The stimulation of H3K4 trimethylation (H3K4me3) by H2B monoubiquitination (H2Bub) has been widely studied, and multiple mechanisms have been proposed for this form of histone cross-talk. Thornton et al. combine biochemical, structural, and in vivo approaches to provide a novel mechanism for the role of H2B ubiquitination machinery in the regulation of histone H3K4 methylation by COMPASS. This study demonstrates that the H2Bub machinery and Cps35/Swd2 function to focus the H3K4me3 activity of COMPASS at promoter-proximal regions in a context-dependent manner.
Keywords: chromatin, gene expression, histone, transcription
Abstract
The stimulation of trimethylation of histone H3 Lys4 (H3K4) by H2B monoubiquitination (H2Bub) has been widely studied, with multiple mechanisms having been proposed for this form of histone cross-talk. Cps35/Swd2 within COMPASS (complex of proteins associated with Set1) is considered to bridge these different processes. However, a truncated form of Set1 (762-Set1) is reported to function in H3K4 trimethylation (H3K4me3) without interacting with Cps35/Swd2, and such cross-talk is attributed to the n-SET domain of Set1 and its interaction with the Cps40/Spp1 subunit of COMPASS. Here, we used biochemical, structural, in vivo, and chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) approaches to demonstrate that Cps40/Spp1 and the n-SET domain of Set1 are required for the stability of Set1 and not the cross-talk. Furthermore, the apparent wild-type levels of H3K4me3 in the 762-Set1 strain are due to the rogue methylase activity of this mutant, resulting in the mislocalization of H3K4me3 from the promoter-proximal regions to the gene bodies and intergenic regions. We also performed detailed screens and identified yeast strains lacking H2Bub but containing intact H2Bub enzymes that have normal levels of H3K4me3, suggesting that monoubiquitination may not directly stimulate COMPASS but rather works in the context of the PAF and Rad6/Bre1 complexes. Our study demonstrates that the monoubiquitination machinery and Cps35/Swd2 function to focus COMPASS's H3K4me3 activity at promoter-proximal regions in a context-dependent manner.
Eukaryotic chromatin is highly modified, with many diverse post-translational modifications. How these modifications influence each other to regulate DNA-templated processes has been widely studied (Smith and Shilatifard 2010; Gardner et al. 2011). One of the oldest and best-characterized examples of cross-talk between two histone modifications is the requirement of monoubiquitinated H2B for higher levels of H3K4 trimethylation (H3K4me3) and H3K79me3 (Briggs et al. 2002; Dover et al. 2002; Ng et al. 2002; Sun and Allis 2002; Wood et al. 2003; Lee et al. 2010). Evidence for the requirement for histone H2B monoubiquitination (H2Bub) for histone methylation includes (1) the observed loss of H3K4me3 when the RAD6 and BRE1 genes, which encode the enzyme for H2Bub, are deleted (Wood et al. 2003) and (2) mutation of the monoubiquitinated residue Lys123 to arginine, which also leads to a reduction in H3K4me3 levels (Dover et al. 2002; Sun and Allis 2002). Later, in vitro studies were performed that demonstrated that H2Bub directly stimulates the enzymes mediating H3K4 and H3K79 methylations (McGinty et al. 2008; Kim et al. 2009, 2013).
In yeast, all H3K4 monomethylation (H3K4me1), H3K4 dimethylation (H3K4me2), and H3K4me3 are catalyzed by the Set1 enzyme within the macromolecular COMPASS (complex of proteins associated with Set1) (Miller et al. 2001; Krogan et al. 2002; Shilatifard 2012). COMPASS is composed of seven subunits in addition to Set1, which, ordered by molecular weight, are Cps60/Bre2, Cps50/Swd1, Cps40/Spp1, Cpd35/Swd2, Cps30/Swd3, Cps25/Sdc1, and Cps15/Shg1 (Miller et al. 2001). Three independent groups have demonstrated a connection between the Cps35/Swd2 subunit of COMPASS and its interaction with monoubiquitinated chromatin and cross-talk to H3K4me3 (Lee et al. 2007; Zheng et al. 2010; Soares and Buratowski 2012). In the absence of the Rad6 or Bre1 ubiquitin ligase or in strains bearing the K123R mutant form of H2B, Cps35/Swd2 is not properly recruited to chromatin (Lee et al. 2007; Zheng et al. 2010; Soares and Buratowski 2012). Furthermore, Cps35/Swd2's association with COMPASS was reduced threefold in yeast mutants lacking H2Bub (Lee et al. 2007; Zheng et al. 2010). However, Cps35/Swd2 is in at least one other complex in addition to COMPASS, and complicated genetic interactions between these complexes leave it unclear which are direct and which are indirect effects (Soares and Buratowski 2012).
