Significance
Helicobacter pylori is the greatest risk factor for gastric adenocarcinoma and has been classified as a carcinogen by the World Health Organization. Cytotoxin associated gene A (CagA) is the primary virulence determinant of H. pylori and is sufficient to induce tumor formation in animal models. We show here that the host tumor suppressor Apoptosis-stimulating Protein of p53-2 (ASPP2) binds robustly to an N-terminal domain of CagA and elucidate the crystal structure of this complex, revealing the details of the CagA–ASPP2 interaction. Structure-based mutagenesis disrupts this complex in vitro and in cells. Furthermore, we show that the CagA–ASPP2 interaction modulates critical ASPP2 functions, such as p53-binding and apoptosis of H. pylori-infected cells.
Abstract
The Cytotoxin associated gene A (CagA) protein of Helicobacter pylori is associated with increased virulence and risk of cancer. Recent proteomic studies have demonstrated an association of CagA with the human tumor suppressor Apoptosis-stimulating Protein of p53-2 (ASPP2). We present here a genetic, biochemical, and structural analysis of CagA with ASPP2. Domain delineation of the 120-kDa CagA protein revealed a stable N-terminal subdomain that was used in a yeast two-hybrid screen that identified the proline-rich domain of ASPP2 as a host cellular target. Biochemical experiments confirm this interaction. The cocrystal structure to 2.0-Å resolution of this N-terminal subdomain of CagA with a 7-kDa proline-rich sequence of ASPP2 reveals that this domain of CagA forms a highly specialized three-helix bundle, with large insertions in the loops connecting the helices. These insertions come together to form a deep binding cleft for a highly conserved 20-aa peptide of ASPP2. ASPP2 forms an extended helix in this groove of CagA, burying more than 1,000 Å2 of surface area. This interaction is disrupted in vitro and in vivo by structure-based, loss-of-contact point mutations of key residues in either CagA or ASPP2. Disruption of CagA and ASPP2 binding alters the function of ASPP2 and leads to the decreased survival of H. pylori-infected cells.
Infecting nearly 50% of the human population, Helicobacter pylori (Hp) is the only pathogen known to colonize the stomach and is linked to duodenal and gastric ulcers, adenocarcinomas, and mucosa associated lymphoid tissue (MALT) lymphomas (1). The pathogen itself is listed as a group 1 carcinogen by the World Health Organization and is the strongest risk factor for the development of gastric cancer (1).
Hp translocates through a type IV secretion system Cytotoxin associated gene A (CagA), a protein composed of more than 1,200 aa that is injected into the epithelial cells lining the stomach (2–8). CagA interacts with more than 20 host proteins and has been shown to manipulate many host cellular functions, including cytoskeletal structure, cell-to-cell adhesion, and intracellular signal transduction (9). The expression of CagA in transgenic mice is tumorigenic (10). Furthermore, mongolian gerbils challenged with type I Hp strains develop gastric dysplasia and adenocarcinoma at 12 wk after infection in a CagA-dependent manner (11).
Many of the prooncogenic activities of CagA are believed to be mainly exerted through its C-terminal domain. Host-phosphorylated CagA associates with many SH2-containing host proteins (such as SHP2, Grb2, SHP1, Csk, and Crk) through its C terminus (9, 12–14). For example, binding to the tyrosine phosphatase oncoprotein SHP-2 contributes to the activation of signaling cascade that ultimately causes CagA-induced dramatic alterations in cultured cell morphology and cytoskeletal structure that is associated with epithelial to mesenchymal transition (15–17). A critical phosphorylation-independent interaction occurs between CagA and the PAR1/MARK family of serine/theronine kinases (18). A short segment of CagA that functions as a pseudosubstrate mimic of these kinases is critical for this interaction (19), leads to the loss of PAR1/MARK kinase activity in cells, and is required for the SHP-2 promoted hummingbird phenontype and maintenance of cell polarity.
More recently, the N terminus of CagA has become the focus of research interest and several binding partners have been uncovered (Runx3, ASPP2, β-integrin, TAK1, and TRAF) (20–23). Two groups have determined the crystal structure of a large, 800-aa N-terminal portion of CagA (24, 25). These structures revealed a set of unique folds, comprised of three structurally distinct domains [labeled variously I–III (24) and D1–D3 (25)]. A part of domain II, encompassing a single-layer β-sheet and a conserved surface-exposed patch is implicated in the specific binding to β1 integrin, suggesting a unique mechanism of CagA translocation. Structural and mutational analysis identified key residues within one of the basic surface patches of domain II responsible for binding to phosphatidyl serine (24).
Oncogenesis is considered to occur as a multifactorial set of events. Two key events in most known cancers are common: the activation of oncogenes and the inactivation of tumor suppressor genes (26). CagA is now known to activate cellular protooncogenes (see above). Recent data has identified two important tumor suppressors as potential binding targets of CagA, thereby raising the possibility that the oncogenic potential of this virulence factor is also due to manipulation of cellular tumor suppressors. One of these targets is RUNX3 (20, 27, 28). The second is ASPP2 (21).
ASPP2 was originally identified as a p53 binding protein and has been shown to be a haplo-sufficient tumor suppressor that cooperates with p53 (and its family members p63 and p73) to suppress tumor growth in vivo (29, 30). Moreover, increasing evidence expands ASPP2 cellular functions to include the formation of tight junctions, maintenance of cell polarity of epithelial cells, the control of autophagy, and oncogene-induced senescence (31–34). ASPP2 is often down-regulated in many aggressive malignancies and gastric cancers (35–37). Infection with Hp and delivery of CagA increases the level of ASPP2 (21, 38). Furthermore, after Hp infection, CagA coimmunoprecipitates with ASPP2 and alters its proapoptotic function (21).
