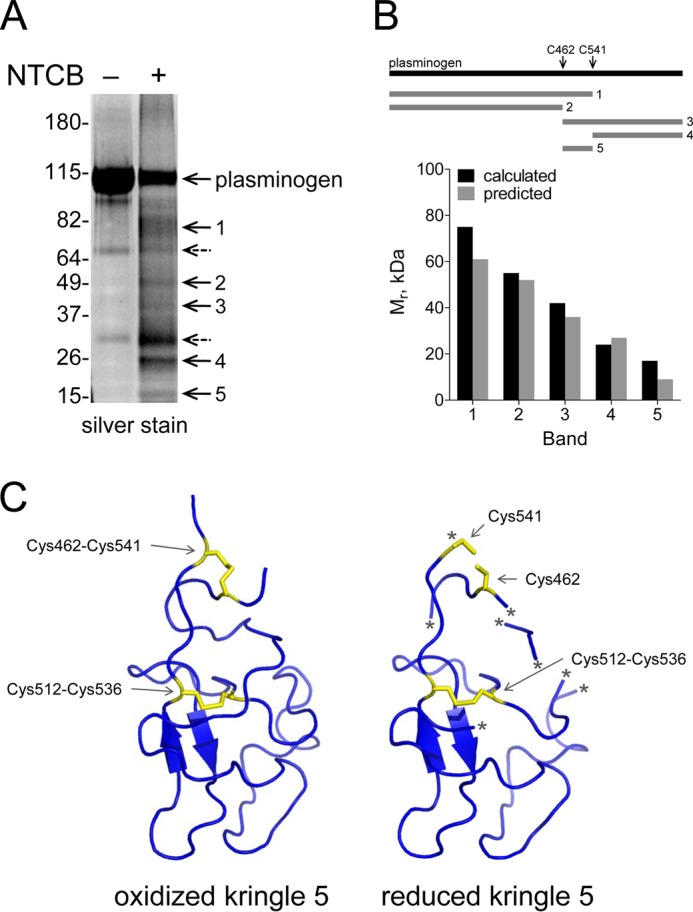
Alkylation of the Cys462 and Cys541 plasmin thiols inhibits PGK-mediated angiostatin formation. A, plasminogen (20 μg) was untreated or alkylated with 20 mm iodoacetamide (IAM) for 24 h. The plasminogen was converted to plasmin, and 20 μg was incubated with 80 μg of PGK in 0.5 ml of Hepes-buffered saline for 30 min at 37 °C. Plasmin products were separated on SDS-PAGE and blotted with polyclonal anti-plasminogen antibodies. The angiostatin fragments are indicated. The positions of molecular mass markers are shown at left. B, plasminogen (6 μg) was incubated with 50 mm iodoacetamide for 8 h and then digested with chymotrypsin. Tandem mass spectrum of the chymotryptic fragment, LPNVETPSEEDCMF, shows Cys462 (underlined) labeled with the iodoacetamide adduct, carboxyamidomethyl. The accurate mass spectrum of the peptide is shown in the inset (observed [M+2H]2+ = 834.3497 m/z; expected [M+3H]3+ = 834.35 m/z).
The reduced Cys462 and Cys541 thiols in plasminogen were alkylated with iodoacetamide and the disulfide-bonded cysteines with methyl methanethiolsulfonate following reduction with dithiothreitol. The ratio of carboxyamidomethyl to methyldisulfide labeling represents the fraction of the Cys462-Cys541 disulfide bond in the population that is in the reduced state. A peptide containing Cys462 was resolved, and Cys-carboxyamidomethyl (Fig. 4B) and Cys-methyldisulfide adducts were identified, which provided for an estimate of the redox state of the disulfide bond in the plasminogen preparation. No peptides containing Cys541 were resolved in the analysis. 26% of Cys462 in the population of plasminogen molecules was labeled with iodoacetamide and the remainder with methyl methanethiolsulfonate (Table 3). This result implies that approximately one quarter of the Cys462-Cys541 disulfide bond is reduced in this preparation of plasminogen, which is the range observed from labeling with DTNB (Table 2) or NTCB (Fig. 3).
TABLE 3.