Ideally, being able to connect the powerful genetics and biochemistry of yeast with in vitro experiments could be helpful in identifying the mechanism for how H2Bub facilitates H3K4me3. To this end, a recent study described the use of reconstituted complexes to demonstrate that Cps40/Spp1 and the n-SET domain of Set1 are required for H2Bub stimulation of H3K4me3 (Kim et al. 2013). However, no direct physical interaction between Cps40/Spp1 or the n-SET domain and H2Bub chromatin could be found. Furthermore, there is no evidence for a monoubiquitination-dependent interaction of Cps40/Spp1 with chromatin, leaving the mechanism of this stimulation of H3K4me3 undetermined. In order to demonstrate a role for the n-SET domain and Cps40 in H2Bub-dependent stimulation of H3K4me3 in vivo, a strain with a truncated version of Set1 that was unable to interact with Cps35/Swd2, but was still capable of interacting with Cps40/Spp1, was generated (Kim et al. 2013). This amino acid 762–1080 form of Set1 could achieve wild-type levels of H3K4me3, as determined by Western blotting (Kim et al. 2013). Furthermore, this methylation required Cps40/Spp1, as deletion of Cps40/SPP1 in a strain expressing the truncated Set1 resulted in a loss of H3K4me3 in vivo (Kim et al. 2013).
Here, we explore possible mechanisms for Cps40/Spp1 in stimulating H3K4me3. We investigated the nature of the 762-Set1 form of COMPASS in vivo and found that without Cps40/Spp1, the 762-Set1 protein levels are reduced. Attempts to reconstitute 762-Set1 with core COMPASS subunits failed unless Cps40/Spp1 was included due to degradation of yeast 762-Set1 in the Sf9 insect cells. Also, single-particle electron microscopy (EM) studies suggest that Cps40/Spp1 interacts with the n-SET domain of Set1 and appears to stabilize a particular conformation of Set1 from what would otherwise be a flexible region before the SET domain that would be degraded in the absence of Cps40/Spp1. In order to understand the nature of the robust methylation by 762-Set1, we performed H3K4me3 chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) and found that, unlike full-length Set1, the truncated version implemented this modification in a deviant manner, with reduced H3K4me3 over promoter-proximal regions and increased H3K4me3 over gene bodies, suggesting that Cps35/Swd2 and the H2Bub machinery could play a role in focusing H3K4me3 on the correct location. We also performed detailed screens in yeast and identified mutants with very low levels of H2Bub, however, still demonstrating normal levels of H3K4me3. Based on our findings, we propose a model in which the H2Bub machinery itself, but not the ubiquitinated H2B, is critical for setting up protein–protein interactions, which are important for COMPASS function in implementing high levels of H3K4me3 at gene promoters.
Results and Discussion
Cps40/Spp1 stabilizes a truncated form of Set1 in vivo
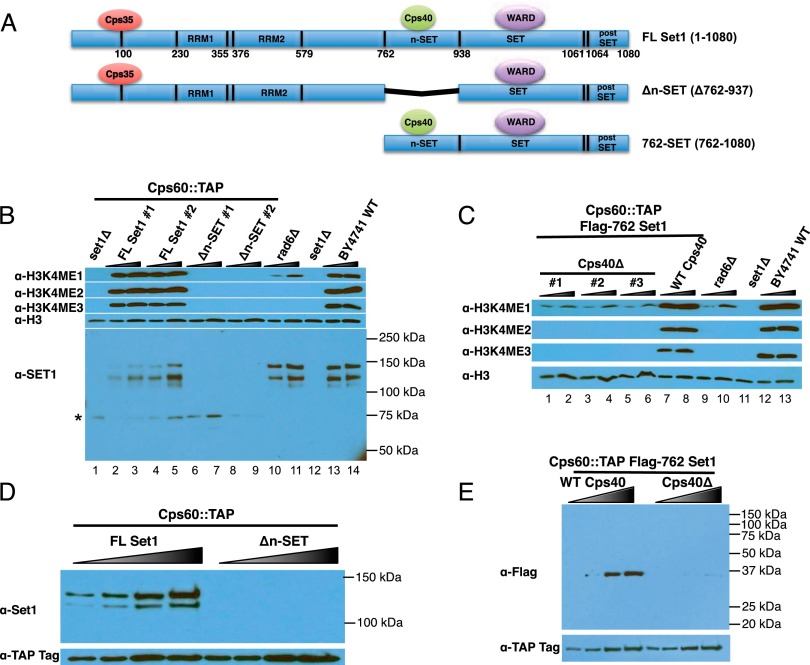
To investigate Cps40/Spp1's contribution to cross-talk between H2Bub and H3K4me3, we replaced the Set1 ORF with truncated versions at the endogenous SET1 locus (Fig. 1A). The Δn-SET has a deletion of the n-SET domain, the Cps40/Spp1-interacting region, but contains the Cps35/Swd2 interaction domain (Takahashi et al. 2011; Kim et al. 2013). Strains were also constructed to express a Flag-tagged Set1 with just the n-SET through post-SET regions (Flag-762-Set1) (Fig. 1A). Each of these constructs stably expresses TAP-tagged Cps60/Bre2 to facilitate analysis of the structural integrity of COMPASS in these mutants (Fig. 1B–E). We found that H3K4 methylation is at a very low or undetectable level in the Δn-SET strains, comparable with a set1 deletion (Fig. 1B; cf. lanes 2–5 and 6–9). Western blotting with antibodies generated toward Set1's C terminus demonstrates that the Δn-SET protein levels are similarly reduced (Fig. 1B). In contrast, the 762-Set1 enzyme implements wild-type levels of H3K4 methylation (Fig. 1C, lanes 7,8). However, deleting CPS40/SPP1 results in a severe loss of H3K4me1, H3K4me2, and H3K4me3 (Fig. 1C, lanes 1–6).
