Abstract
We combined systematic bioinformatics analyses and molecular dynamics simulations to assess the conservation patterns of Ser and Thr motifs in membrane proteins, and the effect of such motifs on the structure and dynamics of α-helical transmembrane (TM) segments. We find that Ser/Thr motifs are often present in β-barrel TM proteins. At least one Ser/Thr motif is present in almost half of the sequences of α-helical proteins analyzed here. The extensive bioinformatics analyses and inspection of protein structures led to the identification of molecular transporters with noticeable numbers of Ser/Thr motifs within the TM region. Given the energetic penalty for burying multiple Ser/Thr groups in the membrane hydrophobic core, the observation of transporters with multiple membrane-embedded Ser/Thr is intriguing and raises the question of how the presence of multiple Ser/Thr affects protein local structure and dynamics. Molecular dynamics simulations of four different Ser-containing model TM peptides indicate that backbone hydrogen bonding of membrane-buried Ser/Thr hydroxyl groups can significantly change the local structure and dynamics of the helix. Ser groups located close to the membrane interface can hydrogen bond to solvent water instead of protein backbone, leading to an enhanced local solvation of the peptide.
Keywords: Bioinformatics, Molecular dynamics, Molecular transporters and receptors, Ser/Thr motifs, Transmembrane proteins
Hydrophilic amino acids of transmembrane (TM) proteins can have important structural and functional roles. For example, hydrophilic amino acids can contribute to the association of TM segments via hydrogen-bonding or salt-bridging interactions (Hermansson and von Heijne 2003; Zhou et al. 2000, 2001), influence the boundary of the insertion of membrane proteins into the lipid bilayer (Krishnakumar and London 2007), reduce (via salt bridging) the free energy of membrane partitioning of a helix (Chin and von Heijne 2000), and can influence ligand binding (Junne et al. 2007) or participate in chemical reactions of the TM protein (Metz et al. 1991). Ser and Thr groups are distinguished from other polar side chains by their ability to compete with the backbone groups for intrahelical hydrogen bonding. Such hydrogen bonding may have important consequences for the structure and function of the membrane protein. To understand the conservation patterns of Ser/Thr groups in membrane proteins, and the effect of Ser/Thr groups on the local protein structure and dynamics, here we combined extensive bioinformatics analyses with molecular dynamics simulations.
Regardless of whether the residue is buried or solvent-accessible, the side chains of the polar amino acids Ser and Thr have a high propensity for intrahelical hydrogen bonding (Gray and Matthews 1984); this intrahelical hydrogen bonding can be either with a backbone amide or with a backbone carbonyl group (Gray and Matthews 1984; Presta and Rose 1988; Richardson and Richardson 1988). Hydrogen bonding to the backbone amide is observed when Ser/Thr are located in the position immediately preceding the N terminus of α-helices (N-cap) (Doig et al. 1997; Kumar and Bansal 1998; Vijayakumar et al. 1999) and in membrane proteins (Eilers et al. 2000).
The interior location preferred by Ser/Thr in membrane proteins (Pilpel et al. 1999) was associated with the high packing values of these amino acids (Eilers et al. 2000); statistical analysis suggested that Ser/Thr whose hydroxyl groups hydrogen bond to the backbone of the TM helix can induce a small local bending of the helix (Ballesteros et al. 2000). When associated with Pro in the TM segment sequence, a Ser/Thr can modulate significantly the structural deformation of the helix induced by Pro (Deupi et al. 2004). The preference of Ser/Thr amino acids for intrahelical hydrogen bonding with the backbone could also be responsible for the lack of a significant contribution of Ser/Thr to the association of TM segments (Zhou et al. 2001). But, when specific Ser/Thr motifs are present at the interface of TM helices, they can drive oligomerization of the helices via interhelical hydrogen bonding (Dawson et al. 2002).
The competition with backbone groups for hydrogen bonding may be related to the low propensity of these amino acids in the middle of α-helices (Vijayakumar et al. 1999). Nevertheless, the frequency of occurrence of Thr/Ser at buried sites within TM segments of helical is higher than that of other polar and charged amino acids (Gratkowski et al. 2001). At buried sites in proteins, Ser/Thr tend to hydrogen bond (Worth and Blundel 2008).
Given the expectation that Ser/Thr are relatively infrequent in a TM α-helical segment, we were intrigued by our observations that multiple Ser/Thr are present within TM segments of membrane proteins with different function, particularly in functionally important regions of several molecular transporters and receptors. In what follows we discuss briefly several such examples.
The analysis of sequences of retinal proteins indicates the presence of a TT motif at the heart of the protein; the TT motif is largely conserved as TT, ST, or TC (Nack et al. 2012). In the bacteriorhodopsin proton pump the two Thr are present at position 89 and 90 (i.e., close to the primary proton acceptor D85), and the Thr90Ala mutation has profound effects on the reaction cycle (Perálvarez et al. 2001). G-protein-coupled receptors squid and bovine rhodopsin contain several Ser/Thr motifs along the TM segments—for example, S79DFT82, S122IMT125 and S263IVIVSQFLSWS275 in squid rhodopsin (pdb 2Z73, Murakami and Kouyama 2008), and T58LIVT62, T92TTLYTS98, and T297S298 in bovine rhodopsin (pdb 1U19, Okada et al. 2004). S122 of squid rhodopsin is part of the hydrogen-bonded network that may be involved in signal relay (Murakami and Kouyama 2008, Jardon-Valadez et al. 2010), and bovine rhodopsin T94 is involved in controlling the protonation state of the retinal Schiff base (Buss et al. 2003). The conserved squid rhodopsin Y315 makes potentially important hydrogen bonds with water (Sugihara et al. 2011); Y315 is located on the cytoplasmic side of a TM helix, near the loop segment S316VS318.
In the GlpG intramembrane protease, helix TM3 that interconnects distant hydrogen-bonding clusters contains the sequence S171GKLIVITLISALS185 (Bondar et al. 2009); another intramembrane protease, FlaK, also has a TM helix that contains three closely spaced Thr/Ser groups (T710LSYLVT716) (pdb 3S0X, Hu et al. 2011). In the Thermotoga maritima SecY TM7, a helix thought to be critical for the translocon function (Plath et al. 1998; van den Berg et al. 2004; Du Plessis et al. 2009), contains the sequence S277AIVSIPSAIASIT290; the hydroxyl groups of the four Ser amino acids and of the Thr hydrogen bond to backbone carbonyl groups of other TM7 amino acids (Bondar et al. 2010). The corresponding region of the Escherichia coli translocon contains three Ser and one Thr groups in the sequence S281S282IILFPAT288IAS291 (Bondar et al. 2010). Mutation of the E. coli S282 to Arg leads to the prlA401 phenotype (Osborne and Silhawy 1993) thought to be characterized by the destabilization of the closed state of the translocon (Smith et al. 2005); adding one more Thr that replaces I290 reduced export of staphylokinase by the translocon (Sako 1991; Osborne and Silhavy 1993). Arrangements of Ser/Thr separated by two or three residues, with the Ser/Thr side chains largely on one side of the TM helix, are also observed in the P-type proton pump AHA2 from Arabidopsis thaliana—T686IMT689, S762IIS765, or S827IVT830 (pdb 3B8C, Pedersen et al. 2007); the T686IMT689 segment within TM6 is very close to the primary proton donor/acceptor D684. The membrane-embedded region of the α subunit of the nicotinic acethylcholine receptor consists of four helices, each with at least one Ser/Thr motif; helix M2 contains three Thr and six Ser groups (Unwin 2005). Finally, the TM segment of the amyloid precursor protein has two Thr amino acids separated by two VI pairs, the N-terminal Thr being at the γ42 cleavage site (see e.g., Munter et al. 2007).
