Abstract
Background
Wood cell walls are rich in cellulose, hemicellulose and lignin. Hence, they are important sources of renewable biomass for producing energy and green chemicals. However, extracting desired constituents from wood efficiently poses significant challenges because these polymers are highly cross-linked in cell walls and are not easily accessible to enzymes and chemicals.
Results
We show that aspen pectate lyase PL1-27, which degrades homogalacturonan and is expressed at the onset of secondary wall formation, can increase the solubility of wood matrix polysaccharides. Overexpression of this enzyme in aspen increased solubility of not only pectins but also xylans and other hemicelluloses, indicating that homogalacturonan limits the solubility of major wood cell wall components. Enzymatic saccharification of wood obtained from PL1-27-overexpressing trees gave higher yields of pentoses and hexoses than similar treatment of wood from wild-type trees, even after acid pretreatment.
Conclusions
Thus, the modification of pectins may constitute an important biotechnological target for improved wood processing despite their low abundance in woody biomass.
Keywords: Populus, Wood development, Secondary cell wall, Lignocellulose, Biofuel, Pectin
Background
There is high interest in cultivating Populus species as energy crops because they grow rapidly, producing abundant lignocellulosic biomass that can be used as feedstock for biofuel and biomaterial production [1]. However, a major challenge hindering large-scale commercial use of Populus lignocellulose is its recalcitrance to degradation by bacterial and fungal enzymes, which complicates the separation of non-crystalline carbohydrate polymers and cellulose contents from lignin, and their subsequent saccharification. The constitutive lignocellulose polymers are non-randomly arranged within cell-wall layers that each have a distinct composition and architecture [2]. The outermost thin layer of middle lamella and primary cell wall, frequently referred to as the compound middle lamella, is rich in pectin and xyloglucan (XG) and is heavily lignified, whereas the inner thicker secondary-wall layers are enriched in cellulose and in hemicelluloses such as xylans and mannans, but are thought to contain relatively less lignin. Secondary-wall layers comprise the bulk of wood biomass and are traditionally believed to govern wood properties. However, hydrolysis of XG, a primary wall-layer hemicellulose in Populus, has been unexpectedly shown to affect solid wood traits [3] and accelerate the lignocellulose saccharification [4]. Moreover, a recent study has shown that alleles of the Eucalyptus genes PME6 and PME7, encoding pectin methyl esterases, are associated with solid-wood quality properties, suggesting that pectin structure significantly influences wood properties [5].
Pectins in wood cell walls are composed mainly of homogalacturonan (HG) and rhamnogalacturonan I (RGI), which are concentrated in a thin outermost coat of the compound middle lamella, and have not been reported in xylan-containing secondary-wall layers [2]. The backbones of different pectin polymers are thought to be covalently connected, forming a supramolecular network linked to other cell-wall polymers via unknown bonds [6]. A recent study has identified a low abundance proteoglycan covalently linking pectins, arabinogalactan (AG) and arabinoxylan [7]. The final structure of the pectin network with its associated polymers is determined by activities of wall-residing trans-glycosylases, esterases, hydrolases and pectate lyases (PELs) [8].
Plant PELs (EC 4.2.2.2), which belong to polysaccharide lyase family 1 (PL1) (http://www.cazy.org), cleave the α-1,4 glycosidic bond between the galacturonic acid units of HG by β-elimination and release unsaturated oligogalacturonides. Genome-wide expression analyses in Arabidopsis have suggested that this gene family is involved in growth, cell adhesion and primary cell wall decomposition [9,10]. A PEL, encoded by ZePel, also appears to be involved in the differentiation of tracheary elements in Zinnia elegans[11]. In Populus, PEL genes have been assigned the CAZy family name PL1 and consecutive numbers PL1-1 to PL1-28[12]. Many of these genes are highly expressed during xylem cell expansion [13]. PtxtPL1-27 has been found to be highly upregulated in transgenic hybrid aspen (Populus tremula L. x tremuloides Michx.; Ptxt) with enhanced secondary growth caused by overexpression of GA20 oxidase [14]. Here we present a biochemical characterization of the recombinant PtxtPL1-27 protein and an analysis of the effects of its overexpression in hybrid aspen. The results indicate that PtxtPL1-27 can substantially modify the extractability of polymers from xylem cell walls, enhancing saccharification. Although pectin degradation has been shown previously to enhance saccharification of stems in herbaceous plants [15], it was surprising to find that it also affects saccharification in the case of aspen wood. Our analyses showed however that these effects in aspen are qualitatively and quantitatively different than those reported for herbaceous plants.
Results and discussion
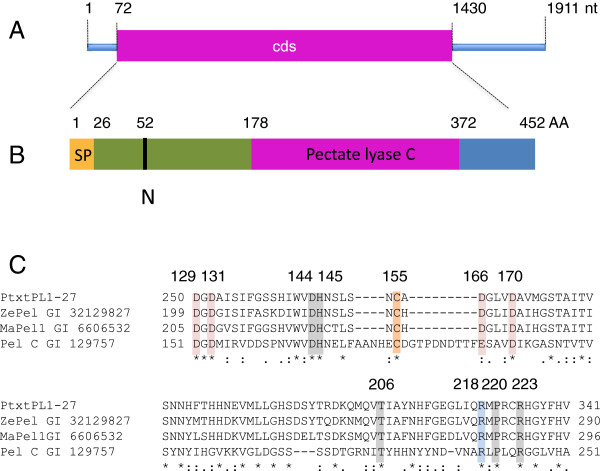
An EST clone, A053p77u [GenBank: AI164054], of hybrid aspen corresponding to the Populus trichocarpa gene PtPL1-27[12] represented the most upregulated gene in a microarray study of fast-growing GA20 oxidase overexpressing lines [14]. The corresponding 1911 nucleotide (nt)-long transcript was isolated by 5′ RACE [GenBank: EU379971.1.] and its length was confirmed by northern blot analysis (Additional file 1). The isolated transcript contains an open reading frame of 1359 nt and has 99.6% identity at the amino acid level to PtPL1-27 (Figure 1A). Thus, we named this cDNA PtxtPL1-27. The deduced mature protein encoded by the cloned cDNA has a molecular mass of 47 kDa, an isoelectric point of 7.75 (Figure 1B,C), and contains all of the conserved residues of PELs known to be involved in Ca2+-binding, substrate binding and catalysis [16].
Figure 1.
Structural features of PtxtPL1-27. (A) Structure of mRNA [GenBank: EU379971.1]. Cds, coding sequence; nt, nucleotides. (B) Structure of the deduced peptide showing the signal peptide (SP), the pectate lyase C catalytic domain, and the predicted N-glycosylation site (N). AA, amino acids. (C) A multiple alignment of the catalytic site of PtxtPL1-27 and pectate lyases of known enzymatic activities, showing the conserved residues involved in Ca+2 binding (pink), disulfide bonds (orange), catalysis (blue), and substrate binding (gray) with numbering according to Pel C structure as in [16]. ZePel, Zinnia elegans pectate lyase; MaPel1, Musa acuminata pectate lyase 1; Pel C, Erwinia chrysanthemi pectate lyase C.