Figure 1.
Importance of the n-SET domain for Set1 levels and Cps40/Spp1-dependent H3K4me3. (A) Domain structure of full-length (FL) Set1, Δn-SET, and 762-Set1, indicating sites of COMPASS subunit binding. WARD is an abbreviation for the core complex subunits, named after the four mammalian homologs Wdr5 (Cps30), Ash2 (Cps60), Rbbp5 (Cps50), and Dpy30 (Cps25). (B) H3K4me1, H3K4me2, H3K4me3, and loss of Set1 in Δn-SET strains. Whole-cell extracts were fractionated by SDS-PAGE and probed with the indicated antibodies. The asterisk indicates a nonspecific band that is seen even in set1 deletion strains. (C) H3K4me1, H3K4me2, and H3K4me3 are lost in cps40Δ 762-Set1 strains. Western blotting with the indicated antibodies of whole-cell extracts, as in B. (D) Set1 is missing from Cps60/Bre2 purifications in the Δn-SET strain. TAP-tagged Cps60/Bre2 was isolated from strains with wild-type or Δn-SET Set1 with IgG Sepharose and elution with TEV protease. Antibodies recognizing the Set1 C terminus were used to assess Set1 levels. TAP tag antibody indicates the level of purified Cps60/Bre2. (E) Protein levels of the Flag-762 version of Set1 are reduced in the absence of Cps40/Spp1. TAP-tagged Cps60/Bre2 was isolated from strains coexpressing Flag-762-Set1 in either the presence or absence of Cps40/Spp1. The M2 Flag monoclonal antibody was used to assess 762-Set1 levels. TAP tag antibody indicates the level of purified Cps60/Bre2.
Previous studies demonstrated that deletion of CPS40/SPP1 from strains expressing full-length Set1 have wild-type levels of mono- and dimethylation and only show reduced trimethylated H3K4 (Schneider et al. 2005; Takahashi et al. 2009). This finding suggests that the Flag-762 Set1 construct (Fig. 1C, lanes 1–8) is unusually dependent on the Cps40/Spp1 subunit of COMPASS. To assess the structural integrity of COMPASS in the Δn-SET and Flag-762-Set1 strains, cellular lysates were purified using the protein-A moiety of the TAP tag on Cps60/Bre2. Antibodies recognizing the TAP tag show equal levels of Cps60/Bre2 in these purifications, while the Δn-SET protein levels in the presence of Cps40/Spp1 (Fig. 1D) and the Flag-762-Set1 protein levels in the absence of Cps40/Spp1 (Fig. 1E) are each greatly reduced. Together, these findings suggest that Cps40/Spp1 functions to stabilize Set1 in vivo. The observed loss of H3K4me3 in the absence of Cps40/Spp1 in the 762-Set1 strain could be explained by the loss of Set1's stability and therefore is not necessarily ascribable to the misregulation of the H2Bub cross-talk pathway.