Because Thr/Ser have positive free energies of insertion in the membrane hydrophobic core (Hessa et al. 2005; Moon and Fleming 2011), and weakly inhibit membrane insertion (Xie et al. 2007)—although the context of the amino acid in the TM segment is also very important for its recognition by the protein translocon (Hessa et al. 2005), it is indeed plausible, as some of the examples above may suggest, that the presence of Ser/Thr motifs in TM segments may be important for the structural and/or functional role of the protein.
We revisit here the presence of Ser/Thr amino acids in TM proteins and their effect on the local structure and dynamics of model TM protein segments. We performed systematic bioinformatics analyses of a data set consisting of 339 unique sequences of protein chains that contain α-helical and β-barrel TM segments. We find that, within the data sets analyzed here, about half of the sequences of the α-helical membrane proteins have at least one Ser/Thr motif, although sequences containing a large number of motifs are a minority. In the α-helical membrane proteins, Ser/Thr motifs can be present not only along the TM helices, but also in solvent-exposed regions of the protein—which may contain β-strands. On the basis of the bioinformatics analyses and visual inspection of protein structures we could identify membrane transporters that have a remarkable number of Ser/Thr groups along TM helices (see examples in Fig. S1). To assess how various Ser/Thr motifs affect the local structure and dynamics of α-helical TM segments, we carried out molecular dynamics (MD) simulations of four model single-spanning α-helical TM segments, including TM7 of T. maritima SecY. The MD simulations indicate that presence of Ser/Thr motifs can affect the local structure, dynamics, and solvation of TM helices. We thus conclude that the presence of multiple Ser/Thr motifs along the TM helices of a protein, although a relatively infrequent event, likely has significant implications for the structure and dynamics of the protein.
Methods
Bioinformatics Analyses
TM protein sequence chains were selected from the Protein Data Bank of Transmembrane Proteins (PDB_TM, Tusnády et al. 2005a), which is based on scanning all PDB entries with the TMDET algorithm (Tusnády et al. 2005b). Only nonredundant sequences were selected from the complete PDB_TM data set. The sequences are classified into two subsets: chains containing α-helical TM segments, and chains containing β-barrel TM segments; these two data sets are denoted here as, respectively, data-α (291 unique sequences) and data-β (48 unique sequences).
For the protein sequences in the both data-α and data-β sets, we analyzed the presence of the following specific motifs (denoted in what follows as Signatures) containing Ser/Thr amino acids: SS, ST, TT, SxxS, SxxxS, SxxT, SxxxT, TxxT, and TxxxT, where “x” denotes any amino acid. For the Signature analysis of the protein sequences, we used the program Preg (Rice et al. 2000). The search for Signatures included the entire protein chain—that is, regions of the protein that may not be within the lipid membrane region are also included in the search.
The statistical analysis of the sequences was performed using the R software (R Development Core Team 2010). Using R, we first tested the normality of the data-α and data-β samples using the Shapiro–Wilk normality test (Shapiro and Wilk 1965; Royston and Remark 1995). To compare the results for the different Signatures in data-α and data-β, we used the nonparametric (i.e., distribution-free) statistical hypothesis Mann–Whitney U-test, also called the Wilcoxon rank-sum test (Mann and Whitney 1947).
To cluster the data-α and data-β Sequences according to the number and type of Signatures, we proceeded as follows. The results of the Signature analysis described above were used to derive the Signature number information per sequence (that is, the number of SS, ST, TT, SxxS, SxxS, TxxT, TxxxT, SxxT and SxxxT per sequence) for each data set. For each Signature, all counts were normalized between 0 (no Signature) and 1 (the highest number of Signatures in each column). The Signature data were analyzed using unsupervised cluster learning methods. We used two distinct clustering methods: hierarchical clustering (Mitchell 1997; Jain et al. 1999), and k-means clustering (MacQueen 1967). The hierarchical clustering was performed using the Euclidean distance and the complete linkage approach. The number of clusters was calculated using the inconsistency threshold and coefficient (Bezdek and Pal 1998) as validity indices. The Euclidian space was used for the k-means clustering. In order to reduce the sensitivity of the algorithm to the initial random cluster centroids, we repeated each of the k-means runs 10 times and chose the best solution. We used the silhouette method (Rousseeuw 1987) to estimate the number of clusters. The potentially optimal number of k-means clusters was then chosen in order to maximize the average distance between silhouette means. Unless indicated otherwise, all functions used are part of the Matlab Statistics Toolbox (Jones 1993).
Molecular Dynamics Simulations
We investigated the structure and dynamics of four model peptides that contain different types and numbers of Ser/Thr Signatures (Table 1). The peptide in Sim1 consists of the seventh TM helix of the SecYEG protein translocon from T. maritima (Zimmer et al. 2008), and a fragment of the loop that connects the seventh and eighth TM helices of the T. maritima translocon (for a total of 27 amino acids); there are four Ser groups and one Thr in this peptide, separated by one, two, or three amino acids that are hydrophobic (Ile, Phe, or Val), or mildly hydrophobic (Ala). Sim2 and Sim3 are on peptides that contain two (Sim2) or four (Sim3) SerLeu stretches. In Sim4 we consider a model peptide that has four Ser groups within three SLLS repeats. The sequences of all peptides used in the MD simulations are given in Table 1. The simulation systems contained the TM peptide embedded in the center of hydrated palmitoyloleoyl phosphatidylethanolamine (POPE) hydrated lipid membrane, for a total of ~72,600 atoms (~280 lipid molecules, and ~12670 water molecules).
Table 1.
Summary of the MD simulations performed
| Simulation | Peptide sequence | Simulation length (ns)a | Average RMSD (Å)b |
|---|---|---|---|
| Sim1 | VIPIIPAS277AIVS281IPS284A IAS288IT290AAET294LK |
34 | 2.7 ± 0.1c |
| Sim2 | (L)3-(SL)2-(L)13 | 49 | 2.0 ± 0.1 |
| Sim3 | (L)3-(SL)4-(L)9 | 34 | 1.8 ± 0.1 |
| Sim4 | L-(SLL)4-(L)7 | 34 | 1.8 ± 0.1 |
The simulation length denotes the length of the trajectory without any constraints
The average RMSD and standard deviation values were computed from the last 20 ns of the trajectory without constraints (2000 frames). Time series of the RMSD values computed from Sim1–Sim4 are provided in Fig. S10
For the TM helical segment of the peptide in Sim1, the average RMSD is 1.3 ± 0.2 Å (see also Fig. S2A)
Coordinates for the peptide in Sim1 were taken from the crystal structure of Zimmer et al. (2008). The helical model peptides investigated in Sim2–Sim4 were constructed using the CHARMM software (Brooks et al. 1983). The protonatable amino acid residues in Sim1 were considered in their standard protonation states (Glu—negatively charged, and Lys—positively charged).
MD simulations were performed using the NAMD software (Kalé et al. 1999; Phillips et al. 2005) with the CHARMM22 force field for the protein atoms (MacKerell et al. 1998), CHARMM27 for lipids (Feller and MacKerell 2000), and the TIP3P model for the water molecules (Jorgensen et al. 1983). We cut off the short-range real-space interactions at 12 Å using a switching function between 8 and 12 Å. The smooth particle mesh Ewald summation (Darden et al. 1993; Essmann et al. 1995) was used to compute the Coulombic interactions. To maintain a constant temperature of 310 K and the pressure at 1 barr we used a Langevin dynamics scheme and a Nosé-Hoover Langevin piston (Feller et al. 1995; Martyna et al. 1994).
In the initial stages of the MD simulation the system was subject to weak harmonic constraints, as follows. During minimization, heating, and the first 1.5 ns of the equilibration, we used harmonic constraints of 5 kcal mol−1 Å−2 for the peptide atoms, and 2 kcal mol−1 Å−2 for the lipid and water molecule atoms. The constraints on the lipid atoms were then switched off, and we continued with 1 ns of equilibration with the constraints on the peptide and water atoms unchanged. The constraints on the peptide atoms were set to 2 kcal mol−1 Å−2 for the subsequent 1 ns of the equilibration. We then switched off the constraints on the peptide, and continued the equilibration with a constraint of 2 kcal mol−1 Å−2 on the water atoms only. All harmonic constraints were switched off for the remaining part of the simulations.
