Abstract
Biofilm formation of Campylobacter jejuni, a major cause of human gastroenteritis, contributes to the survival of this pathogenic bacterium in different environmental niches; however, molecular mechanisms for its biofilm formation have not been fully understood yet. In this study, the role of oxidative stress resistance in biofilm formation was investigated using mutants defective in catalase (KatA), superoxide dismutase (SodB), and alkyl hydroperoxide reductase (AhpC). Biofilm formation was substantially increased in an ahpC mutant compared to the wild type, and katA and sodB mutants. In contrast to the augmented biofilm formation of the ahpC mutant, a strain overexpressing ahpC exhibited reduced biofilm formation. A perR mutant and a CosR-overexpression strain, both of which upregulate ahpC, also displayed decreased biofilms. However, the introduction of the ahpC mutation to the perR mutant and the CosR-overexpression strain substantially enhanced biofilm formation. The ahpC mutant accumulated more total reactive oxygen species and lipid hydroperoxides than the wild type, and the treatment of the ahpC mutant with antioxidants reduced biofilm formation to the wild-type level. Confocal microscopy analysis showed more microcolonies were developed in the ahpC mutant than the wild type. These results successfully demonstrate that AhpC plays an important role in the biofilm formation of C. jejuni.
Introduction
Campylobacter jejuni is a leading bacterial cause of human gastroenteritis and accounts for 400 million–500 million cases of diarrhea worldwide per year [1]. C. jejuni is also implicated in the onset of approximately a quarter of cases of Guillain–Barré syndrome, an autoimmune disorder characterized by acute and progressive neuromuscular paralysis [2]. While C. jejuni is isolated from a variety of domestic animals and wildlife, poultry is considered to be the major reservoir for C. jejuni [3]. Campylobacter infection is zoonotic and most cases of human campylobacteriosis are associated with consumption of undercooked poultry [4]. Although C. jejuni is known to be fastidious to culture because of complex nutrient and growth requirements [5], this bacterium is isolated from diverse environmental sources, such as surface water, sewage and farms [6], [7], suggesting that C. jejuni may possess unique survival mechanisms to persist in the environment. However, mechanisms for stress resistance and survival in the environment have not been well understood in C. jejuni.
Oxidative stress resistance is an important defense mechanism. As a microaerophilic bacterium, C. jejuni possesses unique oxidative defense systems. C. jejuni has a sole catalase (KatA) and a sole superoxide dismutase (SodB) for the detoxification of H2O2 and superoxide, respectively [8]. In Escherichia coli, alkyl hydroperoxide reductase consists of AhpC and AhpF [9] and plays an important role in scavenging endogenous H2O2 [10], whereas an ahpF homolog is absent from the C. jejuni genome [11]. Although the substrates of AhpC have not yet been identified in C. jejuni, an ahpC mutation increases susceptibility to aerobic stress and cumene hydroperoxide, but not to H2O2 [11]. As to the regulation of oxidative stress response, C. jejuni lacks homologs of the oxidative stress regulators OxyR and SoxRS, one or both of which are usually present in many bacterial species [8]. Instead, PerR, Fur and CosR regulate genes of oxidative stress resistance in C. jejuni [12]–[15].
Biofilms are a mode of bacterial growth which is often found in natural settings [16]. High numbers of Campylobacter spp. are isolated from biofilms in nature [17], implying that the ability of Campylobacter to form biofilms contributes to its prevalence in the environment [17]–[19]. C. jejuni forms biofilms on various abiotic surfaces, such as glass, plastics and stainless steel [20]–[22]. The biofilm formation of C. jejuni is affected by nutritional and environmental conditions. Cultivation with Mueller Hinton media at 37°C under 10% CO2 enhanced biofilms, whereas nutrient-rich media (e.g., Brucella broth) or high osmolarity (e.g, >0.05M NaCl) decreases biofilm formation of Campylobacter [22]. As motility is an important factor of biofilm formation in many bacterial species [23], mutations of genes associated with bacterial motility significantly affect the biofilm formation of C. jejuni [20], [21], [24]. Surface polysaccharides influence biofilm formation as mutations of genes involved in the synthesis of capsular polysaccharide (CPS) or lipooligosaccharide (LOS) increase biofilm formation [20], [25]. Interestingly, the pgp1 gene encoding a peptidoglycan DL-carboxypeptidase affects the corkscrew morphology of C. jejuni, and a pgp1 mutation results in defects in motility and biofilms [26]. Quorum sensing is also involved in biofilm formation as a luxS mutant that is defective in the production of autoinducer-2 (AI-2) exhibited reduced biofilms [22].
Owing to the aggregated bacterial growth in biofilms, bacterial cells in biofilms may encounter a series of nutritional and physiological stress. Thus, bacterial resistance to stress may significantly affect biofilm formation. For example, the stringent response is an important stress resistance mechanism associated with bacterial survival under unfavorable conditions. The stringent response of C. jejuni is mediated by SpoT, a bifunctional enzyme that synthesizes and hydrolyzes guanosine teteraphosphate (ppGpp) [27]. In contrast to the stringent response mutants in other bacteria which usually show defects in biofilms, interestingly, the spoT mutation significantly increases biofilm formation and produces more mature biofilms compared with the wild type [28]. Oxidative stress resistance significantly impacts C. jejuni’s aerotolerance, freeze-thaw resistance, antibiotic resistance, intracellular survival and chicken colonization [11], [14], [29]–[31]. However, nothing is known about the role of oxidative stress resistance in the biofilm formation of C. jejuni. In this study, we compared the biofilm formation of mutants defective in key enzymes of oxidative stress resistance, including KatA, SodB, and AhpC, and show that AhpC plays a pivotal role in the biofilm formation of C. jejuni.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains used in this study are listed in Table 1. All C. jejuni strains were grown at 42°C with Mueller-Hinton (MH) media (Oxoid, Canada) under a microaerobic condition (5% O2, 10% CO2, and 85% N2). Occasionally, MH media were supplemented with kanamycin (50mg/L) and/or chloramphenicol (25mg/L). E. coli DH5α harboring plasmids was grown at 37°C with Luria-Bertani (LB) media (Difco, US) that were supplemented with ampicillin (100mg/L), chloramphenicol (25mg/L), or kanamycin (50mg/L), where required.