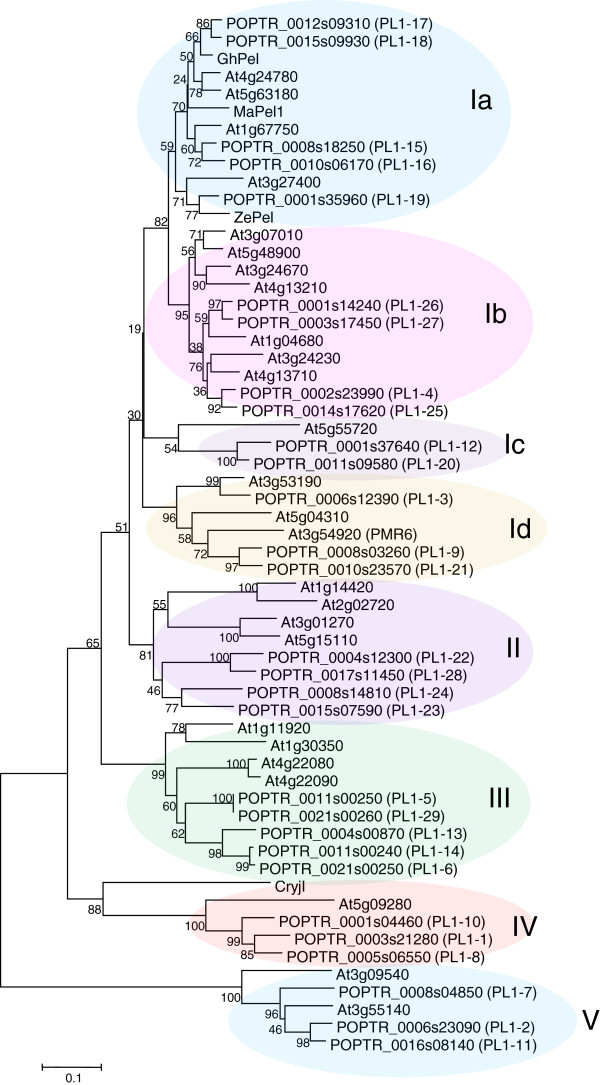
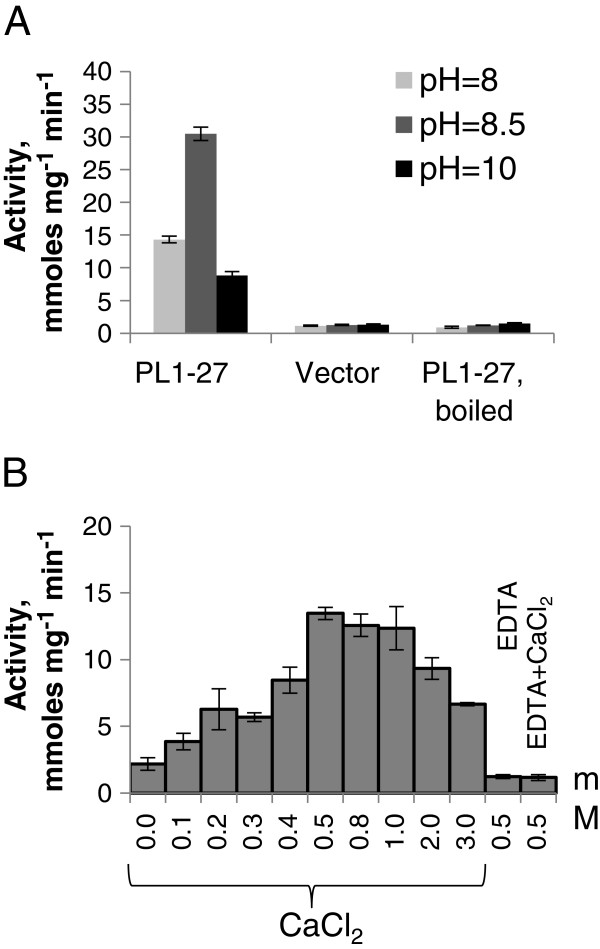
Phylogenetic analysis of 26 Arabidopsis thaliana[10] and 29 Populus trichocarpa PL1 members (Additional file 2) revealed five major clades, with clade I further divided into four sub-clades (Figure 2). PL1-27 belongs to clade Ib. Plant PEL-like genes encoding proteins with experimentally validated PEL activity are members of clades Ia (ZelPel [11]), MaPel1 [17], GhPel [18]) or clade IV (Cry j I [19]). To characterize the enzymatic activity of PtxtPL1-27, we expressed the mature protein in Escherichia coli and partially purified it by affinity chromatography (Additional file 3). The purified protein displayed PEL activity (4.2.2.2) towards polygalacturonic acid substrate, releasing 30 mmol of unsaturated uronides min-1 mg-1 at pH 8.5 (Figure 3). This is comparable to the specific activity of cotton GhPel [18], and much higher than that of ZelPel [11], CryJ I [19] and MaPel1 [17]. As is typical for PELs of family PL1 [20], the PEL activity of PtxtPL1-27 is optimal at basic pH, stimulated by Ca+2 and inhibited by ethylenediaminetetraacetic acid (EDTA).
Figure 2.
Phylogenetic tree of pectate lyases in Arabidopsis thaliana, Populus trichocarpa, and other plants. The tree was constructed by the neighbor-joining method using MEGA5. The Populus gene numbering is consistent with [12] and family designation (I-V) is consistent with [10]. The optimal tree with the sum of branch length = 5.9 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. Cryj1, Cryptomeria japonica pectate lyase [GenBank: BAA05542]; GhPel, Gossypium hirsutum pectate lyase [GenBank: ADB90478]; MaPel1, Musa acuminata pectate lyase 1 [GenBank: AAF19195]; ZePel, Zinnia elegans pectate lyase [GenBank: Y09541].
Figure 3.
PEL activity of PtxtPL1-27 expressed in Escherichia coli. PEL activity was measured as the formation of unsaturated uronides. Effect of pH (A) and Ca2+ ions (B). Means of five replicates ± standard error. Vector denotes the proteins extracted from the bacteria expressing the vector without PtxtPL1-27. EDTA, ethylenediaminetetraacetic acid.
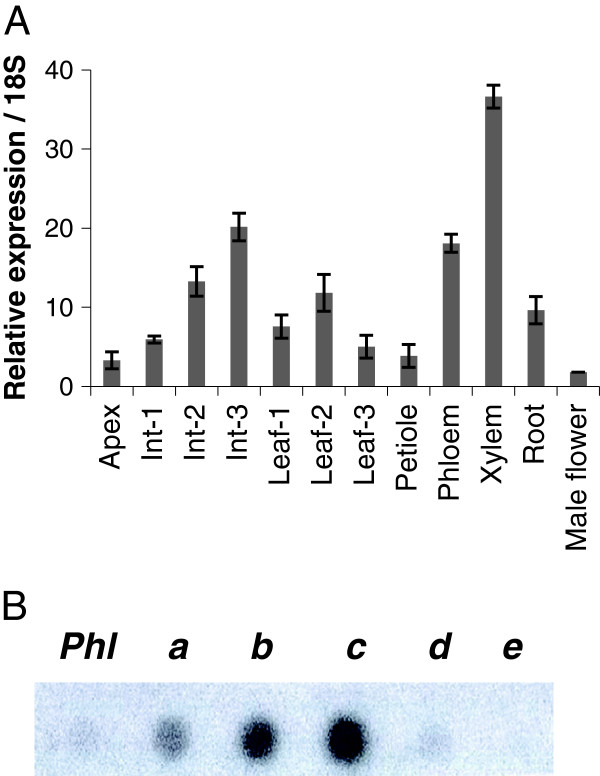
Real time RT-PCR analysis was used to determine PtxtPL1-27 transcript distribution in different organs and tissues of hybrid aspen. The analysis showed the highest expression in developing secondary xylem (Figure 4A). In the stem, the expression was higher in fully elongated internodes than in partially elongated internodes. Transcript profiling of the cambial region by tangential sectioning revealed that expression was highest in the transition zone, where xylem cell expansion declines and secondary cell-wall deposition begins (Figure 4B). These data indicate that PtxtPL1-27 is expressed at a later stage of xylem cell development than other known pectin-degrading enzymes [13], outside the region of the IAA peak [21] where major cell expansion takes place and cell walls are expected to have low pH as a result of IAA action [22]. This expression pattern, along with the high pH optimum for PtxtPL1-27 enzymatic activity (Figure 3A), suggests that the enzyme may be involved in cell-wall modification during the late stage of xylem differentiation and even the secondary walled stage rather than in cell expansion.
Figure 4.
PtxtPL1-27 expression analysis in aspen. (A) Real time RT-PCR analysis of PtxtPL1-27 expression in different tissues and organs of hybrid aspen indicates the highest expression of this gene in developing xylem and organs containing this tissue. Data shown are means of two biological replicates ± standard error. (B) Expression of PtxtPL1-27 in different wood development zones isolated by tangential sectioning. The same amount of total RNA from each zone was blotted onto a membrane and hybridized with a radiolabeled probe corresponding to PtxtPL1-27 cDNA. Phl, phloem; a, vascular cambium; b, expansion zone; c, transition zone; d, secondary wall formation zone; e, cell death zone.