Quantitation of the redox state of the plasminogen Cys462-Cys541 disulfide bond
The fraction of the LPNVETPSEEDCMF peptide containing Cys462 (underlined) that labeled with either iodoacetamide or methyl methanethiolsulfonate is shown.
| Alkylator | Abundance | Percentage |
|---|---|---|
| Iodoacetamide adduct | 51,481 | 26 |
| Methyl methanethiolsulfonate adduct | 144,319 | 74 |
DISCUSSION
Both the protein chemical studies described here and the recent description of the crystal structure of plasminogen (28) indicate that the Cys462-Cys541 disulfide bond is cleaved in a fraction of the plasma protein. Law et al. (28) suggested that the bond may have been cleaved by the synchrotron radiation used to reveal the structure. The findings presented here indicate that the bond is naturally cleaved in a fraction of plasminogen, what we term reduced plasminogen, and it is the reduced plasminogen that is the substrate for PGK-mediated angiostatin formation.
These and previous results (3, 4, 10, 11) are consistent with the following sequence of molecular events in plasmin that lead to angiostatin formation (Fig. 5). The plasminogen Cys462-Cys541 disulfide bond is presumably reduced by a reductase in blood. This event is likely regulated as different amounts of reduced plasminogen were observed in different plasminogen preparations from plasma. A candidate reductase is thioredoxin, which is secreted by immune cells during inflammation (29, 30), resulting in elevated serum levels in patients with asthma (31), rheumatoid arthritis, and heart failure (32). Binding of tumor-derived PGK to reduced plasmin induces a conformational change in kringle 5 that leads to attack by the Cys541 thiolate anion on the Cys536 sulfur atom of the Cys512-Cys536 disulfide bond, resulting in reduction of the bond. Cleavage of the Cys512-Cys536 disulfide bond leads to a conformational change in plasmin and exposure of the peptide backbone to proteolysis on the C-terminal side of residues 468, 474, and 530.
FIGURE 5.
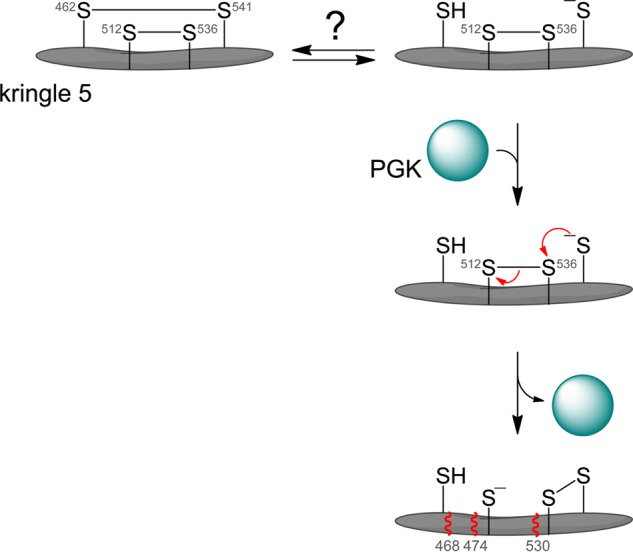
Model for the mechanism of angiostatin formation from plasmin(ogen). The Cys462-Cys541 disulfide bond is reduced in a fraction of plasminogen in blood or in the injured vascular wall. We propose that binding of PGK to plasmin induces a conformational change in kringle 5 that leads to attack by the Cys541 thiolate anion (present as a certain percentage of all thiols by action of the buffer) on the Cys536 sulfur atom of the Cys512-Cys536 disulfide bond, resulting in reduction of the bond. Cleavage of the disulfide allows further conformational change and exposure of the peptide backbone to proteolysis C-terminal of residues 468, 474, and 530 (represented by wavy red lines).
