Abstract
Spinocerebellar ataxia-3 (SCA3) (also known as Machado Joseph Disease) is an incurable neurodegenerative disorder caused by expression of a mutant variant of ataxin-3 protein (ATX3). Inhibiting expression of ATX-3 would provide a therapeutic strategy, but indiscriminant inhibition of both wild-type and mutant ATX3 might lead to undesirable side-effects. An ideal silencing agent would block expression of mutant ATX3 while leaving expression of wild-type ATX3 intact. We have previously observed that peptide nucleic acid (PNA) conjugates targeting the expanded CAG repeat within ATX3 mRNA block expression of both alleles. We now identify additional PNAs capable of inhibiting ATX3 expression that vary in length and in the nature of the conjugated cation chain. We can also achieve potent and selective inhibition using duplex RNAs containing one or more mismatches relative to the CAG repeat. Anti-CAG antisense bridged nucleic acid (BNA) oligonucleotides that lack a cationic domain are potent inhibitors but are not allele-selective. Allele-selective inhibitors of ATX-3 expression provide insights into the mechanism of selectivity and promising lead compounds for further development and in vivo investigation.
Keywords: Ataxin-3, Spinocerebellar ataxia-3, allele-selective, siRNA, peptide nucleic acid
Introduction
Spinocerebellar ataxia type 3 (SCA3, Machado Joseph Disease) is a progressive neurological disorder (Paulson, 2007a; Paulson, 2007b). SCA3 is caused by heterozygous mutations within one allele of the ataxin 3 (ATX3) gene. The ATX3 gene contains a tract with multiple copies of the trinucleotide CAG. In unaffected individuals, this tract is typically less than 31 repeats. Individuals with 45 to 51 repeats sometimes show symptoms but disease penetrance is incomplete. When greater than 52 repeats are present, there is full penetrance. Patients can have as many as 86 CAG repeats within the mutant allele. Clinical symptoms are affected by repeat size and mean repeat length can vary from 73 to 80 repeats in different patient populations (Sasaki, 1995).
The symptoms of SCA3 are severe (Paulson 2007). Typically these symptoms begin to be observed in patients over fifty, but are also noted in younger individuals and age of onset correlates with the number of mutant repeats. Patients can have problems with walking, speech, and blurred vision. Symptoms worsen over ten to fifteen years and patients may require wheelchairs or other devices to maintain mobility. The patient’s conditions deteriorate over time and death from pulmonary complications can occur.
One approach to treating SCA3 would be to inhibit ATX3 protein expression, removing the cause of the disease and slowing or preventing its progression. Supporting this conclusion, in a conditional mouse model of SCA3 turning off ATX3 expression early in the disease state yielded a phenotype that was indistinguishable from wild-type mice (Boy et al., 2009).
One approach to reducing levels of ATX3 protein is to use duplex RNAs or antisense oligonucleotides complementary to ATX3 mRNA. Researchers have identified antisense oligonucleotides and duplex RNAs targeting mRNAs for huntingtin (HTT) (Huntington’s Disease), ATX3, and other triplet repeat-containing genes (Gonzalez-Alegre and Paulson, 2007; Denovan-Wright and Davidson 2006; Scholefield and Wood, 2010). Other elegant studies using antisense oligonucleotides have shown that blocking the long (>500 repeat) CUG repeat in the DMPK (Myotonic Dystrophy) gene can limit aberrant muscleblind protein binding to the expanded repeat region (Mulders et al., 2009; Wheeler et al., 2009).
Most trinucleotide repeat expansion diseases are autosomal dominant conditions caused by expression of a mutant allele. A key consideration for nucleic acid-based therapy is whether inhibition of both alleles can be achieved without undue toxicity due to reduced expression of wild-type protein. For ATX3, one recent report suggests that inhibiting expression of both the mutant and wild-type alleles did not cause observable toxicity, suggesting that approaches for therapy that reduce expression of both alleles might be feasible (Alves et al., 2010).
There is no guarantee, however, that successfully inhibiting both alleles in mice will translate into successful treatments for humans. Preferential inhibition of the mutant allele may be beneficial and allele-selective strategies have the potential for fewer side effects in patients. To achieve allele-selective inhibition, Paulson targeted a duplex RNA to a single-nucleotide polymorphism linked to SCA3 (Miller et al., 2003). Subsequently, Pereira de Almeida (Alves et al., 2008) observed that targeting siRNAs to a SNP found in 70 % of patients with SCA3 led to allele-selective inhibition.
A fundamental difference between the wild-type and mutant alleles of all patients is the number of CAG repeats. CAG repeats are known to form hairpin structures when analyzed in cell free systems (Sobczak et al. 2003; Kiliszek et al., 2010). In the context of a complete cellular mRNA, these hairpins might differ significantly in structure depending on the number of CAG repeats present. We reasoned that short single-stranded oligomers complementary to CAG repeats might take advantage of differences in RNA structure between wild-type and mutant CAG repeat tracts, selectively recognize the mutant repeat region, and block expression of the mutant protein while leaving expression of the wild-type protein relatively unchanged.
We tested this hypothesis by targeting peptide nucleic acids (PNAs) to CAG repeat tracts in fibroblast cell lines derived from patients with SCA3 and a related triplet expansion disease, Huntington’s Disease (HD) caused by expanded CAG repeats within the huntingtin (HTT) gene (Hu et al., 2009a; Hu et al. 2009b; Hu et al., 2009c). PNA is a DNA/RNA mimic in which nucleotide bases are linked by an amide backbone (Nielsen et al., 1991). PNA is known to be able to recognize complementary sequences within structured RNAs (Marin and Armitage, 2005), making it a promising starting point for studies investigating recognition of CAG hairpins. Anti-CAG PNAs inhibited expression of both mutant genes and did not cause toxicity or affect expression of other cellular genes containing triplet repeats. We subsequently observed that single-stranded oligonucleotides (Hu et al., 2009a; Gagnon et al., 2010) and mismatch- containing duplex RNAs (Hu et al., 2010) could also be allele-selective inhibitors of HTT expression. We now report inhibition of ATX-3 by additional PNA derivatives and by double-stranded RNAs designed to achieve allele-selective inhibition by shifting the mechanism used during RNAi.
Results
Experimental Design
Our experiments employ a patient-derived fibroblast cell line GM06151 that is heterozygous for mutant ATX3. The wild-type allele contains 24 repeats and the mutant allele has 74 repeats. The 74 repeats within the mutant allele are typical of the repeat number found in SCA3 patients (Sasaki et al., 1995) making GM06151 cells a good test for the applicability of the approach to most patients.