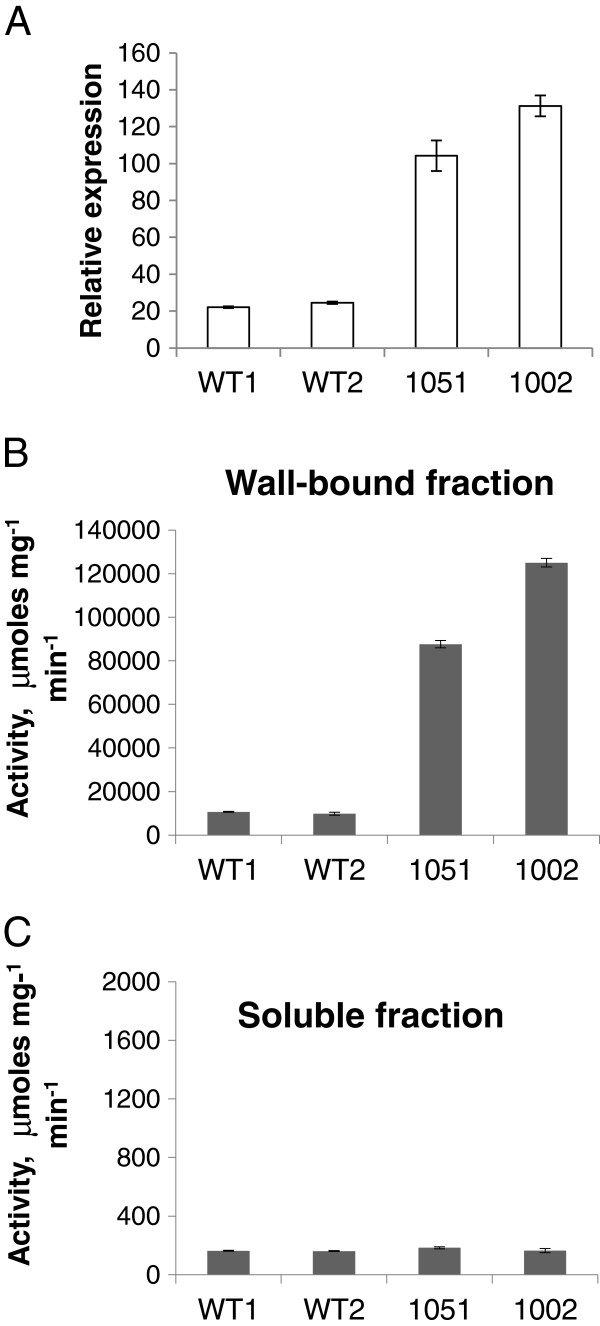
To investigate effects of PtxtPL1-27 on developing wood, its coding sequence was overexpressed in aspen under the control of the CaMV 35S. The two lines that showed the highest expression levels out of the 20 lines that were obtained were selected for chemical wood analyses. Compared with wild-type (WT) counterparts, PtxtPL1-27 transcript levels were five to six times higher in the secondary-walled developing xylem (Figure 5A), and PEL activity was ten to twelve times higher in the wall-bound protein fraction in samples from these lines, whereas no change in the soluble fraction was observed (Figure 5B,C). In addition, the ectopic expression of PtxtPL1-27 resulted in growth inhibition (Additional file 4) similar to that observed after ectopic overexpression of polygalacturonase [15]. To account for differences in growth rates between the lines, wood samples were taken from the same internodes (40 to 44) to compare samples of the same cambial age in plastochrons. Basic wood chemical analyses did not reveal any major changes in the transgenic lines, only minor increases in lignin and trifluoroacetic acid (TFA)-soluble Glc contents, and a small decrease in Man contents (Table 1). Contrary to our expectations based on the activity of PtxtPL1-27, we did not observe significant reduction in the uronic acid (UA) contents of the wood cell walls in the transgenic lines (Table 1). It is possible that there is a substantial fraction of glucuronic acid from glucuronoxylan in total wood uronic acids [23], and therefore changes in galacturonic acid in the pectin fraction of the transgenic lines are proportionally small and remain undetected. On the other hand, it is also possible that HG, after being cleaved by PEL, remains in cell walls and is held by interacting polymers. In apple leaves, the overexpression of PG did not lead to overall decrease in UAs but to increased HG extractability [24]. Therefore, we examined UA contents in pectin fractions sequentially extracted from cell walls by water and 1,2-cyclohexylenedinitrilotetraacetic acid (CDTA) 1 M potassium hydroxide (KOH) and 4 M KOH, and their contents remaining in the residual wall pellets in the transgenic lines and in WT (Figure 6). Significantly higher content of UAs in the CDTA extract and a lower content in the 1 M KOH extract were recorded in the transgenic lines compared to WT. These results suggest that the overexpressed PEL induced important changes in extractability of uronic acid-containing polymers.
Figure 5.
Overexpression of PtxtPL1-27 in developing wood of aspen. Relative PtxtPL1-27 transcript level determined by RT-qPCR and calibrated to 18S RNA (A) and PEL activity in wall-bound and soluble protein fractions (B, C) in developing xylem in the most highly expressing independent transgenic lines carrying 35S: PtxtPL1-27 (1051, 1002) and wild-type (WT). Means of two experiments, each with three biological replicates. WT1 and WT2 represent two sets of randomly selected WT trees. Bars represent standard error.
Table 1.
Lignin, hydrolyzable sugar anhydrate content and crystalline cellulose content of aspen wood
| Wild-type | 1051 | 1002 | |
|---|---|---|---|
| Klason lignin |
16.8 ± 0.21* |
17.7 ± 0.3** |
17.1 ± 0.3 |
| Acid soluble lignin |
2.1 ± 0.0 |
2.1 ± 0.0 |
2.0 ± 0.1 |
| Cellulose |
364 ± 28 |
346 ± 62 |
363 ± 28 |
| Rha |
11.7 ± 1.2 |
9.2 ± 0.6 |
9.0 ± 1.2 |
| Ara |
5.6 ± 0.1 |
6.8 ± 0.4 |
6.1 ± 0.5 |
| Xyl |
312.4 ± 5.7 |
304.1 ± 6.4 |
317.8 ± 2.1 |
| Man |
20.5 ± 0.4** |
18.8 ± 0.6 |
16.1 ± 0.9** |
| Gal |
11.5 ± 0.9 |
13.3 ± 0.5 |
13.1 ± 1.0 |
| Glc |
178.8 ± 0.4* |
187.2 ± 2.3 |
191.9 ± 4.8 |
| Total sugar |
540.4 ± 7.4 |
539.4 ± 5.3 |
554.1 ± 6.8 |
| Uronic acids | 31.8 ± 2.4 | 30.4 ± 2.0 | 27.9 ± 1.5 |
Lignin content is expressed in % (w/w). Sugar content is expressed in μg/mg of wood de-starched alcohol-insoluble residue (AIR). Means ± standard error, n = 10 or 2 biological replicates for lignin and sugars, respectively.
1Averages for the transgenic lines in bold are significantly different from wild-type (WT) (post analysis-of-variance (ANOVA) t-test). Averages for WT in bold are significantly different from both transgenic lines taken together (post-ANOVA contrast). *P ≤10%; **P ≤5%.
Figure 6.
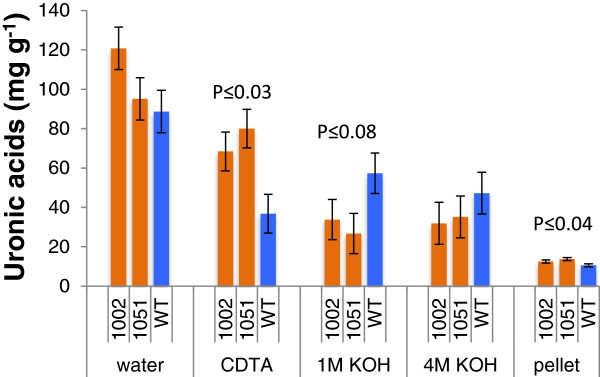
Overexpression of PtxtPL1-27 affects extractability of acidic polymers from wood. Wood alcohol-insoluble residues (AIRs) were sequentially extracted with water, 1,2-cyclohexylenedinitrilotetraacetic acid (CDTA) 1 M potassium hydroxide (KOH) and 4 M KOH, and the amounts of uronic acids extracted by each solvent and remaining in the pellet was determined by the biphenyl assay and expressed per weight of extracted cell wall material. Data are means of three biological replicates and standard error. Probability refers to the post analysis-of-variance analysis by contrast between transgenic lines and wild-type (WT).