Table 1. Strains and plasmids used in this study.
| Strains and Plasmids | Description | Sources or references |
| E.coli | ||
| DH5α | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+)phoA supE44 λ– thi-1 gyrA96 relA1 | Life Technologies |
| C.jejuni | ||
| NCTC 11168 | Wild type, a human isolate | [37] |
| ΔahpC | ahpC mutant, ahpC::aphA-3 | This study |
| ΔkatA | katA mutant, katA:: aphA-3 | [31] |
| ΔsodB | sodB mutant, sodB::cat | [31] |
| ΔperR | perR mutant, perR::cat | [57] |
| ahpC over | C. jejuni NCTC 11168 harboring an extra copy of ahpC in rRNA region | This study |
| cosR over | C. jejuni NCTC 11168 harboring an extra copy of cosR in rRNA region | [13] |
| ΔperR &ΔahpC | ahpC (ahpC::aphA-3) and perR (perR::cat) double mutant | This study |
| cosR over&ΔahpC | ahpC mutant (ahpC::aphA-3) and CosR- overexpression mutant | This study |
| ahpC comp | ahpC complementation | This study |
| katA comp | katA complementation | [31] |
| sodB comp | sodB complementation | [31] |
| perR comp | perR complementation | [57] |
| Plasmids | ||
| pUC19 | Cloning vector used for suicide vector in C. jejuni; Ampr | New England Biolabs |
| pFMBcomCM | pUC19 derivative carrying an rRNA gene cluster; Cmr | [13] |
| pMW10 | E.coli-C. jejuni shuttle vector | [32] |
Construction of the ahpC mutant, and the perR&ahpC and cosR over&ahpC double mutants
The ahpC gene and its flanking region were amplified by PCR with DahpC-F and DahpC-R primers (Table 2). After digestion with EcoRI and BamHI, the PCR product was ligated to pUC19 that had been treated with the same enzymes. The pUC19::ahpC plasmid was digested with EcoRV and ligated with the kanamycin resistance cassette which had been amplified from pMW10 [32] with Kan-F and Kan-R primers (Table 2). The constructed suicide plasmid was introduced to C. jejuni NCTC 11168 by electroporation and mutants were selected on MH agar plates supplemented with kanamycin (50 mg/L). The ahpC mutation was confirmed by PCR with mahpC-F and mahpC-R primers (Table 2). The ahpC complementation strain was constructed by chromosomal integration of ahpC as described previously [33]. Briefly, ahpC was PCR-amplified with com_ahpC-F and com_ahpC-R primers (Table 2). After digestion with XbaI, ahpC was ligated with pFMBcomCM [13]. The pFMBcomCM::ahpC plasmid was transformed into the ahpC mutant strain by electroporation and a complementation strain was selected by growing on MH agar plates containing kanamycin (50mg/L) and chloramphenicol (25mg/L). In addition, pFMBcomCM::ahpC was introduced into C. jejuni NCTC 11168 as described above to construct an ahpC-overexpression strain. Increased transcription of ahpC in the ahpC-overexpression strain was confirmed by qRT-PCR (Fig. S1). Chromosomal DNA extracted from the ahpC mutant strain was introduced into the perR mutant and the CosR-overexpression strain by electroporation to construct the perR&ahpC and cosR over&ahpC double mutants. Transformants were selected on MH agar plates supplemented with kanamycin (50mg/L) and chloramphenicol (25mg/L). Transfer of the ahpC mutation was confirmed by PCR.
Table 2. Primers used in this study.
| Primers | Sequences (5′-3′) |
| DahpC-F | GGAATTCCTCCCCACTTCTCATATC |
| DahpC-R | GGGATCCCAATAGCTGCCGCATCTTG |
| Kan-F | GCGATGAAGTGCGTAAG |
| Kan-R | CGGCTCCGTCGATACTATG |
| mahpC-F | CATGATAGTTACTAAAAAAGCTTTAG |
| mahpC-R | GTTAAAGTTTAGCTTCGTTTTTGCC |
| com_ahpC-F | GTCTCTAGAAGCTGCCGCATCTTGAGACTTTG |
| com_ahpC-R | CGTTCTAGACACCTTCTGGATTGTTAGTATCAT |
Biofilm Assay
Biofilm assay was performed as described previously [25] with some modifications. Briefly, overnight cultures of C. jejuni strains were harvested from MH agar plates and resuspended in MH broth to an OD600 of 0.07. After culturing 5 h at 42°C with shaking (200rpm) under a microaerobic condition, the bacterial suspension was diluted with fresh MH broth to an OD600 of 0.07 and inoculated into 96-well plates (Nunc, US). Biofilms were cultivated under the same growth condition without shaking. The 96-well plates were washed twice with PBS (pH 7.4) and dried 20 min at room temperature after discarding supernatants. Fifty microliter of 1% crystal violet was added to each well. After staining 15 min at room temperature, crystal violet solution was removed and the plates were washed 3 times with PBS (pH 7.4). Stained biofilm was eluted with a solution of 10% acetic acid and 30% methanol, and OD595 was measured with a spectrophotometer (Thermo Scientific, US). Occasionally, biofilm assay was carried out in the presence of 1 µM CosR-PNA [34], which was commercially synthesized by PANAGENE (Daejeon, South Korea), or antioxidants, including L-proline, L-cysteine, β-carotene and N-acetyl cysteine, which were purchased from Sigma (St. Louis, US).