Plasmin-binding proteins other than PGK can apparently facilitate the same thiol/disulfide exchange events in plasmin kringle 5. Incubation of annexin A3-S100A10 heterotetramer (24), plasma-membrane associated β-actin (25), or a truncated porcine plasminogen activator inhibitor 1 (residues 80–265) (26) with plasmin results in generation of kringle-containing angiostatin fragments that are indistinguishable from those generated from incubation of plasmin with PGK. Plasmin also undergoes autoproteolysis at alkaline pH, producing kringle fragments and microplasmin that has a Lys531 N terminus (33–35). We have suggested that plasmin reduction and proteolysis at alkaline pH are mediated by the same chemical events facilitated by PGK at neutral pH (4). Reduction of disulfide bonds by oxidoreductases or by thiol/disulfide exchange is influenced by the pKa and electrostatic milieu of the attacking cysteine thiol (36, 37). It is possible that alkaline pH facilitates angiostatin formation by simply increasing the fraction of Cys541 in the thiolate anion state, which is the state that attacks the Cys512-Cys536 disulfide bond leading to angiostatin release.
Recombinant C462A mutant plasminogen was expressed in insect cells to further test this mechanism. The mutant was predicted to represent reduced plasminogen and, therefore, should be fully converted to the kringle fragments. However, the protein degraded prior to purification from the conditioned medium in three separate attempts. The instability of this mutant is perhaps not surprising considering the range of other factors that can facilitate attack of the Cys512-Cys536 disulfide bond by the Cys541 thiolate (Fig. 5).
The two labile plasmin disulfide bonds have configurations that are shared by allosteric disulfides in other proteins. The geometry of a disulfide bond is defined by the six atoms linking the two α-carbons of the cysteine residues (6). These six atoms plus the amide nitrogen attached to the α-carbons are used to define the five χ angles of the cystine residue, which are calculated by the rotation around the bonds linking the atoms. There are 20 different disulfide bond configurations based on the sign of each χ angle (6, 7). The three main types of bond classification are the spirals, hooks, and staples, and disulfides are either right- or left-handed (RH or LH) depending on whether the sign of the central χ angle is positive or negative, respectively. The −RHStaple is the most common allosteric configuration to date (8, 9).
The plasminogen Cys462-Cys541 and Cys512-Cys536 disulfide bonds have −/+RHHook and −LHHook configurations, respectively. There are three other known allosteric disulfides with a −/+RHHook configuration and three with a −LHHook configuration (Table 4). The −/+RHHooks are in β2-glycoprotein I, angiotensinogen, and factor XI, whereas the −LHHooks are in interleukin receptor subunit γ, XRCC1, and vascular endothelial growth factors C and D. There are parallels between the plasminogen allosteric bonds and the functional roles of these other allosteric disulfides.
β2-Glycoprotein I is a blood protein involved in thrombosis and is the autoantigen in the antiphospholipid syndrome (38). The thrombosis in this syndrome is associated with accelerated atherosclerosis and recurrent miscarriages. The Cys288-Cys326 −/+RHHook disulfide bond in the fifth complement module domain is reduced in a fraction of the protein in blood (39, 40). There is more oxidized β2-glycoprotein I in the plasma of patients with antiphospholipid syndrome, which is associated with increased immunogenicity of the protein and increased thrombosis (41). Blood angiotensinogen is cleaved by renin and angiotensin-converting enzyme to produce angiotensin peptides that control blood pressure and fluid homeostasis (42). The Cys18-Cys138 −/+RHHook disulfide bond is reduced in a fraction of plasma angiotensinogen (43). There is more oxidized angiotensinogen in the plasma of pregnant females with preeclampsia (43), which results in increased cleavage by renin and elevated blood pressure. Proteolytic activation of factor XI to XIa initiates the intrinsic/consolidation phase of blood coagulation. The Cys362-Cys482 −/+RHHook disulfide bond is reduced in a fraction of plasma factor XI (44). The reduced protein is more efficiently activated by thrombin, factor XIIa, or factor XIa than the oxidized protein. Higher levels of reduced factor XI are found in the blood of patients with antiphospholipid syndrome, which may contribute to the thrombosis in this syndrome. These examples mirror the situation with plasminogen described here. The kringle 5 −/+RHHook disulfide bond is reduced in a fraction of plasma plasminogen, and the reduced protein has properties different from those of the oxidized protein.