Given the importance of ASPP2 in suppressing the tumor formation and its potential role in CagA-induced carcinogenesis, we took a biochemical, structural, and cellular approach to better understand the nature of this interaction. We identified a minimal, CagA-interacting subdomain of ASPP2 and solved the crystal structure of this complex to 2.0-Å resolution. Based on the structure-guided mutagenesis, we identified residues critical for the stability of this interaction. Finally, we show that the specific interaction between CagA and ASPP2 is key for the survival of Hp-infected cells.
Results
Recruitment of ASPP2 to the Sites of Hp Infection and Identification of CagA and ASPP2 Interacting Domains.
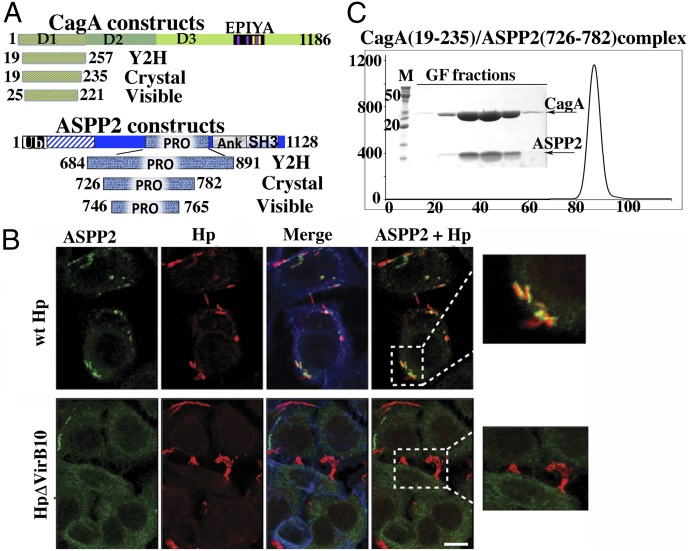
Using a combination of bioinformatic and biochemical analysis (SI Experimental Procedures), we were able to identify and produce a soluble, proteolytically resistant subdomain of CagA (residues 19–257) that is N-terminal to the well-studied repeats region of the molecule and corresponds well to structural subdomain I (D1) in recently published crystal structures of the first 800 aa of CagA (Fig. 1A) (24, 25).
Fig. 1.
ASPP2 binding and recruitment by CagA. (A) Domain delineation of a CagA–ASPP2 minimal complex. The schematic diagram of constructs of CagA and ASPP2 used in this study are as follows: Crystal, constructs that were coexpressed in E. coli, copurified, and used in crystallization trials; Visible, domains that are visible in crystal structure; Y2H, constructs used in Y2H screen. (B) Relocalization of endogenous ASPP2 during the course of Hp infection is CagA dependent. AGS cells were infected with the indicated Hp strains (moi 1:50) or left uninfected. After 7 h, cells were fixed and stained with anti-ASPP2 (green) and anti-CagA (red) antibodies and phalloidin for F-actin (blue). Insets show that only wild-type Hp (wt Hp, Upper Inset) strongly associates with ASPP2, whereas there is no association with Hp ΔVirb10 mutant (Lower Inset). (Scale bar: 10 μm.) (C) Gel filtration profile of CagA (19–235) and ASPP2 (726–782) complex. The final step of purification of CagA (19–235) and ASPP2 (726–782) complex used in crystallization experiments is shown. The elution profile from gel filtration column (Superdex 200; GE Health) and SDS/PAGE of peak fractions are presented.
To identify host proteins that might interact with this domain, CagA (19–257) was used as bait in a yeast two-hybrid (Y2H) screen against a universal human cDNA library. Among several potential interacting proteins identified in this screen, we obtained 15 independent clones of the protein ASPP2. This screen was an independent genetic confirmation of the proteomic results that showed that the first 800 aa of CagA could pull down ASPP2 (21). Furthermore, the Y2H screening enabled us to narrow down CagA and ASPP2 interacting domains. All of the ASPP2 clones identified by Y2H screening overlapped in a region spanning residues 684–891, containing the so-called “proline-rich domain” (Fig. 1A).
Helicobacter infection and CagA delivery causes a strong relocalization of endogenous ASPP2 to the sites of bacteria attachment to cells, clearly suggesting an important role for ASPP2 during Hp infection (Fig. 1B and Fig. S1) and, thereby, establishing the biological relevance of previous studies that relied on exogenously expressed ASPP2 and CagA. Based on this observation and on the association between the CagA and ASPP2 in proteomic and genetic assays (21), these results provided a strong mandate to pursue structural studies and examine this interaction in more detail. To identify an optimal construct for crystallographic studies, we screened numerous sequences of ASPP2 based on the 15 clones obtained from Y2H and coexpressed them recombinantly in Escherichia coli where they formed a stable complex with the bait subdomain of CagA as shown in Fig. 1C. The purified complex of CagA (19–235) and ASPP2 (726–782) was crystallized, and the high-resolution crystal structure was solved (Table S1 and Experimental Procedures).
Crystal Structure of a Minimal CagA–ASPP2 Complex.