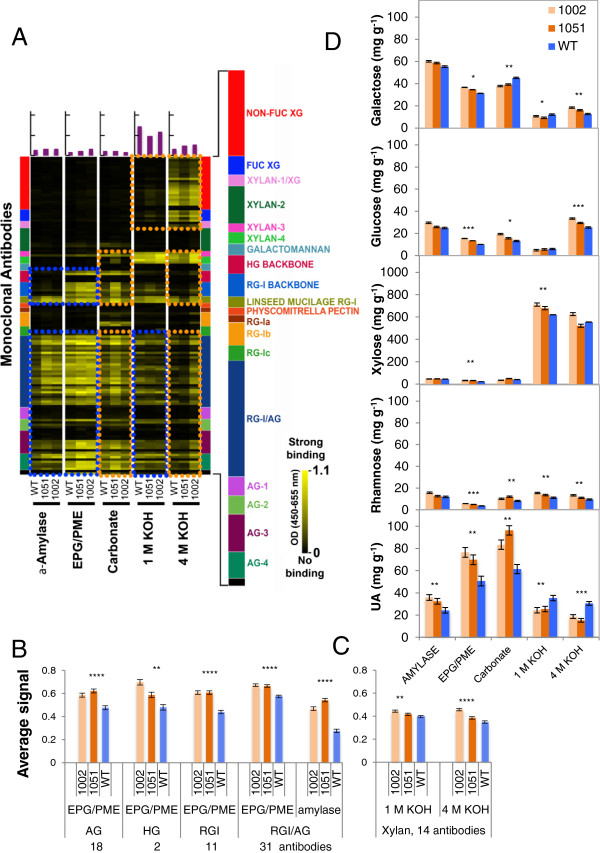
To identify the polymers with extractability that was affected by PEL overexpression, we applied antibody-based glycome profiling [25-28] to the sequential extracts of wood from PtxtPL1-27-overexpressing and WT saplings, prepared using α-amylase, endo-polygalacturonase (EPG)/pectin methylesterase (PME), CaCO3, 1 M KOH and 4 M KOH. Signals from most of 150 monoclonal antibodies used in the profiling ([26]; Additional file 5) showed clear qualitative differences between genotypes (Figure 7A). The main differences between the two overexpressing lines and WT plants were the increased signals for many typical epitopes of pectins and AGs in the α-amylase and/or EPG/PME extracts (and in some cases, weaker or stronger in 1 M KOH fractions) of the transgenics (blue boxes; Figure 7A), strongly indicating that overexpression of PtxtPL1-27 increased the extractability of these polymers. The signals stronger than optical density (OD) = 0.1 from these polymers were statistically analyzed, and significant changes are shown in Figure 7B. The increased amounts of HG, RGI and AG epitopes in EPG/PME extracts are supported by the increased contents of UAs, Rha and Gal in this fraction (Figure 7D). Other differences were noted in comparing the glycome profiles of the overexpressing lines with WT, particularly in the carbonate and 4 M KOH extracts (orange boxes, Figure 7A). In particular, higher signals with xylan-directed antibodies in line 1002 were noted in the 4 M KOH extract. However, these differences were not always consistent between the two overexpressing lines, making it difficult to correlate these changes directly with the overexpression of PtxtPL1-27. It was also evident that few of the anti-xylan antibodies included in these initial studies reacted strongly with aspen hemicelluloses. To address the question of xylan extractability, we expanded the panel of xylan-directed antibodies (http://www.wallmabdb.net), and analyzed aspen 1 M KOH and 4 M KOH extracts. Fourteen of these xylan-directed antibodies bound strongly to WT aspen hemicellulose extracts (OD ≥0.1) (Additional file 5). Average ELISA signals of these monoclonal antibodies showed increases in transgenic lines in both 1 M KOH and 4 M KOH extracts compared to the corresponding extracts from WT walls (Figure 7C), indicating increased extractability of xylan. These results are in agreement with the changes seen in xylose content in the 1 M KOH extract (Figure 7D and Additional file 6). In summary, the glycome profiling of sequential wall extracts indicated increased solubility of pectins, AGs and xylans in the transgenic lines.
Figure 7.
Overexpression of PtxtPL1-27 affects extractability of many polymers of wood. Sequential cell wall extracts, obtained using α-amylase, endo-polygalacturonase (EPG)/ pectin methylesterase (PME), sodium carbonate, 1 M potassium hydroxide (KOH) and 4 M KOH, were probed with a panel of 150 monoclonal antibodies (A-C) and analyzed for the monosaccharide composition (D). (A-C) Extracts were screened against an array of plant glycan-directed monoclonal antibodies using ELISA. (A) Antibody binding (average of two biological replicates) is depicted as colored heat maps; bright yellow, maximal binding; black, no binding. The colored panel (right) depicts groups of antibodies used, identified according to polysaccharides predominantly recognized by each group [26] (list of antibodies provided in Additional file 5). Colored boxes indicate regions of the glycome profiles that were altered in both transgenic lines (blue boxes) or predominantly in one line (orange boxes) compared to wild-type (WT). (B) Signals ≥0.1 from pectin- and arabinogalactan (AG)-directed antibodies recognizing epitopes in α-amylase and EPG/PME extracts. (C) Signals ≥0.1 from an expanded set of xylan-directed antibodies specifically recognizing epitopes in aspen wood hemicelluloses in 1 M KOH and 4 M KOH extracts. (D) Monosaccharide composition of sequential extracts. Only monosaccharides that differed in content in extracts between PtxtPL1-27 overexpressing lines and the WT are presented (full dataset in Additional file 6). Data in B-D are means of two biological replicates ± standard error. Asterisks indicate probability for significance of difference between transgenic lines and WT (post analysis-of-variance contrast: *P ≤10%, **P ≤5%, ***P ≤1%, ****P ≤0.1%).
In conclusion, the above cell-wall analyses revealed that PEL overexpression induced no major changes in the composition of cell walls in the wood, but substantially altered extractabilities of pectins, AGs and xylans in the wood. These results concerning the cells with secondary cell-walls are reminiscent of the previously published data on primary-walled tissues. In strawberry fruits, the downregulation of PEL expression was found to reduce the solubility of UAs and neutral sugars [29]. Thus, in both primary and secondary walls, the integrity of an enzyme-accessible HG domain appears to influence the mobilization of major cell-wall matrix components (HG, RGI, xylan and AG). Several previous studies have suggested that pectins and hemicelluloses are cross-linked in primary walls [28-34]. A covalent linkage between RGI, HG, and arabinoxylan involving an AG protein has been identified recently in the Arabidopsis cell suspension culture medium [7]. It is possible that similar covalent linkages bind HG to AG and glucuronoxylan in the wood cell walls of aspen, and therefore the fragmentation of HG by PEL increases solubility of xylan and AG (Figure 7). Another possibility is that PEL activity alters pectin structure in a way that influences lignin polymerization, either by affecting lignin nucleation sites or activities of laccases or peroxidases [35,36]. Slightly increased Klason lignin contents were seen in the transgenic lines (Table 1) and might have affected polysaccharide extractability. Yet another possibility is that PEL increases cell wall porosity, thus facilitating access of native cell-wall endohydrolases to their substrates [8], thereby making these polymers more soluble.
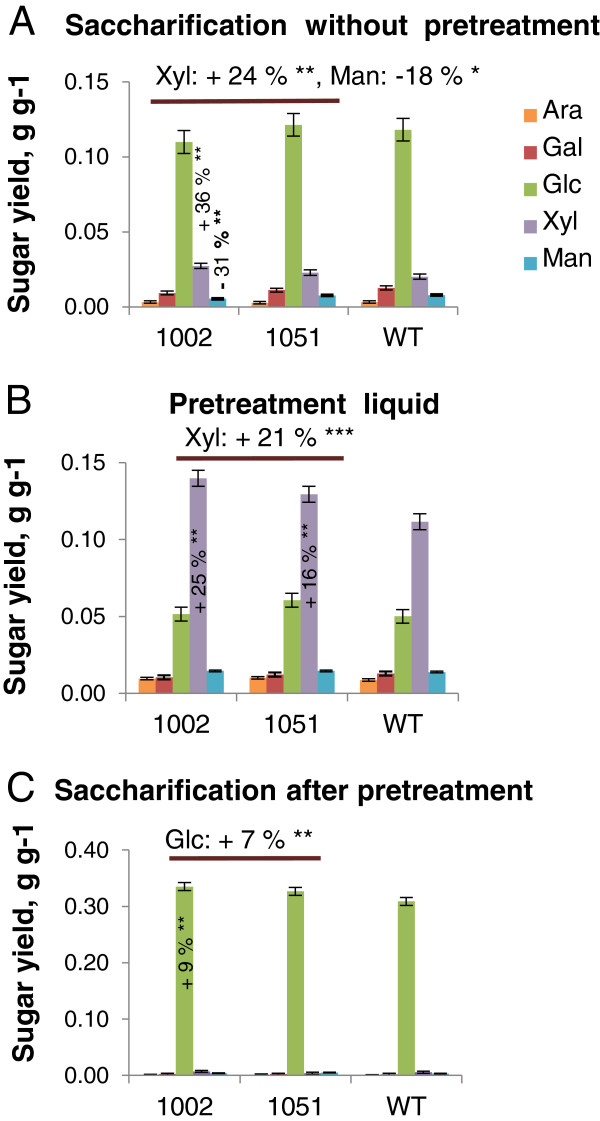
To test the possibility that altered extractability of xylan and other matrix polymers improves the digestibility of lignocellulose, milled wood of the transgenic and WT lines was enzymatically hydrolyzed with and without acid pretreatment. Without pretreatment, glucose yields from the transgenic lines were not altered, but their xylose yields were 24% higher than WT yields (Figure 8). An 18% reduction in mannose yield was also observed, probably reflecting a similar reduction in mannose levels in the starting material (Table 1) rather than a change in mannan digestibility. Acid pretreatment resulted in the release of 21% more xylose, and subsequent enzymatic digestion yielded 7% more glucose from transgenic lines than from WT. As the liquid fraction obtained after pretreatment was removed before enzymatic hydrolysis of cellulose and as no increase in the cellulose content of the transgenic lines was detected (Table 1), the increased glucose yield after pretreatment (Figure 8C) could mainly be attributed to improved digestibility of cellulose. Overall, the combined yields of pentoses, hexoses and all monosaccharides from acid pretreatment and enzymatic saccharification were increased by up to 24 ± 0.08%, 8 ± 0.03% and 11 ± 0.02%, respectively (Table 2). These results contrast with those obtained with herbaceous plants that ectopically express fungal polygalacturonase or plant pectin methyl esterase inhibitor [15], in which saccharification efficiency was increased without pretreatment, but not when acid pretreatment was applied. In further contrast to our results, the increased saccharification (without pretreatment) in material from herbaceous plants was due to increases in glucose yields whereas xylose yields were not affected. It is likely that in primary walls, the removal of pectin increases accessibility of cellulose to enzymes. The accessibility of cellulose in secondary walls has been shown to be strongly influenced by their xylan and lignin contents [37-39], and xylan acid hydrolysis yields are reportedly inversely correlated to the syringyl/guaiacyl (S/G) ratio [40]. However, the effects of pectin structure and composition on these variables have not been studied previously in woody plants. This study shows the positive effect of pectin degradation on saccharification in woody tissues (Figure 8, Table 2).
