Abstract
DNA barcodes were obtained for 81 butterfly species belonging to 52 genera from sites in north-central Pakistan to test the utility of barcoding for their identification and to gain a better understanding of regional barcode variation. These species represent 25% of the butterfly fauna of Pakistan and belong to five families, although the Nymphalidae were dominant, comprising 38% of the total specimens. Barcode analysis showed that maximum conspecific divergence was 1.6%, while there was 1.7–14.3% divergence from the nearest neighbour species. Barcode records for 55 species showed <2% sequence divergence to records in the Barcode of Life Data Systems (BOLD), but only 26 of these cases involved specimens from neighbouring India and Central Asia. Analysis revealed that most species showed little incremental sequence variation when specimens from other regions were considered, but a threefold increase was noted in a few cases. There was a clear gap between maximum intraspecific and minimum nearest neighbour distance for all 81 species. Neighbour-joining cluster analysis showed that members of each species formed a monophyletic cluster with strong bootstrap support. The barcode results revealed two provisional species that could not be clearly linked to known taxa, while 24 other species gained their first coverage. Future work should extend the barcode reference library to include all butterfly species from Pakistan as well as neighbouring countries to gain a better understanding of regional variation in barcode sequences in this topographically and climatically complex region.
Keywords: COI, endemism, Lepidoptera, mtDNA, Pakistan
Introduction
DNA barcoding has emerged as a useful tool for the identification and discovery of animal species. It employs sequence diversity in a 648 base pair fragment near the 5′ end of the mitochondrial cytochrome c oxidase subunit I (COI) gene as a tool for species discrimination (Hebert et al. 2003a). Barcoding has been shown to discriminate species across the animal kingdom (Tyagi et al. 2010; Virgilio et al. 2010) including fishes, mammals, birds, insects, crustaceans and many other groups (Hebert et al. 2004a; Foottit et al. 2008; Hastings et al. 2008; Hubert et al. 2008; Hou et al. 2009; Wong et al. 2009; Clare et al. 2011). Reflecting the rapid growth in barcode coverage (Jinbo et al. 2011), BOLD, the Barcode of Life Data System (Ratnasingham & Hebert 2007), now includes records for more than 261K animal species. The order Lepidoptera has received particular attention (Hajibabaei et al. 2006; Silva-Brandao et al. 2009; Hebert et al. 2010; Kim et al. 2010) with 691K barcode records on BOLD (Feb 3, 2013), including data for 9124 named butterfly (Papilionoidea, Hesperioidea) species from 194 countries.
The gap between maximum intraspecific and minimum interspecific distances has been used for species delimitation in various animal groups (Hebert et al. 2004a; Meyer & Paulay 2005; Meier et al. 2006, 2008; Puillandre et al. 2012). This approach has helped to resolve cryptic species complexes (Hebert et al. 2004b; Burns et al. 2007; Park et al. 2011; Deng et al. 2012) and has aided ecological studies (Valentini et al. 2009; Pramual & Kuvangkadilok 2012). For example, Vaglia et al. (2008) used DNA barcodes to reveal cryptic species of sphingid moths, while van Nieukerken et al. (2012) discriminated cryptic species of leaf-mining Lepidoptera. Likewise, Carletto et al. (2009) discriminated sibling species of Aphis gossypii.
The effectiveness of DNA barcoding has spurred efforts to construct DNA barcode reference libraries for various animal groups (Ekrem et al. 2007; Guralnick & Hill 2009; Janzen et al. 2009; Lee et al. 2011; Zhou et al. 2011; Webb et al. 2012). These libraries not only aid the documentation of biodiversity (Janzen et al. 2005; Naro-Maciel et al. 2010) including endangered species (Elmeer et al. 2012; Vanhaecke et al. 2012), but can disclose endemism (Bossuyt et al. 2004; Quilang et al. 2011; Sourakov & Zakharov 2011). Because Lepidoptera have been selected as a model group for intensive analysis, the order is well represented on BOLD, but some regions such as South-East Asia have seen little investigation. Barcode records are available for a significant fraction of the Central Asian butterfly fauna (Lukhtanov et al. 2009) and for a smaller number of species from Western India (Gaikwad et al. 2012). However, these studies fail to provide coverage for many species known from Pakistan (Roberts 2001). The current study had the primary goals of testing the effectiveness of DNA barcodes in the identification of butterfly species from Pakistan and comparing these records with those from other regions to gain a better sense of the extent of intraspecific variation.
Materials and methods
Specimen sampling
Butterflies were collected at 107 locations across central and northern Pakistan (Fig. 1) during 2009–2012. These sites included three different climatic zones: tropical, subtropical and temperate, with altitudes ranging from 127 to 2660 m, and both agricultural and forested environments. Each specimen was labelled, assigned a code number and deposited in the arthropod collection at the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, for subsequent morphological and molecular analysis. Using standard guides to the fauna (Malik 1973; Hasan 1994; Roberts 2001), the 407 specimens were assigned to 81 species belonging to 52 genera. Two species (Lasiommata sp. MA01 and Polycaena sp. MA01) could only be identified to a generic level, but were included in the analysis. Specimen data and images are available on BOLD (Ratnasingham & Hebert 2007) in the project MABUT (Barcoding Butterflies of Pakistan). Fifty-nine of the 81 species were represented by more than one specimen (range 2–20). All sequences generated in this study are available on BOLD (Process IDs: MABUT001-10 to MABUT312-12; MABUT326-13 to MABUT388; MAIMB133-09 to MAIMB137-09, 166-09, 167-09, 169-09, 170-09, 178-09, 179-09) and on GenBank under the following accession nos: KC158311–KC158471, HQ990321–HQ990449, HQ990705, HQ990728–HQ990729, GU681850–GU681851, GU681855–GU681856, GU681859, GU681870 and GU681872–GU681875.
Fig 1.