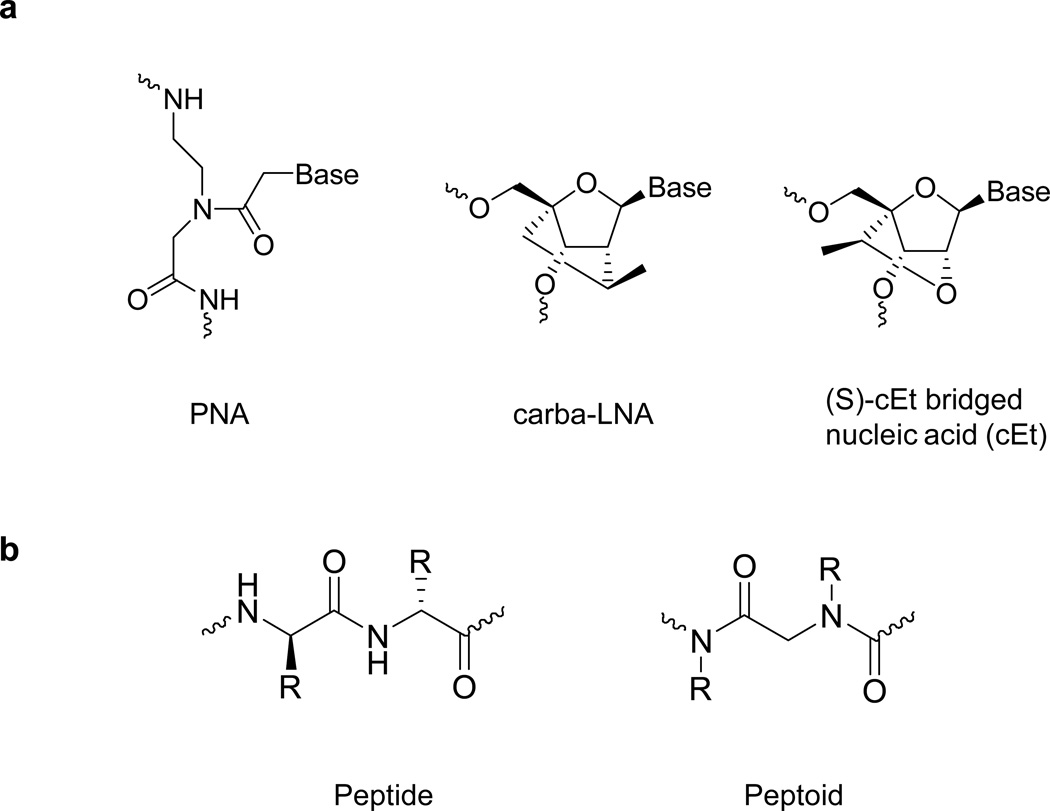
PNAs do not spontaneously enter cultured cells so cationic moieties were attached to them to facilitate uptake. Several different peptide or peptoid conjugates were tested. Single-stranded bridged nucleic acid (BNA) oligonucleotides were introduced using PepMute, a peptide-based transfection reagent. Duplex RNAs were transfected into cells using cationic lipid by standard protocols (Janowski et al., 2006). Structures of PNA, BNA, and peptide and peptoid transporters are shown in Figure 1.
Figure 1.
Chemical structures of (a) PNA, carba-LNA and cEt; (b) peptide and peptoid.
Effect of changing PNA length
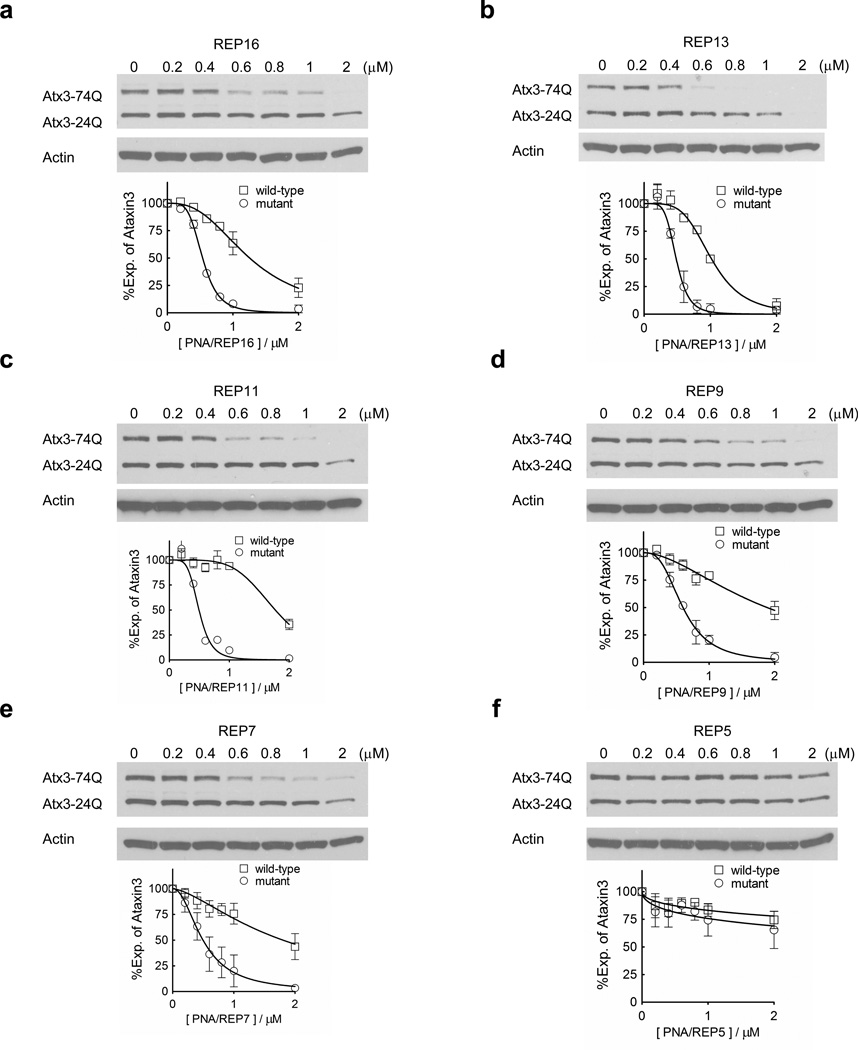
Inhibition by shorter PNAs would provide additional lead compounds for therapeutic development and reveal important details of the mechanism of selective inhibition. We synthesized PNAs of varied lengths and assayed inhibition of ATX3 expression. All PNAs used for these initial comparisons were conjugated to the peptide d-K8 (eight lysines in the d configuration) at the PNA C-terminus by a standard peptide amide linkage (unlike nucleic acids, PNAs are made by peptide synthesis through amide bonds and possess N- and C- termini). The d-K8 peptide was chosen because previous studies had shown that it combined efficient cellular uptake and a simple synthesis using a single relatively inexpensive amino acid (Hu and Corey, 2007).
PNAs of varied lengths achieved potent and selective inhibition. PNAs that were 16, 13, 11, 9, and 7 bases long inhibited ATX3 expression with similar IC50 values of 0.5–0.6 µM (Figure 2, Table 1). These IC50 values are similar to the value for the parent 19-base PNA conjugate (0.36 µM). Selectivities for inhibiting the mutant versus the wild-type allele ranged from 2.4 to 3.6 fold, also similar to the value of allele selectivity for the parent 19 base PNA conjugate (2.7 fold). A 5 base PNA did not affect ATX3 expression, establishing a minimum size for allele-selective inhibition by PNA-peptide conjugates.