The interleukin receptor subunit γ (CD132) is the common γ chain of the cytokine receptors for IL-2, -4, -7, -9, -15, and -21 on the surface of lymphocytes (45). The Cys183-Cys232 −LHHook disulfide bond in CD132 is reduced on the surface of activated thymocytes in mice following an inflammatory challenge (46). Cleavage of the disulfide inhibits IL-2 binding to the receptor complex and, therefore, receptor signaling and cell proliferation. XRCC1 is a DNA repair protein that forms a complex with DNA polymerase β and mediates DNA base excision repair. The Cys12-Cys20 −LHHook disulfide bond can be reduced in XRCC1, which results in higher affinity for DNA polymerase β (47). Vascular endothelial growth factor C and D (VEGF-C, -D) binding to the tyrosine kinase receptor VEGFR-3 is involved in lymphangiogenesis in normal and pathological conditions (48). VEGF-C and -D form disulfide-linked antiparallel homodimers that is important for their productive interaction with their receptors. Dimer formation of these VEGFs is regulated by an unpaired cysteine residue (Cys137 in VEGF-C) that is only a few Å from the interdimer −LHHook disulfide bond (Cys156-Cys165 in VEGF-C) (49, 50). The current evidence implies that the unpaired cysteine residue cleaves the interdimer disulfide bond of VEGF-C and -D by thiol/disulfide exchange, which inhibits the activity of these VEGFs (51, 57). The chemical events in VEGF-C and -D are similar to what is occurring in plasmin. In both cases, an unpaired cysteine thiol is cleaving the −LHHook allosteric bond by thiol/disulfide exchange.
This analysis indicates that the −/+RHHook or −LHHook bonds, in addition to the −RHStaple, are potential allosteric configurations. This information will facilitate the identification of allosteric disulfides in other protein structures.
Acknowledgments
We thank Jason Wong for assistance with the design and analysis of the mass spectrometry experiments. Protein structure figures were generated using PyMOL.
This work was supported by grants from the National Health and Medical Research Council and the Cancer Council New South Wales.
- PGK
- phosphoglycerate kinase
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid)
- ITC
- 2-iminothiazolidine-4-carboxylyl
- K
- kringle
- LH
- left-handed
- MPB
- 3-(N-maleimidylpropionyl)biocytin
- NTCB
- 2-nitro-5-thiocyanobenzoic acid
- RH
- right-handed.
REFERENCES
- 1. Lijnen H. R. (2001) Elements of the fibrinolytic system. Ann. N.Y. Acad. Sci. 936, 226–236 [DOI] [PubMed] [Google Scholar]
- 2. O'Reilly M. S., Holmgren L., Shing Y., Chen C., Rosenthal R. A., Moses M., Lane W. S., Cao Y., Sage E. H., Folkman J. (1994) Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 79, 315–328 [DOI] [PubMed] [Google Scholar]
- 3. Stathakis P., Fitzgerald M., Matthias L. J., Chesterman C. N., Hogg P. J. (1997) Generation of angiostatin by reduction and proteolysis of plasmin: catalysis by a plasmin reductase secreted by cultured cells. J. Biol. Chem. 272, 20641–20645 [DOI] [PubMed] [Google Scholar]
- 4. Stathakis P., Lay A. J., Fitzgerald M., Schlieker C., Matthias L. J., Hogg P. J. (1999) Angiostatin formation involves disulfide bond reduction and proteolysis in kringle 5 of plasmin. J. Biol. Chem. 274, 8910–8916 [DOI] [PubMed] [Google Scholar]
- 5. Hogg P. J. (2003) Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28, 210–214 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt B., Ho L., Hogg P. J. (2006) Allosteric disulfide bonds. Biochemistry 45, 7429–7433 [DOI] [PubMed] [Google Scholar]
- 7. Schmidt B., Hogg P. J. (2007) Search for allosteric disulfide bonds in NMR structures. BMC Struct. Biol. 7, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cook K. M., Hogg P. J. (2013) Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 18, 1987–2015 [DOI] [PubMed] [Google Scholar]