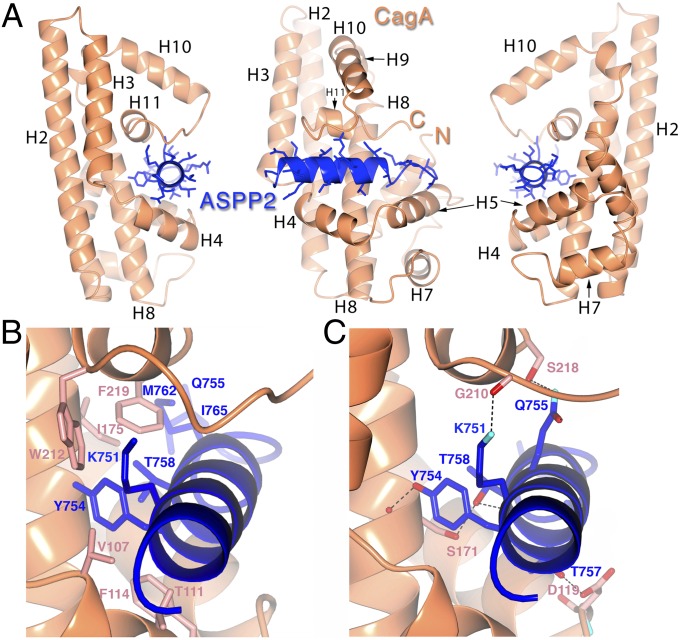
CagA(19–235) is an entirely α-helical structure based on a specialized version of a generic antiparallel, three-helix bundle architecture (Fig. 2A). The bundle consists of helices H2, H4, and H8 (with the third helix distorted and broken by a kink). Two loops between helices have been augmented dramatically with insertions that adopt small subfolds (residues 105–150 and 185–221): The first is a loop that, in a classic bundle, would simply connect two of the helices, and the second follows the last (third) helix. These two small subdomains are on the same face of the molecule, and together form a deep and extended groove that is the binding cleft for the ASPP2 helix (Fig. 2A). The first insertion presents helices H4 and H5 as the “bottom” of the binding cleft for ASPP2, whereas the second insertion forms the “top” of the binding groove with helix H11.
Fig. 2.
Overall structure of the CagA–ASPP2 complex. (A) Ribbon diagrams of CagA (orange) and ASPP2 (blue) in three different orientations related by rotations of 90° about a vertical axis. C, COOH terminus; N, NH2 terminus. Graphics were generated with CCP4 QtMG (55). (B) Details of the CagA–ASPP2 protein–protein interface. The focus is on the hydrophobic interactions between CagA and ASPP2. The protein main chain is shown as in Fig. 2A, with corresponding side chains of CagA and ASPP2 in salmon and blue, respectively. (C) Focus on the hydrogen-bonding interactions between CagA and ASPP2. The protein main chain is shown as in Fig. 2A, with corresponding side chains of CagA and ASPP2 in salmon and blue, respectively. Hydrogen bonds are indicated by black dotted lines, and the atoms of nitrogen and oxygen are show in blue and red, respectively.
The interaction between the CagA N-terminal domain and ASPP2 can thus be thought of as a clamp on a pipe or bar: The tumor-suppressor sequence from 746 to 765 forms four turns of a helical structure with a short tail that is surrounded by the deep groove formed from the two insertions in the CagA three-helix bundle. Overall, ASPP2 buries roughly 1,100 Å2 of surface area in the complex, with more than half of the helical surface buried in the CagA groove.
A large complementary hydrophobic patch in the CagA cleft interacts with hydrophobic residues in the ASPP2 sequence (Fig. 2B and Fig. S2B). In the midst of this predominantly hydrophobic interface are a number of significant hydrogen bond interactions, primarily involving side-chain to main-chain contacts (Fig. 2C and SI Results). The fragment of the proline-rich domain of ASPP2, which encompasses these CagA-binding residues, is one of two regions within the ASPP protein family that are strongly conserved, although only few of the CagA-contacting residues are conserved (Lys751, Tyr754, and Gln755) (Fig. S2A). This observation nonetheless raised the question of whether other ASPP family members could interact with CagA. ASPP1 shares more similarity with ASPP2 than iASPP, but neither was able to form a stable complex with CagA as assayed by Ni-NTA pull-down assay, cation-exchange and size exclusion chromatography (Fig. S3).
Structure-Based Mutagenesis of the CagA–ASPP2 Interface.