Figure 2. Effects of PNA length on ATX3 expression.
All PNAs are conjugated to peptide d-K8 at their C-terminus. (a–f) PNAs with varied length were tested in patient fibroblast cell line GM06151 (CAG 74 mutant repeats/24 wild-type repeats) at increasing concentrations. Representative western blot images are presented. Quantification and a nonlinear fitting curve of ATX3 expression is plotted from multiple experiments. Error bars are standard error of the mean (SEM).
Table 1.
PNA-peptide conjugates targeting the ATXN3 CAG repeat region.
| Name | Sequence (length) | MS Cal./Obs. |
Tm (°C) |
mut IC50 (µM) |
wt IC50 (µM) |
Selectivity (fold) |
|---|---|---|---|---|---|---|
| REP19 | K-GCTGCTGCTGCTGCTGCTG-K8 (19) | 6315/6320 | 85 | 0.36 ± 0.06 | 0.99 ± 0.09 | 2.7‡ |
| REP16 | K-GCTGCTGCTGCTGCTG-K8 (16) | 5506/5507 | 80 | 0.5 ± 0.1 | 1.2 ± 0.2 | 2.4 |
| REP13 | K-GCTGCTGCTGCTG-K8 (13) | 4697/4698 | 72 | 0.5 ± 0.1 | 1.1 ± 0.1 | 2.2 |
| REP11 | K-GCTGCTGCTGC-K8 (11) | 4139/4137 | 64 | 0.5 ± 0.1 | 1.7 ± 0.1 | 3.4 |
| REP9 | K-GCTGCTGCT-K8 (9) | 3597/3599 | 52 | 0.6 ± 0.1 | 1.8 ± 0.3 | 3.0 |
| REP7 | K-GCTGCTG-K8 (7) | 3079/3080 | 44 | 0.5 ± 0.3 | 1.8 ± 0.4 | 3.6 |
| REP5 | K-GCTGC-K8 (5) | 2522/2518 | - | ni | ni | - |
| REP19NK | K8-GCTGCTGCTGCTGCTGCTG-K (19) | 6315/6317 | 84 | 1.1 ± 0.5 | 3.8 ± 1.2 | 3.4 |
| REP19R | K-GCTGCTGCTGCTGCTGCTG-R8 (19) | 6538/6540 | >87 | 0.5 ± 0.1 | 0.8 ± 0.3 | 1.6 |
| REP19NR | R8-GCTGCTGCTGCTGCTGCTG-K (19) | 6538/6538 | >87 | 2.2 ± 0.2 | 5.9 ± 1.9 | 2.7 |
| REP7NK | K8-GCTGCTG-K (7) | 3079/3080 | 38 | 3.5 ± 0.9 | >6 | >1.7 |
| REP7R | K-GCTGCTG-R8 (7) | 3303/3304 | 46 | 0.7 ± 0.1 | >2 | 2.8 |
| REP7NR | R8-GCTGCTG-K (7) | 3303/3301 | 45 | 1.5 ± 0.3 | >2 | 1.3 |
| REP13K6 | K-GCTGCTGCTGCTG-K6 (13) | 4441/4442 | 71 | 0.8 ± 0.2 | 4.0 ± 0.4 | 5.0 |
| REP13K4 | K-GCTGCTGCTGCTG-K4 (13) | 4184/4182 | 71 | 2.3 ± 0.3 | 5.4 ± 0.9 | 2.3 |
| REP13KK | K8-GCTGCTGCTGCTG-K8 (13) | 5594/5593 | 74 | 0.08 ± 0.03 | 0.15 ± 0.08 | 1.8 |
| REP19S | K-GCTGCTGCTGCTGCTGCTG-Spacer-K8 (19) | 6605/6602 | >87 | 0.4 ± 0.1 | 0.7 ± 0.2 | 1.7 |
| REP19P | K-GCTGCTGCTGCTGCTGCTG-(Kpeptoid)8 (19) | 6314/6314 | >87 | 0.9 ± 0.4 | >2 | >2.2 |
PNAs are listed from N to C terminal. All PNAs contain a lysine residue at the N or C termini and a polypeptide attached at the other end. d-amino acids are used in all peptide conjugates. Selectivity is calculated by comparing the IC50 for inhibition of wild-type versus the IC50 for inhibition of mutant ataxin-3 protein. Error is standard deviation. The Tm value is measured from PNA with its complementary DNA sequence. Spacer is — (NCH2CH2OCH2CH2OCH2CO)2—. ni: no inhibition observed.
Values from Hue et al., 2009a.
We measured melting temperature (Tm) values to evaluate the relative affinities of the PNA conjugates for complementary sequences (Table 1). Tm values were determined for association with complementary DNA strands. Strikingly, REP7 and REP19 have similar potencies and selectivities even though the Tm value for REP7 is 44°C (Tm), approximately half the Tm value observed for REP19. A Tm value could not be measured for the inactive 5 base PNA conjugate, suggesting that its lack of potency was due to poor binding to its target sequence.
Because the amino acids are in the d configuration, it is likely that they are not efficiently hydrolyzed inside cells and may be present when the PNA recognizes its mRNA target. It is possible, therefore, that the attached cationic peptide may affect recognition of ATX mRNA. Interactions between the peptide domain and ATX3 mRNA may compensate for the lower binding potential of the relatively few PNA bases found in REP7 and be responsible for the observation that REP7 is as potent and selective as the much longer conjugate REP19.
The importance of the attached peptide for allele-selective inhibition by PNA conjugates is supported by our previous results that the nature of the peptide domain can alter selectivity even when the PNA domain is held constant (Hu et al., 2009a). When arginine, rather than lysine was used as a peptide domain, allele selectivity was lost. When peptide d-K8 is attached at the N-terminus rather than the C-terminus, selectivity is greatly enhanced. Taken together, data from inhibiting expression of ATX3 and HTT demonstrate that selectivity is a property of the entire conjugate, not just the domain designed to base-pair with the CAG repeat target.