Cps40/Spp1 stabilizes a truncated form of Set1 in reconstituted COMPASS
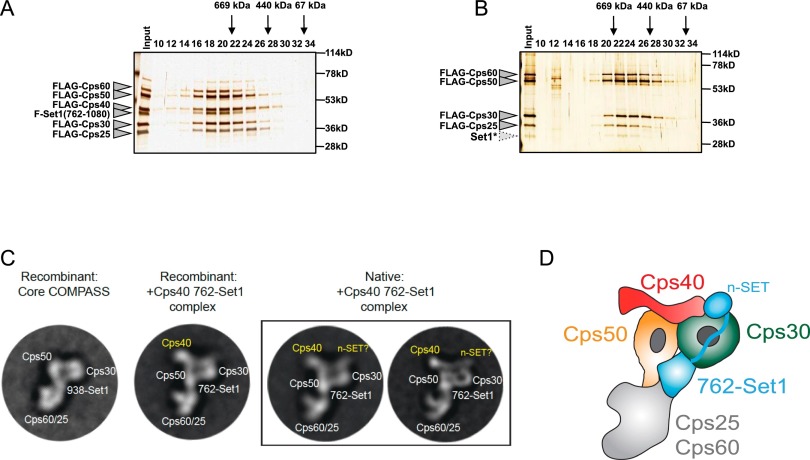
To further investigate the relationship of Cps40/Spp1 with the Flag-762-Set1-containing COMPASS, we used the baculovirus system for reconstitution of COMPASS. Viruses encoding Flag-tagged versions of Cps60/Bre2, Cps50/Swd1, 762-Set1, Cps30/Swd3, and Cps25/Sdc1 with or without Cps40/Spp1 were coinfected into Sf9 cells. Protein lysates were used for Flag affinity purification followed by size exclusion chromatography and silver staining. Flag-762-Set1 peaks in fraction 18 in the presence of Cps40/Spp1, eluting at an apparent size of ∼800 kDa (Fig. 2A). However, attempts at reconstituting the Flag-762-Set1 protein with COMPASS subunits were unsuccessful in the absence of Cps40/Spp1 (Fig. 2B). The complex without Cps40/Spp1 elutes in fractions 22–23 with an apparent size of ∼600 kDa, similar to the elution behavior of the core COMPASS consisting of the minimal SET domain of Set1 with the four core COMPASS subunits (Takahashi et al. 2011). This observation can be attributed to the lack of intact Flag-762-Set1 (degradation of n-SET) in the reconstituted COMPASS in the absence of Cps40/Spp1. In subsequent fractions, the rest of the core COMPASS subunits elute together, as the previously described “WARD” complex (named after the mammalian homologs of WDR5 [Cps35], Ash2 [Cps60], Rbbp5 [Cps50], and Dpy30 [Cps25]). These findings are consistent with our in vivo studies, suggesting that Cps40/Spp1 has a strong stabilizing influence on 762-Set1.
Figure 2.
Structural basis for Cps40/Spp1 stabilization of 762-Set1. (A,B). Flag-tagged 762-Set1 and other Flag-tagged COMPASS subunits were coexpressed in Sf9 cells using the baculovirus system. (A) When Flag-Cps40/Spp1 is included in the reconstitution, the assembled complex is intact, as determined by Superose 6 gel filtration chromatography, eluting at ∼800 kDa (peaking at fraction 18). (B) When Flag-Cps40/Spp1 is left out of the reconstitution, the Flag-purified complex displays an altered Superose 6 elution profile, peaking at ∼600 kDa (fraction 22). Molecular weight markers and COMPASS subunits are indicated. A truncated version of Set1 that coelutes with the COMPASS core subunits in fraction 22 is labeled as Set1* and likely corresponds to the minimal SET domain. We predict that this form of Set1 (core Set1) is generated when the n-SET domain of 762-Set1 is degraded in the absence of Cps40/Spp1. (C,D) Representative 2D class averages of COMPASS with Cps40/Spp1. (C) Representative 2D class averages of the recombinant “core” COMPASS (Cps60/Bre2, Cps50/Swd1, Cps30/Swd3, Cps25/Sdc1, and 938-Set1) are compared with class averages of recombinant “+Cps40/Spp1, 762-Set1” COMPASS (Cps60/Bre2, Cps50/Swd1, Cps40/Spp1, Cps30/Swd3, Cps25/Sdc1, and 762-Set1) and with native “+Cps40/Spp1, 762-Set1” COMPASS. Additional densities attributed to Cps40/Spp1 and n-SET are observed on top of Cps50/Swd1 and Cps30/Swd3. (D) Model of Cps60/Bre2-, Cps50/Swd1-, Cps40/Spp1-, Cps30/Swd3-, Cps25/Sdc1-, and 762-Set1-containing COMPASS with subunits and domains indicated.
Single-particle EM analysis of Cps40/Spp1 within a Flag-762-Set1-containing COMPASS
The baculovirus-reconstituted COMPASS with Flag-762-Set1 and Cps40/Spp1 was purified by size exclusion chromatography, and peak fractions were visualized by negative stain EM followed by two-dimensional (2D) particle classification and averaging (see Supplemental Figs. 1, 2). For comparison, Flag-762-Set1 COMPASS was purified from yeast cells using TAP purification of Cps60/Bre2. Both the reconstituted and native Flag-762-Set1/Cps40/Spp1 COMPASS complexes reveal views similar to the recombinant “core” COMPASS complex previously characterized (Fig. 2C, left; Takahashi et al. 2011). Compared with “core” COMPASS, native and reconstituted Flag-762-Set1/Cps40/Spp1 COMPASS reveals a significant rod-like density, which we attribute to Cps40/Spp1 attached on top of Cps50/Swd1. Furthermore, the 2D averages reveal a smaller globular density attached on top of Cps30/Swd3. We postulate that this small globular density corresponds to n-SET, which has been previously shown to engage Cps30/Swd3. The fact that the densities attributed to Cps40/Spp1 and n-SET are peripheral to the complex might explain the difference in elution profile on gel filtration analysis of the reconstituted COMPASS assembled with and without Cps40/Spp1 (Fig. 2A,B). Interestingly, several 2D averages of the native complex reveal that the Cps40/Spp1 and n-SET densities are connected, resulting in a long rod that sits tightly on top of the Cps50/Swd1–Cps30/Swd3 module (Supplemental Figs. 1, 2). The association of Cps40/Spp1 with n-SET might protect this region from attracting proteolytic machinery, explaining why the absence of Cps40/Spp1 could lead to proteolysis of Set1 and the major shift in the molecular weight, as observed on size exclusion chromatography (Fig. 2A,B).