During the equilibration with harmonic constraints, and for the first 1 ns of the MD simulation without constraints, we used an integration step of 1 fs. For the remaining part of the simulation we used the reversible multiple time-step algorithm (Grubmüller et al. 1991; Tuckerman and Berne 1992) with time steps of 1 fs for the bonded forces, 2 fs for the short-range nonbonded forces, and 4 fs for the long-range electrostatic forces. The lengths of the bonds to hydrogen atoms were constrained using SHAKE (Ryckaert et al. 1977).
We used VMD (Humphrey et al. 1996) for molecular graphics, inspection of selected structures (e.g., Fig. S1), and trajectory analysis.
Results
Bioinformatics Analysis of the Sequences of TM Proteins
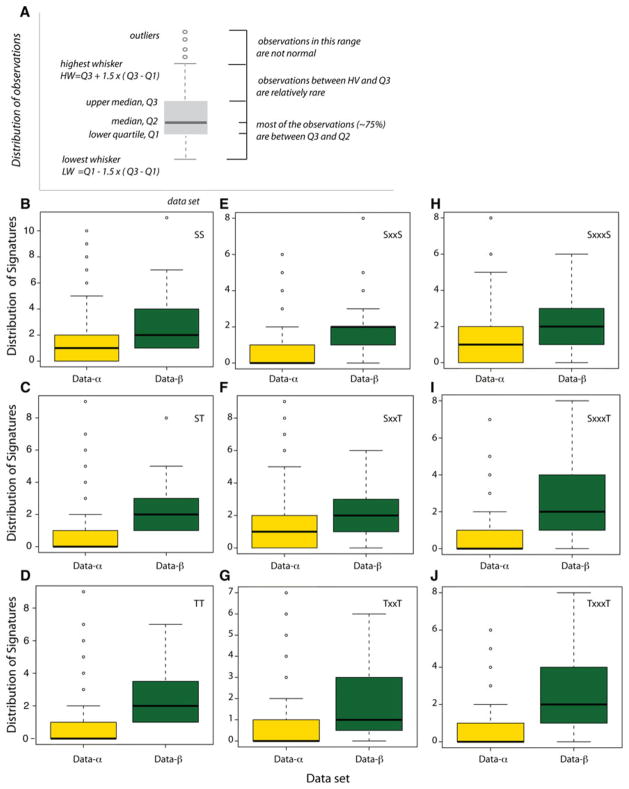
The statistical analysis of the sequences showed that they mostly did not follow a normal distribution; this made necessary the use of nonparametric tests to address the differences in the number of Signatures in data-α and data-β. The nonparametric tests showed significant differences in the number of Signatures appearing in data-α and data-β when using the Wilcoxon rank-sum test (Table 2, Figs. 1, S2–S4). The meaning of the box plots is illustrated in Fig. 1a. A summary of the statistical analyses of the sequences is given in Fig. S4. The Shapiro test is summarized in Fig. S5.
Table 2.
Summary of the percentage of data-α and data-β sequences that contain given numbers of specific Signatures a
| Motif | No. of Signatures |
|||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |
| Data-α | ||||||
| SS | 47.4 | 23 | 13.4 | 8.9 | 1.7 | 3.1 |
| SxxS | 55 | 22 | 9 | 8 | 4 | 1 |
| SxxxS | 45 | 29 | 13 | 7 | 3 | 1 |
| ST | 52.9 | 25.1 | 12 | 4.8 | 3.1 | 0.7 |
| SxxT | 49 | 22 | 17 | 7 | 3 | 1 |
| SxxxT | 58 | 23 | 12 | 4 | 2 | 1 |
| TT | 55.3 | 25.1 | 13.1 | 3.8 | 0.3 | 0.7 |
| TxxT | 53 | 30 | 11 | 4 | 1 | 1 |
| TxxxT | 57 | 25 | 9 | 6 | 2 | 0 |
| Data-β | ||||||
| SS | 5 | 16 | 11 | 23 | 23 | 9 |
| SxxS | 20 | 25 | 35 | 12 | 2 | 4 |
| SxxxS | 23 | 21 | 15 | 9 | 8 | 8 |
| ST | 7 | 21 | 16 | 12 | 21 | 15 |
| SxxT | 19 | 25 | 17 | 21 | 14 | 2 |
| SxxxT | 15 | 21 | 29 | 6 | 13 | 8 |
| TT | 12 | 12 | 19 | 14 | 11 | 14 |
| TxxT | 25 | 31 | 6 | 15 | 11 | 4 |
| TxxxT | 19 | 21 | 13 | 21 | 8 | 6 |
For a graphical representation of the Signature analysis here summarized, and for further details, see Figs. S2 and S3
Fig. 1.
Box-and-whisker plots illustrating the distribution of Signatures in data-α and data-β. Box-and-whisker plots are nonparametric representations of the distribution of the data within the sample assessed. The principles of a box plot are illustrated in (a). For example, the yellow box in (b) (for SS Signatures in data-α) indicates that (1) the median number of SS Signatures in the data-α set is 1; (2) approximately half of the sequences have no Signature; (3) about a quarter of the sequences have one SS Signature; and (4) outliers more than 5 SS Signatures are not normal within the data-α set. For information about the number of specific Signatures for each of the data-α and data-β sets, see Figs. S2–S5. Details of the statistical analyses and the results of the Shapiro test are given in Figs. S6 and S7
On average, most proteins from data-β contain Ser/Thr Signatures (Table 2, Figs. S2 and S3). Only 15–25 % of these sequences do not have any Signature (Table 2). We note significant differences between the distributions of Ser/Thr Signatures separated by two amino acids. Two SxxS Signatures are present in a large part of the sample (35 %), whereas TxxT is present in just 6 %; the presence of SxxT has an intermediate value of 17 % (Table 2; Fig. S3B, E, H).
The number of Signatures in data-α is significantly smaller than in data-β (Fig. 1). In data-α SxxxS (55 %) and SxxT (51 %) remain prevalent, although most often just one Signature is present (Fig. S2C, E); TxxT, SxxxT, TxxxT, and SxxS are found in, respectively, 47, 42, 43, and 45 % of the sequences (Table 2; Fig. S2H, F, I, B).
For a certain Signature, the amino acid sequence encompassed by Ser/Thr can be different for the two classes of TM proteins sequences considered here. Whereas in data-α SxxxS appears most often as SLxxS or SVxxS—that is, with a bulky hydrophobic amino acid (Fig. S6g–i), in data-β the preferred SxxxS sequences are SYxxS and SAxxS (Fig. S7b). Leu and Val are also associated with most of the TxxxT Signatures in data-α (Fig. S6p–s); in data-β, most common TxxxT motifs are TGxxT and TSxxT (Fig. S7e). Signatures SxxxT, SxxT, and SxxS have the same motifs in the both data sets, respectively, SAxxT and SLxxT, SVxT and SGxT, and SSxS and STxS (Figs. S6d–f, j–e, a–c, S7a, c, f) (Fig. 2).