Effect of changing the attached peptide
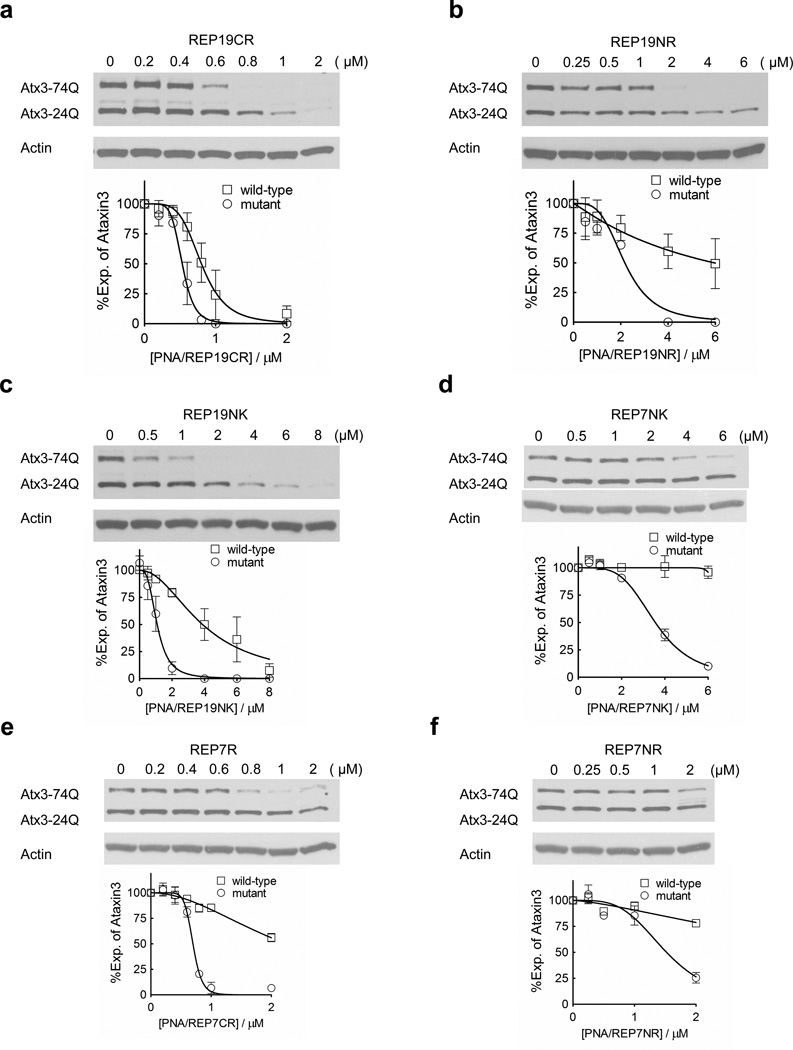
To further examine the effect of peptide attachment on potency and selectivity we varied the peptide domain of the PNA conjugate. We first tested the introduction of arginine residues in place of lysine. When an R8 peptide domain was attached to the C-terminus of a 19-base PNA (REP19R), we observed an IC50 value of 0.5 µM, similar to the IC50 value for the analogous lysine-containing domain (REP19) (Figure 3a). Selectivity for the REP19R, 1.6-fold, was reduced relative to REP19, 2.7-fold. As noted above, we had previously observed a similar reduction in selectivity for inhibition of HTT expression (Hu et al. 2010a). REP19R8 containing the R8 peptide at the N-terminus showed a decreased potency, 2.2 µM, but also a higher selectivity, 2.7-fold (Figure 3b).
Figure 3. Effects of peptide orientation and chemistry on ATX3 expression.
19-base PNAs (a–c) or 7-base PNAs (d–f) with 8 lysines or arginines at their N or C-terminus were tested in patient fibroblast cell line GM06151 (CAG 74 mutant repeats/24 wild-type repeats) at increasing concentrations. Representative western blot images are presented. Quantification and a nonlinear fitting curve of ATX3 expression is plotted from multiple experiments. Error bars are SEM.
When peptide d-K8 was attached to the N terminus, selectivity was 3.4 fold (Figure 3c). For HTT, the effect of switching the orientation of the peptide from the C to N terminus was much greater, >8-fold (Hu et al., 2009a). The finding that attaching peptides yields different results for ATX3 versus HTT expression suggests that they may be forming different interactions with the two mRNAs.
We hypothesized that short PNAs might be more sensitive to the identity of the peptide because the peptide domain makes a greater share of the conjugates’ interactions with its mRNA target. To test this hypothesis we synthesized conjugates between different peptides and a seven base PNA (Figure 3d–f, Table 1). The N-terminal lysine conjugate (REP7NK) had a lower potency (3.5 µM) than the C terminal lysine conjugate (REP7) (1.7 µM), the N-terminal arginine (REP7NR) (1.5 µM), or the C-terminal arginine (REP7R) (0.7 µM) conjugates. Conjugate REP7NK did not inhibit expression of wild-type ATX3 at any of the concentrations tested, suggesting that use of short PNAs might offer a route to higher selectivity.
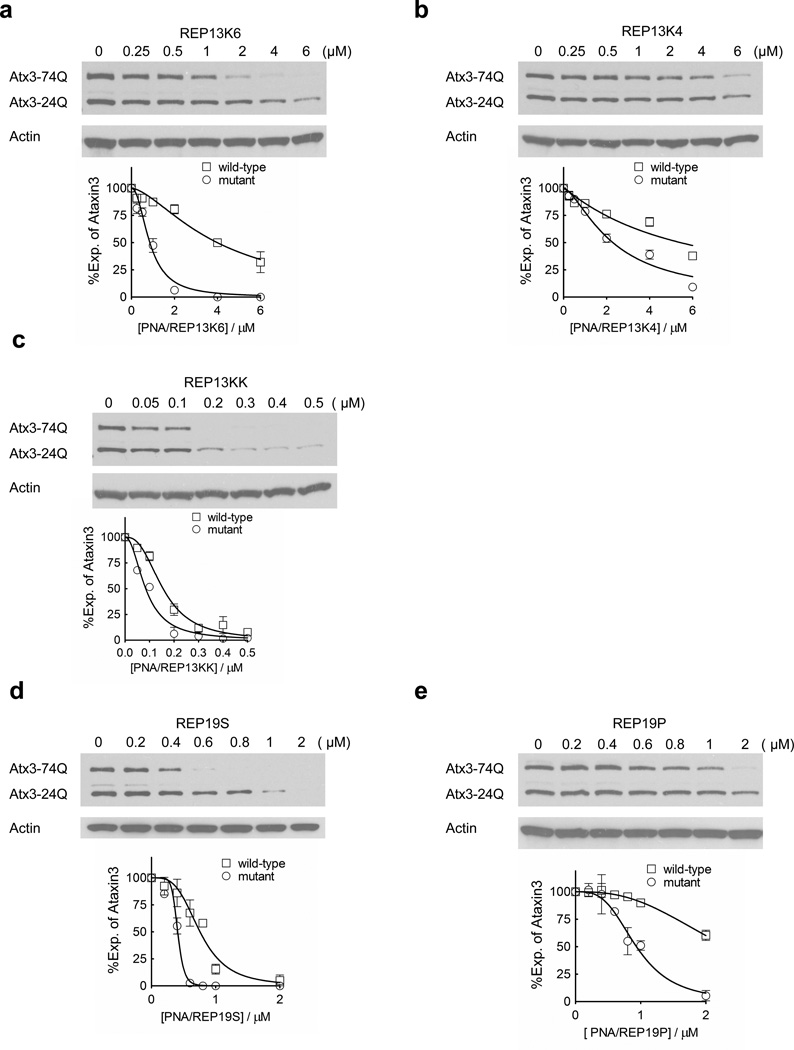
We also examined the effect of the number of positive charges or the chemical composition of the charged domain. Conjugate REP13K6 showed slightly reduced potency, 0.8 µM, but increased selectivity, 5-fold (Figure 4a). Potency for REP13K4 was further reduced to 2.3 µM (Figure 4b). The addition of positive charges had the opposite effect. REP13KK containing two d-K8 domains at the N and C termini was the most potent compound tested, 0.08 µM, but had reduced selectivity, 1.8-fold (Figure 4c). These data indicate that selectivity and potency can be adjusted by varying the length of the lysine conjugate.