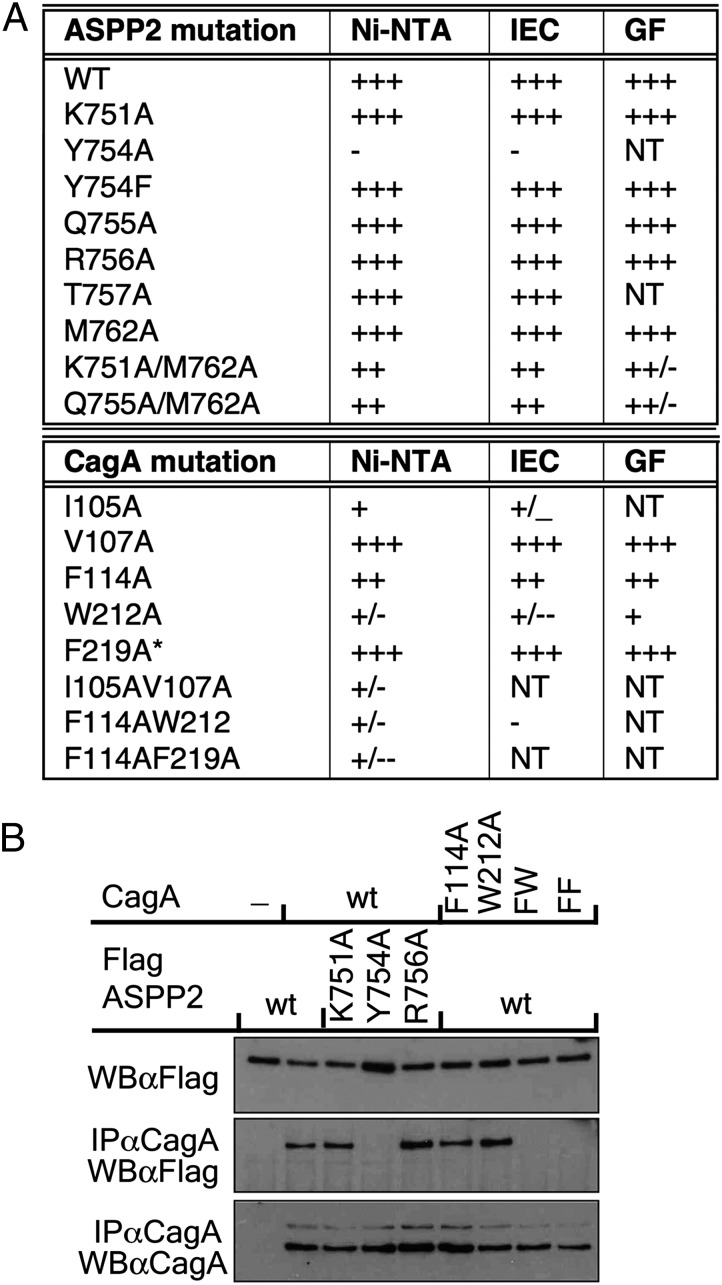
To determine which residues are most important for the interaction between CagA and ASPP2, we used structure-guided mutagenesis. Mutant proteins were coexpressed with wild-type partner proteins in bacteria. Complex formation and integrity was tested by affinity, ion exchange, and size exclusion chromatography. The results are summarized in Fig. 3A, and more details are presented in Figs. S4–S6.
Fig. 3.
Structure-based mutagenesis of residues in the CagA–ASPP2 interface. (A) Summary of in vitro binding assays. GF, gel filtration, size exclusion chromatography of complexes; IEC, ion-exchange chromatography of complexes on SP Sepharose; Ni-NTA, affinity chromatography on Ni-NTA Sepharose of bacterial cell lysates coexpressing wild-type and mutant proteins as indicated; NT, not tested; *, additional mutation I35A/V37A (not involved in binding). (B) Contribution of different residues of ASPP2 and CagA to their interaction in cultured cells. HEK 293-T cells were cotransfected with wild-type or mutant CagA along with an expression construct of FLAG-tagged wild-type or mutant ASPP2, as indicated. Cells were lysed in Nonidet P-40 containing buffer, cellular extract was immunoprecipitated with anti-CagA antibody, and proteins were detected with indicated antibodies.
Several ASPP2 residues making contacts with CagA were targeted for mutagenesis. Creating single, loss-of-contact mutations of these residues (through mutation to alanine) generally did not disrupt complex formation (Fig. 3A) as determined by affinity (Fig. S4A), ion exchange (Fig. S5), and size exclusion chromatography (Fig. S6). Only one mutation, Tyr754Ala, completely abolished binding of ASPP2 to CagA. When Tyr754 was mutated into Phe, the binding to CagA was not disrupted, revealing that it is the hydrophobic contacts of the aromatic ring, and not the hydrogen bonding of the tyrosine hydroxyl, that is most critical in anchoring ASPP2 to CagA. Complexes that contain double mutants of ASPP2 (Lys751Ala/Met762Ala or Asn755Ala/Met762Ala) were slightly destabilized on gel filtration (Fig. 3A and Fig. S6), which suggests that hydrogen bonds of those residues also contribute to the complex stability, but to the lower extent.
Hydrophobic residues of CagA that are deeply buried inside the CagA-binding cleft were mutated into alanine and tested for binding to the proline-rich domain of ASPP2. Single mutations such as Val107Ala and Phe219Ala had no effect on CagA–ASPP2 complex formation (Fig. 3A and Fig. S4B), whereas the Phe114Ala mutation only marginally weakened it. However, the mutations Ile105Ala, and especially Trp212Ala, noticeably destabilized the complex (Fig. 3A and Fig. S4B). The complex between wild-type ASPP2 and these two mutants of CagA completely fell apart on cation exchange chromatography (Fig.3A and Fig. S5), leaving unbound mutant CagA (19–235) exclusively present in the flow-through fraction. Double mutants of CagA (Ile105Ala/Val107Ala; Phe114Ala/Trp212Ala; Phe114Ala/Phe219Ala) exhibit very weak binding to wild-type ASPP2 (Fig. 3A). Only small amounts, if any, of wild-type ASPP2 was coeluted with those CagA mutants on Ni-NTA pull-down (Fig. S4B). If there was any complex assembled, it was further subjected to cation exchange chromatography, where it completely fell apart (Fig. S5). This analysis strongly suggests that hydrophobic contacts between CagA and ASPP2 are crucial for their interaction.
To examine the contribution of different contacts of CagA and ASPP2 in the cellular context, we cotransfected HEK293 cells with different combinations of mutant and wild-type constructs of full-length Flag-tagged ASPP2 and full-length CagA, and tested complex formation by coimmunoprecipitation (Fig. 3B). Replacing Tyr754 of ASPP2 with Ala completely eliminated interaction, whereas a Lys751Ala mutant of ASPP2 could still bind to wild-type CagA at levels comparable to wild-type ASPP2. An Arg756Ala mutation was used as a negative control because this residue, as observed in the structure, is not involved in the binding to CagA. As expected, this mutation had no effect on complex formation. These results entirely mirrored our findings obtained in vitro.