Measurement of Total Reactive Oxygen Species (ROS)
The ROS level in biofilms was measured using CM-H2DCFDA (Life Technologies, US), a general oxidative stress indicator. After discarding supernatants from 96-well plates, biofilms were washed twice with PBS (pH7.4) and then treated with 100 µl PBS containing 10 µM CM-H2DCFDA for 30 min. Fluorescence was measured with a multi-well plate reader (Varioskan Flash; Thermo Scientific). Protein concentrations of each sample were measured with Bradford protein assay (Bio-Rad, US) to normalize the ROS level.
Lipid Hydroperoxide (LPO) Assay
LPO levels were measured using a commercial kit (Cayman Chemical Co., US) according to the manufacturer’s instructions. Briefly, LPOs were extracted from biofilms with chloroform and methanol. The LPO extract (500 µl) was mixed with 50 µl of Chromogen reagent (4.5mM ferrous sulfate in 0.1M HCl and 3% methanolic solution of ammonium thiocyanate) and incubated at room temperature for 5 min. A portion (300 µl) of each sample was transferred into a 96-well glass plate to measure OD500. A standard curve was generated with 13-hydroperoxy-octadecadienoic acid that was provided by the manufacturer.
Confocal Microscopy Analysis
Biofilms were fixed with 4% paraformaldehyde and stained with SYTO9 and propidium iodide from the LIVE/DEAD Biofilm Viability kit (Life Technologies, US) according to the manufacturer’s instructions. Confocal microscopy was performed with an inverted confocal microscope (IX-81, Olympus, Japan) and Volocity 3D Image Analysis Software (PerkinElmer Inc., US).
Statistical Analysis
Data analysis was performed with GraphPad Prism 6 (GraphPad Software Inc., US).
Results and Discussion
Enhanced Biofilm Formation in the ahpC Mutant
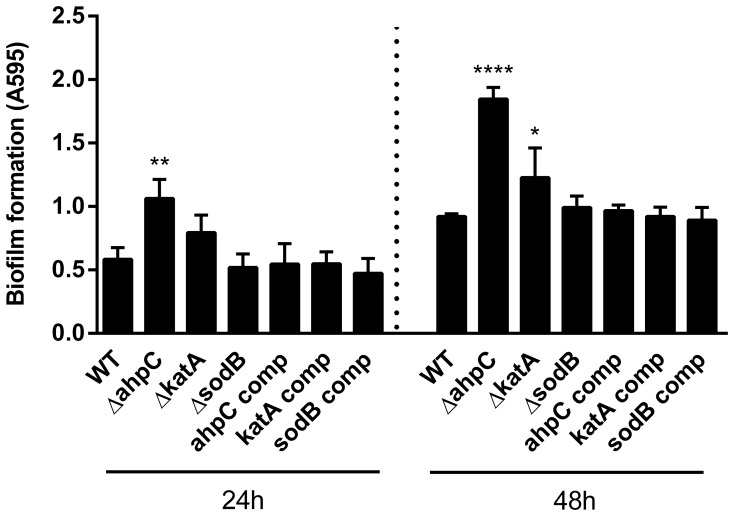
To investigate the role of oxidative stress defense in the biofilm formation of C. jejuni, we measured the biofilm formation levels of three oxidative stress resistance mutants defective in AhpC, SodB and KatA. These enzymes were chosen because of their critical roles in oxidative stress resistance, and C. jejuni possesses only a single gene copy encoding each enzyme. The ahpC mutant displayed most significant increases in the level of biofilm formation compared with the wild type and katA and sodB mutants, and complementation of the ahpC mutant restored biofilm formation to the wild-type level (Fig. 1). The katA mutation slightly increased biofilm formation, whereas the sodB mutant produced biofilms as comparably as the wild type (Fig. 1). Extended (i.e., 48 h) incubation of biofilms increased the levels of biofilm formation in all the tested strains; however, the ahpC mutant developed biofilms to a greater extent than the wild type and katA and sodB mutants both 24 h and 48 h (Fig. 1). SodB is known to be important for Campylobacter’s survival in environmental conditions, such as freeze-thaw stress in foods [35], [36]; however, the sodB mutation did not affect biofilm formation (Fig. 1). Bacterial growth is not likely to be associated with the enhanced biofilm formation of the ahpC mutant because the ahpC, katA, and sodB mutants grew as comparably as the wild type (data not shown). The ahpC mutation slightly reduced the motility (Fig. S2); however, this does not account for the substantial increase in biofilms observed in the ahpC mutant, because mutations resulting in a motility defect impair the biofilm formation of C. jejuni [20], [21], [24], whereas the ahpC mutation substantially increased biofilm formation despite the partial reduction in motility. The results show that AhpC significantly affects the biofilm formation of C. jejuni.
Figure 1. Biofilm formation of oxidative stress resistance mutants.
Biofilm formation was measured using crystal violet staining. The results show the means and standard deviations of a representative assay with triplicate samples. The experiment was repeated six times and all produced similar results. Statistical significance was analyzed using one-way analysis of variance (ANOVA). *P≤0.05, **P≤0.01, ****P≤0.0001.