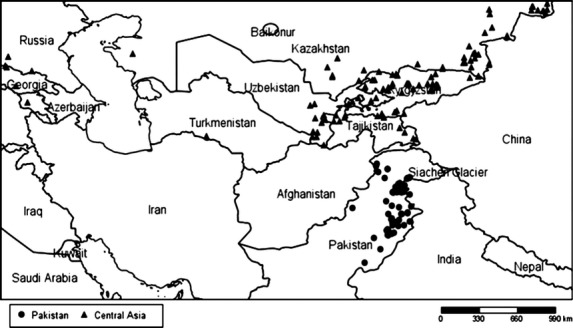
Map of Pakistan and neighbouring nations showing collection localities for this study as well those for specimens examined in a prior study (Lukhtanov et al. 2009).
DNA extractions and PCR amplifications
A single leg was removed from each specimen with a sterile forceps and transferred to a 96-well microplate preloaded with 30 μL of 95% ethanol in each well. DNA extraction, PCR amplification and sequencing were performed at the Canadian Centre for DNA Barcoding (CCDB) following standard protocols (Ivanova et al. 2006, 2007; Ivanova & Grainger 2007a,b2007b,c2007c). DNA extractions were performed by following the protocols developed for invertebrate barcoding (Ivanova et al. 2006). Amplification of the COI-5′ barcode region was performed with primer pair LepF1 (ATTCAACCAATCATAAAGATATTGG)/LepR1 (TAAACTTCTGGATGTCCAAAAAATCA) (Hebert et al. 2004b) using the following PCR conditions: 94 °C (1 min); 5 cycles of 94 °C (30 s), 45 °C (40 s), 72 °C (1 min); 35 cycles of 94 °C (30 s), 51 °C (40 s), 72 °C (1 min); and final extension of 72 °C (10 min). PCRs were carried out in 12.5 μL reactions containing standard PCR ingredients and 2 μL of DNA template. PCR products were analysed on 2% agarose E-gel® 96 system (Invitrogen Inc.). Amplicons were sequenced bidirectionally using BigDye Terminator Cycle Sequencing Kit (v3.1) on an ABI 3730XL DNA Analyzer. The forward and the reverse sequences were assembled and aligned using CodonCode Aligner (CodonCode Corporation, USA). Sequences were also inspected and translated in mega V5 (Tamura et al. 2011) to verify that they were free of stop codons and gaps.
Data analysis
The sequence from each specimen was compared with barcode sequences on GenBank using ‘Blast’ and with sequences on BOLD using the ‘Identification Request’ function. Prior studies have revealed that most different species of Lepidoptera show >2% sequence divergence at CO1 (Hebert et al. 2003b), and researchers have used a 2% pairwise distance threshold for species delimitation (Strutzenberger et al. 2011). For the barcode-based identity analysis, we also used a threshold of 2% divergence. DNA barcodes for 9124 butterfly species from 194 countries are currently available on BOLD, all readily available for sequence comparisons. In addition, the results were compared with those of prior studies in Central Asia (353 butterfly species) (Lukhtanov et al. 2009), Korea (83 species) (Kim et al. 2010) and India (40 species) (Gaikwad et al. 2012). ClustalW nucleotide sequence alignments (Thompson et al. 1994) and NJ clustering analysis were performed using mega V5 (Tamura et al. 2011). The Kimura-2-Parameter (K2P) (Kimura 1980) distance model was used, along with pairwise deletion of missing sites, with nodal support estimated using 500 bootstrap replicates. The online version of Automatic Barcode Gap Discovery (ABGD) (Puillandre et al. 2012) was used for both pairwise distance analyses and to generate distance histograms and distance ranks. The presence or absence of a ‘barcode gap’ (Meyer & Paulay 2005) was also determined for each species as a test of the reliability of its discrimination. Using the barcode gap criterion, a species is distinct from its nearest neighbour (NN) if its maximum intraspecific distance is less than the distance to its NN sequence. The ‘Barcode Gap Analysis’ (BGA) was performed using BOLD. Species identification success by ‘Best Match’ and cluster analysis was performed using TaxonDNA (Meier et al. 2006). The relationship between geographical distance and intraspecific genetic distance was analysed separately for each species (with at least three individuals and three locations) using the Mantel test (Mantel 1967) and by linear regression using xlstat (version 2013.3.02; Addinsoft, Inc., NY, USA).
Results
Barcode sequences greater than 500 base pairs (bp) were recovered from 374 of the 407 specimens (92%), providing at least one sequence for each of the 81 butterfly species. When these sequences were compared with those in the BOLD and NCBI databases, close sequence matches (<2% divergence) were detected for 55 of the species from Pakistan, while 26 lacked a match. The highest number of matches involved records from India (15), Central Asia (11) and Korea (10).
Figure 2 presents results from the ABGD and BGA analyses. Distance values show a gap between the intraspecific and the interspecific distances (Fig. 2A). As well, both the maximum and mean distances to NN are higher than the respective intraspecific distances for all species (Fig. 2B). Nearest neighbour distances were more than 3% for all but three species pairs: Tarucus balkanicus vs. T. rosaceus (1.70%), Junonia orithya vs. J. hierta (2.49%) and Celastrina huegelii vs. C. argiolus (2.64%). Intraspecific distances could not be determined for the 22 species with just a single representative, but NN distances were greater than 4% for 21 of them.
Fig 2.
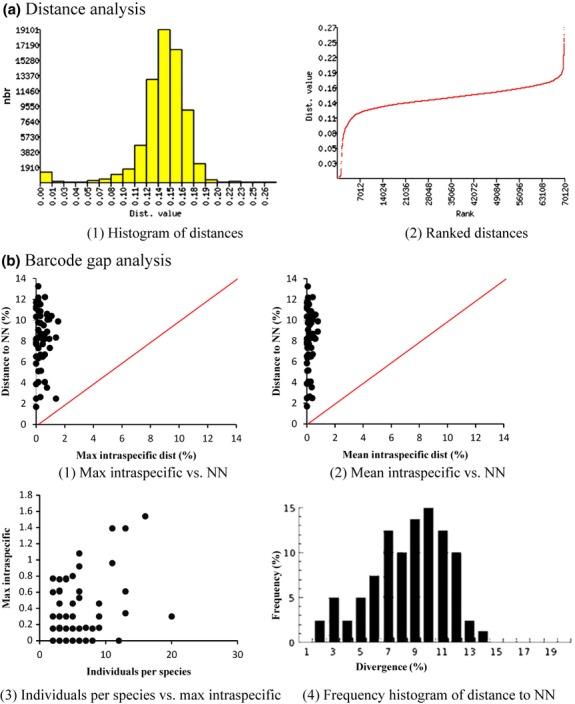
Pairwise distance divergence (%) (a) and barcode gap analysis (b) for butterflies from Pakistan as generated by Automatic Barcode Gap Discovery (Puillandre et al. 2012) and by BOLD (Ratnasingham & Hebert 2007), respectively. NN = nearest neighbour.