The Phe114Ala substitution of CagA slightly diminished interaction with ASPP2 (Fig. 3B), similar to the in vitro data, but the Trp212Ala mutation, which seems to be more detrimental in vitro, did not destabilize the complex in the cellular assay. This difference could be explained by enhanced stability of the Trp212Ala mutant of CagA in HEK293 cells compared with that in vitro. Double mutants of CagA, Phe114Ala/Trp212Ala and Phe114Ala/Phe219Ala, were not able to make any detectable complex with wild-type ASPP2 in cells (Fig. 3B).
Dominant Negative Effect of CagA-Binding Domain on CagA-Induced ASPP2 Function.
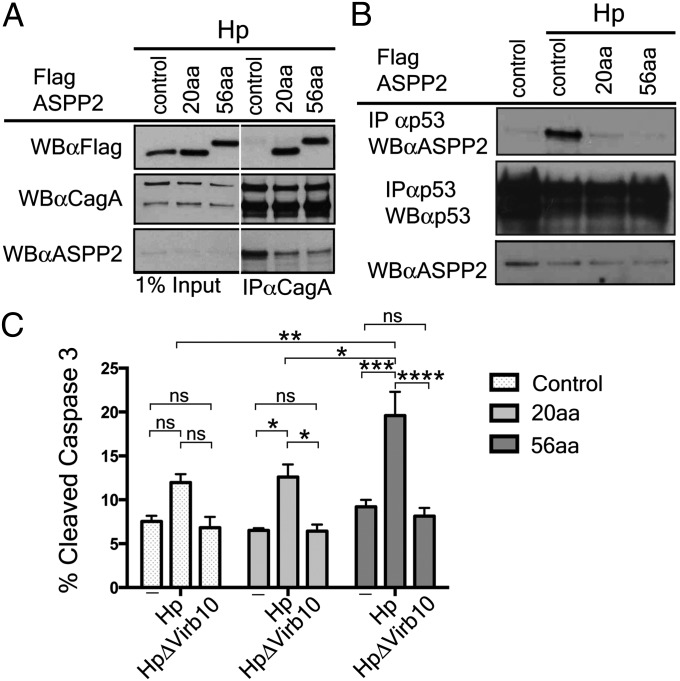
To examine the CagA effect on ASPP2 downstream activities, we used AGS cells to generate stable clones expressing two overlapping constructs of the proline-rich domain of ASPP2: (i) 726–782 (56aa), which was used in the crystallization experiments, and (ii) 746–765 (20aa), which contains only those residues visible in the structure. We were able to demonstrate that even the smaller construct, only 20 aa long, was sufficient for a stable interaction in cells during infection because it coimmunprecipitated with CagA after Hp infection (Fig. 4A). Furthermore, the expression of the 56aa construct and, to a lesser extent, the 20aa construct, reduced the binding between CagA and the endogenous ASPP2 (Fig. 4A). This realization allowed us to use these constructs as dominant negative mutants to examine any changes in ASPP2 activities that specifically depend on the interaction with CagA.
Fig. 4.
Expression of the CagA-binding domain of ASPP2 blocks the downstream pathway activated by delivery of CagA. (A) The CagA-binding domain of ASPP2 is sufficient to bind to CagA during Hp infection and inhibit interaction with endogenous ASPP2. AGS cells stably expressing FLAG-mCherry, FLAG-mCherry ASPP2 56aa, or FLAG-mCherry-ASPP2 20aa were infected with wild-type Hp (moi 1:50). Seven hours after infection, cells were lysed, and lysates were immunoprecipitated or directly immunoblotted with the indicated antibodies. CagA was retrieved with an anti-CagA antibody. Immunoprecipitates were separated by SDS/PAGE and detected by immunoblotting using the indicated antibodies. (B) Dominant negative effect of the CagA-binding domain of ASPP2 on the ASPP2–p53 interaction following Hp infection. AGS cells stably transfected with the indicated constructs were infected with wt Hp or left uninfected. Seven hours after infection, cells were harvested and processed as indicated in A. Uninfected cells were also used as a control. (C) Increased apoptosis of Hp-infected cells upon disruption of CagA–ASPP2 binding. Levels of cleaved Caspase-3 were assayed as a measure of apoptotic response by flow cytometry. AGS cells stably transfected with the indicated constructs (control, 20aa, and 56aa), were infected for 24 h with the indicated Hp strains (moi 1:50) or left uninfected. Error bars ± SEM (n = 4); ns, not significant; ****P < 0.0001; ***P < 0.001; **P < 0.01;*P < 0.05. Significance was tested by using two-way ANOVA Tukey multiple-comparison test.
The association between ASPP2 and p53 is enhanced by Hp in a CagA-dependent manner (21). We therefore sought to test whether the expression of the 20aa and 56aa fragments of ASPP2 would interfere with this interaction. In agreement with the previously published data, Hp infection promotes the association between ASPP2 and p53 in the AGS cells, but the expression of the 56aa ASPP2 fragment abolishes it (Fig. 4A). This result strongly suggests that binding of ASPP2 to CagA is responsible for increased association with p53. The expression of the shorter ASPP2 fragment (20aa) had a lower effect on the ASPP2-p53 association upon Hp challenge. Altogether, these observations confirm that the ASPP2 fragment identified in the crystal structure is sufficient to stably interact with CagA and its expression has a dominant negative effect on the downstream events promoted by the CagA–ASPP2 association.