Truncated Set1 (762-Set1) is a rogue methylase with altered intragenic specificity
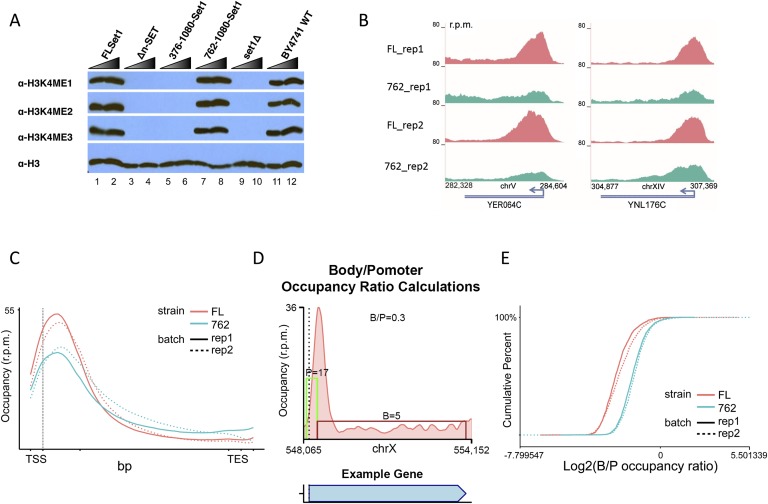
Having demonstrated that the apparent inability of 762-Set1 to implement H3K4me3 in the absence of Cps40/Spp1 is due to degradation of the enzyme and not the proposed loss of cross-talk between Cps40/Spp1 and H2Bub, we set out to understand the nature of the robust bulk levels of H3K4me3 observed in strains containing both Set1-762 and Cps40/Spp1 (Kim et al. 2013). We first compared H3K4 methylation levels in various truncations of COMPASS (Fig. 3A; data not shown). Deletion of the n-SET domain (Δn-SET) or deletion of the first 375 residues that remove the Cps35/Swd2 interaction domains and the first RRM domain of Set1 (leaving amino acids 376–1080-Set1) results in a severe loss of all forms of H3K4 methylation (Fig. 3A, cf. lanes 1–2 and 5–6). The loss of H3K4me3 levels in Set1 N-terminal deletion mutants is suppressed by the further truncation of Set1 to the 762–1080-Set1 strain as assessed by Western blotting of bulk histones (Fig. 3A, cf. lanes 1–2 and 3–8). We therefore performed ChIP-seq in cells expressing full-length Set1 or 762-Set1. We observed that the pattern of H3K4me3 is frequently reduced over the promoter-proximal regions and increased over the gene bodies in the presence of the truncated version of Set1, and this alteration was reproducible in biological replicates (Fig. 3B).
Figure 3.
Wild-type levels of H3K4me3 in the 762-Set1 strain have altered occupancy. (A) The 762-Set1 version of COMPASS is unique among other truncations to sustain high levels of H3K4 methylation when tested by Western blotting (Kim et al. 2013). The Δn-SET and amino acids 376–1080 forms of Set1, both of which lose the ability to interact with Cps35/Swd2, show severely reduced levels of H3K4 methylation. However, deleting even more of Set1, the first 761 amino acids, appears to restore wild-type levels of H3K4 methylation in these cells, including H3K4me3. (B–E) The H3K4me3 implemented by 762-Set1 is not as focused on promoter-proximal regions. (B) Genome browser track of examples of genes with reduced H3K4me3 body/promoter occupancy ratios in a 762-Set1 strain. (C) Metagene analysis of the 1625 genes that are large (>800 bp), isolated (>200 bp from another gene), and enriched for H3K4me3 in the full-length Set1 strain. (D) Schematic for how the ratio of promoter-proximal versus gene body H3K4me3 occupancy is measured. (E) Cumulative distribution analysis of body/promoter occupancy ratio for the 1625 genes that are long, isolated, and enriched for H3K4me3 (Kolmogorov-Smirnov test, two-sided, D = 0.243; P-value < 2.2 × 10−16): (FL) full-length Set1; (762) amino acids 762–1080-Set1. Batches 1 and 2 represent biological replicates.