Overexpression of ahpC Reduced Biofilm Formation
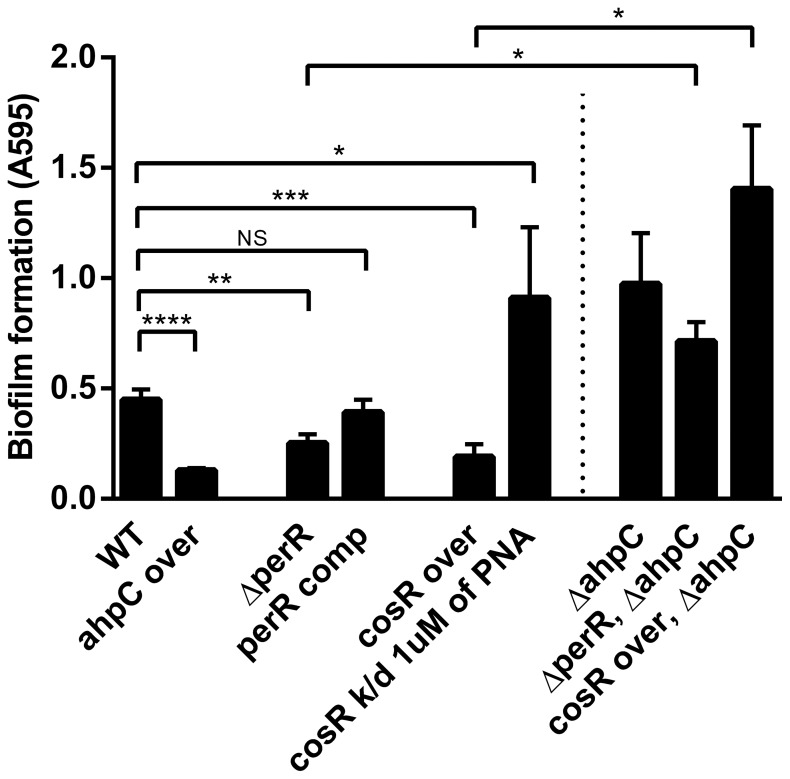
Since AhpC is an enzyme detoxifying ROS, we hypothesized that ahpC overexpression would reverse the phonotype observed in the ahpC mutant if biofilm formation is directly associated with the enzymatic function of AhpC. The ahpC overexpression was achieved in two different ways by: 1) incorporating an extra copy of ahpC to the chromosome of C. jejuni, and 2) modulating the expression of AhpC-controlling regulators, such as PerR and CosR [15], [34]. C. jejuni lacks homologs of OxyR and RpoS [37], which regulate AhpC in E. coli and Salmonella [38], [39]. Instead, PerR negatively regulates AhpC in C. jejuni [15]. CosR is an essential response regulator and positively regulates AhpC in C. jejuni [34]. Thus, ahpC overexpression was alternatively achieved using a perR knockout mutant and a CosR-overexpression strain. In contrast to the stimulated biofilm formation of the ahpC mutant (Fig. 1), interestingly, the ahpC-overexpression strain exhibited significantly reduced biofilm levels compared to the wild type (Fig. 2), and the perR mutant and the CosR-overexpression strain also displayed substantial reductions in the level of biofilm formation (Fig. 2). The perR complementation restored biofilm formation to the wild-type level (Fig. 2). CosR is an essential regulator and its knockout mutant cannot be constructed because of bacterial lethality [40], [41]. Thus, a gene knockdown strategy was used to reduce CosR expression with CosR-specific PNA as described in our previous studies [13], [34]. Interestingly, CosR knockdown with CosR-PNA significantly increased biofilm formation to the level of the ahpC mutant (Fig. 2).
Figure 2. Effect of increased ahpC expression on biofilm formation.
Biofilm formation levels were determined with crystal violet staining after 24 h incubation of samples. The results show the means and standard deviations of a representative experiment with triplicate samples. The experiment was repeated six times and all produced similar results. The statistical differences between the wild type and each mutant were determined by t-test. NS: non-significant, *P≤0.05, **P≤0.01, ***P≤0.001, ****P≤0.0001.
PerR and CosR are involved in the regulation of genes other than ahpC [13], [14], [34]. At least 104 genes belong to the PerR regulon [14], and CosR regulates 93 genes involved in various cellular functions, such as energy production, motility, drug efflux, and oxidative stress resistance [13]. An ahpC mutation was introduced to the perR mutant and the CosR-overexpression strain to clarify the role of AhpC in the reduced biofilm formation of the perR mutant and the CosR-overexpression strain. Interestingly, harboring the ahpC mutation abrogated the inhibitory effect of the perR mutation and the CosR-overexpression on biofilm formation and substantially increased biofilm formation similar to the ahpC mutant (Fig. 2), suggesting that CosR and PerR affect biofilms mainly through ahpC.
Kalmokoff et al. showed that proteins of oxidative stress resistance, including AhpC, Tpx (thiol peroxidase), and CosR (Cj0355c), are upregulated in C. jejuni biofilms [21]. Consistently, in this study, we demonstrated that both AhpC and CosR significantly affect the biofilm development of C. jejuni (Figs. 1 and 2). Given the positive regulation of AhpC by CosR [34], upregulated CosR will increase AhpC expression, consequently promoting the detoxification of ROS in biofilms. Since AhpC overexpression reduced the biofilm formation (Fig. 2), upregulated AhpC will reduce biofilm formation presumably to alleviate oxidative stress in aggregated bacterial cells by decreasing C. jejuni’s capability to develop biofilms. Regulators of oxidative stress resistance are also often involved in the modulation of biofilm formation in C. jejuni. A mutation of csrA (carbon starvation regulator) attenuated both oxidative stress resistance and biofilm formation [42]. CprS is the cognate sensor kinase of the essential response regulator CprR, and the cprS mutation enhances biofilm formation and affects the expression of oxidative stress defense proteins, such as AhpC, KatA, CosR, SodB, and TrxB [43]. Based on the previous reports and our findings, the two important defense/survival mechanisms may be functionally related to each other in C. jejuni.