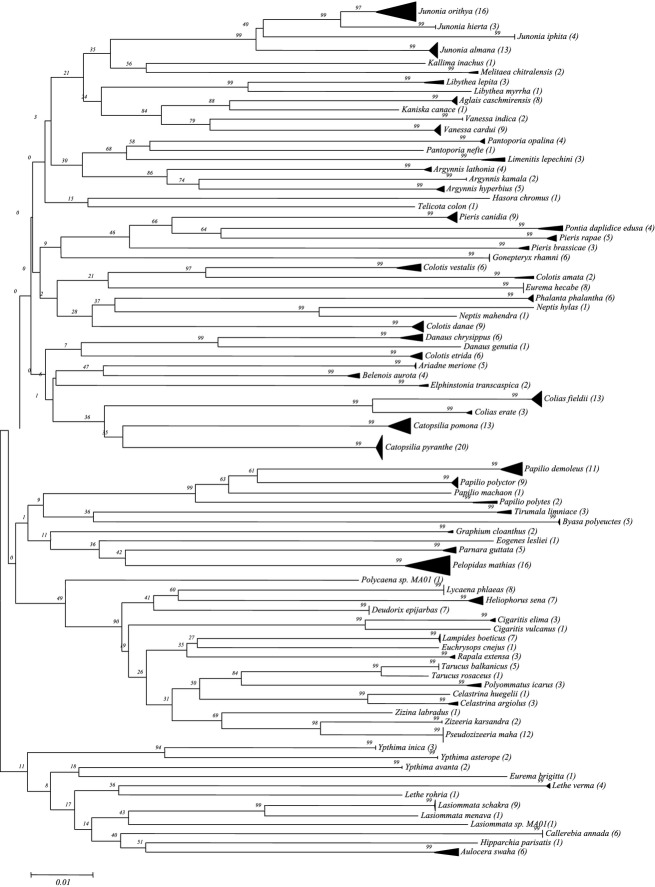
NJ clustering analysis showed that each of the 81 species formed a monophyletic cluster (Fig. 3). Species with two or more barcode sequences were analysed for species identification using TaxonDNA. When a 3% threshold was employed, 100% of the species were correctly identified using the ‘Best Match or Best Close Match’ criterion. Analysis of the 374 sequence records using TaxonDNA led to the recognition of 78 clusters at a 3% threshold and 80 clusters at a 2% threshold. At the 3% threshold, 75 of the 78 clusters were comprised of a single species, with the largest pairwise intraspecific distance being 2.88%, while 79 of the 80 clusters were a single species at the 2% threshold with the largest pairwise intraspecific distance being 1.67%.
Fig 3.
NJ cluster analysis of butterfly species from north-central Pakistan based on the analysis of 374 sequences from 81 species. Bootstrap values (500 replicates) are shown above the branches. The scale bar shows K2P distances. The node for each species with multiple specimens was collapsed to a vertical line or triangle, with the horizontal depth indicating the level of intraspecific divergence. Bracketed numbers next to each species name indicate the number of individuals analysed. Analyses were conducted in mega5.
Genetic divergences increased with taxonomic rank (Table 1; Fig 2) with little overlap between conspecific and congeneric distances. Intraspecific divergences ranged from 0.0 to 1.6% with a mean of 0.2%, while divergences for the species in a genus ranged from 1.7 to 14.3% with a mean of 8.0%. The distances within families ranged from 3.9 to 19.2% with a mean of 13.1%. Fifty-five species were represented by at least one conspecific from another country, but in most cases, there was little increase in intraspecific divergence when they were included in the analysis (Table 2). Seventeen species showed a three-fold or more increase in intraspecific distances (Table 2, bold-faced numbers), but their maximum intraspecific divergence remained <3%, and mean divergence was <1% in all cases except Colotis amata (max = 3.20%, mean = 1.17%) (Table 2). The relationship between geographical and genetic distances was quantified by plotting geographical distances against intraspecific variation (K2P). Table 2 provides species-wise Mantel correlation statistics, while Fig. 4 shows the overall trend between geographical distance and intraspecific genetic divergence. Some species showed a strong correlation between the two parameters, as genetic distances increased with geographical distance, but others did not show a significant relationship between the two variables (Table 2). Overall, this analysis showed a weak relationship (R2 = 0.22; y = 8E-05x + 0.250) between the geographical extent of a species and its maximum intraspecific divergence (Fig. 4).
Table 1.
Percentage K2P sequence divergence at the COI barcode region among the 59 butterfly species with >2 specimens, among the 19 genera with two or more species and among the five families with two or more genera
| Distance class | n | Taxa | Comparisons | Min (%) | Mean (%) | Max (%) |
|---|---|---|---|---|---|---|
| Intraspecific | 352 | 59 | 1349 | 0 | 0.2 | 1.6 |
| Congeners | 233 | 19 | 1274 | 1.7 | 8.0 | 14.3 |
| Confamilial | 372 | 5 | 16 200 | 3.9 | 13.1 | 19.2 |
Table 2.