Figure 8.
Overexpression of PtxtPL1-27 increases sugar yields from saccharification. Sugar yields after 72 h of saccharification without pretreatment (A), after acid pretreatment (B), and after 72 h saccharification following acid pretreatment (C). Means of five biological replicates ± standard error. Percentage differences are shown for individual lines significantly differing from wild-type (WT) (post analysis-of-variance (ANOVA) t-test), and average differences for both transgenic lines when significantly different from WT (post ANOVA contrast between transgenic lines and WT) *P ≤10%, **P ≤5%, ***P ≤1%.
Table 2.
Total sugar yields for transgenic aspen lines and wild-type after acidic pretreatment
| Line |
Total sugar yields (pretreatment liquid and enzymatic hydrolysate) |
||
|---|---|---|---|
| Y Hexoses/W (g g -1 ) a | Y Pentoses/W (g g -1 ) b | Y Total/Wv (g g -1 ) c | |
|
Wild-type |
0.390 ± 0.01 (100%)** |
0.126 ± 0.01 (100%)*** |
0.517 ± 0.01 (100%)*** |
|
1051 |
0.421 ± 0.01 (108%)** |
0.144 ± 0.01 (114%)** |
0.565 ± 0.01 (109%)** |
| 1002 | 0.418 ± 0.03 (107%) | 0.157 ± 0.01 (124%)** | 0.574 ± 0.01 (111%)** |
The % values represent the sugar yield in comparison with wild-type (WT). Means ± standard error, n = 5 biological replicates. aYield of hexoses (g of Gal, Glc, and Man) per g of wood after pretreatment and after 72 h of enzymatic hydrolysis. bYield of pentoses (g of Ara and Xyl) per g of wood after pretreatment and after 72 h of enzymatic hydrolysis. cYield of monosaccharides (g of Ara, Gal, Glc, Man and Xyl) per g of wood after pretreatment and after 72 h of enzymatic hydrolysis. Asterisks (by means for individual lines) indicate yields significantly different from WT when analyzed by post analysis-of-variance (ANOVA) t-test or means (for WT) that were significantly different from both lines by post ANOVA contrast. **P ≤5%, ***P ≤1%.
Since the transgenic lines gave higher enzymatic glucose yields and had slightly higher TFA-soluble glucose contents, we hypothesized that cellulose crystallinity may be affected by PtxtPL1-27 overexpression. To test this hypothesis, crystallite structures in the wood of transgenic and WT lines were examined by x-ray diffraction. However, no major differences in either the crystallite width or overall crystallinity were observed (Table 3). Thus, the higher glucose yields could be partly explained by an increase in TFA-soluble glucan contents and partly by increased accessibility of cellulose due to better xylan extraction during pretreatment (Figure 8B). The latter effect might be directly related to increased solubility of xylan observed in the transgenic lines (Figure 7). Another likely possibility is that PEL activity erodes the outer pectin coat of middle lamella, thereby increasing access of chemicals and enzymes to secondary walls. Interestingly, the relative increase in glucose yields was smaller compared to that on xylose yields. Direct application of pectin-degrading enzymes (EPG/PME) to woody lignocellulose from poplar has been also shown to solubilize xylan, but the subsequent cellulose digestion was not affected by this treatment [41]. These results suggest that the effects of pectin degradation might differ between in vitro and in vivo conditions.
Table 3.
Cellulose crystallite structure in transgenic lines and wild-type determined by x-ray diffraction
| Line | Crystallinity % | Crystallite width Å |
|---|---|---|
| Wild-type |
28.7 ± 1.5 |
29.9 ± 0.1 |
| 1051 |
29.4 ± 1.8 |
30.0 ± 0.2 |
| 1002 | 30.7 ± 1.7 | 30.2 ± 0.1 |
Means ± standard error, n = 5 to 8 biological replicates. Å – ångström.
Conclusions
In overall conclusion, we have shown that PtxtPL1-27 from clade Ib of the PL1 family is a PEL, which is expressed in differentiating xylem at the onset of secondary wall formation. When overexpressed, it is capable of loosening several matrix components from secondary-walled xylem cells without significantly affecting their overall cell wall composition. PEL activity is beneficial for lignocellulose saccharification, increasing yields of monosaccharides, especially xylose, with and without pretreatment. Thus, pectin structure may affect wood saccharification even though it is a minor wood component. These findings could be exploited in several ways. First, our results could be used to design more efficient saccharification cocktails for deconstruction of woody biomass. Second, our results suggest that targeting pectin in the genetic improvement programs for woody feedstocks might prove beneficial for subsequent processing and deconstruction of woody biomass. Third, genetic engineering targeted toward pectin-modification in woody plants might improve their suitability as feedstock for biofuel production. They also show that the genetic engineering strategy would need to be targeted to mature woody tissues in order to avoid negative effects on plant growth and development that have been observed in this and other studies [15,42].
Materials and methods
Cloning of PtxtPL1-27
Full-length cDNA was cloned from the hybrid aspen cDNA from elongating shoots by 5′ RACE using Marathon (Clontech Laboratories Inc., Mountain View, CA, USA), and the gene-specific primer Race1 (Tab. S3) based on AI164054. The complete cDNA was reconstructed by PCR using Advantage PCR Cloning Kit (Clontech) with F1 and R1 primers that included BamHI restriction sites (Additional file 7) and hybrid aspen cDNA. The PCR product was sub-cloned, sequenced and named PtxtPL1-27 [GenBank: EU379971.1].
Heterologous expression and enzyme assays
Mature PtxtPL1-27 peptide (Δ1-26PtxtPL1-27) was expressed in E. coli (ER2566) using the pTYB11 vector (IMPACT™-CN System, New England Biolabs Inc., Ipswich, MA, USA) carrying the PCR product obtained with the primers MF1 and MR1 (Additional file 7). Cells containing the recombinant plasmid were grown in LB medium with 100 μg/ml carbenicillin to an OD of A600 = 0.5. Expression of the recombinant gene was then induced by incubating the cells with 0.3 mM isopropyl β-D-thiogalactoside (IPTG) at 14°C for 15 h. The cells were resuspended in 20 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) 500 mM NaCl and 1 mM EDTA, pH 8.0, lysed by sonication on ice, and centrifuged at 20,000 g for 30 minutes at 4°C. The pellet was dissolved in the same buffer and purified using an IMPACT™-CN chitin beads column (New England Biolabs). PEL activity (EC 4.2.2.2) was assayed in the column-purified protein extract according to Collmer et al.[43] using 0.24% w/v of polygalacturonic acid (Sigma-P3889) in 60 mM Tris–HCl, pH 8.5 (unless otherwise indicated), 0.5 mM CaCl2, and 0.3 ml of protein extract in 1.5 ml reaction mixtures. The unsaturated uronide products were measured by absorbance at 235 nm over 180 minutes at 30°C using the molar extinction coefficient 4.6 × 103 M-1 cm-1. Specific activity was expressed per mg of protein as determined by a Bio-Rad Assay kit (Life Science, Sunbyberg, Sweden) using BSA as a protein standard.
Generation of transgenic aspen lines
The full-length PtxtPL1-27 cDNA clone lacking 5′ and 3′ untranslated regions (UTRs) was released by digestion with XbaI and SalI and ligated in sense orientation into the plant binary vector pMH1.kana (a gift from Mattias Holmlund, SLU, Umeå, Sweden) under control of the CaMV 35S promoter. The construct was transformed into hybrid aspen clone T89 by Agrobacterium-mediated transformation, and plants were regenerated, planted in the greenhouse and grown for approximately three months, as previously described [42]. Briefly, the growing conditions were: 18-h photoperiod, a temperature regime of 22°C/17°C (day/night), and a relative humidity of 70%.