Metagene analysis shows that H3K4me3 is generally reduced from the promoter region and increased in the gene body in the 762-Set1 strain as compared with a full-length Set1 strain (Fig. 3C). To increase the accuracy of our analyses, we only considered genes that were long (>800 base pairs [bp]), isolated (at least 200 bp from any other gene), and enriched for H3K4me3 in the full-length Set1-containing strain (n = 1625). To quantitate the redistribution of H3K4me3 from its normal marking at transcription start sites (TSSs), we calculated the ratio of gene body regions to promoter-proximal regions, taking the −100- to +300-bp region around the TSSs as the promoter and the rest of the gene as the body (Fig. 3D). The distribution of the body/promoter occupancy ratio between the wild-type and 762-Set1 strain was contrasted (Fig. 3E) and found to be significant (Kolmogorov-Smirnov test, two-sided, D = 0.243; P-value < 2.2 × 10−16). Thus, H3K4me3 in the 762-Set1 strain is far from wild type.
A role for monoubiquitination machinery but not monoubiquitination in H3K4me3: context dependency in monoubiquitination/methylation cross-talk
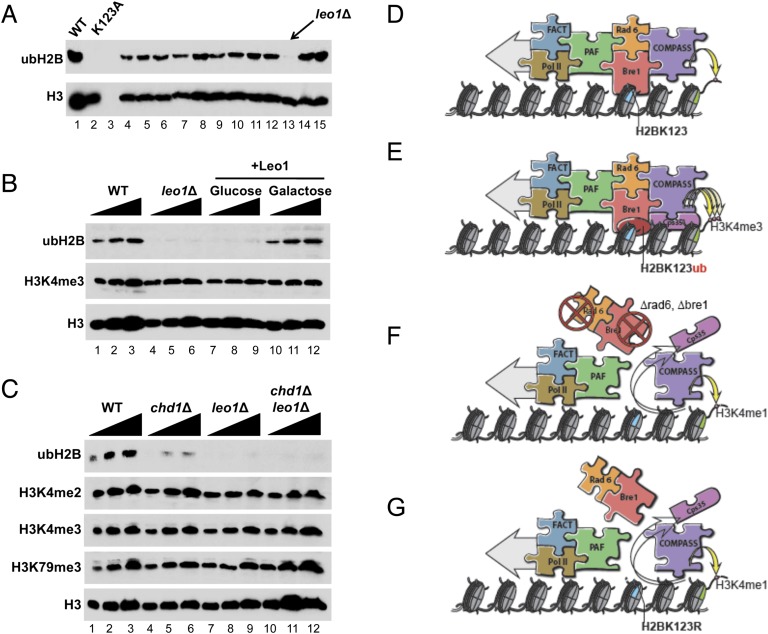
The clear difference in H3K4me3 patterns by Set1 that can interact with Cps35/Swd2 and by Set1 that cannot interact with Cps35/Swd2 could mean that the H2Bub might be important for focusing H3K4me3 at promoter-proximal regions. However, we recently used global proteomic screening (GPS) to identify Chd1 as a factor required for H2Bub (Lee et al. 2012). While earlier GPS studies showed that Rad6 and Bre1, the E2 and E3 ligases implementing H2Bub, also affected H3K4me3 (Dover et al. 2002), the loss of Chd1 had no global effect on H3K4me3 (Lee et al. 2012). Since Chd1 loss resulted in ∼90% reduction in H2Bub levels, we could not conclude that H3K4me3 could occur in the absence of H2Bub. Therefore, we used GPS to identify additional factors that are required for H2Bub without affecting H3K4me3 levels. One of the hits identified is the PAF complex subunit Leo1 (Fig. 4A, lane 13). As seen with Chd1, loss of Leo1 had no effect on bulk levels of H3K4me3 (Fig. 4B) despite the loss of significant levels of H2Bub. We confirmed a requirement for Leo1 in H2Bub by complementing the leo1Δ strain with LEO1 expressed under the GAL1 promoter. In the presence of glucose, H2Bub levels are greatly reduced, while switching to galactose restores H2Bub to wild-type levels (Fig. 4B). Histone H3K4me3 levels are unaffected in all conditions (Fig. 4B).
Figure 4.