Removal of ASPP2 in AGS cells results in the increased CagA-dependent apoptosis of Hp-infected cells (21). We therefore asked whether we could observe a similar phenotype when blocking the CagA–ASPP2 interaction. After 24 h of infection with the wild-type Hp (CagA+), the apoptotic response was determined by measuring the level of cleaved Caspase-3. The apoptotic level was significantly enhanced in infected AGS cells expressing the 56aa ASPP2 fragment, compared with infected AGS control cells, uninfected or cells infected with CagA-translocation mutant of Hp (Fig. 4C). However, the level of apoptosis in infected AGS expressing 20aa fragment was comparable to controls, most likely due to incomplete block of CagA interaction with endogenous ASPP2. Thus, these data confirm that one of the mechanisms put in place by CagA to prevent the apoptotic response in the host cell is to complex with ASPP2 and to misregulate its proapoptotic function. To achieve this modulation, a physical complex between CagA and ASPP2 is required to block the cell death otherwise induced by Hp infection.
Discussion
Infection with Hp is the single greatest risk factor associated with gastric cancer (39). The virulence factor CagA of Hp has been shown to possess oncogenic properties in cells and animals, as well as being associated epidemiologically with an elevated risk of cancer (1, 10, 15, 40). Many of the interactions of CagA with proteins involve promitogenic signaling molecules in critical regulatory cascades (e.g., SHP-2), and these interactions have been the focus of models for the oncogenic potential of this bacterial protein. The recent identification of human tumor suppressors as potential targets of CagA (20, 21) has raised the possibility that it may drive oncogenesis both through the stimulation of growth and the suppression of growth inhibitory pathways.
Our results show that CagA is highly evolved to specifically recognize a stretch of roughly 20 aa in the tumor suppressor ASPP2, which form a conserved helix buried deeply in a conserved pocket of CagA. A single, loss-of-contact point mutation in ASPP2 (Tyr754Ala) was sufficient to disrupt the complex in vitro and in vivo. The CagA interface did not possess such an Achilles’ heel, however, and disruption of the complex required the mutation of multiple residues (e.g., Phe114Ala/Trp212Ala, Phe114Ala/Phe219Ala, and Ile105Ala/Val107Ala). Targeted disruption of intermolecular hydrogen bonding did not disrupt the complex, establishing that the interaction between CagA and ASPP2 is dominated by hydrophobic contacts.
Two crystal structures of large, N-terminal regions of CagA have been published (24, 25). The structure of subdomain I (D1), encompassing residues (24–221) and corresponding to the CagA subdomain used in this study (19–235), in both structures exhibited significant disorder relative to remaining subdomains. It appears from our structure that binding to ASPP2 stabilized the subdomain of CagA. We aligned our structure of the ASPP2 binding domain of CagA to the Hayashi et al. (24) structure of subdomain I of CagA to examine potential ASPP2-induced conformational changes (Fig. S7). Overall, the structures are very similar, with differences centered on the ASPP2 binding cleft. The two insertions that form the cleft both shift toward the bound helix like pinchers in a clamp (Fig. S7).
ASPP2 is a well-known tumor suppressor that activates the p53-mediated apoptotic response upon cellular stress. However, ASPP2 has pleiotropic activities in maintaining the homeostasis of epithelial cells, such as regulation of tight junctions formation, maintenance of cell polarity, as well as autophagy and the oncogene-induced senescence (31–34). Although most of the ASPP2-binding proteins that are critical for its proapoptotic function (p53, NF-κB, Bcl-2) interact with the ankyrin-SH3 domain of ASPP2, the plasma membrane localization and the prosenescence activity of ASPP2 depend on cellular partners that bind ASPP2 at its N-terminal region (41–44). It was shown (41, 45) that peptides generated from the proline-rich domain (e.g., 693–746) could bind to the ankyrin-SH3 domain of ASPP2 and compete for binding with NF-κB or p53. This result raised the possibility that an autoinhibitory intramolecular mechanism exists to regulate associations with ankyrin-SH3 domain. In binding ASPP2, the N-terminal domain of CagA partially overlaps with this region and, indeed, increases association with p53. In favor of this autoinhibitory hypothesis, we also show that the 56aa fragment of ASPP2 (726–782) inhibits association with p53 during the course of the Hp infecion, by competing with endogenous ASPP2 for binding to CagA and possibly by directly blocking the ankyrin-SH3 binding site of p53. Further experiments are required to better understand this mechanism.
Furthermore, Hp-induced ASPP2 plasma membrane localization could stimulate untimely interaction with Par3, and interfere with cell polarity, because it has also been suggested that CagA interacts with Par3 (21). Taken together, along with the MARK2–CagA interaction, these complexes could contribute to a loss of cell polarity, a signature event of Hp infection. This depolarization could present an advantage to Hp that would enable it to replicate and grow on a wider area of epithelial cells (46, 47).
Several studies suggest that CagA has an antiapoptotic effect during the course of Hp infection (48, 49). Buti et al. (21) have shown that the CagA–ASPP2 interaction contributes to this effect. We confirm this finding here and provide evidence that direct interaction between CagA and ASPP2 is necessary to prevent apoptosis and allow survival of Hp-infected cells. Using structural information from this study, it should be possible to address this and other questions even more directly to ascertain the contribution of the CagA–ASPP2 interaction to CagA-induced oncogenesis.
Experimental Procedures
Purification of the Hp 26695 CagA and ASPP2 Complex.
His-tagged N-terminal domain of Hp 26695 CagA and GST-tagged proline-rich domain of human ASPP2 were coexpressed in BL-21 E. coli and purified through Ni-NTA Sepharose (Qiagen). The CagA–ASPP2 complex was further purified by cation-exchange and gel-filtration chromatography. More detailed purification steps can be found in SI Experimental Procedures. Mutagenesis was performed as described, (19) and mutant complexes were purified the same way as wild type. Complexes of CagA (19–235) with ASPP1 (694–755) or iASPP (511–555) were purified and analyzed in the same manner as CagA–ASPP2 complexes.
