Abstract
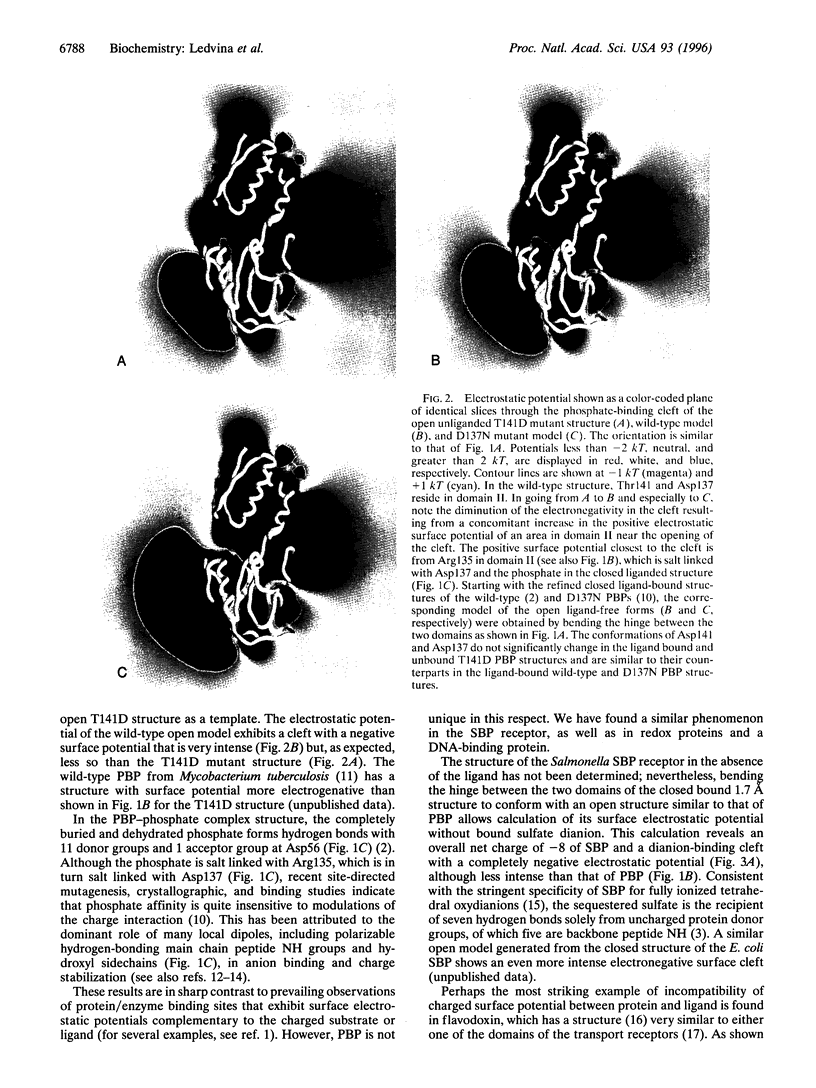
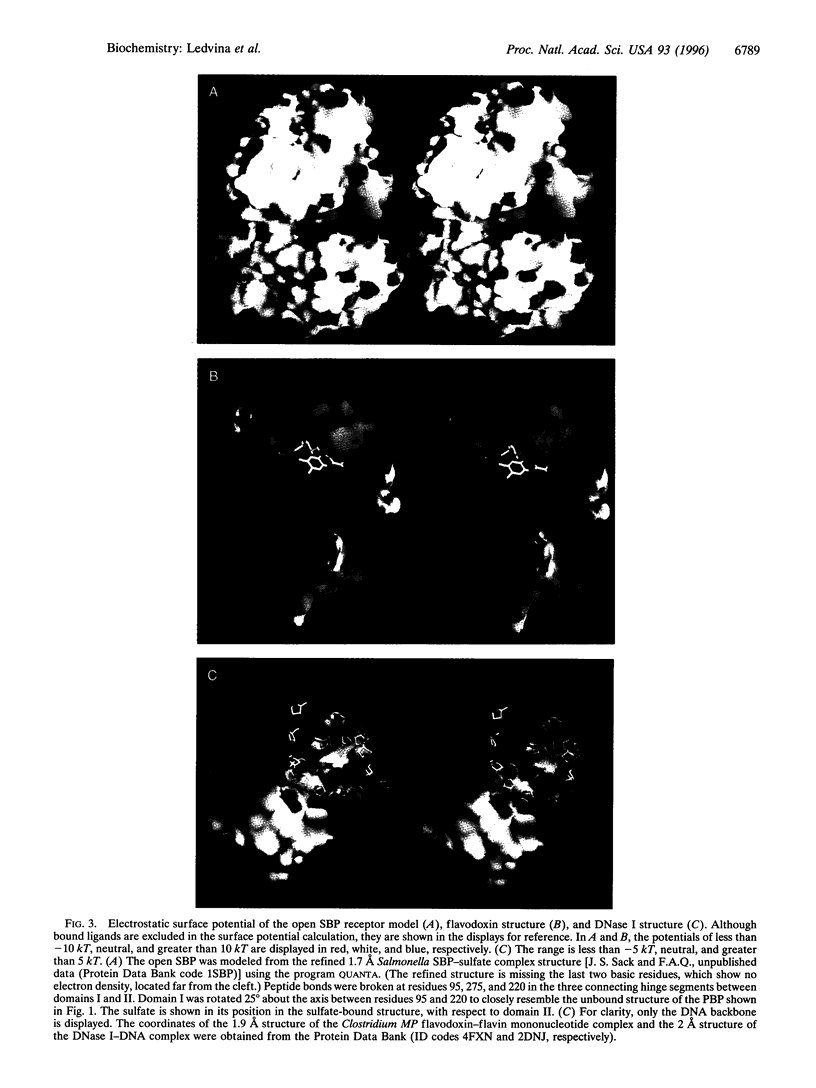
Determination of the crystal structure of an "open" unliganded active mutant (T141D) form of the Escherichia coli phosphate receptor for active transport has allowed calculation of the electrostatic surface potential for it and two other comparably modeled receptor structures (wild type and D137N). A discovery of considerable implication is the intensely negative potential of the phosphate-binding cleft. We report similar findings for a sulfate transport receptor, a DNA-binding protein, and, even more dramatically, redox proteins. Evidently, for proteins such as these, which rely almost exclusively on hydrogen bonding for anion interactions and electrostatic balance, a noncomplementary surface potential is not a barrier to binding. Moreover, experimental results show that the exquisite specificity and high affinity of the phosphate and sulfate receptors for unions are insensitive to modulations of charge potential, but extremely sensitive to conditions that leave a hydrogen bond donor or acceptor unpaired.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aqvist J., Luecke H., Quiocho F. A., Warshel A. Dipoles localized at helix termini of proteins stabilize charges. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):2026–2030. doi: 10.1073/pnas.88.5.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Choudhary A., Lathigra R., Quiocho F. A. The immunodominant 38-kDa lipoprotein antigen of Mycobacterium tuberculosis is a phosphate-binding protein. J Biol Chem. 1994 Jan 21;269(3):1956–1958. [PubMed] [Google Scholar]
- Gilliland G. L., Quiocho F. A. Structure of the L-arabinose-binding protein from Escherichia coli at 2.4 A resolution. J Mol Biol. 1981 Mar 5;146(3):341–362. doi: 10.1016/0022-2836(81)90392-2. [DOI] [PubMed] [Google Scholar]
- He J. J., Quiocho F. A. Dominant role of local dipoles in stabilizing uncompensated charges on a sulfate sequestered in a periplasmic active transport protein. Protein Sci. 1993 Oct;2(10):1643–1647. doi: 10.1002/pro.5560021010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995 May 26;268(5214):1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- Jacobson B. L., Quiocho F. A. Sulfate-binding protein dislikes protonated oxyacids. A molecular explanation. J Mol Biol. 1988 Dec 5;204(3):783–787. doi: 10.1016/0022-2836(88)90369-5. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 A resolution. J Mol Biol. 1989 Nov 5;210(1):163–180. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- Lahm A., Suck D. DNase I-induced DNA conformation. 2 A structure of a DNase I-octamer complex. J Mol Biol. 1991 Dec 5;222(3):645–667. doi: 10.1016/0022-2836(91)90502-w. [DOI] [PubMed] [Google Scholar]
- Luecke H., Quiocho F. A. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature. 1990 Sep 27;347(6291):402–406. doi: 10.1038/347402a0. [DOI] [PubMed] [Google Scholar]
- Mandel-Gutfreund Y., Schueler O., Margalit H. Comprehensive analysis of hydrogen bonds in regulatory protein DNA-complexes: in search of common principles. J Mol Biol. 1995 Oct 20;253(2):370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Pardee A. B. Purification and properties of a sulfate-binding protein from Salmonella typhimurium. J Biol Chem. 1966 Dec 25;241(24):5886–5892. [PubMed] [Google Scholar]
- Pflugrath J. W., Quiocho F. A. Sulphate sequestered in the sulphate-binding protein of Salmonella typhimurium is bound solely by hydrogen bonds. Nature. 1985 Mar 21;314(6008):257–260. doi: 10.1038/314257a0. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Ledvina P. S. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996 Apr;20(1):17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Sack J. S., Vyas N. K. Stabilization of charges on isolated ionic groups sequestered in proteins by polarized peptide units. Nature. 1987 Oct 8;329(6139):561–564. doi: 10.1038/329561a0. [DOI] [PubMed] [Google Scholar]
- Smith W. W., Burnett R. M., Darling G. D., Ludwig M. L. Structure of the semiquinone form of flavodoxin from Clostridum MP. Extension of 1.8 A resolution and some comparisons with the oxidized state. J Mol Biol. 1977 Nov 25;117(1):195–225. doi: 10.1016/0022-2836(77)90031-6. [DOI] [PubMed] [Google Scholar]
- Wang Z., Choudhary A., Ledvina P. S., Quiocho F. A. Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies. J Biol Chem. 1994 Oct 7;269(40):25091–25094. doi: 10.2210/pdb1pbp/pdb. [DOI] [PubMed] [Google Scholar]
- Yao N., Ledvina P. S., Choudhary A., Quiocho F. A. Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor. Biochemistry. 1996 Feb 20;35(7):2079–2085. doi: 10.1021/bi952686r. [DOI] [PubMed] [Google Scholar]