Analysis of RNA in the transgenic and WT lines
RNA was isolated from developing xylem scraped from the wood surface after peeling the bark, cambium and phloem scraped from the exposed bark surface, root tips approximately 0.5 cm long, internodes up to 50%, 50 to 80%, and 100% of their final length, designated internodes 1, 2 and 3, leaves subtending these internodes (leaf 3 was fully expanded), the apical bud and petioles from internodes 1 and 2, pooled. In addition, we extracted RNA from a series of samples of tangentially sectioned developing wood. Total RNA was extracted by hexadecyl-trimethylammonium bromide (CTAB) and purified as previously described [42].
cDNA was synthesized from the total RNA samples using a blend of oligo (dT) and random primers and an iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Dot blot was prepared as described previously [42] and probed under stringent conditions with a probe based on AI164054. Primers for real-time PCR were designed using Beacon Designer (v 2.1, Premier Biosoft International, Palo Alto, CA, USA) and tested for efficiency and specificity. The selected primers are listed in Additional file 7. PCR reactions were performed in triplicate using a Bio-Rad iCycler MyiQ Real-Time PCR Detection System and products were detected with SYBR Green (iQ SYBR Green Supermix). The threshold cycle (Ct) was calculated using iCycler MyiQ software 1.0 (Bio-Rad, Hercules, CA, USA). The PCR efficiencies (E) were determined using 4-fold serial dilutions of pooled cDNA originating from the assayed tissues. Relative transcript levels were then calculated as:
Protein extraction and PEL activity in aspen
Proteins were extracted from scraped xylem from internodes 44 to 60: 5 g of tissue were ground in liquid N2 to a fine powder, stirred for 30 minutes at 4°C in 25 ml of buffer A (50 mM sodium phosphate, pH 7.0, 2 mM EDTA, 4% polyvinylpyrolidone (PVP) mw 360 000, 1 mM dithiothreitol (DTT), and centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant was collected as the soluble fraction and the pellet was re-suspended in 25 ml of buffer A supplemented with 1 M NaCl, stirred for 30 minutes at 4°C, centrifuged as above, and the supernatant was collected as the wall-bound fraction. Saturated ammonium sulfate (AS) was added to reach 20% and 19% of saturation, to the soluble- and wall-bound fractions, respectively, stirred for 30 minutes at 4°C, and centrifuged as above to remove PVP from solution. Solid AS was added to the supernatants, increasing the AS saturation to 90% and 80% for the soluble- and wall-bound fractions, respectively, and stirred for 30 minutes at 4°C to precipitate proteins. Following the centrifugation as above, the pellets were dissolved in 1 ml TE buffer, pH 8.0. The dissolved proteins were desalted with PD10 columns and protein concentrations were determined by the Bradford method using the Bio-Rad Assay kit (Life Science). Pectate lyase activity was assayed as described above using 0.3 ml of extracted proteins.
Wood chemical analyses
Wood of internodes 40 to 44 was freeze-dried and ground using an A11 Basic Analytical Mill (IKA, Staufen, Germany) and then using an Ultra Centrifugal Mill ZM 200 equipped with a 0.5 mm ring sieve (Retsch) at 30 Hz for 150 sec. The contents of acid-resistant lignin (Klason lignin) and acid-soluble lignin were determined according to Theander and Westerlund [44].
Alcohol-insoluble residue (AIR) was obtained by washing the ground wood sequentially in 70% ethanol, methanol:chloroform 1:1 (v/v) and acetone, then drying overnight under vacuum. Starch was removed by treatment with Bacillus α-amylase (Sigma - A6380) at 5,000 units per g of AIR, 16 h at 37°C. Crystalline cellulose was purified by hydrolysing non-cellulosic polysaccharides with acetic acid:nitric acid:water (8:1:2, v/v/v) and removing the supernatant. The resulting pellet was washed four times with acetone, dried, hydrolysed in 72% sulphuric acid, and the glucose content was determined with the anthrone method [45].
Sequential extraction of the wood cell walls followed a previously published procedure [25]. The neutral glycosyl residue composition of non-fractionated and fractionated cell wall polysaccharides was analyzed by the alditol acetate method [46]. Briefly, after preparing alditol acetates, 1 μL samples were injected, splitless, into a Hewlett-Packard chromatograph 5890 (Ramsey, MI, USA) equipped with an SP 2330 (30 m × 0.25 mm, 0.25 μm film thickness, 24019 Supelco) column using helium as carrier gas, and the analytes were detected by a coupled mass spectrometer. UA contents of the cell wall polysaccharides were determined according to Filisetti-Cozzi and Carpita [47].
Cell wall extracts were diluted to 60 μg mL - 1 and 50 μL samples were used for glycome profiling, as previously described [25]. α-Amylase (type II-A from Bacillus) was obtained from Sigma (A6380), and EPG (I and II from Aspergillus niger) [48] and PME (from A. niger) [49] were donated by Carl Bergmann (Complex Carbohydrate Research Center, University of Georgia), and used at approximately 1.0 units/100 mg AIR in 50 mM sodium acetate, pH 5.0. Plant cell wall glycan-directed monoclonal antibodies [26] were obtained as hybridoma cell-culture supernatants either from laboratory stocks maintained by the Complex Carbohydrate Research Center (CCRC, JIM and MAC series; available from CarboSource Services (http://www.carbosource.net)) or from Fabienne Guillon (AX1 [27] (INRA, Nantes)).
Saccharification
Freeze-dried wood from internodes 21 to 39 was milled as above, followed by sieving with an Analytical Sieve Shaker AS 200 (Retsch) to obtain 0.1- to 0.5-mm particles. Pre-pretreatment (if applied) consisted of incubation of 50 mg milled and sieved wood in 1% (w/w) sulfuric acid at 165˚C for 10 minutes under stirring in a single-mode microwave system (Initiator Exp, Biotage, Uppsala, Sweden). After acid pretreatment, the suspension was centrifuged, the liquid phase (the pretreatment liquid) was removed, and the pellet was washed. The remaining solid (pretreated wood), and the untreated wood powder (50 mg), were treated with 50 mg of a liquid enzyme mixture consisting of equal proportions of Celluclast 1.5 L (a liquid enzyme preparation from Trichoderma reesei and the main source of cellulase) and Novozyme 188 (a liquid enzyme preparation from A. niger and the main source of β-glucosidase) (both from Sigma-Aldrich, St. Louis, MO, USA) in sodium citrate buffer (50 mM, pH 5.2), using 1,000 mg as the total size of the reaction mixture, at 45°C for 72 h in an Ecotron orbital shaker (Infors, Bottmingen, Switzerland) at 170 rpm.
Concentrations of monosaccharides (Ara, Gal, Glc, Man, and Xyl) in the samples were then determined using an HPAEC system (ICS3000, Dionex, Sunnyvale, CA, USA) equipped with a Pulsed Amperometric Detection system and a CarboPac PA20 column (3 × 150 mm) with a CarboPac PA20 guard column (3 × 30 mm) (Dionex).
Abbreviations
AG: arabinogalactan; AIR: alcohol-insoluble residue; ANOVA: analysis of variance; CDTA: 1,2-cyclohexylenedinitrilotetraacetic acid; EDTA: ethylenediaminetetraacetic acid; ELISA: enzyme-linked immunosorbent assay; EPG: endo-polygalacturonase; HG: homogalacturonan; KOH: potassium hydroxide; nt: nucleotide; OD: optical density; PEL: pectate lyase; PME: pectin methylesterase; RGI: rhamnogalacturonan I; SP: single nucleotide; TFA: trifluoroacetic acid; UA: uronic acid; WT: wild-type; XG: xyloglucan.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AKB participated in plant phenotyping, expression studies and cell wall analyses, carried out enzymology analyses, sequence alignment and helped to draft the manuscript. KS participated in generation of transgenic plants, phenotyping, and expression analyses. MLG participated in saccharification analyses. PI participated in cell wall analyses. SP participated in glycome profiling. JL participated in x-ray diffraction analysis. RS participated in x-ray diffraction analyses and in the design of research. MGH participated in glycome profiling and in the design of research. TM participated in the generation of transgenic lines and in the design of research. LJJ participated in saccharification analyses and in the design of research. MIN participated in generation of transgenic lines, phenotyping, expression analyses and in the design of research. EJM conceived the study, carried out statistical analyses, participated in the design, coordinated research and helped to draft the manuscript. All authors read, edited and approved the final manuscript.
Supplementary Material
Detection of PtxtPL1-27 transcript (EU379971.1) by northern blot. Northern blot analysis showing the size of PtxtPL1-27 transcript in the wild-type (WT) at 1.9 kb as compared to transcript size in four independent transgenic lines overexpressing PtxtPL1-27 ORF (without untranslated regions (UTRs)) under control of the CaMV 35S promoter at 1.4 kb. A polyubiquitin probe was used as a reference control. The RNA used in the northern blot analysis originated from developing xylem tissue.
Populus trichocarpa PL1 family genes. The complete list of Populus PL1 family gene models based on Populus trichocarpa assembly version 2.2, including current names, former models, closest Arabidopsis homologues, corresponding Populus ESTs from Populus DB, and protein domains.