The H2Bub machinery, but not the modification alone, is required for H3K4me3 by COMPASS in vivo. (A) GPS of the nonessential gene collection in Saccharomyces cerevisiae for genes required for H2Bub. Leo1, a member of the PAF complex, was identified in this screen. (B) Complementation of the leo1Δ strain with LEO1 expressed under the control of the GAL1 promoter. In the presence of galactose, H2Bub levels return to wild-type levels. H3K4me3 levels remain largely unchanged in all conditions. (C) CHD1, encoding a previously identified factor required for full levels of H2Bub in cells, was knocked out singly and in combination with LEO1. Western blotting of extracts from the chd1Δ leo1Δ strain has greatly reduced H2Bub without a major effect on H3K4me3 or H3K79me3. (D–G) A model for the context dependency for proper H3K4 methylation by COMPASS. (D) The PAF and FACT complexes are recruited to promoters along with RNA polymerase II and help recruit COMPASS and the ubiquitin E2 and E3 ligases Rad6 and Bre1, respectively. (E) Rad6 and Bre1 recruitment results in the monoubiquitination of H2B and the recruitment of Cps35/Swd2, leading to the transition from H3K4me1 to H3K4me2 and H3K4me3. (F) In strains in which the gene encoding Rad6 or Bre1 is deleted, COMPASS can only monomethylate histone H3K4 due to the absence of Cps35/Swd2 on chromatin. (G) Mutation of H2BK123 to arginine (H2BK123R) can result in the loss of interaction of Rad6 and Bre1 and/or PAF with chromatin and therefore the loss of association of Cps35/Swd2 with COMPASS on chromatin, resulting in a reduction in the level of H3K4me2 and H3K4me3.
In an attempt to achieve a greater reduction in H2Bub levels, we made a double deletion strain, chd1Δ leo1Δ. Even with the substantial reduction of H2Bub seen in the leo1Δ and chd1Δ leo1Δ strains, H3K4me3 and H3K79me3 levels remain unchanged (Fig. 4C). Importantly, Handa and colleagues (Chen et al. 2009) have previously observed that RNAi-mediated knockdown of Leo1 in mammalian cells leads to a loss of H2Bub but not H3K4me3, suggesting that monoubiquitination of H2B is also not a prerequisite for H3K4me3 by the COMPASS family in mammals. ChIP-seq analysis shows that the wild-type levels of H3K4me3 seen by Western blotting in these mutants have the expected distribution over the TSS regions, indicating proper regulation of H3K4me3 by COMPASS in these mutants (Supplemental Fig. 3).
A model for the context dependency for proper H3K4me3 by COMPASS
The study presented here supports a model in which the H2Bub machinery (i.e., Rad6/Bre1 and the interacting factors such as PAF and FACT complexes), but not the monoubiquitin modification itself, could function as a prerequisite for H3K4me3. In this model, the PAF complex and the FACT complex go to all active genes, and the Rad6 and Bre1 H2Bub ligases are corecruited to chromatin by PAF and FACT (Fig. 4D; Pavri et al. 2006). If Rad6 and Bre1 are present, then COMPASS's trimethylase function, which requires Cps35/Sdw2, can be properly focused to the promoter-proximal region for H3K4me3 (Fig. 4E). In the rad6 and bre1 deletion strains, Cps35/Swd2 is not recruited to chromatin, resulting in monomethylation of H3K4 by COMPASS (Fig. 4F). We speculate that Rad6 and Bre1 recruitment is dependent on the identity of the K123 residue and is not substitutable by the similarly charged arginine residue (Fig. 4G).
One way to reconcile the seemingly contradictory claims of whether it is Cps35/Swd2 or Cps40/Spp1 that mediates H2Bub cross-talk would be that both factors are needed but at different steps. In wild-type yeast, Cps35/Swd2's recruitment to chromatin in an H2Bub machinery-dependent manner could change the conformation of COMPASS such that Cps40/Spp1 directly stimulates H3K4me3 by COMPASS. Since loss of Cps40 leads to reductions in trimethylation, but not dimethylation (Schneider et al. 2005), and since mutation of the monoubiquitination machinery loses both di- and trimethylation, under this model, the conformations and interactions facilitated by Cps35/Swd2 would be enough for dimethylation but not trimethylation in the absence of Cps40/Spp1.
An unexpected lesson from our study is that many of the assumptions on which our model and others have been built were obtained from Western blotting and by performing ChIP at a few genes. What was considered wild-type H3K4me3 levels in 762-Set1 cells by Western blotting (Kim et al. 2013) was in fact mislocalized H3K4me3, as evidenced by detailed ChIP-seq studies. This finding indicates that many alterations of this model could occur as modern techniques are applied to this now decade-old phenomenon of histone cross-talk between H2Bub and H3K4me3.
Materials and methods
Western analysis and immunoprecipitation
Whole-cell extracts were prepared and subjected to Western blotting. IgG Sepharose was used to isolate Cps60/Bre2 from nuclear extracts. Immunoprecipitations were assayed by Western blotting with antibodies to Set1, the Flag epitope on Set1, or the TAP tag on Cps60/Bre2. TAP purification of Cps60 was performed as previously described (Miller et al. 2001; Takahashi et al. 2011).