Figure 4. Effects of PNAs with different conjugate designs on ATX3 expression.
13-base PNA conjugates (a–c) or 19-base PNA conjugates (d,e) were tested in patient fibroblasts GM06151 (CAG 74 mutant repeats/24 wild-type repeats) at increasing concentrations. Representative western blot images are presented. Quantification and a nonlinear fitting curve of ATX3 expression is plotted from multiple experiments. Error bars are SEM.
We altered the chemical structure of the cationic domain by introducing a spacer between the PNA and peptide (Figure 4d) or replacing the peptide with a peptoid (Figure 4e). Introducing a spacer between the peptide and PNA yielded a conjugate that was less selective, 1.7 fold. Attachment of peptoid (Kwon and Kodadek, 2007) with eight lysine-like residues yielded a conjugate that was less potent (0.9 µM) and possessed a selectivity of >2.2 fold. These data show that substantial alterations in how the positive charge is displayed yield relatively modest effects on potency and selectivity.
Effect of bridged nucleic acids
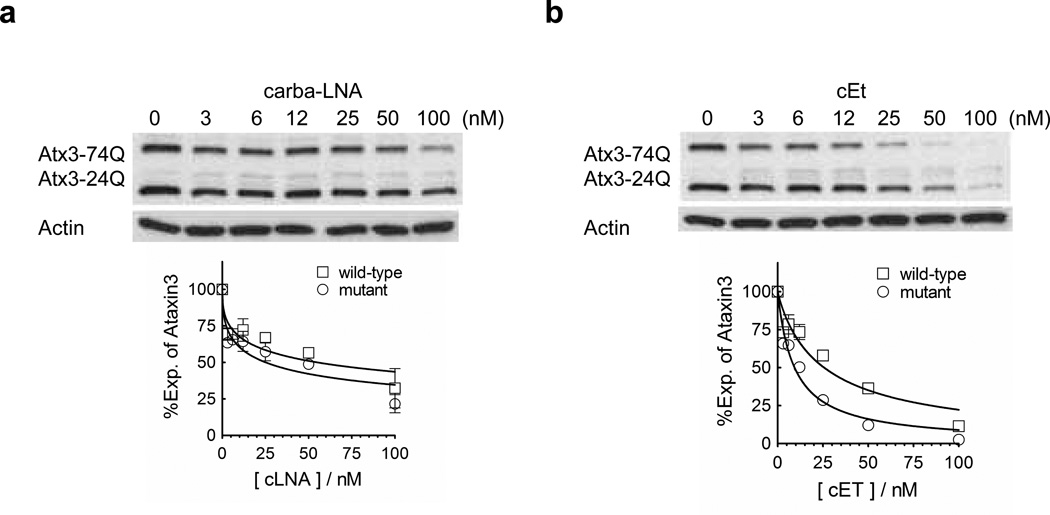
Bridged nucleic acids (BNAs) include nucleotides that contain a covalent linkage constraining the ribose ring between the 2’-O and 4’-C positions. We have reported that the introduction of locked nucleic acid (LNA) (Kumar et al. 1998; Obika et al. 1998; Braasch and Corey 2002) , carba LNA (Srivastava et al., 2007), and cET (Prakash et al., 2010) into anti-CAG oligonucleotides leads to potent and selective inhibition of HTT expression (Hu et al. 2009a; Gagnon et al. 2010). To test whether expression of ATX3 could be inhibited by anti-CAG oligonucleotides we transfected carba LNA and cET oligonucleotides into cells. In contrast to allele-selective inhibition of HTT, we observed potent but nonselective inhibition of both alleles of ATX3 (Figure 5a,b).
Figure 5. Effect of single stranded BNA oligonucleotides.
Effects on ATX3 protein levels after adding increasing amount of (a) carba-LNA, or (b) cET in fibroblasts GM06151 (CAG 74 mutant repeats/24 wild-type repeats). The BNAs were introduced into cells by PepMute transfection reagent. Representative western blot images are presented. Quantification and a nonlinear fitting curve of ATX3 expression is plotted from multiple experiments. Error bars are SEM.
Allele-selective inhibition by duplex RNAs
Double-stranded RNA is an alternate approach to gene silencing. We had previously shown that duplex RNAs that were fully complementary to the CAG repeat tract were potent but non-selective inhibitors of ATX3 expression (Hu et al., 2009a). Double-stranded RNA, however, can silence gene expression through two distinct mechanisms (Filipowicz et al., 2008; Kurreck et al., 2009). When duplex RNA is fully complementary to its mRNA target it is likely that the mRNA will be cleaved by the protein Argonaute 2 (AGO2) (Liu et al., 2004). However, if the RNA is imperfectly complementary, interactions at the AGO2 active site are disrupted (Du et al., 2005; Wang et al., 2008) and cleavage of the RNA is less likely. Reduced levels of protein will be due to inhibition of translation or increased degradation of mRNA. This strategy yielded potent and allele-selective inhibitors of HTT (Hu et al., 2010).
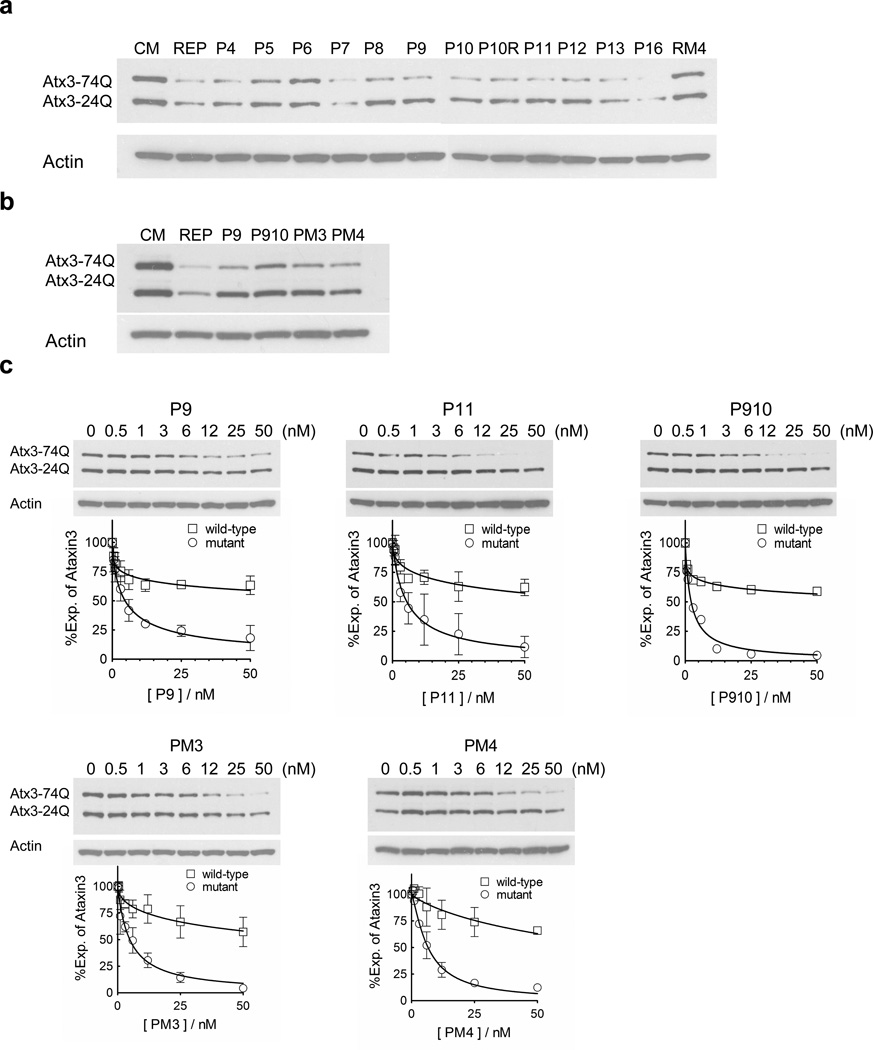
To test whether introducing mismatched bases might enhance allele-selectivity we tested anti-CAG duplexes containing mismatches at positions 4–13 and 16 (Figure 6a). With the exception of duplex RNA P6 (mutated at position 6), the duplex RNAs inhibited expression of ATX3. Duplex RNAs P8, P9, P10, P910, 10R, P11, and P12 appeared to be selective for inhibition of the mutant allele. Interestingly, P6 also failed to inhibit HTT expression, indicating that position 6 in the seed sequence is a critical determinant for recognition of the CAG target sequence. Mutations at other positions in the seed sequence are much less important.