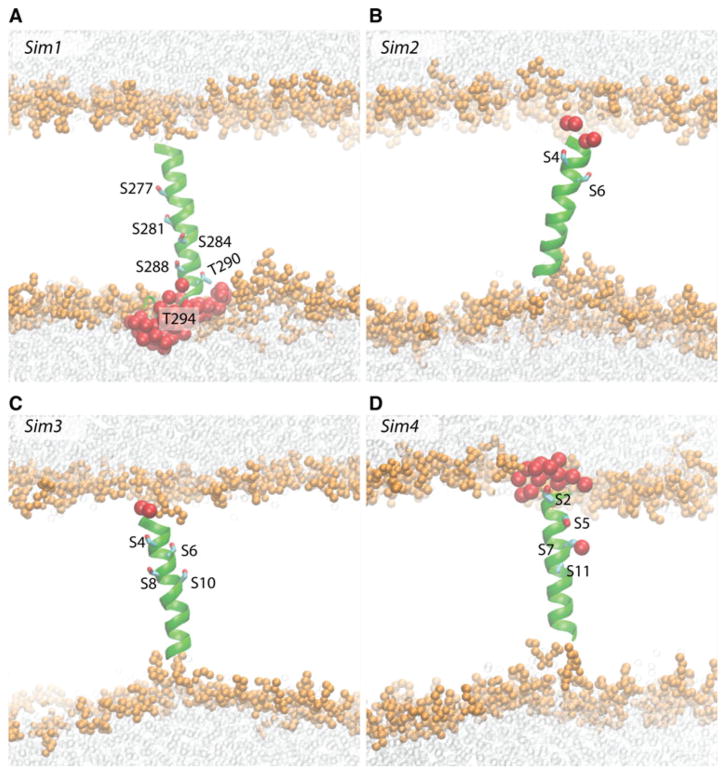
Fig. 2.
Schematic representations of Sims1–4. Coordinate snapshots from the end of Sim1 (a), Sim2 (b), Sim3 (c), and Sim4 (d) are shown as cutaway views with heavy atoms of lipid glycerol, phosphate, and choline groups depicted as van der Waals spheres scaled down by 40 %, and solvent water oxygen atoms depicted as transparent van der Waals spheres (also scaled down by 40 %). Water oxygen atoms within 6 of any Ser/Thr atom are shown as large spheres (van der Waals spheres enlarged by 20 %). The Ser/Thr side chains are shown as bonds. VMD (Humphrey et al. 1996) was used to prepare molecular graphics images
Our preliminary analysis of a possible relationship between noticeable numbers of Signatures and the molecular function of the protein indicates that, within the data sets considered here, the proteins with large numbers of Signatures tend to be transporters or receptors. Below we discuss briefly several such examples.
Identification of Molecular Transporters and Receptors with Significant Numbers of Ser/Thr Motifs
Visual inspection of data-α molecular transporters with a large number of Signatures (Fig. S8) indicates that the Signatures can be present not only in the membrane-embedded region of the protein, but also on solvent-exposed regions—loops, or larger soluble domains. The inclusion in the analysis of solvent-exposed parts of TM proteins is a limitation of the data sets used for the current analysis that leads to an over-estimation of the number of Ser/Thr Signatures. Below we discuss briefly examples of molecular transporters that contain large numbers of Signatures in the TM and/or solvent-exposed domains. In the discussion and in the Supplementary Information files the proteins are identified by the Protein Data Bank (PDB, Berman et al. 2003) chain used in the bioinformatics analysis; for example, 1f6g_A indicates that we used chain A from PDB 1f6g.
The AMPA subtype ionotropic glutamate receptor (Sobolevsky et al. 2009) has numerous Ser/Thr groups that give rise to sixteen SxxxS Signatures, nine SxxT, and smaller numbers of other Signatures—although most of the groups are solvent exposed (3kg2_A, Fig. S1A). The full-length potassium channel KcsA (1f6g_A; Cortes et al. 2001) (Fig. S1B), and the P-type ATPases AHA2–proton pump (3b8c_A, Pedersen et al. 2007) (Fig. S1C), sarcoplasmic reticulum Ca2+ pump (Obara et al. 2005) (2agv_A, Fig. S1D), and the Neurospora proton pump (1mhs_A; Kühlbrandt et al. 2002) (Fig. S1E) have Ser/Thr Signatures in the both TM and solvent-exposed regions.
Solvent-exposed domains of the TM protein may contain β-strand domains where most of the Ser/Thr Signatures are found, such as entry 3h9v_A, corresponding to the ATP-gated P2X4 ion channel from Kawate et al. 2009) (Fig. S1F). The structure of the KirBac3.4 potassium channel from Gulbis et al. (n.d.) (pdb entry 1XL4) has Signatures in the both TM helices and solvent-exposed β-strands (Fig. S1G). The polysaccharides translocon Wza (2j58_A; Dong et al. 2006) has 12 Signatures, of which three (two TxT and one SxxxT) are within the C-terminal D4 helical segment thought to be embedded in the outer membrane, and the remaining Signatures are in solvent-exposed α-helical or β-strand segments (Fig. S1H). A stunning example of a molecular transporter with solvent-exposed helical and β-strand segments and a large number of Signatures is mouse P-glycoprotein (3g5u_A; Aller et al. 2009), which has numerous SxxxS and ST Signatures, and at least one from each of the other Signatures (Figs. S1I, S8–J).
There are other examples of data-α transporters containing a remarkable number of Signatures within the TM region. In the putative ammonium channel Nitrosomonas europaea Rh50 (data-α entry 3b9w_A; Lupo et al. 2007), some of the Ser/Thr amino acid residues are part of TT (4), SxxS (2), TxxT (16), or TxxxT (1) Signatures, and a single TM helix can have up to five Ser/Thr groups (Fig. S1R). Alignment of the sequences of N. europaea Rh50 and the E. coli ammonium transporter AmtB (Lupo et al. 2007) indicates that some of the Ser/Thr Signatures are conserved, whereas others are present only in N. europaea Rh 50. For example, the S44AT46T47GT49YLV Rh50 segment of TM2 corresponds to S43MLT46QVT49VT51 in AmtB. In Rh50, F86 and F194 are thought to form a gate in the substrate transport path (Lupo et al. 2007). F194 is part of a SF194AT segment in TM6, and this sequence is present as SFNS in human Rhesus RhD and RhAG—two membrane proteins whose molecular function is not entirely clear, though RhAG may participate in ammonium transport (van Kim et al. 2006). TM6 of Rh50 also contains the SxxxS Signature S180MLGS184 (Lupo et al. 2007) S180 being part of the pore-lining amino acid residues (Hub et al. 2010). The arginine-agmatine antiporter (AdiC, data-α entry 3hqk_A; Fang et al. 2009) has a total of 14 Signatures, of which four are SS, and the remaining are one or two of the ST, TT, T/SxxxT/S or SxxT Signatures (Fig. S1J). Aquaporin AQP1 (data-α entry 1J4N_A; Sui et al. 2001) has Ser/Thr Signatures (mostly with Ser) both along TM helices and solvent-exposed loops (Fig. S1K).
We also identified membrane proteins with signal-transducing activities that have Ser/Thr Signatures in regions of the protein known to be critical for function, or have numerous Signatures. Thus, we found that bovine rhodopsin has 14 Signatures, four of which TT (Fig. S1L; 3cgl_A, Stenkamp 2008). The muscarinic M3 receptor (2amk_A, Han et al. 2005; Li et al. 2005), has numerous Ser/Thr along the TM helices (Fig. S1M).
From the data-β set, the outer membrane heme transporter ShuA (data-β entry 3fhh_A, Brillet et al. n.d.) is loaded with Ser and Thr groups: the structure depicted in Fig. S1N has 111 Ser/Thr groups, which is approximately 18 % of the 621 amino acids. This significant number of Ser/Thr gives rise to 11 SS Signatures and 29 other Signatures, SxxT being the least represented—just one Signature. Another example of a data-β protein with numerous Signatures is the hemophore receptor (and transporter) HasR (Krieg et al. 2009) (data-β entry 3csl_A, Figs. S1O, S9); this protein structure has 115 Ser/Thr groups, accounting for 15 % of the total amino acids. The protein has seven SS Signatures, eight SxxS, and between three and six from each of the other Signatures. At the other extreme in terms of number of Signatures for a data-β protein we mention the outer membrane porin OmpG (data-β entry 2f1c_X; Subbarao and van der Berg 2006). Out of the 286 amino acids, 27 (~9 %) are Ser/Thr (Fig. S1P), but there are only six Signatures: one ST, one TT, two TxxxT, one SxxxT, and one TxxT Signature (Fig. S9).