Maximum intraspecific distances for 55 butterfly species with barcode records from Pakistan and other nations
| Maximum intraspecific distance (individuals) | Mantel correlation statistics for geographical vs. genetic distances | ||||
|---|---|---|---|---|---|
| No. | Species | Pakistan | Combined | Countries with matches | (α = 0.05) |
| 1 | Aglais caschmirensis | 0.15 (8) | 0.2 (12) | Kyrgyzstan, Mongolia, Nepal, Russia, Uzbekistan | r = 0.8; P = 0.038 |
| 2 | Argynnis kamala | 0.0 (2) | 0.79 (3) | Nepal | r = 1.0; P = 0.333 |
| 3 | Argynnis hyperbius | 0.46 (5) | 0.96 (14) | Australia, Japan, South Korea | r = 0.28; P = 0.005 |
| 4 | Ariadne merione | 0.15 (6) | 0.3 (12) | India | r = 0.55; P = 0.239 |
| 5 | Aulocera swaha | 0.92 (6) | 0.96 (7) | India | r = 0.74; P = 0.000 |
| 6 | Belenois aurota | 0.46 (4) | 0.76 (7) | Kenya | * |
| 7 | Byasa polyeuctes | 0.15 (5) | 0.48 (6) | Taiwan | r = 0.71; P = 0.064 |
| 8 | Catopsilia pomona | 1.39 (13) | 1.93 (34) | Australia, China, Papua New Guinea, Thailand, Taiwan | r = 0.76; P = 0.000 |
| 9 | Catopsilia pyranthe | 0.30 (20) | 0.36 (13) | Australia, Malaysia | r = 0.82; P = 0.0001 |
| 10 | Celastrina argiolus | 0.31 (3) | 2.2 (76) | Armenia, Canada, Cyprus, Finland, France, Georgia, Germany, Iran, Italy, Kazakhstan, Mexico, Morocco, Romania, Russia, Spain, South Korea, United States | r = 0.81; P = 0.0001 |
| 11 | Colias erate | 0.15 (3) | 0.15 (4) | Kyrgyzstan | r = −0.27; P = 0.708 |
| 12 | Colias fieldii | 0.61 (13) | 0.64 (16) | China | r = 0.93; P = 0.0001 |
| 13 | Colotis amata | 0.6 (2) | 3.2 (44) | Angola, Iran, Kenya, Madagascar, Namibia, Oman, South Africa, Somalia, Tanzania, Yemen | r = 0.27; P = 0.042 |
| 14 | Colotis danae | 0.64 (9) | 1.53 (6) | Iran | * |
| 15 | Colotis etrida | 0.35 (6) | 0.35 (8) | India | * |
| 16 | Colotis vestalis | 0.92 (6) | 1.4 (21) | Algeria, Ethiopia, Iran, Israel, Oman, Sudan, Yemen | r = −0.17; P = 0.39 |
| 17 | Danaus chrysippus | 1.08 (6) | 1.47 (48) | Egypt, India, Italy, Kenya, Madagascar, Morocco, Philippines, Spain, South Africa, Taiwan, Tanzania | r = 0.007; P = 0.941 |
| 18 | Danaus genutia | – (1) | 0.8 (6) | India, Malaysia, Taiwan | r = −0.96; P = 0.0001 |
| 19 | Deudorix epijarbas | 0.0 (7) | 0.0 (8) | Taiwan | Genetic distances are ‘zero’ |
| 20 | Eurema hecabe | 0.0 (8) | 1.2 (34) | Australia, China, India, Japan, Korea, Malaysia, Papua New Guinea, Sri Lanka, Thailand | r = 0.14; P = 0.559 |
| 21 | Hasora chromus | – (1) | 0.64 (8) | Australia, Papua New Guinea | r = −0.52; P = 0.001 |
| 22 | Hipparchia parisatis | – (1) | 1.22 (4) | Iran | * |
| 23 | Junonia almana | 0.34 (13) | 1.02 (14) | India, Malaysia | r = 0.77; P = 0.008 |
| 24 | Junonia hierta | 0.0 (3) | 2.05 (41) | India, Kenya, Madagascar, South Africa, Tanzania | r = −0.46; P = 0.186 |
| 25 | Junonia iphita | 0.0 (4) | 0.92 (8) | India | r = 1.0; P = 0.333 |
| 26 | Junonia orithya | 1.39 (16) | 2.0 (19) | Australia, India, Malaysia, Taiwan | r = 0.21; P = 0.55 |
| 27 | Kallima inachus | – (1) | 0.31 (3) | India | * |
| 28 | Kaniska canace | – (1) | 0.8 (8) | Malaysia, South Korea | r = −0.97; P = 0.0001 |
| 29 | Lampides boeticus | 0.16 (7) | 2.63 (126) | Australia, Cyprus, Germany, Egypt, Iran, Israel, Italy, Kenya, Madagascar, Morocco, Papua New Guinea, Portugal, Romania, Spain, Taiwan, Tanzania | r = 0.35; P = 0.075 |
| 30 | Lasiommata menava | – (1) | 1.22 (4) | Iran, Tajikistan | r = 0.89; P = 0.167 |
| 31 | Lasiommata schakra | 0.16 (9) | 0.16 (15) | Nepal | r = 0.42; P = 0.707 |
| 32 | Lethe rohria | – (1) | 1.07 (2) | China | * |
| 33 | Lethe verma | 0.31 (4) | 1.12 (6) | China | r = 0.93; P = 0.039 |
| 34 | Libythea lepita | 0.61 (3) | 0.92 (8) | South Korea, Taiwan | r = 0.13; P = 0.789 |
| 35 | Limenitis lepechini | 0.77 (3) | 0.77 (7) | Uzbekistan | r = 0.0; P = 0.0001 |
| 36 | Lycaena phlaeas | 0.0 (8) | 1.12 (107) | Armenia, Canada, Cyprus, Finland, France, Germany, Iran, Italy, Morocco, Nepal, Norway, Portugal, Romania, Russia, Spain, Tunisia, USA | r = 0.7; P = 0.0001 |
| 37 | Neptis hylas | – (1) | 1.7 (6) | India | * |
| 38 | Papilio demoleus | 0.96 (11) | 1.02 (11) | Taiwan | r = 0.31; P = 0.331 |