- 9. Hogg P. J. (2013) Targeting allosteric disulphide bonds in cancer. Nat. Rev. Cancer 13, 425–431 [DOI] [PubMed] [Google Scholar]
- 10. Lay A. J., Jiang X. M., Daly E., Sun L., Hogg P. J. (2002) Plasmin reduction by phosphoglycerate kinase is a thiol-independent process. J. Biol. Chem. 277, 9062–9068 [DOI] [PubMed] [Google Scholar]
- 11. Lay A. J., Jiang X. M., Kisker O., Flynn E., Underwood A., Condron R., Hogg P. J. (2000) Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature 408, 869–873 [DOI] [PubMed] [Google Scholar]
- 12. Daly E. B., Wind T., Jiang X. M., Sun L., Hogg P. J. (2004) Secretion of phosphoglycerate kinase from tumour cells is controlled by oxygen-sensing hydroxylases. Biochim. Biophys. Acta 1691, 17–22 [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Wang J., Dai J., Jung Y., Wei C. L., Wang Y., Havens A. M., Hogg P. J., Keller E. T., Pienta K. J., Nor J. E., Wang C. Y., Taichman R. S. (2007) A glycolytic mechanism regulating an angiogenic switch in prostate cancer. Cancer Res. 67, 149–159 [DOI] [PubMed] [Google Scholar]
- 14. Wang J., Ying G., Jung Y., Lu J., Zhu J., Pienta K. J., Taichman R. S. (2010) Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 70, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Zieker D., Königsrainer I., Tritschler I., Löffler M., Beckert S., Traub F., Nieselt K., Bühler S., Weller M., Gaedcke J., Taichman R. S., Northoff H., Brücher B. L., Königsrainer A. (2010) Phosphoglycerate kinase 1 a promoting enzyme for peritoneal dissemination in gastric cancer. Int. J. Cancer 126, 1513–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castellino F. J., Powell J. R. (1981) Human plasminogen. Methods Enzymol. 80, 365–378 [DOI] [PubMed] [Google Scholar]
- 17. Lay A. J., Hogg P. J. (2002) Measurement of reduction of disulfide bonds in plasmin by phosphoglycerate kinase. Methods Enzymol. 348, 87–92 [DOI] [PubMed] [Google Scholar]
- 18. Riddles P. W., Blakeley R. L., Zerner B. (1983) Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 [DOI] [PubMed] [Google Scholar]
- 19. Sottrup-Jensen L., Claeys H., Zajdel M., Petersen T. E., Magnusson S. (1978) in Progress in Chemical Fibrinolysis and Thrombolysis (Davidson J. F., Rowan R. M., Samama M. M., Desnoyers P. C., eds) Vol. 3, pp. 191–209, Raven Press, New York [Google Scholar]
- 20. Jacobson G. R., Schaffer M. H., Stark G. R., Vanaman T. C. (1973) Specific chemical cleavage in high yield at the amino peptide bonds of cysteine and cystine residues. J. Biol. Chem. 248, 6583–6591 [PubMed] [Google Scholar]
- 21. Wu J., Gage D. A., Watson J. T. (1996) A strategy to locate cysteine residues in proteins by specific chemical cleavage followed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 235, 161–174 [DOI] [PubMed] [Google Scholar]
- 22. Ganderton T., Wong J. W., Schroeder C., Hogg P. J. (2011) Lateral self-association of VWF involves the Cys2431-Cys2453 disulfide/dithiol in the C2 domain. Blood 118, 5312–5318 [DOI] [PubMed] [Google Scholar]
- 23. Wong J. W., Hogg P. J. (2010) Analysis of disulfide bonds in protein structures. J. Thromb. Haemost. 8, 2345. [DOI] [PubMed] [Google Scholar]
- 24. Kwon M., Caplan J. F., Filipenko N. R., Choi K. S., Fitzpatrick S. L., Zhang L., Waisman D. M. (2002) Identification of annexin II heterotetramer as a plasmin reductase. J. Biol. Chem. 277, 10903–10911 [DOI] [PubMed] [Google Scholar]
- 25. Wang H., Schultz R., Hong J., Cundiff D. L., Jiang K., Soff G. A. (2004) Cell surface-dependent generation of angiostatin4.5. Cancer Res. 64, 162–168 [DOI] [PubMed] [Google Scholar]
- 26. Mulligan-Kehoe M. J., Wagner R., Wieland C., Powell R. (2001) A truncated plasminogen activator inhibitor-1 protein induces and inhibits angiostatin (kringles 1–3), a plasminogen cleavage product. J. Biol. Chem. 276, 8588–8596 [DOI] [PubMed] [Google Scholar]