Structure and Dynamics of Model TM Peptides
The root mean squared differences (RMSD) relative to the starting coordinates has reached plateau values for all four model peptides considered here (Table 1; Fig. S10). The MD simulations (Sim1-Sim4) indicate that the presence of Ser/Thr amino acids within the membrane-embedded peptide segment has a significant effect on the structure and dynamics of the TM helix.
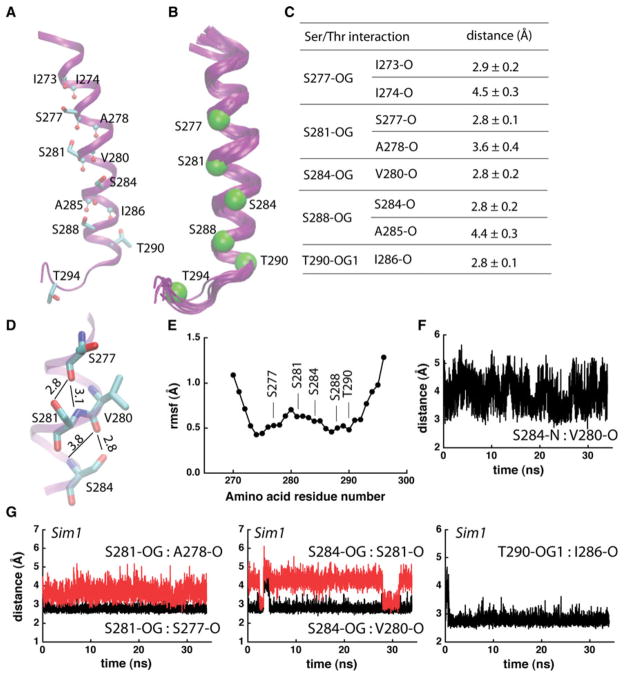
In Sim1, T290 and each of the four Ser amino acids hydrogen bond to the carbonyl group of the fourth amino acid downstream the sequence; in addition to the stable i-4 hydrogen bond, each of the two central Ser groups have a transient hydrogen bond with the carbonyl group of amino acid i-3 (Fig. 3a, c–e, g). Competition of the Ser hydroxyls with the amide groups for hydrogen bonding to the carbonyl groups (Fig. 3d, g) induces local kinks in the helix (Fig. 3b, d), and an enhanced dynamics of this region. Indeed, the root-mean-squared-fluctuation (rmsf) profile indicates high mobility for the groups close to S281 and S284, in particular upstream the sequence where the backbone hydrogen bonding is most perturbed (Fig. 3d–f).
Fig. 3.
Intrahelical hydrogen bonding in Sim1. a Each of the Ser amino acids from the TM helical part of peptide 1 (Table 1) engages in at least 1 intrahelical hydrogen bond. Ser/Thr groups are shown as bonds, and Leu backbone groups that hydrogen bond with Ser/Thr are shown with atoms as small spheres. b Overlap of 10 snapshots of the last 10 ns of Sim1 taken at every 1 ns. Cα atoms of the Ser/Thr amino acid residues at the end of Sim1 are shown as Van der Waals spheres. c Average distances between Ser/Thr hydroxyl oxygen atoms and their interaction partners. Average and standard deviation values were computed from the last 10 ns of Sim1 (1000 coordinate sets). d Close view of hydrogen bonding in the middle segment of peptide 1. Thin lines with numbers indicate hydrogen bonding and the corresponding distances in Å. The standard deviations for the side chain distances are given in (c); for the backbone S281-N:S277-O, and S284-N:V280-O distances, the standard deviations are ±0.3 Å and ±0.5 Å, respectively. e Cα-rmsf profile of peptide 1, computed from the last 10 ns segment of Sim1. The rmsf profile and the overlap of structural snapshots in (b) would suggest an enhanced flexibility of the Ser/Thr-rich TM helix. f Time series of the S284-N:V280-O distance. g Examples of distances (in Å) between Ser/Thr hydroxyl groups and backbone carbonyl groups monitored during Sim1. For all time series presented here and in Fig. 4, the origin of the time axis corresponds to all harmonic constraints being switched off
SecY-TM7, the TM peptide investigated in Sim1, contains four Ser/Thr Signatures in which Ser/Thr are separated by 1, 2, or 3 amino acid residues (Table 1; Fig. 3a); of the amino acids that separate Ser/Thr in these four Signatures, Ile and Ala are each present in three Signatures (Ile: S273xxxS277, S281xxS284, S284xxxS288, and S288 xS290; Ala: S273xxxS277, S284xxxS288, and T290xxx T294) (Table 1; Fig. 3a). The presence of Ile in the Ser Signatures of SecY-TM7 is consistent with the observation from the bioinformatics analysis above that in data-α; these Signatures are often associated with a bulky hydrophobic amino acid. In Sim2–Sim4 we explore further the structure and dynamics of Ser-containing TM segments by considering peptides with different numbers of Ser amino acid residues, and with different numbers of Leu separating the Ser groups (Table 1).
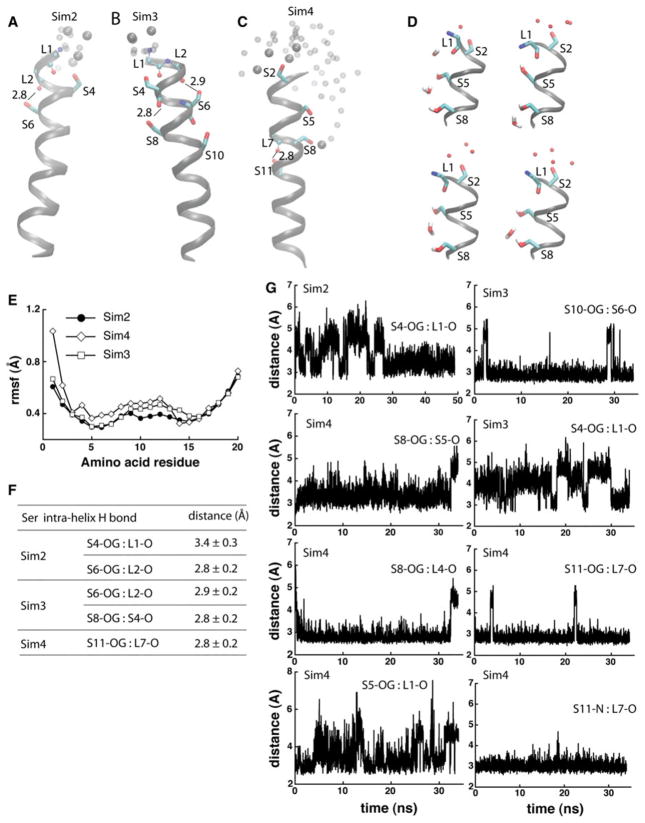
The results on the three Ser-containing poly-Leu model peptides tested in Sim2–Sim4 indicate that the dynamics and hydrogen-bonding interactions of the peptides depend on the number and location of the Ser groups. In Sim2, the two Ser at positions 4 and 6 in the sequence (Table 1; Fig. 4a) hydrogen bond to, respectively, the i-3 and i-4 backbone carbonyls (Fig. 4f, g); S4 can also interact with water (Fig. 4a).
Fig. 4.