| 39 | Papilio machaon | – (1) | 2.9 (110) | Canada, Finland, France, Germany, Israel, Italy, Japan, Morocco, Nepal, United States, Russia, Spain, Romania, South Korea | r = 0.23; P = 0.001 |
| 40 | Papilio Polyctor | 0.31 (9) | 1.95 (18) | China | r = 0.99; P = 0.046 |
| 41 | Papilio polytes | 0.8 (2) | 1.67 (10) | Malaysia, Thailand | r = 0.51; P = 0.347 |
| 42 | Pelopidas mathias | 1.6 (16) | 2.6 (23) | Indonesia, Madagascar, South Africa, UAE | r = 0.47; P = 0.177 |
| 43 | Phalanta phalantha | 0.15 (6) | 0.2 (10) | India | * |
| 44 | Pieris brassicae | 0.31 (3) | 1.53 (63) | Armenia, Austria, Finland, France, Germany, Italy, Kyrgyzstan, Morocco, Portugal, Romania, Russia, Spain | r = 0.02; P = 0.895 |
| 45 | Pieris canidia | 0.31 (9) | 0.8 (13) | Kyrgyzstan, Uzbekistan | r = 0.78; P = 0.161 |
| 46 | Pieris rapae | 0.30 (5) | 0.31 (6) | Nepal, South Korea | r = 0.39; P = 0.000 |
| 47 | Pontia daplidice edusa | 0.77 (4) | 1.25 (41) | Armenia, Austria, Finland, Georgia, Germany, Iran, Israel, Italy, Kazakhstan, Romania, Russia, UAE | r = 0.04; P = 0.689 |
| 48 | Pseudozizeeria maha | 0.0 (12) | 0.19 (18) | Japan, South Korea, Taiwan | r = 0.97; P = 0.068 |
| 49 | Tarucus balkanicus | 0.0 (5) | 2.23 (22) | Cyprus, Egypt, Israel, Morocco, Tunisia, Turkey, UAE | r = −0.69; P = 0.963 |
| 50 | Telecota colon | – (1) | 0.77 (5) | Australia | * |
| 51 | Tirumala limniace | 0.62 (3) | 2.09 (11) | India, Kenya, Tanzania | r = 0.85; P = 0.133 |
| 52 | Vanessa cardui | 0.49 (9) | 1.61 (115) | Algeria, Armenia, Australia, Canada, Eritrea, Finland, France, Germany, Israel, India, Italy, Japan, Kazakhstan, Kenya, Morocco, Romania, Russia, South Africa, South Korea, Spain, Taiwan, Tanzania, UAE, USA | r = 0.09; P = 0.329 |
| 53 | Vanessa indica | 0.0 (2) | 0.66 (6) | South Korea, Taiwan | r = 0.11; P = 0.932 |
| 54 | Zizeeria karsandra | 0.0 (2) | 1.53 (14) | Algeria, Australia, Cyprus, Egypt, UAE | r = 0.4; P = 0.247 |
| 55 | Zizina labradus | – (1) | 2.42 (86) | Australia, Kenya, New Zealand, Papua New Guinea, Tanzania | r = 0.29; P = 0.2 |
Species from Pakistan with no matches in the databases (n = 26): Argynnis lathonia,Callerebia annada,Celastrina huegelii,Cigaritis elima,Cigaritis vulcanus,Elphinstonia transcaspica,Eogenes lesliei,Euchrysops cnejus,Eurema brigitta,Gonepteryx rhamni,Graphium cloanthus,Heliophorus sena,Lasiommata sp. MA01,Libythea myrrha,Melitaea chitralensis,Neptis mahendra,Pantoporia nefte,Pantoporia opalina,Parnara guttata,Polycaena sp. MA01,Polyommatus icarus,Rapala extensa,Tarucus rosaceus,Ypthima avanta,Ypthima sakra,Ypthima inica
The number of individuals of a species included in the analysis is indicated in brackets. A double dash indicates that a given species was presented by only one specimen, and thus, maximum intraspecific divergence is not presented, while bold highlighting is used to indicate those species that exhibit a three-fold or greater increase in intraspecific variation when records outside of Pakistan were included.
Insufficient data to run the Mantel test.
Fig 4.
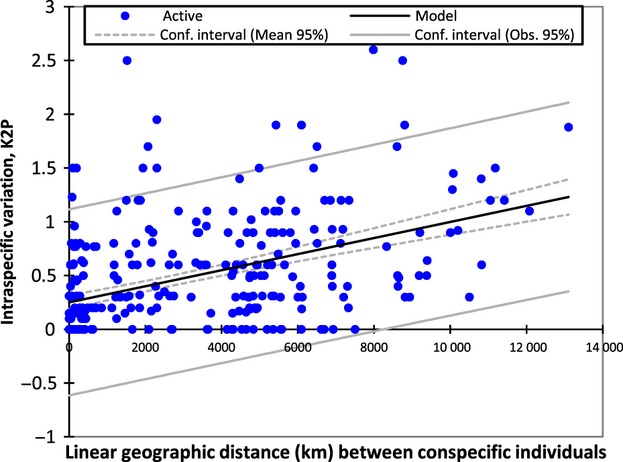
Intraspecific variation (K2P) against geographical extent (km) of butterflies from Pakistan and their conspecifics from other regions (linear regression, y = 8E-05x + 0.250; R2 = 0.22).
Discussion
Identification success for the butterflies of Pakistan
This study has begun the construction of a DNA barcode reference library for the butterflies of Pakistan. Cluster analysis revealed that all 81 species examined in the study formed a monophyletic cluster which corresponded perfectly with the taxa recognized on morphological criteria. Although three species pairs showed limited divergence (<3%), maximum intraspecific divergence was always lower than the NN distance, enabling the separation of all species. Even the most closely related (1.70%) species pair, Tarucus balkanicus and T. rosaceus, was separated with strong bootstrap support in the NJ tree. Our results confirm the usefulness of DNA barcoding in identifying the butterflies of Pakistan, but the sample size was low for some species and 75% of the fauna awaits analysis.