Y2H screen and delineation of CagA-binding domain of ASPP2.
Y2H screening was performed by Genomics and Proteomics Core Facility at German Cancer Research Center. The N-terminal domain of CagA (19–257) was used as a bait to screen a Mate and Plate Universal Human (normalized) Library (Clonthech). Clones encoding ASPP2 were picked up 15 times during the screen under moderate stringency conditions (0.4 mM 3-aminotriazole). Based on those clones, limited proteolyisis with subtilisin, and N-terminal sequencing of digested products, the minimal binding domains were defined as ASPP2 (721–782) and CagA (19–235). Further deletion of 5 aa of ASPP2 (726–782) was necessary to obtain diffracting crystals of ASPP2–CagA complex. Details are included in SI Experimental Procedures.
Crystallization and Structural Determination.
For crystallization, both native and selenomethionine-substituted (SeMet) protein complexes were purified as described above. Crystals were grown by vapor diffusion by using hanging drops as detailed in SI Experimental Procedures. Higher quality crystals were obtained from SeMet protein complexes, and they were used for the final refinement.
Data were collected from SeMet protein crystals at Brookhaven National Synchrotron light source beamline X29 at the selenium absorption edge and processed by using HKL2000 (50). The crystals belonged to the space group C2, with unit-cell parameters a = 118.64 Å, b = 120.24 Å, c = 100.66 Å, and α, β, γ = 90.00°, 115.64°, and 90.00°. There were four heterodimers of CagA–ASPP2 in the asymmetric unit. Phases were determined by using SHELX (51), and 90% of the final model was built by ARP/WARP (52). Cycles of manual building with COOT (53) and refinement with REFMAC5 (54) resulted in a model with an R/Rfree of 19.1%/23.8% to 2.05-Å resolution. The crystallographic statistics are summarized in Table S1.
Cells and Transfection.
HEK293T and AGS cells were transfected by using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. FLAG-mCherry, FLAG-mCherry-ASPP2 20aa (751–765), and FLAG-mCherry-ASPP2 56aa (726–782) were all cloned in the retroviral expression vector pLHCX (Clontech) and used to generate stably expressing AGS cell lines under HygromycinB (0.4 μg/mL) selection. For transient expression in HEK 293-T cells full-length, wild type or mutant, ASPP2 were cloned into pREV-TRE (Clontech), whereas full-length wild-type or mutant CagA were cloned into pcDNA5FRT/TO (Invitrogen). Mutant ASPP2 and CagA genes were generated as above described.
Bacterial Strains and Infections.
Hp strain G27 and the isogenic mutants ΔVirb10 were used for the infection of AGS cells as described (21) and in SI Experimental Procedures.
In Vitro Binding Assays.
Pull-down on Ni-NTA Sepharose columns was used to determine binding of wild-type or mutant His-tagged Cag A (19–235) to wild-type or mutant GST-tagged ASPP2 (726–782). This step was followed by cation-exchage (SP Sepharose) and size-exclusion chromatography (Superdex 200) as detailed in SI Experimental Procedure.
Immunoprecipitation.
Cells were treated as indicated and lysed in lysis buffer (0.5% Nonidet P-40, 5 mM EDTA, 50 mM Tris⋅HCl at pH 7.5, and 100 mM NaCl). An equal amount of each sample was used for immunoprecipitation overnight at 4 °C with anti-CagA (sc-b300; Santa Cruz Biotechnologies). Immunoprecipitates were resolved by SDS/PAGE and subjected to Western blotting by using either anti-FLAG M2 (F1804; Sigma) or anti-CagA (sc-b300) antibodies.
Immunofluorescence.
Cells were grown on coverslip and, following Hp infection, were fixed in 4% (vol/vol) paraformaldehyde and permeabilized in 0.1% Triton X-100. After blocking in 2% (wt/vol) BSA, the cells were incubated with the indicated primary antibodies for 1 h at room temperature followed by Alexa Fluorochrome secondary antibodies or phalloidin-647 for actin, and mounted onto glass slides for the confocal image acquisition. Confocal images were taken under LSM 710 Zeiss confocal microscope.
Supplementary Material
Acknowledgments
We thank D. Oren and W. Shi for access to and assistance with crystallographic equipment. This work was funded in part by National Institutes of Health (NIH) Grant AI098109 and The Rockefeller University (to C.E.S.) The use of the Rigaku/MSC microMax 007HF in The Rockefeller University Structural Biology Resource Center was made possible by Grant Number 1S10RR022321-01 from the National Center for Research Resources of the NIH. L.B. and X.L. are supported by Ludwig Institute for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates for the CagA–ASPP2 complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4IRV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320631111/-/DCSupplemental.