Effect of ROS Accumulation on Biofilm Formation in the ahpC Mutant
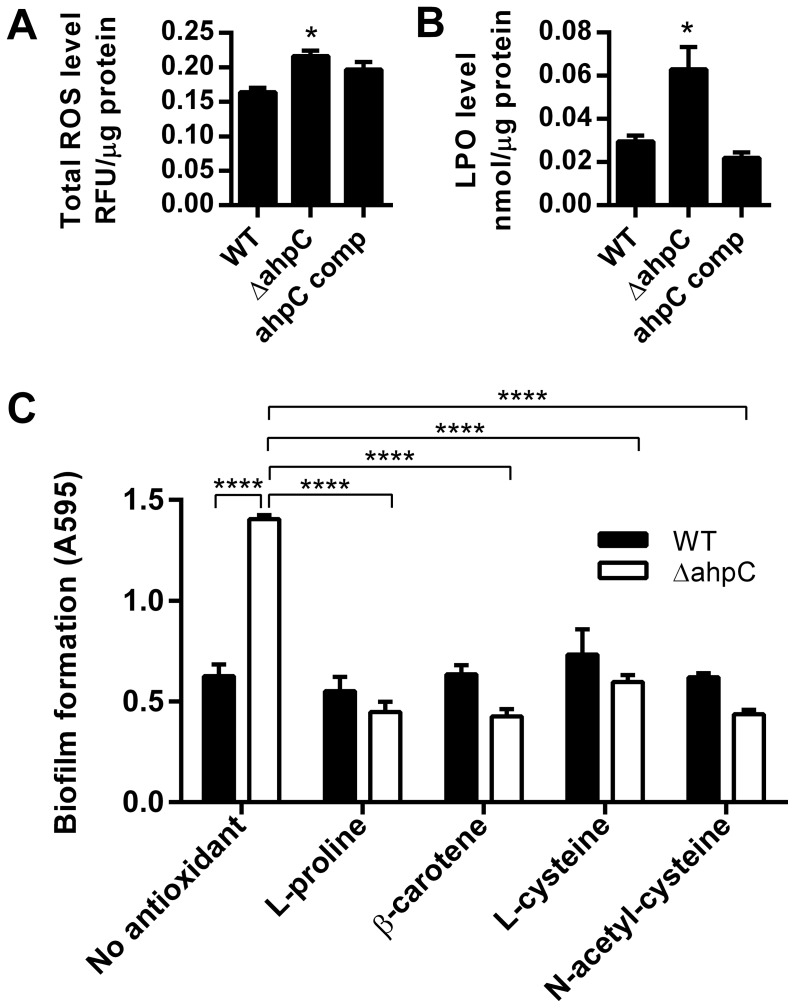
AhpC scavenges endogenous H2O2 at the physiological level in E. coli [10]. The substrates of the Salmonella Typhimurium AhpC are small hydroperoxides [44] and organic hydroperoxides, including alkyl hydroperoxides which can be produced intracellularly from unsaturated fatty acids and nucleic acids [9]. However, the enzymatic function and substrate of AhpC have not been defined in C. jejuni. In this study, total ROS and lipid hydroperoxide (LPO) were measured and compared between the ahpC mutant and the wild type (Fig. 3A and B). Consistent with the enzymatic function of AhpC in ROS detoxification, the ahpC mutant showed increased accumulation of total ROS and LPO compared to the wild type (Fig. 3A and B). In particular, the LPO level of the ahpC mutant was 2-fold higher than that of the wild type (Fig. 3B), suggesting that AhpC may be involved in the detoxification of LPO in C. jejuni. The results prompted us to hypothesize that the accumulation of endogenous ROS may be involved in the augmented biofilm formation of the ahpC mutant. To examine this possibility, biofilm assay was performed in the presence of antioxidants to reduce the ROS level. Interestingly, addition of antioxidants reduced the biofilm of the ahpC mutant to the wild-type level (Fig. 3C). Bacterial growth of the wild type and the ahpC mutant was not affected by the treatment with antioxidants (data not shown). The results strongly suggest that the accumulation of ROS enhanced biofilm formation in the ahpC mutant.
Figure 3. Determination of total ROS level (A) and lipid hydroperoxide (LPO; B) in the ahpC mutant, and reduced biofilm formation in the ahpC mutant by treatment with antioxidants (C).
The assays were carried out with 24 h old samples. Antioxidants were treated to a final concentration of 1nM. The results show the means and standard deviations of a representative experiment with triplicate samples. The assays were repeated at least three times and similar results were reproducible in all the experiments. Statistical significance was analyzed with t-test (Fig. 3A and 3B) and two-way ANOVA (Fig. 3C). *P≤0.05, ****P≤0.0001.