When sequences for butterfly species from Central Asia (Lukhtanov et al. 2009) were included, eight species pairs formed paraphyletic clusters. Among these pairs, the NN distance between Aglais caschmirensis (from Pakistan) and A. nixa (from Uzbekistan) was 0.2%, while that between A. caschmirensis and A. urticae (from Kazakhstan) was 1.4%. Although NN distances for these sister species pairs were small, barcode-based identifications were possible as reported by Tavares & Baker (2008) in their study on sister species of birds.
‘Barcode Gap Analysis’ showed that NN distance for all the species was higher than the maximum intraspecific distance. The Barcode Index Number (BIN) system (Ratnasingham & Hebert 2013) provided further evidence of the genetic distinctiveness of the species as it assigned the 81 species to 80 BINs with only T. balkanicus and T. rosaceus sharing a BIN. When identity analysis was performed using Best Match/Best Close Match at a 3% threshold, all the species were correctly identified. Other studies have generally reported similar results (Janzen et al. 2005; Lukhtanov et al. 2009; Gaikwad et al. 2012) with a few exceptions. For example, Gaikwad et al. (2012) found that intraspecific divergence was higher (7.8%) in the butterfly Lethe europa than the distance to its NN (7.4%). Such cases can, of course, arise through a failure to discriminate sibling taxa. Bortolus (2008) has emphasized the importance of detailed taxonomic study in cases where DNA barcode results are discordant with taxonomic assignments. Costa et al. (2012) have reinforced this conclusion, noting the need for a ranking system to register the certainty of identifications for specimens used to develop reference barcode libraries. These suggestions reinforce the importance of an integrative approach to species delimitation by considering morphological, genetic, ecological and geographical information, rather than considering taxonomic identifications as facts against which to ‘test’ DNA barcoding (e.g. Smith et al. 2008). Nevertheless, focusing on one region of the genome is useful to the community for generating a comparable set of sequences across a large number of diverse taxa and geographical regions.
Genetic divergence patterns with increasing geographical distance: a regional Asian perspective
The within-species divergence values for most species in the study were under the 2%. In most cases, the addition of conspecific sequences from other countries increased the intraspecific distance, but the relationship between geographical distance and the level of intraspecific divergence was not strong. In a few cases, substantial intraspecific distances were observed between specimens from the same region. For example, Pelopidas mathias collected from sites in Pakistan <250 km apart showed 1.54% divergence. On the other hand, Deudorix epijarbas from Pakistan and Taiwan (4832 km) lacked barcode divergence. Other species showed regional variation that was not linked to distance. For example, specimens of Lampides boeticus from Pakistan and Queensland Australia were just 0.4% divergent, but specimens from Papua New Guinea were 1.9% divergent. These results reinforce previous conclusions that geographical distance is often associated with an increased genetic divergence, but that the increase is too small to impede the identification of species (Lukhtanov et al. 2009; Bergsten et al. 2012; Gaikwad et al. 2012).
Diversity hotspots and endemism in Asia underscores the need for regional barcode libraries
Although Pakistan and neighbouring Central Asia are only 700 km apart, prior studies have indicated that there is little overlap in their butterfly faunas. In fact, just 42 species (14%) are shared among the 320 butterfly species from Pakistan (Roberts 2001) and the 353 species from Central Asia (Lukhtanov et al. 2009). Their distinctive faunas undoubtedly reflect the effectiveness of the Pamir mountain chain, which rises to more than 5000 m, as a dispersal barrier. This limited overlap suggests the presence of multiple regions of endemism in this segment of Asia, mirroring a pattern of low overlap between the biodiversity hotspots in the Western Ghats (India) and Sri Lanka (Bossuyt et al. 2004). Although India and Sri Lanka are on the same continental shelf, and the strait separating them does not exceed 70 m in depth, limited biotic interchanges have left the two areas with an unexpectedly large number of endemics. This fact highlights the need to expand barcode coverage for all animal groups from the various subregions in southern Asia. Certainly, barcode reference libraries based on species from other nations will only permit the identification of a fraction of Pakistan's biodiversity.
Acknowledgments
This research was enabled by grant HEC No. 20-1403/R& D/09, Sequencing DNA Barcodes of Economically Important Insect Species from Pakistan, from the Higher Education Commission of Pakistan. It was also supported by grant 106106-001, Engaging Developing Nations in iBOL, from IDRC. Sequence analysis was made possible by a grant from the Government of Canada through Genome Canada and the Ontario Genomics Institute in support of the International Barcode of Life (iBOL) project. We thank staff at the CCDB for aid with sequence analysis.
M.A. designed and performed experiment, analyzed data and wrote the paper. S.A. and A.M.K. collected and identified butterflies. S.J.A. provided help with analysis and writing the paper. P.D.N.H. designed the methods, contributed reagents and wrote the paper.
Data Accessibility
Specimen data, images and DNA sequences: BOLD project MABUT (Barcoding Butterflies of Pakistan).
DNA sequences: BOLD IDs and GenBank accession nos for each butterfly specimen listed in Table S1 (Supporting information).
Sequence alignments: Supporting information.
Supporting information
Additional Supporting Information may be found in the online version of this article:
ClustalW alignment of barcode sequences of 81 butterfly species from Pakistan.
BOLD IDs and GenBank accessions of butterfly specimens included in the study.