SDS-PAGE showing the degree of purification of Δ 1-26 Ptxt PL1-27 expressed in E. coli. PtxtPL1-27 protein was expressed in E. coli using pTYB11 vector and partially purified by affinity chromatography. Proteins were separated by gel electrophoresis and stained by Coomassie. Lane 1: proteins extracted from E. coli expressing recombinant protein before induction. Lane 2: proteins extracted from E. coli expressing recombinant protein after induction by 0.3 mM isopropyl β-D-thiogalactoside (IPTG). The induced product can be seen at 98.8 kDa polypeptide representing PtxtPL1-27 fused to intein-chitin binding domain. Lane 3: partially purified recombinant protein is shown as 41.2 kDa band. The preparation is contaminated mainly by the 56.0 kDa polypeptide containing intein and chitin binding domain, which is the cleavage product of 98.8 kDa recombinant polypeptide.
Ectopic overexpression of PtxtPL1-27 inhibits plant growth. Data are relative stem increments and PtxtPL1-27 expression levels in elongating stem internodes of independent transgenic lines. Stem-length increments were determined as an increase in stem length during 64 days of growth in the greenhouse. Between two and eight plants per line were measured. Expression was determined by RT-q-PCR using 18S as a reference gene.
Antibodies used for the ELISA-based glycome profiling. The list of all monoclonal antibodies used in Figure 7.
Monosaccharide composition of wood sequential extracts and the resulting pellets. Sugar composition of wood extracts; means ± standard error, n = 2 biological replicates. Asterisks beside means for individual lines in bold type indicate values significantly different from wild-type (WT) by post analysis-of-variance (ANOVA) t-test. Asteriks beside the means for WT in bold type indicate values significantly different from both lines by post ANOVA contrast; *P ≤10%, **P ≤5%, ***P ≤1%.
List of PCR primers used in the study. Sequences of all forward and reverse PCR primers used in this study.
Contributor Information
Ajaya K Biswal, Email: biswalajaya@ccrc.uga.edu.
Kazuo Soeno, Email: soeno@affrc.go.jp.
Madhavi Latha Gandla, Email: gmadhavilatha7@gmail.com.
Peter Immerzeel, Email: Peter.Immerzeel@slu.se.
Sivakumar Pattathil, Email: siva@ccrc.uga.edu.
Jessica Lucenius, Email: jessica.lucenius@helsinki.fi.
Ritva Serimaa, Email: ritva.serimaa@helsinki.fi.
Michael G Hahn, Email: hahn@ccrc.uga.edu.
Thomas Moritz, Email: Thomas.Moritz@slu.se.
Leif J Jönsson, Email: leif.jonsson@chem.umu.se.
Maria Israelsson-Nordström, Email: maria.nordstrom@slu.se.
Ewa J Mellerowicz, Email: Ewa.Mellerowicz@slu.se.
Acknowledgements
We thank Mattias Holmlund for a gift of pMH1.kana vector and Jesper Harholt for fruitful discussions. The research was funded by FORMAS, the Swedish Research Council, UPSC Berzelii Center of Forest Biotechnology (funded by VR and Vinnova), Biomime Center (funded by the Swedish Foundation for Strategic Research, the Knut & Alice Wallenberg Foundation), Bio4Energy and SNS. The glycome profiling was supported by the BioEnergy Science Center administered by Oak Ridge National Laboratory and funded by a grant (DE-AC05-00OR22725) from the United States Department of Energy. Generation of the CCRC series of plant glycan-directed monoclonal antibodies was supported by the NSF Plant Genome Program (DBI-0421683).
References
- Somerville C, Youngs H, Taylor C, Davis SC, Long SP. Feedstocks for lignocellulosic biofuels. Science. 2010;329:790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Gorshkova TA. Tensional stress generation in gelatinous fibres: a review and possible mechanism based on cell wall structure and composition. J Exp Bot. 2012;63:551–565. doi: 10.1093/jxb/err339. [DOI] [PubMed] [Google Scholar]
- Park YW, Baba K, Furuta Y, Iida I, Sameshima K, Arai M, Hayashi T. Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett. 2004;564:183–187. doi: 10.1016/S0014-5793(04)00346-1. [DOI] [PubMed] [Google Scholar]
- Kaida R, Kaku T, Baba K, Oyadomari M, Watanabe T, Nishida K, Kanaya T, Shani Z, Shoseyov O, Hayashi T. Loosening xyloglucan accelerates the enzymatic degradation of cellulose in wood. Mol Plant. 2009;2:904–909. doi: 10.1093/mp/ssp060. [DOI] [PubMed] [Google Scholar]
- Sexton TR, Henry RJ, Harwood CE, Thomas DS, McManus LJ, Raymond C, Henson M, Shepherd M. Pectin methylesterase genes influence solid wood properties of Eucalyptus pilularis. Plant Physiol. 2012;158:531–541. doi: 10.1104/pp.111.181602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008;11:1–12. doi: 10.1016/j.pbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, Baldwin D, Pham C, Orlando R, Darvill A, Hahn MG, Kieliszewski MJ, Mohnen D. An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell. 2013;25:270–287. doi: 10.1105/tpc.112.107334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 2004;161:641–675. doi: 10.1111/j.1469-8137.2004.00980.x. [DOI] [PubMed] [Google Scholar]
- Palusa SG, Golovkin M, Shin SB, Richardson DN, Reddy AS. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007;174:537–550. doi: 10.1111/j.1469-8137.2007.02033.x. [DOI] [PubMed] [Google Scholar]
- Sun L, van Nocker S. Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 2010;22:152. doi: 10.1186/1471-2229-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Roberts K, Stacey NJ, Connerton I, Ruiz-Teran F, McCann MC. A pectate lyase from Zinnia elegans is auxin inducible. Plant J. 1998;13:17–28. doi: 10.1046/j.1365-313x.1998.00002.x. [DOI] [PubMed] [Google Scholar]
- Geisler-Lee J, Geisler M, Coutinho PM, Segerman B, Nishikubo N, Takahashi J, Aspeborg H, Djerbi S, Master E, Andersson-Gunnerås S, Sundberg B, Karpinski S, Teeri TT, Kleczkowski LA, Henrissat B, Mellerowicz EJ. Poplar carbohydrate-active enzymes (CAZymes). Gene identification and expression analyses. Plant Physiol. 2006;140:946–962. doi: 10.1104/pp.105.072652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Sundberg B. Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr Opin Plant Biol. 2008;11:293–300. doi: 10.1016/j.pbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Israelsson M, Eriksson ME, Hertzberg M, Aspeborg H, Nilsson P, Moritz T. Changes in gene expression in the wood-forming tissue of transgenic hybrid aspen with increased secondary growth. Plant Mol Biol. 2003;52:893–903. doi: 10.1023/A:1025097410445. [DOI] [PubMed] [Google Scholar]
- Lionetti V, Francocci F, Ferrari S, Volpi C, Bellincampi D, Galletti R, D’Ovidio R, de Lorenzo G, Cervone F. Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci USA. 2010;107:616–621. doi: 10.1073/pnas.0907549107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron SR, Scavetta RD, Garrett M, Legner M, Jurnak F. Characterization and implications of Ca2+ binding to pectate lyase C. J Biol Chem. 2003;278:12271–12277. doi: 10.1074/jbc.M209306200. [DOI] [PubMed] [Google Scholar]
- Marín-Rodríguez MC, Smith DL, Manning K, Orchard J, Seymour GB. Pectate lyase gene expression and enzyme activity in ripening banana fruit. Plant Mol Biol. 2003;51:851–857. doi: 10.1023/A:1023057202847. [DOI] [PubMed] [Google Scholar]
- Wang H, Guo Y, Lv F, Zhu H, Wu S, Jiang Y, Li F, Zhou B, Guo W, Zhang T. The essential role of GhPEL gene, encoding a pectate lyase, in cell wall loosening by depolymerization of the de-esterified pectin during fiber elongation in cotton. Plant Mol Biol. 2010;72:397–406. doi: 10.1007/s11103-009-9578-7. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Ono A, Sawatani M, Nanba M, Kohno K, Usui M, Kurimoto M, Matuhasi T. Cry j I, a major allergen of Japanese cedar pollen, has a pectate lyase enzyme activity. Allergy. 1995;50:90–93. doi: 10.1111/j.1398-9995.1995.tb02489.x. [DOI] [PubMed] [Google Scholar]
- Garron M-L, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- Tuominen H, Puech L, Fink S, Sunberg B. A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 1997;115:577–585. doi: 10.1104/pp.115.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. Role of the plasma membrane H + -ATPase in auxin-induced elongation growth: historical and new aspects. J Plant Res. 2003;116:483–505. doi: 10.1007/s10265-003-0110-x. [DOI] [PubMed] [Google Scholar]
- Bose SK, Barber VA, Alves EF, Kiemle DJ, Stipanovic AJ, Francis RC. An improved method for the hydrolysis of hardwood carbohydrates to monomers. Carbohydr Pol. 2009;78:396–401. doi: 10.1016/j.carbpol.2009.04.015. [DOI] [Google Scholar]