Figure 6. Western analysis of ATX3 expression in fibroblasts GM06151 (CAG 74 mutant repeats/24 wild-type repeats) after treating with mismatch-containing siRNAs.
(a) Effects of 25 nM siRNAs on ATX3 expression. (b) Comparison of 25 nM siRNAs with different numbers of centrally mismatched bases. (c) siRNAs P9, P11 P910, PM3, and PM4 selectively inhibit mutant ATX3 expression. Representative gel images are presented. Quantification and a nonlinear fitting curve of ATX3 expression is plotted from multiple experiments. Error bars are SEM.
RNA duplexes that had two (P910), three (PM3), or four mismatched bases (PM4) also appeared to be selective (Figure 6b). IC50 values were similar regardless of whether the duplexes possessed two, three, or four mismatched bases (Figure 6c) and fold selectivities were all greater than 8-fold (Table 1). Potent and selective inhibition by a several different mismatch-containing duplexes provides many lead compounds for further development. If one compound is toxic or has unfavorable properties in vivo, another compound may substitute.
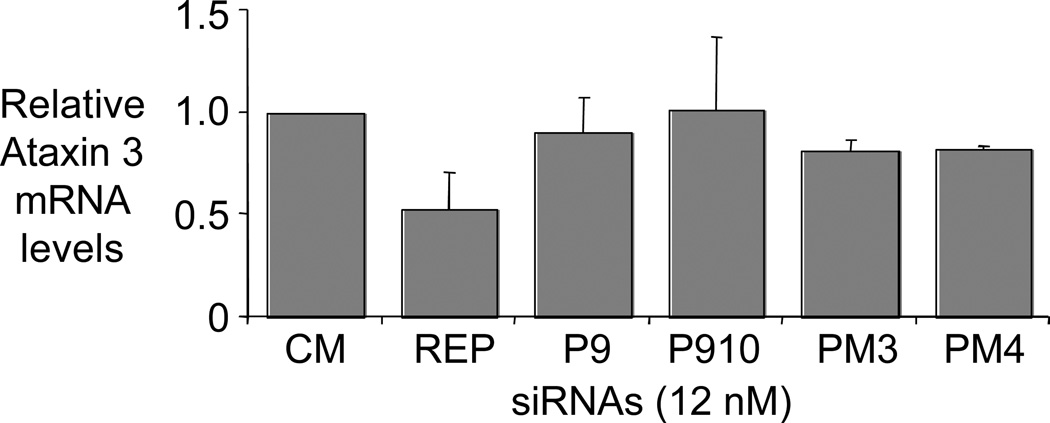
To test our hypothesis that the mismatch-containing duplex RNAs function by a mechanism that does not involve RNA cleavage we used quantitative PCR (qPCR) to determine the levels of ATX-3 mRNA (Figure 7). Duplex RNAs P9, P910, PM3, and PM4 caused little or no reduction in amounts of measured ATX-3 mRNA. Fully complementary duplex RNA REP reduced ATX-3 levels by 50 %. These data support the hypothesis that mismatch-containing mRNAs act by blocking translation rather than reducing RNA levels through cleavage of the mRNA target or by blocking transcription.
Figure 7. Selective siRNAs have little effects on ATX3 mRNA levels.
qPCR analysis of ATX3 mRNA levels after treatment with siRNAs at 12 nM.
Discussion
There are at least 16 diseases caused by expanded trinucleotide repeats (Orr and Zogbhi, 2007). These diseases are currently incurable and usually have severe neurological consequences for patients. The lack of adequate therapies makes development of effective drugs a priority and innovative approaches will be necessary to identify safe and efficacious agents.
One strategy for reducing levels of mutant genes is use of synthetic antisense oligonucleotides or duplex RNAs to silence their expression (Gonzalez-Alegre and Paulson, 2007; Denovan-Wright and Davidson 2006; Scholefield and Wood, 2010). These compounds are much larger than traditional small molecule (<700 daltons molecular weight) drugs but advances in delivery protocols are making them a viable approach for developing drugs to treat neurological disease. A Phase I trial using an antisense oligonucleotide designed to silence superoxide dismutase and delivered directly into the central nervous system has recently been initiated for amyotrophic lateral sclerosis (ALS) (isispharm.com). Gene silencing is an especially promising approach for neurological diseases like SCA3 or HD because they are caused by a single autosomal dominant mutation.
Here we examine three strategies for silencing ATX3 expression that target the expanded CAG repeat. Two strategies, mismatch-containing duplex RNAs and PNA-peptide conjugates led to potent and allele-selective inhibition of ATX3 expression. One strategy, antisense oligonucleotides, achieved potent inhibition without allele-selectivity.
We have now shown that two different genes with expanded trinucleotide repeats, ATX3 and HTT, can be selectively silenced by two different anti-CAG strategies, PNA-peptide conjugates and mismatch-containing siRNAs. For PNA-peptide conjugates the design of the peptide domain is critical for achieving selective inhibition. The length of the PNA is a relatively unimportant, with a wide range of lengths yielding the same result. This finding demonstrates that interactions beyond simple base-pairing at the CAG repeat are critical for optimizing allele-selectivity. The ability of the same compounds to silence two different disease genes suggests that it may be possible to develop a single agent that can treat multiple diseases.