Bacterial cells in biofilms are physiologically different from planktonic cells and often exhibit increased resistance to environmental stress and antimicrobials [45]. The aggregate growth of bacteria in biofilms reduces the penetration of nutrients into inner layers and limits the diffusion of metabolic wastes, resulting in nutritional limitation and physiological stress [46]. The oxidative stress generated from endogenous ROS in biofilm cells enhances bacterial mutability and diversity [47], [48], demonstrating that ROS substantially impacts bacterial physiology in biofilms. Genes involved in general and oxidative stress response are often induced in biofilms, possibly to alleviate the stress generated in biofilms [21], [48]–[50]. The nutritional starvation of Pseudomonas aeruginosa in biofilms confers increased antibiotic tolerance in association with oxidative stress defense as the inactivation of stringent response significantly impaired the activities of catalase and superoxide dismutase [51]. The expression levels of Zn-superoxide dismutase (SodC) and thiol peroxidase (Tpx) are increased in biofilms of E. coli O157:H7, and sodC and tpx mutations impaired biofilm formation [52]. Iron stimulates the formation of rugose biofilms in E. coli in connection with ROS. Generation of superoxide stress by adding a superoxide generator or mutating sodA and sodB promotes the development of rugose biofilms [53]. The biofilm formation of S. aureus is stimulated by exposure to cigarette smoke that contains bioactive compounds such as free radicals and ROS. Addition of antioxidant (e.g., N-acetyl cysteine) eliminated the enhanced biofilm formation caused by cigarette smoke exposure [54].
Enhanced Development of Microcolonies in the ahpC Mutant
Biofilm development consists of multiple stages, starting with the attachment of bacteria, formation of microcolonies, maturation of biofilms forming mushroom-like structures and water channels, and dispersion of cells from biofilms [55]. C. jejuni forms microcolonies prior to the development of biofilms, and flagellar mutants are defective in forming microcolonies and biofilms [56]. Biofilms were observed with a confocal microscope to investigate the biofilm formation stage that is affected by the ahpC mutation. According to the results of confocal microscopy analysis, the ahpC mutant showed increased development of microcolonies compared to the wild type (Fig. 4). This is consistent with the results of biofilm assays, showing that the difference in biofilm formation between the wild type and the ahpC mutant was obvious even after one day (Fig. 1). The results suggest that AhpC is involved in the development of microcolonies at the early stage of biofilm formation.
Figure 4. Confocal microscopy analysis of biofilms of the wild type (A), the ahpC mutant (B), and the complementation strain (C).
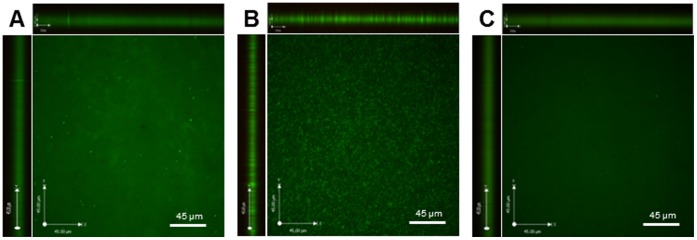
Biofilms were grown 24 h and stained with the LIVE/DEAD Biofilm Viability kit (Life Technologies, US).
Physiological heterogeneity exits in biofilms due to the concentration gradients between layers inside and outside [46]. Reduced diffusion of oxygen into biofilms will expose cells in outside layers to more oxygen and oxidative stress compared to bacteria inside biofilms. The impact of heterogeneity in biofilms would not be significant in the ahpC mutant because AhpC affects biofilm formation even at an early stage when biofilms are not fully developed (Fig. 4).
To the best of our knowledge, this is the first report showing that AhpC affects biofilm formation. The kind of ROS responsible for the enhanced biofilm formation of the ahpC mutant is still unknown. We speculate that the ROS would be neither H2O2 nor superoxide, because mutations of C. jejuni’s sole catalase and superoxide dismutase did not affect biofilm formation significantly, albeit the katA mutant increased biofilm formation at substantially lower levels than the ahpC mutant (Fig. 1). Although AhpC scavenges endogenous H2O2 at physiological levels in E. coli [10], such a function has not been demonstrated in the C. jejuni AhpC, and addition of exogenous H2O2 rather reduced the biofilm formation (data not shown). Based on the extrapolation of the enzymatic function of AhpC in other bacteria and the increased accumulation of LPO in the ahpC mutant, the ROS involved in biofilm formation in the ahpC mutant would be an organic peroxide(s) endogenously generated within the cell. Validation of this possibility first of all requires the identification of ROS substrates of AhpC in C. jejuni and still awaits future studies.
Supporting Information
Increased levels of ahpC transcription in the ahpC overexpression strain. qRT-PCR was carried out with primer pairs; qPCR_ahpC-F (GGTATTGGTCAGGTTAAATTCCC) and qPCR_ahpC-R (GGTAAATCATTAACCACAGCATG). The results were normalized to the expression level of 16S rRNA (Cjr01) as described previously [34]. The assay was repeated three times. **P≤0.01.
(TIFF)
Motility of the wild type (WT), the ahpC mutant (ΔahpC), the ahpC complementation strain (ahpC comp), and the ahpC overexpression strain (ahpC over). (A) The ahpC mutation resulted in a slight reduction in motility with a full restoration to the wild-type level by complementation. The assay was performed with MH medium containing 0.4% agar, and the motility agar plate was incubated microaerobically at 42°C for 2 days. The result is a representative of three independent experiments with similar results. (B) Comparison of the size of motility zones. The results show the means and standard deviations. NS: non-significant.
(TIF)
Acknowledgments
We thank Drs. Lynn McMullen and Michael Gänzle (University of Alberta) for sharing their laboratory facilities.