References
- 1.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Censini S, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93(25):14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi M, et al. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med. 2000;191(4):593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert S, et al. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol. 2000;2(2):155–164. doi: 10.1046/j.1462-5822.2000.00043.x. [DOI] [PubMed] [Google Scholar]
- 5.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 6.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA. 2000;97(3):1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covacci A, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90(12):5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: Evidence of linkage to cytotoxin production. Infect Immun. 1993;61(5):1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr Opin Microbiol. 2008;11(1):30–37. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Ohnishi N, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105(3):1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco AT, et al. Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68(2):379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002;277(9):6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 13.Poppe M, Feller SM, Römer G, Wessler S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene. 2007;26(24):3462–3472. doi: 10.1038/sj.onc.1210139. [DOI] [PubMed] [Google Scholar]
- 14.Stein M, et al. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol Microbiol. 2002;43(4):971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 15.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4(9):688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 16.Higashi H, et al. EPIYA motif is a membrane-targeting signal of Helicobacter pylori virulence factor CagA in mammalian cells. J Biol Chem. 2005;280(24):23130–23137. doi: 10.1074/jbc.M503583200. [DOI] [PubMed] [Google Scholar]
- 17.Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci USA. 2005;102(45):16339–16344. doi: 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saadat I, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447(7142):330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 19.Nesić D, et al. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17(1):130–132. doi: 10.1038/nsmb.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsang YH, et al. Helicobacter pylori CagA targets gastric tumor suppressor RUNX3 for proteasome-mediated degradation. Oncogene. 2010;29(41):5643–5650. doi: 10.1038/onc.2010.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buti L, et al. Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc Natl Acad Sci USA. 2011;108(22):9238–9243. doi: 10.1073/pnas.1106200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez-Soto LF, et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5(12):e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamb A, et al. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10(11):1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, et al. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe. 2012;12(1):20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan-Türköz B, et al. Structural insights into Helicobacter pylori oncoprotein CagA interaction with β1 integrin. Proc Natl Acad Sci USA. 2012;109(36):14640–14645. doi: 10.1073/pnas.1206098109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1(2):157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 27.Katoh M. Dysregulation of stem cell signaling network due to germline mutation, SNP, Helicobacter pylori infection, epigenetic change and genetic alteration in gastric cancer. Cancer Biol Ther. 2007;6(6):832–839. doi: 10.4161/cbt.6.6.4196. [DOI] [PubMed] [Google Scholar]
- 28.Li QL, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 29.Samuels-Lev Y, et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell. 2001;8(4):781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan A, Lu X. ASPP: A new family of oncogenes and tumour suppressor genes. Br J Cancer. 2007;96(2):196–200. doi: 10.1038/sj.bjc.6603525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong W, et al. ASPP2 regulates epithelial cell polarity through the PAR complex. Curr Biol. 2010;20(15):1408–1414. doi: 10.1016/j.cub.2010.06.024. [DOI] [PubMed] [Google Scholar]
- 32.Wang XD, et al. SUMO-modified nuclear cyclin D1 bypasses Ras-induced senescence. Cell Death Differ. 2011;18(2):304–314. doi: 10.1038/cdd.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. Autophagic activity dictates the cellular response to oncogenic RAS. Proc Natl Acad Sci USA. 2012;109(33):13325–13330. doi: 10.1073/pnas.1120193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sottocornola R, et al. ASPP2 binds Par-3 and controls the polarity and proliferation of neural progenitors during CNS development. Dev Cell. 2010;19(1):126–137. doi: 10.1016/j.devcel.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Sgroi DC, et al. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59(22):5656–5661. [PubMed] [Google Scholar]
- 36.Tordella L, et al. ASPP2 suppresses squamous cell carcinoma via RelA/p65-mediated repression of p63. Proc Natl Acad Sci USA. 2013;110(44):17969–17974. doi: 10.1073/pnas.1309362110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park SW, An CH, Kim SS, Yoo NJ, Lee SH. Mutational analysis of ASPP1 and ASPP2 genes, a p53-related gene, in gastric and cololorectal cancers with microsatellite instability. Gut Liver. 2010;4(2):292–293. doi: 10.5009/gnl.2010.4.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eftang LL, Esbensen Y, Tannæs TM, Bukholm IR, Bukholm G. Interleukin-8 is the single most up-regulated gene in whole genome profiling of H. pylori exposed gastric epithelial cells. BMC Microbiol. 2012;12:9. doi: 10.1186/1471-2180-12-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wroblewski LE, Peek RM, Jr, Wilson KT. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin Microbiol Rev. 2010;23(4):713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaser MJ, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111–2115. [PubMed] [Google Scholar]
- 41.Rotem S, Katz C, Friedler A. Insights into the structure and protein-protein interactions of the pro-apoptotic protein ASPP2. Biochem Soc Trans. 2007;35(Pt 5):966–969. doi: 10.1042/BST0350966. [DOI] [PubMed] [Google Scholar]
- 42.Naumovski L, Cleary ML. The p53-binding protein 53BP2 also interacts with Bc12 and impedes cell cycle progression at G2/M. Mol Cell Biol. 1996;16(7):3884–3892. doi: 10.1128/mcb.16.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang JP, et al. NF-kappaB subunit p65 binds to 53BP2 and inhibits cell death induced by 53BP2. Oncogene. 1999;18(37):5177–5186. doi: 10.1038/sj.onc.1202904. [DOI] [PubMed] [Google Scholar]
- 44.Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274(5289):1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 45.Rotem-Bamberger S, Katz C, Friedler A. Regulation of ASPP2 interaction with p53 core domain by an intramolecular autoinhibitory mechanism. PLoS ONE. 2013;8(3):e58470. doi: 10.1371/journal.pone.0058470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan S, Noto JM, Romero-Gallo J, Peek RM, Jr, Amieva MR. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7(5):e1002050. doi: 10.1371/journal.ppat.1002050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan S, Tompkins LS, Amieva MR. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 2009;5(5):e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umehara S, Higashi H, Ohnishi N, Asaka M, Hatakeyama M. Effects of Helicobacter pylori CagA protein on the growth and survival of B lymphocytes, the origin of MALT lymphoma. Oncogene. 2003;22(51):8337–8342. doi: 10.1038/sj.onc.1207028. [DOI] [PubMed] [Google Scholar]
- 49.Oldani A, et al. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5(10):e1000603. doi: 10.1371/journal.ppat.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 55.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.