A third anti-CAG approach, antisense oligonucleotides, achieved selective inhibition of HTT (Gagnon et al., 2010) but not ATX3. We observe inhibition of HTT expression but not ATX3 expression even though 1) the same oligonucleotides were used, 2) the target CAG sequence is preserved, and 3) the number of CAG repeats is similar (74 for ATX3, 69 for HTT). The most obvious explanation for the difference in selectivities is that the sequence of RNA surrounding the CAG repeat differs causing different RNA secondary structures and different potentials for contacts. In the future, approaches that optimize base-pairing to the CAG repeat and interactions with surrounding RNA structure may permit oligonucleotides to be selective for reducing mutant ATX3.
Materials and Methods
PNAs, BNAs and siRNAs
PNA-peptide conjugates were synthesized on an Expedite 8909 synthesizer (Applied Biosystems, Foster City, CA) and purified by C-18 reversed phase HPLC. Peptoid residue was manually synthesized first on the resins according to the published protocol (Hernando et al., 2002). The resins were loaded on the machine to continue synthesis of the PNA conjugate. carba-LNA was provided by Glen Research Corporation (Sterling, VA) (Srivastava et al., 2007) and cEt was synthesized by Isis Pharmaceuticals (Seth et al., 2010). siRNAs and DNA oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA).
Thermal Denaturation by UV Melt Analysis
Thermal denaturation analysis of BNA, PNA, or RNA-containing duplexes was carried out using a CARY Varian model 3 UV-Vis spectrophotometer. Absorbance was monitored at 260 nm in a 1 cm quartz cuvette. Oligomers were annealed (1 µM each strand) in 1× Dulbecco’s phosphate-buffered saline (Sigma Aldrich) and melted three times from 18°C to 99°C at a ramp rate of 2°C/min. To evaluate the concentration dependence of Tm, 0.1 cm quartz cuvettes were used with variable concentrations of oligonucleotide ranging from 0.5 to 50.0 µM. Absorbance was collected at one reading per 1°C. Tm was calculated using CARY WinUV Thermal Application software using a baseline fitting method.
Cell Culture and Transfection
Patient-derived fibroblast cell lines GM06151 were obtained from the Coriell Institute (Camden, NJ). Cells were maintained at 37 °C and 5% CO2 in MEM eagle media (Sigma, M4655) supplemented with 10% heat inactivated fetal bovine serum (Sigma) and 0.5% MEM nonessential amino acids (Sigma). Cells were plated in 6-well plates at 70,000 cells/well in supplemented MEM media two days prior to transfection. Stock solutions of PNA-peptide conjugates were heated at 65 °C for 5 min, then diluted to the appropriate concentration using OptiMEM (Invitrogen, Carlsbad, CA) and then added to cells. After 24 h, the media were removed and replaced by fresh supplemented MEM media. Cells were typically harvested 4 days after transfection for protein assay. LNAs were transfected to cells using PepMute transfection reagent (SignaGen, Ijamsville, MD) according to the manufacturer's instructions. LNAs were pre-heated at 65°C for five minutes reduce potential for aggregation before dilution with transfection buffer. siRNAs were introduced into cells by RNAiMAX (Invitrogen) (3 µL for 100 nM siRNA).
Analysis of ataxin-3 expression
Cells were harvested with trypsin-EDTA solution (Invitrogen). The protein concentration was quantified with BCA assay (Thermo Scientific, Waltham, MA). SDS-PAGE (7.5% acrylamide pre-cast gels; Bio-Rad) was used to separate the mutant and wild-type ataxin-3 protein. Primary antibodies for ataxin-3 (MAB5360; 1:10000; Chemicon), anti-β-actin antibody (1:10000; Sigma), HRP conjugate anti-mouse secondary antibody (1:10000; Jackson ImmunoResearch Laboratories, West Grove PA) were used for visualizing proteins by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Protein bands were quantified using ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2007). The percentage of inhibition was calculated relative to a control sample. The GraphPad Prism 4 program was used to generate the fitting curves for inhibition of ataxin-3. The following equation was used for fitting, y = 100-(100×m)/(nm+xm), where y is percentage of inhibition and×is the oligomer concentration, m and n are fitting parameters, where n is taken as the IC50 value. IC50 values were calculated from each experimental replicate and the standard deviation of these values is taken as the erro for the IC50.
Total RNA was extracted using TRIzol (Invitrogen) 3 days after transfection. After DNase I treatment, reverse transcription reactions were done using High Capacity Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Quantitative PCR was performed on a 7500 real-time PCR system (Applied Biosystems) using iTaq SYBR Green Supermix (Bio-rad). Data was normalized relative to levels of 18S mRNA. Primer sequences specific for ataxin-3 are as follows: forward primer, 5′–GGAAATATGGATGACAGTGG–3′; reverse primer, 5′–ATCCTGAGCCTCTGATACTC–3′. Primers specific for 18S were obtained from Applied Biosystems.
Table 2.
siRNAs and BNAs targeting the ATXN3 CAG repeat region.
| siRNA | Sequence | Position of mismatch |
Tm (°C) |
mutIC50 (nM) |
wtIC50 (nM) |
Selectivity (fold) |
|---|---|---|---|---|---|---|
| siREP | GCUGCUGCUGCUGCUGCUGtt | 86.8 | 12 ± 4 | 24 ± 9 | 2 lit | |
| siP4 | GCUACUGCUGCUGCUGCUGtt | 4 | 84.0 | - | - | - |
| siP5 | GCUGAUGCUGCUGCUGCUGtt | 5 | 83.2 | - | - | - |
| siP6 | GCUGCAGCUGCUGCUGCUGtt | 6 | 85.1 | - | - | - |
| siP7 | GCUGCUACUGCUGCUGCUGtt | 7 | 82.7 | - | - | - |
| siP8 | GCUGCUGAUGCUGCUGCUGtt | 8 | 83.8 | - | - | - |
| siP9 | GCUGCUGCAGCUGCUGCUGtt | 9 | 86.7 | 4.6 | >50 | 11 |
| siP10 | GCUGCUGCUACUGCUGCUGtt | 10 | 83.5 | - | - | - |
| siP10R | GCUGCUGCUUCUGCUGCUGtt | 10 | 78.0 | - | - | - |
| siP11 | GCUGCUGCUGAUGCUGCUGtt | 11 | 83.7 | 5.4 ± 3.9 | >50 | 9 |
| siP12 | GCUGCUGCUGCAGCUGCUGtt | 12 | 85.6 | - | - | - |
| siP13 | GCUGCUGCUGCUACUGCUGtt | 13 | 82.8 | - | - | - |
| siP16 | GCUGCUGCUGCUGCUACUGtt | 16 | 76.4 | - | - | - |
| siP910 | GCUGCUGCAACUGCUGCUGtt | 9,10 | 83.5 | 3.1 ± 0.9 | >50 | 16 |
| siPM3 | GCUGCUGCAAAUGCUGCUGtt | 9,10,11 | 79.9 | 5.2 ± 1.1 | >50 | 10 |
| siPM4 | GCUGCUGAAAAUGCUGCUGtt | 8,9,10,11 | 76.4 | 6.5 ± 2.8 | >50 | 8 |
| siRM4 | GCAGCUGUUGCUACUGUUGtt | 3,8,13,17 | 78.0 | - | - | - |
| siCM | GCUAUACCAGCGUCGUCAUtt | - | 80.0 | - | - | - |
| BNAs | ||||||
| carba LNA |
gc[T]gc[T]gc[T]gc[T]gc[T]gc[T]g | 76.5 | 25 ± 6.0 | 55 ± 18 | 2.2 | |
| cET | gcUgcUgcUgcUgcUgcUg | 82.8 | 9.3 ± 0.8 | 25 ± 3.8 | 2.6 | |
All sequences are listed from 5’ to 3’. Only the guide strand of siRNA is shown. RNA bases are in capital letters. Mismatched bases are underlined and in bold letters. For BNA oligomers, carba-LNA modified bases are in square bracket, cEt bases are in italics. DNA bases are in lowercase. Selectivity is calculated by comparing the IC50 for inhibition of wild-type versus the IC50 for inhibition of mutant ataxin-3 protein. Error is standard deviation. siCM is a non-complementary negative control siRNA. The Tm value for BNAs was calculated using complementary DNA sequences. Tm values for siRNAs are values for the duplex The molecular weight (calculated/observed) is 6042/6037 for carba LNA and 5965/5969 for cET.