- Atkinson RG, Schröder R, Hallett IC, Cohen D, MacRae EA. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002;129:122–133. doi: 10.1104/pp.010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Hahn MG. Immunological approaches to plant cell wall and biomass characterization: glycome profiling. Methods Mol Biol. 2012;908:61–72. doi: 10.1007/978-1-61779-956-3_6. [DOI] [PubMed] [Google Scholar]
- Pattathil S, Avci U, Baldwin D, Swennes AG, McGill JA, Popper Z, Bootten T, Albert A, Davis RH, Chennareddy C, Dong R, O’Shea B, Rossi R, Leoff C, Freshour G, Narra R, O’Neill M, York WS, Hahn MG. A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 2010;153:514–525. doi: 10.1104/pp.109.151985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillon F, Tranquet O, Quillien L, Utille JP, Ortiz JJO, Saulnier L. Generation of polyclonal and monoclonal antibodies against arabinoxylans and their use for immunocytochemical location of arabinoxylans in cell walls of endosperm of wheat. J Cereal Sci. 2004;40:167–182. doi: 10.1016/j.jcs.2004.06.004. [DOI] [Google Scholar]
- Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG. Molecular analysis of a family of arabidopsis genes related to galacturonosyltransferases. Plant Physiol. 2011;155:1791–1805. doi: 10.1104/pp.110.163220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Doménech N, Jiménez-Bemúdez S, Matas AJ, Rose JKC, Muñoz-Blanco J, Mercado JA, Quesada MA. Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J Exp Bot. 2008;59:2769–2779. doi: 10.1093/jxb/ern142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming CM, Rizkallah HD, McKendrick KA, Abdel-Massih RM, Baydoun EA-H, Brett CT. Biosynthesis and cell-wall deposition of a pectin–xyloglucan complex in pea. Planta. 2005;222:546–555. doi: 10.1007/s00425-005-1560-2. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Widespread occurrence of a covalent linkage between xyloglucan and acidic polysaccharides in suspension-cultured angiosperm cells. Ann Bot. 2005;96:91–99. doi: 10.1093/aob/mci153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Fry SC. Xyloglucan-pectin linkages are formed intra-protoplasmically, contribute to wall-assembly, and remain stable in the cell wall. Planta. 2008;227:781–794. doi: 10.1007/s00425-007-0656-2. [DOI] [PubMed] [Google Scholar]
- Marcus SE, Verhertbruggen Y, Herve C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WG, Knox JP. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8:60. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C, Rogowski A, Gilbert HJ, Knox PJ. Enzymatic treatments reveal differential capacities for xylan recognition and degradation in primary and secondary plant cell walls. Plant J. 2009;58:413–422. doi: 10.1111/j.1365-313X.2009.03785.x. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Liu CH. Deciphering the enigma of lignification: precursor transport, oxidation, and the topochemistry of lignin assembly. Mol Plant. 2012;5:304–317. doi: 10.1093/mp/ssr121. [DOI] [PubMed] [Google Scholar]
- Chang VS, Holtzapple MT. Fundamental factors affecting biomass enzymatic reactivity. Appl Biochem Biotechnol. 2000;84–86:5–37. doi: 10.1385/abab:84-86:1-9:5. [DOI] [PubMed] [Google Scholar]
- Lee C, Teng Q, Huang W, Zhong R, Ye Z-H. Down-regulation of PoGT47C expression in poplar results in a reduced glucuronoxylan content and an increased wood digestibility by cellulase. Plant Cell Physiol. 2009;50:1075–1089. doi: 10.1093/pcp/pcp060. [DOI] [PubMed] [Google Scholar]
- Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE. A. 2011. pp. 6300–6305. [DOI] [PMC free article] [PubMed]
- Davison BH, Drescher SR, Tuskan GA, Davis MF, Nghiem NP. Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl Biochem Biotechnol. 2006;129–132:427–435. doi: 10.1385/abab:130:1:427. [DOI] [PubMed] [Google Scholar]
- DeMartini JD, Pattathil S, Miller JS, Li H, Hahn MG, Wyman CE. Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energy Environ Sci. 2013;6:898–909. doi: 10.1039/c3ee23801f. [DOI] [Google Scholar]
- Siedlecka A, Wiklund S, Péronne M-A, Micheli F, Leśniewska J, Sethson I, Edlund U, Richard L, Sundberg B, Mellerowicz EJ. Pectin methyl esterase inhibits intrusive and symplastic cell growth in developing wood cells of Populus. Plant Physiol. 2008;146:554–565. doi: 10.1104/pp.107.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A, Ried JL, Mount MS. Assay methods for pectic enzymes. Meth Enzymol. 1988;161:329–335. [Google Scholar]
- Theander O, Westerlund EA. Studies on dietary fiber. 3. Improved procedures for analysis of dietary fiber. J Agric Food Chem. 1986;34:330–336. doi: 10.1021/jf00068a045. [DOI] [Google Scholar]
- Scott AS, Melvin EH. Determination of dextran with anthrone. Anal Chem. 1953;25:1656–1661. doi: 10.1021/ac60083a023. [DOI] [Google Scholar]
- Albersheim P. A method for the analysis of sugars in plant cell-wall polysaccharides by gas–liquid chromatography. Carbohydr Res. 1967;5:340–345. doi: 10.1016/S0008-6215(00)80510-8. [DOI] [Google Scholar]
- Filisetti-Cozzi TMCC, Carpita NC. Measurement of uronic acids without interference from neutral sugars. Anal Biochem. 1991;197:157–162. doi: 10.1016/0003-2697(91)90372-Z. [DOI] [PubMed] [Google Scholar]
- Benen JA, Kester HC, Visser J. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur J Biochem. 1999;259:577–585. doi: 10.1046/j.1432-1327.1999.00080.x. [DOI] [PubMed] [Google Scholar]
- Warren ME, Kester H, Benen J, Colangelo J, Visser J, Bergmann C, Orlando R. Studies on the glycosylation of wild-type and mutant forms of Aspergillus niger pectin methylesterase. Carbohydr Res. 2002;337:803–812. doi: 10.1016/S0008-6215(02)00032-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of PtxtPL1-27 transcript (EU379971.1) by northern blot. Northern blot analysis showing the size of PtxtPL1-27 transcript in the wild-type (WT) at 1.9 kb as compared to transcript size in four independent transgenic lines overexpressing PtxtPL1-27 ORF (without untranslated regions (UTRs)) under control of the CaMV 35S promoter at 1.4 kb. A polyubiquitin probe was used as a reference control. The RNA used in the northern blot analysis originated from developing xylem tissue.
Populus trichocarpa PL1 family genes. The complete list of Populus PL1 family gene models based on Populus trichocarpa assembly version 2.2, including current names, former models, closest Arabidopsis homologues, corresponding Populus ESTs from Populus DB, and protein domains.
SDS-PAGE showing the degree of purification of Δ 1-26 Ptxt PL1-27 expressed in E. coli. PtxtPL1-27 protein was expressed in E. coli using pTYB11 vector and partially purified by affinity chromatography. Proteins were separated by gel electrophoresis and stained by Coomassie. Lane 1: proteins extracted from E. coli expressing recombinant protein before induction. Lane 2: proteins extracted from E. coli expressing recombinant protein after induction by 0.3 mM isopropyl β-D-thiogalactoside (IPTG). The induced product can be seen at 98.8 kDa polypeptide representing PtxtPL1-27 fused to intein-chitin binding domain. Lane 3: partially purified recombinant protein is shown as 41.2 kDa band. The preparation is contaminated mainly by the 56.0 kDa polypeptide containing intein and chitin binding domain, which is the cleavage product of 98.8 kDa recombinant polypeptide.
Ectopic overexpression of PtxtPL1-27 inhibits plant growth. Data are relative stem increments and PtxtPL1-27 expression levels in elongating stem internodes of independent transgenic lines. Stem-length increments were determined as an increase in stem length during 64 days of growth in the greenhouse. Between two and eight plants per line were measured. Expression was determined by RT-q-PCR using 18S as a reference gene.
Antibodies used for the ELISA-based glycome profiling. The list of all monoclonal antibodies used in Figure 7.
Monosaccharide composition of wood sequential extracts and the resulting pellets. Sugar composition of wood extracts; means ± standard error, n = 2 biological replicates. Asterisks beside means for individual lines in bold type indicate values significantly different from wild-type (WT) by post analysis-of-variance (ANOVA) t-test. Asteriks beside the means for WT in bold type indicate values significantly different from both lines by post ANOVA contrast; *P ≤10%, **P ≤5%, ***P ≤1%.
List of PCR primers used in the study. Sequences of all forward and reverse PCR primers used in this study.