For LEO1 complementation, cells containing the pRS315 vector bearing LEO1 under the control of the GAL1 promoter were cultured in SD medium deficient of leucine overnight at 30°C, transferred to synthetic medium containing dextrose and galactose, respectively, and incubated for 6 h at 30°C. Cell extracts were prepared and analyzed by Western blotting. Antibodies were generated by our laboratory except for anti-H3K79me3 (Abcam, ab2621), anti-monoubiquitinated histone H2B (Cell Signaling Technology, #5546), anti-TAP (Thermo Scientific, CAB1001), and the M2 anti-Flag antibody (Sigma, F3165).
Specimen preparation and EM imaging of negative-stained samples
Baculovirus reconstitution was performed as previously described (Takahashi et al. 2011). All COMPASS complexes were prepared for EM using the conventional negative-staining protocol (Ohi et al. 2004) and imaged at room temperature with a Tecnai T12 electron microscope operated at 120 kV using low-dose procedures. Images were recorded at a magnification of 71,138× and a defocus value of ∼1.5 μm on a Gatan US4000 CCD camera. All images were binned (2 × 2 pixels) to obtain a pixel size of 4.16 Å on the specimen level. All particles were manually excised using Boxer (part of the EMAN 1.9 software suite) (Ludtke et al. 1999).
2D classification of negative-stained COMPASS complexes.
2D reference-free alignment and classification of particle projections were performed using SPIDER (Frank et al. 1996). For the recombinant “+Cps40 762-Set1” COMPASS complex (Cps60, Cps50, Cps40, Cps30, Cps25, and 762-Set1), 12,974 particle projections were classified into 100 classes. For the native “+Cps40 762-Set1” COMPASS complex (Cps60, Cps50, Cps40, Cps30, Cps25, and 762-Set1), 7806 particle projections were initially classified into 50 classes. Two-thousand-one-hundred-sixty-four particle projections from classes clearly showing densities for all domains were subsequently subjected to an additional secondary classification into 10 classes.
ChIP
ChIP was performed as previously described (Lee et al. 2012). For body/promoter ratio, the average normalized coverage between body and promoter was computed for every gene using the −100- to +300-bp region around the TSSs as the promoter and the rest of the gene as the body. See the Supplemental Material for additional details. ChIP-seq data have been deposited at the Gene Expression Omnibus (GEO) under the accession number GSE53241.
Competing interest statement
Antibodies toward monoubiquitinated histone H2B were obtained from Cell Signaling Technology (#5546). A.S. is a paid member of the Scientific Advisory Board of Cell Signaling Technology.
Acknowledgments
We thank Austin Carroll and Larry Rudolph for technical assistance; Rhonda Egidy, Kate Malanowski, Allison Peak, Anoja Perera, and the rest of the Stowers Institute Molecular Biology core for help with Illumina sequencing; and Judy Foye for medium preparation. We thank Mark Miller for artwork, and Laura Shilatifard and Lisa Kennedy for editorial assistance. These studies were supported in part by grants from the Canadian Institutes of Health Research (BMA-355900) to J.-F.C., and G.H.W. is supported by the National Institute of General Medical Sciences (NIGMS) Molecular Biophysics Training Grant GM008270-23. G.S. is a Pew Scholar of Biomedical Sciences. The yeast studies in the Shilatifard laboratory are supported by the National Institutes of Health grant R01GM069905.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.232215.113.
References
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD 2002. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 418: 498. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yamaguchi Y, Tsugeno Y, Yamamoto J, Yamada T, Nakamura M, Hisatake K, Handa H 2009. DSIF, the Paf1 complex, and Tat-SF1 have nonredundant, cooperative roles in RNA polymerase II elongation. Genes Dev 23: 2765–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277: 28368–28371 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A 1996. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gardner KE, Allis CD, Strahl BD 2011. Operating on chromatin, a colorful language where context matters. J Mol Biol 409: 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD, Roeder RG 2013. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell 49: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A 2002. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A 2010. The language of histone crosstalk. Cell 142: 682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Garrett AS, Yen K, Takahashi YH, Hu D, Jackson J, Seidel C, Pugh BF, Shilatifard A 2012. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev 26: 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W 1999. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW 2008. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453: 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K 2002. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 277: 34655–34657 [DOI] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T 2004. Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D 2006. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19: 849–856 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A 2010. The chromatin signaling pathway: Diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell 40: 689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LM, Buratowski S 2012. Yeast Swd2 is essential because of antagonism between Set1 histone methyltransferase complex and APT (associated with Pta1) termination factor. J Biol Chem 287: 15219–15231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418: 104–108 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, Trievel RC, Shilatifard A 2009. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: Implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol 29: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G 2011. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci 108: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Krogan NJ, Dover J, Schneider J, Heidt J, Boateng MA, Dean K, Golshani A, Zhang Y, Greenblatt JF, et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell 11: 267–274 [DOI] [PubMed] [Google Scholar]
- Zheng S, Wyrick JJ, Reese JC 2010. Novel trans-tail regulation of H2B ubiquitylation and H3K4 methylation by the N terminus of histone H2A. Mol Cell Biol 30: 3635–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]