Dynamics and hydrogen bonding of Ser-containing poly-Leu peptides. a, b Coordinate snapshots from Sim2–Sim4 showing the peptide and specific amino acids at the end of the Sims. Ser side chains are shown as bonds; atoms of backbone groups involved in Ser hydrogen bonding are shown as small spheres. Oxygen atoms of water molecules within 6 Å from Ser groups are shown as van der Waals spheres for the snapshot at the of the Sims, and as small transparent spheres for 2 coordinate snapshots taken 2 and 1 ns before the end of the Sims, respectively. For simplicity, in panels a–c hydrogen are not shown. In Sim4, water molecules penetrate into the lipid bilayer, forming hydrogen bonds with S8 and/or S5. d Snapshots from the last ~1 ns segment of Sim4 illustrating hydrogen bonding between Ser side chains and water molecules; selected hydrogen atoms are depicted for examples of hydrogen bonding. e Rmsf (Å) computed from the last 10 ns of Sim2–Sim4. f Selected Ser hydrogen-bond distances measured during the last 10 ns segments of Sim2–Sim4. g Examples of time series of Ser hydrogen-bond distances in Sim2–Sim4
Compared to Sim2, the peptide in Sim3 has two additional Ser groups at positions 8 and 10 in the sequence (Table 1; Fig. 4b). Hydrogen bond dynamics of S4 is similar in Sim2 and Sim3—during the first ~30 ns of the simulations the S4 hydroxyl: L1 backbone hydrogen bond breaks and reforms, and then stabilizes to ~3.4 Å. S10, located deep in the membrane core, is mostly hydrogen bonded to the S6 backbone carbonyl (Fig. 4g); S8 hydrogen bonds to the S4 carbonyl (Fig. 4b). The presence of S8 and S10 in Sim3 is associated with the middle segment of the peptide being more flexible than in Sim2 (Fig. 4e).
The 4 Ser groups of peptide in Sim4 are separated by short LL stretches (Table 1; Fig. 4c, d). That is, unlike in Sims 2 and 3, where the Ser side chains are distributed approximately symmetrically around the helix (Fig. 4a, b), in Sim4 the Ser side chains are roughly on the same side of the helix turns, leading to an imbalanced polarity of the helix (Fig. 4c, d). Toward the end of Sim4, we observe water molecules penetrating the lipid bilayer, where they hydrogen bond with S5 and S8 (Fig. 4d, c). That water molecules enter the lipid membrane to solvate the Ser groups in a peptide containing SLLS Signatures (Fig. 4c) is compatible with previous experiments on synthetic (LSSLLSL)3 peptides in diphytanol phosphatodylcholine lipid membranes: such peptides oligomerized and gave rise to ionic currents (Lear et al. 1998).
Upon hydrogen bonding to water S8 no longer hydrogen bonds to the backbone carbonyls of L4 and S5 (Fig. 4d, g); we also observe geometries in which the side chains of S5 and S8 are bridged via a water molecule (Fig. 4d). Backbone hydrogen bonding of S11 is largely stable (Fig. 4c, g). The presence in Sim4 of more Ser side chains that can interact with backbone or water molecules is associated with the flexibility of the middle segment of the peptide being larger than that observed for the peptides in Sim2 and Sim3 (Fig. 4e).
Discussion
We have revisited the conservation patterns of Ser/Thr motifs in α-helical and β-barrel TM proteins using a data set of 339 unique chain sequences. The bioinformatics analyses indicate the noticeable presence of Ser/Thr Signatures in both β-barrel and α-helical membrane proteins (Table 2, Figs. S1–S3). There is, however, a significant difference between these two classes of proteins: whereas just ~20 % of the β-barrel protein sequences analyzed here do not have any Ser/Thr Signature, in the α-helical TM proteins set the percentage of such sequences is 45–58 % (Table 2, Figs. S2, S3). The finding that within the data sets used here, proteins having more than two Ser/Thr Signatures are a minority (Table 2; Fig. 1) is consistent with Ser/Thr being relatively little represented in the single transmembrane helices analyzed in (Landolt-Marticorena et al. 1993), and with the expectation from the kPROT analysis that the probability of finding multiple Ser groups within the same TM segment degreases rapidly from ~20 % for two Ser groups, to ~0 for 5–6 groups (Pilpel et al. 1999). It is important to note here that the results on the frequency of Ser/Thr motifs depend on the protein sequences included in the data sets for bioinformatics analyses. Further work is necessary to understand whether the number and identity of Ser/Thr motifs within the TM domain of a TM protein depend on the structural details of the protein—e.g., on the number of TM helices or β-sheets.
Although multiple Ser/Thr motifs are relatively rare in TM proteins, we identified intriguing examples of α-helical TM proteins that contain multiple Ser/Thr groups within their TM helices (Fig. S1), including in regions that are important for function (Fig. S1R). The bioinformatics analysis further indicates that some Signatures are clearly preferred: compared to other Ser/Thr Signatures, the SxxS Signatures are disfavored in the β-barrel TM proteins set; such Signatures are also infrequent in the α-helical TM proteins. The SxxxS Signature, present in 55 % of the data-α TM proteins analyzed here, is often associated with a bulky hydrophobic side chain (e.g., SLxxS or SVxxS).
The MD simulations on model Ser-containing α-helical TM segments suggest that SxxS motifs may be disfavored because the energetic cost for hydrogen bonding to the helix backbone such buried Ser hydroxyl groups may be too high. In the simulation on the model peptide that contains four Ser groups each separated by two Leu (Sim4), water molecules enter the lipid bilayer and solvate all except for the deepest buried Ser (Fig. 4c, d). Penetration of water molecules into the hydrophobic core of the lipid bilayer had been observed in previous MD simulations on the partitioning of charged and polar amino acid models (MacCallum et al. 2008).
The side chains of all five Ser/Thr groups of the seventh TM helix of the T. maritima translocon hydrogen bond to the carbonyl group of the i-4 amino acid (Fig. 4a, c). In addition to the i-4 hydrogen bond, the two central Ser hydroxyl groups hydrogen bond transiently to the i-3 backbone carbonyls (Fig. 4a, g). Rh50 was identified here as a transporter with Ser/Thr Signatures within the TM region. In the 1.3 Å resolution structure of Rh50 (Lupo et al. 2007), T46 and T47 of the T46T47xT49 motif have the side chains on the membrane-facing side of a TM helix, whereas T49 faces the protein interior. Each of the three threonine hydroxyl groups is hydrogen bonded to backbone carbonyls from the same TM helix: T47 and T49 hydrogen bond to the i-4 carbonyl, whereas T46 has two hydrogen bonds, with the i-4 (2.9 Å) and i-3 (3.2 Å) carbonyl groups. Hydrogen bonding of Ser/Thr hydroxyl groups with the backbone carbonyl of the i-4 amino acid was also observed in previous MD simulations on model GGPG-flanked α-helical TM peptides in hydrated DMPC lipid bilayers (Johansson and Lindahl 2006). Hydrogen bonding to the backbone stabilizes the Ser/Thr hydroxyl groups within the hydrophobic membrane environment without the need of water entering the bilayer. That is, whether or not water molecules penetrate into the membrane to solvate buried hydroxyl groups depends on the sequence context.
The hydroxyl groups of Ser/Thr compete with backbone amide groups for hydrogen bonding to the carbonyls, and their backbone amide groups can hydrogen bond to the i-4 carbonyls (Fig. 3d). In the case of the peptide in Sim1, the maximum perturbation of the backbone amide:carbonyl hydrogen bond is observed for the central S284:V280 interaction (Fig. 3d, f). Perturbation of the backbone hydrogen bonding appears associated with an enhanced flexibility of the peptide (Fig. 4e). Although the details are expected to depend somewhat on how many motifs are present and where these motifs are located within the sequence, the computations on the model peptides considered here support a model in which Ser/Thr intrahelical hydrogen bonding can have significant effects on the local structure, dynamics, and water interactions of a TM α-helix.