Funding Statement
This study was a start-up from the University of Alberta. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ruiz-Palacios GM (2007) The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin Infect Dis 44: 701–703. [DOI] [PubMed] [Google Scholar]
- 2. Hughes RA, Cornblath DR (2005) Guillain-Barré syndrome. Lancet 366: 1653–1666. [DOI] [PubMed] [Google Scholar]
- 3. Allos BM (2001) Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis 32: 1201–1206. [DOI] [PubMed] [Google Scholar]
- 4. Butzler JP (2004) Campylobacter, from obscurity to celebrity. Clin Microbiol Infect 10: 868–876. [DOI] [PubMed] [Google Scholar]
- 5. Snelling WJ, Matsuda M, Moore JE, Dooley JS (2005) Campylobacter jejuni . Lett Appl Microbiol 41: 297–302. [DOI] [PubMed] [Google Scholar]
- 6. Jokinen C, Edge TA, Ho S, Koning W, Laing C, et al. (2011) Molecular subtypes of Campylobacter spp., Salmonella enterica, and Escherichia coli O157:H7 isolated from faecal and surface water samples in the Oldman River watershed, Alberta, Canada. Water Res 45: 1247–1257. [DOI] [PubMed] [Google Scholar]
- 7. Moore JE, Corcoran D, Dooley JS, Fanning S, Lucey B, et al. (2005) Campylobacter . Vet Res 36: 351–382. [DOI] [PubMed] [Google Scholar]
- 8. Atack JM, Kelly DJ (2009) Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol 4: 677–690. [DOI] [PubMed] [Google Scholar]
- 9. Jacobson FS, Morgan RW, Christman MF, Ames BN (1989) An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage. Purification and properties. J Biol Chem 264: 1488–1496. [PubMed] [Google Scholar]
- 10. Seaver LC, Imlay JA (2001) Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli . J Bacteriol 183: 7173–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baillon ML, van Vliet AH, Ketley JM, Constantinidou C, Penn CW (1999) An iron-regulated alkyl hydroperoxide reductase (AhpC) confers aerotolerance and oxidative stress resistance to the microaerophilic pathogen Campylobacter jejuni . J Bacteriol 181: 4798–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Butcher J, Sarvan S, Brunzelle JS, Couture JF, Stintzi A (2012) Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc Natl Acad Sci U S A 109: 10047–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang S, Zhang Q, Ryu S, Jeon B (2012) Transcriptional regulation of the CmeABC multidrug efflux pump and the KatA catalase by CosR in Campylobacter jejuni . J Bacteriol 194: 6883–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palyada K, Sun YQ, Flint A, Butcher J, Naikare H, et al. (2009) Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni . BMC Genomics 10: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Vliet AH, Baillon ML, Penn CW, Ketley JM (1999) Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J Bacteriol 181: 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Branda SS, Vik S, Friedman L, Kolter R (2005) Biofilms: the matrix revisited. Trends Microbiol 13: 20–26. [DOI] [PubMed] [Google Scholar]
- 17. Maal-Bared R, Bartlett KH, Bowie WR, Hall ER (2012) Campylobacter spp. distribution in biofilms on different surfaces in an agricultural watershed (Elk Creek, British Columbia): using biofilms to monitor for Campylobacter . Int J Hyg Environ Health 215: 270–278. [DOI] [PubMed] [Google Scholar]
- 18. Young KT, Davis LM, Dirita VJ (2007) Campylobacter jejuni: molecular biology and pathogenesis. Nat Rev Microbiol 5: 665–679. [DOI] [PubMed] [Google Scholar]
- 19. Murphy C, Carroll C, Jordan KN (2006) Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni . J Appl Microbiol 100: 623–632. [DOI] [PubMed] [Google Scholar]
- 20. Joshua GW, Guthrie-Irons C, Karlyshev AV, Wren BW (2006) Biofilm formation in Campylobacter jejuni . Microbiology 152: 387–396. [DOI] [PubMed] [Google Scholar]
- 21. Kalmokoff M, Lanthier P, Tremblay TL, Foss M, Lau PC, et al. (2006) Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J Bacteriol 188: 4312–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA (2007) Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl Environ Microbiol 73: 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guttenplan SB, Kearns DB (2013) Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 37: 849–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reuter M, Mallett A, Pearson BM, van Vliet AH (2010) Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol 76: 2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naito M, Frirdich E, Fields JA, Pryjma M, Li J, et al. (2010) Effects of sequential Campylobacter jejuni 81–176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J Bacteriol 192: 2182–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, et al. (2012) Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni . PLoS Pathog 8: e1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gaynor EC, Wells DH, MacKichan JK, Falkow S (2005) The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol Microbiol 56: 8–27. [DOI] [PubMed] [Google Scholar]
- 28. McLennan MK, Ringoir DD, Frirdich E, Svensson SL, Wells DH, et al. (2008) Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive polysaccharide. J Bacteriol 190: 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Day WA Jr, Sajecki JL, Pitts TM, Joens LA (2000) Role of catalase in Campylobacter jejuni intracellular survival. Infect Immun 68: 6337–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garénaux A, Ritz M, Jugiau F, Rama F, Federighi M, et al. (2009) Role of oxidative stress in C. jejuni inactivation during freeze-thaw treatment. Curr Microbiol 58: 134–138. [DOI] [PubMed] [Google Scholar]
- 31. Hwang S, Ryu S, Jeon B (2013) Roles of the superoxide dismutase SodB and the catalase KatA in the antibiotic resistance of Campylobacter jejuni . J Antibiot (Tokyo) 66: 351–353. [DOI] [PubMed] [Google Scholar]