References
- Bergsten J, Bilton DT, Fujisawa T, et al. The effect of geographical scale of sampling on DNA barcoding. Systematic Biology. 2012;61:1–19. doi: 10.1093/sysbio/sys037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolus A. Error cascades in the biological sciences: the unwanted consequences of using bad taxonomy in ecology. Ambio. 2008;37:114–118. doi: 10.1579/0044-7447(2008)37[114:ecitbs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bossuyt F, Meegaskumbura M, Beenaerts N, et al. Local endemism within the Western Ghats – Sri Lanka biodiversity hotspot. Science. 2004;306:479–481. doi: 10.1126/science.1100167. [DOI] [PubMed] [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN. DNA barcodes of closely related (but morphologically and ecologically distinct) species of skipper butterflies (Hesperiidae) can differ by only one to three nucleotides. Journal of the Lepidopterists Society. 2007;61:138–153. [Google Scholar]
- Carletto J, Blin A, Vanlerberghe-Masutti F. DNA-based discrimination between the sibling species Aphis gossypii Glover and Aphis frangulae Kaltenbach. Systematic Entomology. 2009;34:307–314. [Google Scholar]
- Clare EL, Lim BK, Fenton MB, Hebert PDN. Neotropical bats: estimating species diversity with DNA barcodes. PLoS ONE. 2011;6:e22648. doi: 10.1371/journal.pone.0022648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FO, Landi M, Martins R, et al. A ranking system for reference libraries of DNA barcodes: application to marine fish species from Portugal. PLoS ONE. 2012;7:e35858. doi: 10.1371/journal.pone.0035858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Yu F, Zhang T-X, et al. DNA barcoding of six Ceroplastes species (Hemiptera: Coccoidea: Coccidae) from China. Molecular Ecology Resources. 2012;12:791–796. doi: 10.1111/j.1755-0998.2012.03152.x. [DOI] [PubMed] [Google Scholar]
- Ekrem T, Willassen E, Stur E. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Molecular Phylogenetics and Evolution. 2007;43:530–542. doi: 10.1016/j.ympev.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Elmeer K, Almalki A, Mohran KA, Al-Qahtani KN, Almarri M. DNA barcoding of Oryx leucoryx using the mitochondrial cytochrome c oxidase gene. Genetics and Molecular Research. 2012;11:539–547. doi: 10.4238/2012.March.8.2. [DOI] [PubMed] [Google Scholar]
- Foottit RG, Maw HEL, Dohlen von CD, Hebert PDN. Species identification of aphids (Insecta: Hemiptera: Aphididae) through DNA barcodes. Molecular Ecology Resources. 2008;8:1189–1201. doi: 10.1111/j.1755-0998.2008.02297.x. [DOI] [PubMed] [Google Scholar]
- Gaikwad SS, Ghate HV, Ghaskadbi SS, Patole MS, Shouche YS. DNA barcoding of nymphalid butterflies (Nymphalidae: Lepidoptera) from Western Ghats of India. Molecular Biology Reports. 2012;39:2375–2383. doi: 10.1007/s11033-011-0988-7. [DOI] [PubMed] [Google Scholar]
- Guralnick R, Hill A. Biodiversity informatics: automated approaches for documenting global biodiversity patterns and processes. Bioinformatics. 2009;25:421–428. doi: 10.1093/bioinformatics/btn659. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SA. Butterflies of Islamabad and the Murree Hills. Islamabad: Asian Study Group; 1994. p. p68. [Google Scholar]
- Hastings JM, Schultheis PJ, Whitson M, et al. DNA barcoding of new world cicada killers (Hymenoptera: Crabronidae) Zootaxa. 2008;1713:27–38. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London, Series B. Biological Sciences. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London, Series B. Biological Sciences. 2003b;270(Suppl):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biology. 2004a;2:1657–1663. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America. 2004b;101:14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, deWaard JR, Landry J-F. DNA barcodes for 1/1000 of the animal kingdom. Biology Letters. 2010;6:359–362. doi: 10.1098/rsbl.2009.0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ZE, Li Z, Li SQ. Identifying Chinese species of Gammarus (Crustacea: Amphipoda) using DNA barcoding. Current Zoology. 2009;55:158–164. [Google Scholar]
- Hubert N, Hanner R, Holm E, et al. Identifying Canadian fresh water fishes through DNA barcodes. PLoS ONE. 2008;3:e2490. doi: 10.1371/journal.pone.0002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova NV, Grainger CM. 2007a. CCDB protocols, COI amplification. Available from http://www.dnabarcoding.ca/CCDB-DOCS/CCDB_amplification.pdf on November 6, 2012.
- Ivanova NV, Grainger CM. 2007b. CCDB protocols, sequencing. Available from http://www.dnabarcoding.ca/CCDB_DOCS/CCDB_Sequencing.pdf on November 6, 2012.
- Ivanova NV, Grainger CM. 2007c. CCDB protocols, primer sets. Available from http://www.dnabarcoding.ca/CCDB_DOCS/CCDB_Amplification.pdf on November 6, 2012.
- Ivanova NV, deWaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Notes. 2006;6:998–1002. [Google Scholar]
- Ivanova NV, DeWaard JR, Hebert PDN. 2007. CCDB protocols, glass fiber plate DNA extraction. Available from http://www.dnabarcoding.ca/CCDB_DOCS/CCDB_DNA_Extraction.pdf on November 6, 2012.