The influence of the Ser/Thr Signatures on the local structure, dynamics, and solvation of the TM helices could be interpreted to suggest that such Signatures may have important functional roles. For example, Ser/Thr motifs may be used by the protein to shape the local structure by inducing local kinks (Fig. 3d), to enhance solvation (Fig. 4a–d) or flexibility (Fig. 4e). Within the data set considered here, proteins having significant numbers of Ser/Thr Signatures include molecular transporters and receptors. Examples of transporters and receptors containing Ser/Thr groups that are part of Signatures and known as functionally important include squid rhodopsin—S122, part of a SxxT motif, which contributes to a hydrogen-bonded network along the signal relay path (Jardon-Valadez et al. 2010); bacteriorhodopsin—the Thr90Ala mutation in the T89T90 Signature has drastic effects on the kinetics of the reaction cycle (Perálvarez et al. 2001); and SecY—the E. coli S282R mutation in the S281S282II-LFPAT288IAS291 sequence destabilizes the translocon (Smith et al. 2005).
As another implication of the results here on the Ser/Thr motifs, we suggest the possibility that the interpretation of serine scanning mutagenesis as tool for assessing the structure–function relationship in membrane proteins may be complicated by the introduction, through mutation, of Ser/Thr Signatures. A recent example of a serine-scanning investigation in which Ser/Thr Signatures have been introduced is the investigation by Miranda et al. (2011) of the structure–function relationship in TM6 of the yeast H+ ATPase Pma1. The yeast Pma1 has a Thr at position 733; the A732S and A735S mutations, which introduce ST and, respectively, TxS Signatures, led to a reduction in the proton pumping activity (Miranda et al. 2011). In the absence of detailed information about the structure, dynamics, and water interactions of the mutant proteins, the molecular origin of the mutation effect is unclear. But the consequences of removing from or inserting a Signature-related Ser/Thr group are likely due to a complex set of factors that would include not only the loss/addition of a hydrogen-bonding hydroxyl group, but also modification of the structure (e.g., releasing/promoting a kink), changes in the local dynamics, and reducing/enhancing the local hydration. In multipass TM proteins, Ser/Thr groups could also be involved in interhelical hydrogen bonds (Adamian and Liang 2002; Dawson et al. 2002); changes in the interhelical hydrogen bonds via mutation could affect significantly the conformational dynamics of the protein.
Supplementary Material
Acknowledgments
This research was supported in part by Grant GM-74637 from the National Institute of General Medical Sciences (to S.H.W), the Spanish Ministerio de Ciencia e Innovación (project TIN-2009-13950), the Consejería de Innovación, Investigación y Ciencia de la Junta de Andalucía (project TIC-02788) (to C.M.D.V.), the Marie Curie International Reintegration Award IRG276920/Biol-Transp-Comput (to A.-N.B), and an allocation of computer time from the National Science Foundation through the TeraGrid resources.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00232-012-9452-4) contains supplementary material, which is available to authorized users.
Contributor Information
Coral del Val, Email: C.DelVal@decsai.ugr.es, Department of Computer Science and Artificial Intelligence, University of Granada, 18071 Granada, Spain. CITIC-UGR, Centro de Investigación en Tecnologías de la Información y de las Comunicaciones de la Universidad de Granada, 18071 Granada, Spain.
Stephen H. White, Department of Physiology and Biophysics, University of California, Irvine, Medical Sciences I, D-374, Irvine, CA 92697-4560, USA
Ana-Nicoleta Bondar, Department of Physiology and Biophysics, University of California, Irvine, Medical Sciences I, D-374, Irvine, CA 92697-4560, USA.
References
- Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Prot Struct Funct Gen. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrel PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Deupi X, Olivella M, Haaksma EEJ, Pardo L. Serine and threonine residues bend a-helices in the χ1 = g− conformation. Biophys J. 2000;79:2754–2760. doi: 10.1016/S0006-3495(00)76514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Heinrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Bezdek JC, Pal NR. Some new indexes of cluster validity. IEEE Trans Syst Man Cybern B Cybern. 1998;28:301–315. doi: 10.1109/3477.678624. [DOI] [PubMed] [Google Scholar]
- Bondar A-N, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar A-N, del Val C, Freites JA, Tobias JA, White SH. Dynamics of SecY translocons with translocation-defective mutations. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillet K, Meksem A, Cobessi D. Crystal structure of the heme/hemoglobin outer membrane transporter ShuA from Shigella dysenteriae. Protein Data Bank. n.d doi: 10.2210/pdb3fhh/pdb. [DOI] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics. J Comput Chem. 1983;4:187–217. [Google Scholar]
- Buss V, Sugihara M, Entel P, Hafner J. Thr94 and wat2b effect protonation on the retinal chromophore in rhodopsin. Angew Chem Int Ed. 2003;42:3245–3247. doi: 10.1002/anie.200351034. [DOI] [PubMed] [Google Scholar]
- Chin CN, von Heijne G. Charge pair interactions in a model transmembrane helix in the ER membrane. J Mol Biol. 2000;303:1–5. doi: 10.1006/jmbi.2000.4122. [DOI] [PubMed] [Google Scholar]
- Cortes DM, Cuello LG, Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J Gen Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- Dawson JP, Weinger JS, Engelman DM. Motifs of serine and threonine can drive association of transmembrane helices. J Mol Biol. 2002;316:799–805. doi: 10.1006/jmbi.2001.5353. [DOI] [PubMed] [Google Scholar]
- Deupi X, Olivella M, Govaerts C, Ballesteros JA, Campillo M, Pardo L. Ser and Thr residues modulate the conformation of Pro-kinked transmembrane α-helices. Biophys J. 2004;86:105–115. doi: 10.1016/S0006-3495(04)74088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig AJ, Malcolm WM, Stapley BJ, Thornton JM. Structures of N-termini of helices in proteins. Prot Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Beis K, Nesper J, Brunkan-LaMontagne AL, Clarke BR, Whitfiled C, Naismith JH. Wza the translocon for E. coli capsular polysaccharides defines a new class of membrane protein. Nature. 2006;444:226–229. doi: 10.1038/nature05267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM. The lateral gate of SecYEG opens during protein translocation. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci U S A. 2000;97:5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- Fang Y, Jayaram H, Shane T, Kolmakova-Partensky L, Wu F, Williams C, Xiong Y, Miller C. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature. 2009;460:1040–1043. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller SE, MacKerell AD., Jr An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci U S A. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TM, Matthews BW. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within α-helices and its relevance to membrane-bound proteins. J Mol Biol. 1984;175:75–81. doi: 10.1016/0022-2836(84)90446-7. [DOI] [PubMed] [Google Scholar]
- Grubmüller H, Heller H, Windemuth A, Schulten K. Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions. Mol Simul. 1991;6:121–142. [Google Scholar]
- Gulbis JM, Kuo A, Smith B, Doyle DA, Edwards A, Arrowsmith C, Sundstrom M. Intermediate gating structure 1 of the inwardly rectifying K+ channel KirBac3.1. Protein Data Bank. n.d doi: 10.2210/pdb1xl4/pdb. [DOI] [Google Scholar]
- Han S-J, Hamdan FF, Kim S-K, Jacobson KA, Brichta L, Bloodworth LM, Li JH, Wess J. Pronounced conformational changes following agonist activation of the M3 muscarinic acethylcho-line receptor. J Biol Chem. 2005;280:24870–24879. doi: 10.1074/jbc.M500379200. [DOI] [PubMed] [Google Scholar]
- Hermansson M, von Heijne G. Inter-helical hydrogen bond formation during membrane protein integration into the ER membrane. J Mol Biol. 2003;334:803–809. doi: 10.1016/j.jmb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hu J, Xue Y, Lee S, Ha Y. The crystal structure of GxxGD membrane protease FlaK. Nature. 2011;475:528–531. doi: 10.1038/nature10218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hub JS, Winkler FK, Merrick M, de Groot BL. Potentials of mean force and permeabilities for carbon dioxide, ammonia, and water flux across the Rhesus protein channel and lipid membranes. J Am Chem Soc. 2010;132:13251–13263. doi: 10.1021/ja102133x. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke W, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31:264–323. [Google Scholar]