- 27. Torchinski Y. M. (1974) in Thiol and Disulfide Groups of Proteins (Dixon H. B., ed.), p. 24, Consultants Bureau, New York [Google Scholar]
- 28. Law R. H., Caradoc-Davies T., Cowieson N., Horvath A. J., Quek A. J., Encarnacao J. A., Steer D., Cowan A., Zhang Q., Lu B. G., Pike R. N., Smith A. I., Coughlin P. B., Whisstock J. C. (2012) The X-ray crystal structure of full-length human plasminogen. Cell Rep. 1, 185–190 [DOI] [PubMed] [Google Scholar]
- 29. Schwertassek U., Balmer Y., Gutscher M., Weingarten L., Preuss M., Engelhard J., Winkler M., Dick T. P. (2007) Selective redox regulation of cytokine receptor signaling by extracellular thioredoxin-1. EMBO J. 26, 3086–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura H. (2005) Thioredoxin and its related molecules: update 2005. Antioxid. Redox Signal. 7, 823–828 [DOI] [PubMed] [Google Scholar]
- 31. Yamada Y., Nakamura H., Adachi T., Sannohe S., Oyamada H., Kayaba H., Yodoi J., Chihara J. (2003) Elevated serum levels of thioredoxin in patients with acute exacerbation of asthma. Immunol. Lett. 86, 199–205 [DOI] [PubMed] [Google Scholar]
- 32. Lillig C. H., Holmgren A. (2007) Thioredoxin and related molecules: from biology to health and disease. Antioxid. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 33. Kassam G., Kwon M., Yoon C. S., Graham K. S., Young M. K., Gluck S., Waisman D. M. (2001) Purification and characterization of A61: an angiostatin-like plasminogen fragment produced by plasmin autodigestion in the absence of sulfhydryl donors. J. Biol. Chem. 276, 8924–8933 [DOI] [PubMed] [Google Scholar]
- 34. Wu H. L., Shi G. Y., Bender M. L. (1987) Preparation and purification of microplasmin. Proc. Natl. Acad. Sci. U.S.A. 84, 8292–8295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu H. L., Shi G. Y., Wohl R. C., Bender M. L. (1987) Structure and formation of microplasmin. Proc. Natl. Acad. Sci. U.S.A. 84, 8793–8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chivers P. T., Raines R. T. (1997) General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry 36, 15810–15816 [DOI] [PubMed] [Google Scholar]
- 37. Hansen R. E., Østergaard H., Winther J. R. (2005) Increasing the reactivity of an artificial dithiol-disulfide pair through modification of the electrostatic milieu. Biochemistry 44, 5899–5906 [DOI] [PubMed] [Google Scholar]
- 38. Giannakopoulos B., Krilis S. A. (2013) The pathogenesis of the antiphospholipid syndrome. N. Engl. J. Med. 368, 1033–1044 [DOI] [PubMed] [Google Scholar]
- 39. Ioannou Y., Zhang J. Y., Passam F. H., Rahgozar S., Qi J. C., Giannakopoulos B., Qi M., Yu P., Yu D. M., Hogg P. J., Krilis S. A. (2010) Naturally occurring free thiols within β2-glycoprotein I in vivo: nitrosylation, redox modification by endothelial cells, and regulation of oxidative stress-induced cell injury. Blood 116, 1961–1970 [DOI] [PubMed] [Google Scholar]
- 40. Passam F. H., Rahgozar S., Qi M., Raftery M. J., Wong J. W., Tanaka K., Ioannou Y., Zhang J. Y., Gemmell R., Qi J. C., Giannakopoulos B., Hughes W. E., Hogg P. J., Krilis S. A. (2010) β2-Glycoprotein I is a substrate of thiol oxidoreductases. Blood 116, 1995–1997 [DOI] [PubMed] [Google Scholar]