- Janzen DH, Hajibabaei M, Burns JM, et al. Wedding biodiversity inventory of a large and complex Lepidopteran fauna with DNA barcoding. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences. 2005;360:1835–1845. doi: 10.1098/rstb.2005.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen DH, Hallwachs W, Blandin P, et al. Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Molecular Ecology Resources. 2009;9:1–26. doi: 10.1111/j.1755-0998.2009.02628.x. [DOI] [PubMed] [Google Scholar]
- Jinbo U, Kato T, Ito M. Current progress in DNA barcoding and future implications for entomology. Entomological Science. 2011;14:107–124. [Google Scholar]
- Kim MII, Wan X, Kim MJ, et al. Phylogenetic relationships of true butterflies (Lepidoptera: Papilionoidea) inferred from COI, 16S rRNA and EF-1α sequences. Molecules and Cells. 2010;30:409–425. doi: 10.1007/s10059-010-0141-9. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lee W, Kim H, Lim J, et al. Barcoding aphids (Hemiptera: Aphididae) of the Korean Peninsula: updating the global data set. Molecular Ecology Resources. 2011;11:32–37. doi: 10.1111/j.1755-0998.2010.02877.x. [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov EV, Hebert PDN. DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Molecular Ecology Resources. 2009;9:1302–1310. doi: 10.1111/j.1755-0998.2009.02577.x. [DOI] [PubMed] [Google Scholar]
- Malik JM. Notes on the butterflies of Pakistan in the collection of Zoological Survey Department Karachi. Part II. Records of the Zoological Survey of Pakistan. 1973;5:11–28. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Meier R, Kwong S, Vaidya G, Ng PKL. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Systematic Biology. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- Meier R, Zhang G, Ali F. The use of mean instead of smallest interspecific distances exaggerates the size of the “barcoding gap” and leads to misidentification. Systematic Biology. 2008;57:809–813. doi: 10.1080/10635150802406343. [DOI] [PubMed] [Google Scholar]
- Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro-Maciel E, Reid B, Fitzsimmons NN, Le M, DeSalle R, Amato G. DNA barcodes for globally threatened marine turtles: a registry approach to documenting biodiversity. Molecular Ecology Resources. 2010;10:252–263. doi: 10.1111/j.1755-0998.2009.02747.x. [DOI] [PubMed] [Google Scholar]
- Nieukerken van EJ, Doorenweerd C, Stokvis FR, Groenenberg DSJ. DNA barcoding of the leaf-mining moth subgenus Ectoedemia s. str. (Lepidoptera: Nepticulidae) with COI and EF1-a: two are better than one in recognizing cryptic species. Contributions to Zoology. 2012;81:1–24. [Google Scholar]
- Park DS, Suh SJ, Hebert PD, Oh HW, Hong KJ. DNA barcodes for two scale insect families, mealybugs (Hemiptera: Pseudococcidae) and armored scales (Hemiptera: Diaspididae) Bulletin of Entomological Research. 2011;101:429–434. doi: 10.1017/S0007485310000714. [DOI] [PubMed] [Google Scholar]
- Pramual P, Kuvangkadilok C. Integrated cytogenetic, ecological, and DNA barcode study reveals cryptic diversity in SimuliumGomphostilbiaangulistylum (Diptera: Simuliidae) Genome. 2012;55:1–12. doi: 10.1139/g2012-031. [DOI] [PubMed] [Google Scholar]
- Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, automated barcode gap discovery for primary species delimitation. Molecular Ecology. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- Quilang JP, Santos BS, Ong PS, et al. DNA barcoding of the Philippine endemic freshwater sardine Sardinella tawilis (Clupeiformes: Clupeidae) and its marine relatives. The Philippine Agricultural Scientist. 2011;94:248–257. [Google Scholar]
- Ratnasingham S, Hebert PDN. BOLD: the Barcode of Life Data System. Molecular Ecology Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. http://www.barcodinglife.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. A DNA-based registry for all animal species: the Barcode Index Number System. PLoS ONE. 2013 doi: 10.1371/journal.pone.0066213. , in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TJ. The Butterflies of Pakistan. Karachi: Oxford University Press; 2001. p. p290. [Google Scholar]
- Silva-Brandao KL, Lyra ML, Freitas AVL. Barcoding Lepidoptera: current situation and perspectives on the usefulness of a contentious technique. Neotropical Entomology. 2009;38:441–451. doi: 10.1590/s1519-566x2009000400001. [DOI] [PubMed] [Google Scholar]
- Smith MA, Rodriguez JJ, Whitfield JB, et al. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12359–12364. doi: 10.1073/pnas.0805319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourakov A, Zakharov EV. “Darwin's butterflies”? DNA barcoding and the radiation of the endemic Caribbean butterfly genus Calisto (Lepidoptera, Nymphalidae, Satyrinae) Comparative Cytogenetics. 2011;5:191–210. doi: 10.3897/CompCytogen.v5i3.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutzenberger P, Brehm G, Fiedler K. DNA barcoding-based species delimitation increases species count of Eois (Geometridae) moths in a well-studied tropical mountain forest by up to 50% Insect Science. 2011;18:349–362. [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares ES, Baker AJ. Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evolutionary Biology. 2008;8:81. doi: 10.1186/1471-2148-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi A, Bag SK, Shukla V, Roy S, Tuli R. Oligonucleotide frequencies of barcoding loci can discriminate species across kingdoms. PLoS ONE. 2010;5:e12330. doi: 10.1371/journal.pone.0012330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglia T, Haxaire J, Kitching IJ, Meusnier I, Rougerie R. Morphology and DNA barcoding reveal three cryptic species within the Xylophanes neoptolemus and loelia species-groups (Lepidoptera: Sphingidae) Zootaxa. 2008;1923:18–36. [Google Scholar]
- Valentini A, Pompanon F, Taberlet P. DNA barcoding for ecologists. Trends in Ecology and Evolution. 2009;24:110–117. doi: 10.1016/j.tree.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Vanhaecke D, Leaniz de CG, Gajardo G, et al. DNA barcoding and microsatellites help species delimitation and hybrid identification in endangered Galaxiid fishes. PLoS ONE. 2012;7:e32939. doi: 10.1371/journal.pone.0032939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgilio M, Backeljau T, Nevado B, Meyer MD. Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics. 2010;11:206. doi: 10.1186/1471-2105-11-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JM, Jacobus LM, Funk DH, et al. A DNA barcode library for North American Ephemeroptera: progress and prospects. PLoS ONE. 2012;7:e38063. doi: 10.1371/journal.pone.0038063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Shivji MS, Hanner RH. Identifying sharks with DNA barcodes: assessing the utility of a nucleotide diagnostic approach. Molecular Ecology Resources. 2009;9(Suppl. 1):243–256. doi: 10.1111/j.1755-0998.2009.02653.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Robinson JL, Geraci CJ, et al. Accelerated construction of a regional DNA-barcode reference library: caddisflies (Trichoptera) in the Great Smoky Mountains National Park. Journal of North American Benthological Society. 2011;30:131–162. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ClustalW alignment of barcode sequences of 81 butterfly species from Pakistan.
BOLD IDs and GenBank accessions of butterfly specimens included in the study.
Data Availability Statement
Specimen data, images and DNA sequences: BOLD project MABUT (Barcoding Butterflies of Pakistan).
DNA sequences: BOLD IDs and GenBank accession nos for each butterfly specimen listed in Table S1 (Supporting information).
Sequence alignments: Supporting information.