- Jardon-Valadez E, Bondar A-N, Tobias DJ. Coupling of retinal, protein, and water dynamics in squid rhodopsin. Biophys J. 2010;99:2200–2207. doi: 10.1016/j.bpj.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ACV, Lindahl E. Amino-acid solvation structure in transmembrane helices from molecular dynamics simulations. Biophys J. 2006;91:4450–4463. doi: 10.1529/biophysj.106.092767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. MATLAB statistics toolbox: computation, visualization, programming: user’s guide. MathWorks; Natick: 1993. [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- Junne T, Schwede T, Goder V, Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 2009;460:592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg S, Huché F, Diederichs K, Izadi-Pruneyre N, Lecroisey A, Wandersman C, Delepelaire P, Welte W. Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc Natl Acad Sci U S A. 2009;106:1045–1050. doi: 10.1073/pnas.0809406106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar SS, London E. The control of transmembrane helix transverse position in membranes by hydrophilic residues. J Mol Biol. 2007;374:1251–1269. doi: 10.1016/j.jmb.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W, Zeelen J, Dietrich J. Structure, mechanism, and regulation of the Neurosphora plasma membrane H+-ATPase. Science. 2002;297:1692–1696. doi: 10.1126/science.1072574. [DOI] [PubMed] [Google Scholar]
- Kumar S, Bansal M. Dissecting α-helices: position-specific analysis of α-helices in globular proteins. Prot Struct Funct Gen. 1998;31:460–476. doi: 10.1002/(sici)1097-0134(19980601)31:4<460::aid-prot12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RAF. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- Lear JD, Wasserman ZR, DeGrado WF. Synthetic peptide models for protein ion channels. Science. 1998;240:1177–1181. doi: 10.1126/science.2453923. [DOI] [PubMed] [Google Scholar]
- Li B, Nowak NM, Kim SK, Jacobson KA, Bagheri A, Schmidt C, Wess J. Random mutagenesis of the M3 muscarinic acetylcholine receptor. J Biol Chem. 2005;280:5664–5675. doi: 10.1074/jbc.M411623200. [DOI] [PubMed] [Google Scholar]
- Lupo D, Li X-D, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, Merrik M, Winkler FK. The 1.3 Å resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Natl Acad Sci U S A. 2007;104:19303–19308. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum JL, Benett WFD, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- MacQueen JB. Some methods for classification and analysis of multivariate observations. Vol. 1. University of California Press; Berkeley: 1967. [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 1947;18:50–60. [Google Scholar]
- Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular dynamics algorithms. J Chem Phys. 1994;101:4177–4189. [Google Scholar]
- Metz G, Siebert F, Engelhard M. Asp85 is the only internal aspartic acid that gets protonated in the M intermediate and the purple-to-blue transition in bacteriorhodopsin. FEBS Lett. 1991;303:237–241. doi: 10.1016/0014-5793(92)80528-o. [DOI] [PubMed] [Google Scholar]
- Miranda M, Pardo JP, Petrov VV. Structure–function relationships in membrane segment 6 of the yeast plasma membrane Pma1 H+-ATPase. Biochim Biophys Acta. 2011;1808:1781–1789. doi: 10.1016/j.bbamem.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell TM. Machine learning. McGraw-Hill Higher Education; New York: 1997. [Google Scholar]
- Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. Proc Natl Acad Sci U S A. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
- Munter L-M, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. GxxxG motifs within the amyloid precursor transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nack M, Radu I, Schultz B-J, Resler T, Schlesinger R, Bondar A-N, del Val C, Abbruzzetti S, Viappiani C, Bamann C, Bamberg E, Heberle J. Kinetics of proton release and uptake by channelrhodopsin-2. FEBS Lett. 2012;586:1344–1348. doi: 10.1016/j.febslet.2012.03.047. [DOI] [PubMed] [Google Scholar]
- Obara K, Miyashita N, Xu C, Toyoshima I, Sugita Y, Inesi G, Toyoshima C. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+ Proc Natl Acad Sci U S A. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar A-N, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Osborne RS, Silhawy TJ. PrlA suppressor mutations cluster in regions corresponding to three distinct topological domains. EMBO J. 1993;12:3391–3398. doi: 10.1002/j.1460-2075.1993.tb06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- Perálvarez A, Barnadas R, Sabés M, Querol E, Padrós E. Thr90 is a key residue of the bacteriorhodopsin proton pumping mechanism. FEBS Lett. 2001;508:399–402. doi: 10.1016/s0014-5793(01)03080-0. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun B, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Ben-Tal N, Lancet D. kPROT: a knowledge-based scale for the propensity of residue orientation in transmembrane segments. Application to membrane protein structure prediction. J Mol Biol. 1999;294:921–935. doi: 10.1006/jmbi.1999.3257. [DOI] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Presta LG, Rose GD. Helix signals in proteins. Science. 1988;240:1632–1641. doi: 10.1126/science.2837824. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richardson JS, Richardson DC. Amino acid preferences for specific location at the ends of helices. Science. 1988;240:1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Rousseeuw P. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- Royston P, Remark AS. R94: a remark on algorithm AS 181: the W test for normality. Appl Stat. 1995;44:547–551. [Google Scholar]
- Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- Sako T. Novel prlA alleles defective in supporting staphylo-kinase processing in Escherichia coli. J Bacteriol. 1991;173:2289–2296. doi: 10.1128/jb.173.7.2289-2296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SS, Wilk MB. An analysis of variance test for normality (complete samples) Biometrika. 1965;52:591–611. [Google Scholar]
- Smith MA, Clemmons WM, Jr, DeMars CJ, Flower AN. Modelling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp RE. Alternative models for two crystal structures of bovine rhodopsin. Acta Crystallogr. 2008;D64:902–904. doi: 10.1107/S0907444908017162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao GV, van der Berg B. Crystal structure of the monomeric porin OmpG. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Sugihara M, Fujibuchi W, Suwa M. Structural elements of the signal propagation pathway in squid rhodopsin and bovine rhodopsin. J Phys Chem B. 2011;115:6172–6179. doi: 10.1021/jp1101785. [DOI] [PubMed] [Google Scholar]
- Sui H, Han B-G, Lee JK, Wallan P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- R Develompment Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. http://www.R-project.org. [Google Scholar]
- Tuckerman M, Berne BJ. Reversible multiple time scale molecular dynamics. J Chem Phys. 1992;97:1990–2001. [Google Scholar]
- Tusnády GE, Dosztányi ZS, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005a;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnády GE, Dosztányi ZS, Simon I. TMDET: web server for detecting transmembrane domains by using 3D structure of proteins. Bioinformatics. 2005b;21:1276–1277. doi: 10.1093/bioinformatics/bti121. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acethylcholine receptor at 4Å resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport T. X-ray structure of a protein conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- van Kim CL, Colin Y, Cartron J-P. Rh proteins: key structural and functional components of the red cell membrane. Blood Rev. 2006;20:93–110. doi: 10.1016/j.blre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Vijayakumar M, Qian H, Zhou H-X. Hydrogen bonds between short polar side chains and peptide backbone: prevalence in proteins and effects on helix-forming pepensities. Prot Struct Funct Gen. 1999;34:497–507. [PubMed] [Google Scholar]
- Worth CL, Blundell TL. Satisfaction of hydrogen-bonding potential influences the conservation of polar sidechains. Proteins. 2008;75:413–429. doi: 10.1002/prot.22248. [DOI] [PubMed] [Google Scholar]
- Xie K, Hessa T, Seppälä S, Rapp M, von Heijne G, Dalbey RE. Features of transmembrane segments that promote the lateral release from the translocase into the lipid phase. Biochem. 2007;46:15153–15161. doi: 10.1021/bi701398y. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat Struct Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc Natl Acad Sci U S A. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.