- 32. Wösten MM, Boeve M, Koot MG, van Nuenen AC, van der Zeijst BA (1998) Identification of Campylobacter jejuni promoter sequences. J Bacteriol 180: 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karlyshev AV, Wren BW (2005) Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni . Appl Environ Microbiol 71: 4004–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hwang S, Kim M, Ryu S, Jeon B (2011) Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni . PLoS One 6: e22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Purdy D, Cawthraw S, Dickinson JH, Newell DG, Park SF (1999) Generation of a superoxide dismutase (SOD)-deficient mutant of Campylobacter coli: evidence for the significance of SOD in Campylobacter survival and colonization. Appl Environ Microbiol 65: 2540–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stead D, Park SF (2000) Roles of Fe superoxide dismutase and catalase in resistance of Campylobacter coli to freeze-thaw stress. Appl Environ Microbiol 66: 3110–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, et al. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403: 665–668. [DOI] [PubMed] [Google Scholar]
- 38. Jung IL, Kim IG (2003) Transcription of ahpC, katG, and katE genes in Escherichia coli is regulated by polyamines: polyamine-deficient mutant sensitive to H2O2-induced oxidative damage. Biochem Biophys Res Commun 301: 915–922. [DOI] [PubMed] [Google Scholar]
- 39. Morgan RW, Christman MF, Jacobson FS, Storz G, Ames BN (1986) Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc Natl Acad Sci U S A 83: 8059–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garénaux A, Guillou S, Ermel G, Wren B, Federighi M, et al. (2008) Role of the Cj1371 periplasmic protein and the Cj0355c two-component regulator in the Campylobacter jejuni NCTC 11168 response to oxidative stress caused by paraquat. Res Microbiol 159: 718–726. [DOI] [PubMed] [Google Scholar]
- 41. Raphael BH, Pereira S, Flom GA, Zhang Q, Ketley JM, et al. (2005) The Campylobacter jejuni response regulator, CbrR, modulates sodium deoxycholate resistance and chicken colonization. J Bacteriol 187: 3662–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fields JA, Thompson SA (2008) Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J Bacteriol 190: 3411–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Svensson SL, Davis LM, MacKichan JK, Allan BJ, Pajaniappan M, et al. (2009) The CprS sensor kinase of the zoonotic pathogen Campylobacter jejuni influences biofilm formation and is required for optimal chick colonization. Mol Microbiol 71: 253–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parsonage D, Karplus PA, Poole LB (2008) Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc Natl Acad Sci U S A 105: 8209–8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108. [DOI] [PubMed] [Google Scholar]
- 46. Stewart PS, Franklin MJ (2008) Physiological heterogeneity in biofilms. Nat Rev Microbiol 6: 199–210. [DOI] [PubMed] [Google Scholar]
- 47. Boles BR, Singh PK (2008) Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105: 12503–12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ryder VJ, Chopra I, O′Neill AJ (2012) Increased mutability of staphylococci in biofilms as a consequence of oxidative stress. PLoS One 7: e47695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mikkelsen H, Duck Z, Lilley KS, Welch M (2007) Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa . J Bacteriol 189: 2411–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood TK (2009) Insights on Escherichia coli biofilm formation and inhibition from whole-transcriptome profiling. Environ Microbiol 11: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, et al. (2011) Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334: 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim YH, Lee Y, Kim S, Yeom J, Yeom S, et al. (2006) The role of periplasmic antioxidant enzymes (superoxide dismutase and thiol peroxidase) of the Shiga toxin-producing Escherichia coli O157:H7 in the formation of biofilms. Proteomics 6: 6181–6193. [DOI] [PubMed] [Google Scholar]
- 53. DePas WH, Hufnagel DA, Lee JS, Blanco LP, Bernstein HC, et al. (2013) Iron induces bimodal population development by Escherichia coli . Proc Natl Acad Sci U S A 110: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kulkarni R, Antala S, Wang A, Amaral FE, Rampersaud R, et al. (2012) Cigarette smoke increases Staphylococcus aureus biofilm formation via oxidative stress. Infect Immun 80: 3804–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Monroe D (2007) Looking for chinks in the armor of bacterial biofilms. PLoS Biol 5: e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haddock G, Mullin M, MacCallum A, Sherry A, Tetley L, et al. (2010) Campylobacter jejuni 81–176 forms distinct microcolonies on in vitro-infected human small intestinal tissue prior to biofilm formation. Microbiology 156: 3079–3084. [DOI] [PubMed] [Google Scholar]
- 57. Kim M, Hwang S, Ryu S, Jeon B (2011) Regulation of perR expression by iron and PerR in Campylobacter jejuni. J Bacteriol. 193: 6171–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Increased levels of ahpC transcription in the ahpC overexpression strain. qRT-PCR was carried out with primer pairs; qPCR_ahpC-F (GGTATTGGTCAGGTTAAATTCCC) and qPCR_ahpC-R (GGTAAATCATTAACCACAGCATG). The results were normalized to the expression level of 16S rRNA (Cjr01) as described previously [34]. The assay was repeated three times. **P≤0.01.
(TIFF)
Motility of the wild type (WT), the ahpC mutant (ΔahpC), the ahpC complementation strain (ahpC comp), and the ahpC overexpression strain (ahpC over). (A) The ahpC mutation resulted in a slight reduction in motility with a full restoration to the wild-type level by complementation. The assay was performed with MH medium containing 0.4% agar, and the motility agar plate was incubated microaerobically at 42°C for 2 days. The result is a representative of three independent experiments with similar results. (B) Comparison of the size of motility zones. The results show the means and standard deviations. NS: non-significant.
(TIF)