- 41. Ioannou Y., Zhang J. Y., Qi M., Gao L., Qi J. C., Yu D. M., Lau H., Sturgess A. D., Vlachoyiannopoulos P. G., Moutsopoulos H. M., Rahman A., Pericleous C., Atsumi T., Koike T., Heritier S., Giannakopoulos B., Krilis S. A. (2011) Novel assays of thrombogenic pathogenicity in the antiphospholipid syndrome based on the detection of molecular oxidative modification of the major autoantigen β2-glycoprotein I. Arthritis Rheum. 63, 2774–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crowley S. D., Coffman T. M. (2012) Recent advances involving the renin-angiotensin system. Exp. Cell Res. 318, 1049–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou A., Carrell R. W., Murphy M. P., Wei Z., Yan Y., Stanley P. L., Stein P. E., Broughton Pipkin F., Read R. J. (2010) A redox switch in angiotensinogen modulates angiotensin release. Nature 468, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giannakopoulos B., Gao L., Qi M., Wong J. W., Yu D. M., Vlachoyiannopoulos P. G., Moutsopoulos H. M., Atsumi T., Koike T., Hogg P., Qi J. C., Krilis S. A. (2012) Factor XI is a substrate for oxidoreductases: enhanced activation of reduced FXI and its role in antiphospholipid syndrome thrombosis. J. Autoimmun. 39, 121–129 [DOI] [PubMed] [Google Scholar]
- 45. Waldmann T. A. (2006) The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 6, 595–601 [DOI] [PubMed] [Google Scholar]
- 46. Metcalfe C., Cresswell P., Barclay A. N. (2012) Interleukin-2 signalling is modulated by a labile disulfide bond in the CD132 chain of its receptor. Open Biol. 2, 110036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cuneo M. J., London R. E. (2010) Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase β binding affinity. Proc. Natl. Acad. Sci. U.S.A. 107, 6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lohela M., Bry M., Tammela T., Alitalo K. (2009) VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr. Opin. Cell Biol. 21, 154–165 [DOI] [PubMed] [Google Scholar]
- 49. Toivanen P. I., Nieminen T., Viitanen L., Alitalo A., Roschier M., Jauhiainen S., Markkanen J. E., Laitinen O. H., Airenne T. T., Salminen T. A., Johnson M. S., Airenne K. J., Ylä-Herttuala S. (2009) Novel vascular endothelial growth factor D variants with increased biological activity. J. Biol. Chem. 284, 16037–16048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leppänen V. M., Prota A. E., Jeltsch M., Anisimov A., Kalkkinen N., Strandin T., Lankinen H., Goldman A., Ballmer-Hofer K., Alitalo K. (2010) Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc. Natl. Acad. Sci. U.S.A. 107, 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davydova N., Streltsov V. A., Roufail S., Lovrecz G. O., Stacker S. A., Adams T. E., Achen M. G. (2012) Preparation of human vascular endothelial growth factor-D for structural and preclinical therapeutic studies. Protein Expr. Purif. 82, 232–239 [DOI] [PubMed] [Google Scholar]
- 52. Passam F. H., Rahgozar S., Qi M., Raftery M. J., Wong J. W., Tanaka K., Ioannou Y., Zhang J. Y., Gemmell R., Qi J. C., Giannakopoulos B., Hughes W. E., Hogg P. J., Krilis S. A. (2010) Redox control of β2-glycoprotein I-von Willebrand factor interaction by thioredoxin-1. J. Thromb. Haemost. 8, 1754–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leppänen V. M., Jeltsch M., Anisimov A., Tvorogov D., Aho K., Kalkkinen N., Toivanen P., Ylä-Herttuala S., Ballmer-Hofer K., Alitalo K. (2011) Structural determinants of vascular endothelial growth factor-D receptor binding and specificity. Blood 117, 1507–1515 [DOI] [PubMed] [Google Scholar]
- 54. LaPorte S. L., Juo Z. S., Vaclavikova J., Colf L. A., Qi X., Heller N. M., Keegan A. D., Garcia K. C. (2008) Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stauber D. J., Debler E. W., Horton P. A., Smith K. A., Wilson I. A. (2006) Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang X., Rickert M., Garcia K. C. (2005) Structure of the quaternary complex of interleukin-2 with its α, β, and γc receptors. Science 310, 1159–1163 [DOI] [PubMed] [Google Scholar]
- 57. Chiu J., Wong J. W., Gerometta M., Hogg P. J. (2013) Mechanism of dimerisation of a recombinant mature vascular endothelial growth factor C. Biochemistry. 10.1021/bi401518b [DOI] [PubMed] [Google Scholar]