Acknowledgements
This work was supported by the US National Institutes of Health (NIGMS 73042) and the Robert A. Welch Foundation (I-1244).
References
- Alves S, Nascimento-Ferreira I, Dufour N, Hassig R, Auregan G, Nobrega C, Brouillet E, Hantraye P, Pedroso de Lima MC, Deglon N, et al. Silencing ataxin-3 mitigates degeneration in a rat model of Machado-Joseph disease: no role for wild-type ataxin-3? Hum. Mol. Genet. 2010;19:2380–2394. doi: 10.1093/hmg/ddq111. [DOI] [PubMed] [Google Scholar]
- Alves S, Nascimento-Ferreira I, Auregan G, Hassig R, Dufour N, Brouillet E, Pedroso de Lima MC, Hantraye P, Pereira de Almeida L, Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS One. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Wolburg H, Mack A, Nuber S, Bottcher M, Schmitt I, Holzmann C, Zimmermann F, Servadio A, et al. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Hum. Mol. Genet. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- Braasch DA, Liu Y, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucl. Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza EB, Cload ST, Pendergrast PS, Sah DW. Novel therapeutic modalities to address nondrugable protein interaction targets. Neuropsychopharmacology. 2009;34:142–158. doi: 10.1038/npp.2008.115. [DOI] [PubMed] [Google Scholar]
- Denovan-Wright EM, Davidson BL. RNAi: a potential therapy for the dominantly inherited nucleotide repeat diseases. Gene Ther. 2006;13:525–531. doi: 10.1038/sj.gt.3302664. [DOI] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucl. Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Pendergraff HM, Deleavey GF, Swayze EE, Potier P, Randolph J, Roesch EB, Chattopadhyaya J, Damha MJ, Bennett CF, et al. Allele-Selective Inhibition of Mutant Huntingtin Expression with Antisense Oligonucleotides Targeting the Expanded CAG Repeat. Biochemistry. 2010;49:2243–2252. doi: 10.1021/bi101208k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Alegre P, Paulson HL. Technology insight: therapeutic RNA interference--how far from the neurology clinic? Nat. Clin. Pract. Neurol. 2007;3:394–404. doi: 10.1038/ncpneuro0551. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Corey DR. Inhibiting gene expression with peptide nucleic acid (PNA)--peptide conjugates that target chromosomal DNA. Biochemistry. 2007;46:7581–7589. doi: 10.1021/bi700230a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009a;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Dodd DW, Hudson RH, Corey DR. Cellular localization and allele-selective inhibition of mutant huntingtin protein by peptide nucleic acid oligomers containing the fluorescent nucleobase [bis-o-(aminoethoxy)phenyl]pyrrolocytosine. Bioorg. Med. Chem. Lett. 2009b;19:6181–6184. doi: 10.1016/j.bmcl.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Matsui M, Corey DR. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. N. Y. Acad. Sci. 2009c;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Corey DR. Allele-selective inhibition of huntingtin expression by switching RNAi mechanisms. Chem. Biol. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Hu J, Corey DR. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protoc. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases. Nucleic Acids Res. 2010;38:8370–8376. doi: 10.1093/nar/gkq700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. The first analogues of LNA (locked nucleic acids): phosphorothioate-LNA and 2'-thio-LNA. Bioorg. Med. Chem. Lett. 1998;8:2219–2222. doi: 10.1016/s0960-894x(98)00366-7. [DOI] [PubMed] [Google Scholar]
- Kurreck J. RNA interference: from basic research to therapeutic applications. Angew. Chem. Int. Ed. Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YU, Kodadek T. Quantitative evaluation of the relative cell permeability of peptoids and peptides. J. Am. Chem. Soc. 2007;129:1508–1509. doi: 10.1021/ja0668623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute 2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Marin VL, Armitage BA. RNA guanine quadruplex invasion by complementary and homologous PNA probes. J. Am. Chem. Soc. 2005;127:8032–8033. doi: 10.1021/ja051102y. [DOI] [PubMed] [Google Scholar]
- Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. U S A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc. Natl. Acad. Sci. U S A. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Semin. Neurol. 2007a;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- Paulson HL. Bird TC, Dolan CR, Stephens K, editors. Spinocerebeller ataxia type 3. Gene Rev. 2007b ( www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=gene&part=sca3).
- Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, Black C, Seth PP, Swayze EE, Bhat B. Antisense oligonucleotides containing conformationally constrained 2',4'-(N-methoxy)aminomethylene and 2',4'-aminooxymethylene and 2'-O,4'-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J. Med. Chem. 2010;53:1636–1650. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Wakisaka A, Fukazawa T, Iwabuchi K, Hamada T, Takada A, Mukai E, Matsuura T, Yoshiki T, Tashiro K. CAG repeat expansion of Machado-Joseph disease in the Japanese: analysis of the repeat instability for parental transmission, and correlation with disease phenotype. J. Neurol. Sci. 1995;133:128–133. doi: 10.1016/0022-510x(95)00175-2. [DOI] [PubMed] [Google Scholar]
- Scholefield J, Wood MJ. Therapeutic gene silencing strategies for polyglutamine disorders. Trends Genet. 2010;26:29–38. doi: 10.1016/j.tig.2009.11.005. [DOI] [PubMed] [Google Scholar]
- Seth PP, Vasquez G, Allerson CA, Berdeja A, Gaus H, Kinberger GA, Prakash TP, Migawa MT, Bhat B, Swayze EE. Synthesis and biophysical evaluation of 2',4'-constrained 2'O-methoxyethyl and 2',4'-constrained 2'O-ethyl nucleic acid analogues. J. Org. Chem. 2010;75:1569–1581. doi: 10.1021/jo902560f. [DOI] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsiger CS, Ward CM, McAlonis-Downes M, Wei H, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava P, Barman J, Pathmasiri W, Plashkevych O, Wenska M, Chattopadhyaya J. Five- and six-membered conformationally locked 2',4'-carbocyclic ribo-thymidines: synthesis, structure, and biochemical studies. J. Am. Chem. Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]