Significance
To establish long-term infections, bacterial pathogens must adapt their gene expression through sensing and responding to changing conditions or selection of genotypic variations within the population. Hypervariable simple sequence repeats (SSRs) in coding sequences or promoters are a major source of phase variation and often associated with genes involved in host interaction. While their impact on gene regulation at the DNA level is established, we now demonstrate a connection between SSRs and small RNA (sRNA)-mediated posttranscriptional regulation. We show that a homopolymeric G-repeat within the leader of a chemotaxis receptor mRNA in Helicobacter pylori is directly targeted by a small RNA. The length of this G-repeat varies among different Helicobacter strains and thereby determines sRNA-mediated translational repression or activation and strain-specific regulation.
Keywords: homopolymeric repeat, noncoding RNA
Abstract
Phase variation of hypermutable simple sequence repeats (SSRs) is a widespread and stochastic mechanism to generate phenotypic variation within a population and thereby contributes to host adaptation of bacterial pathogens. Although several examples of SSRs that affect transcription or coding potential have been reported, we now show that a SSR also impacts small RNA-mediated posttranscriptional regulation. Based on in vitro and in vivo analyses, we demonstrate that a variable homopolymeric G-repeat in the leader of the TlpB chemotaxis receptor mRNA of the human pathogen Helicobacter pylori is directly targeted by a small RNA (sRNA), RepG (Regulator of polymeric G-repeats). Whereas RepG sRNA is highly conserved, the tlpB G-repeat length varies among diverse H. pylori strains, resulting in strain-specific RepG-mediated tlpB regulation. Based on modification of the G-repeat length within one strain, we demonstrate that the G-repeat length determines posttranscriptional regulation and can mediate both repression and activation of tlpB through RepG. In vitro translation assays show that this regulation occurs at the translational level and that RepG influences tlpB translation dependent on the G-repeat length. In contrast to the digital ON–OFF switches through frame-shift mutations within coding sequences, such modulation of posttranscriptional regulation allows for a gradual control of gene expression. This connection to sRNA-mediated posttranscriptional regulation might also apply to other genes with SSRs, which could be targeting sites of cis- or trans-encoded sRNAs, and thereby could facilitate host adaptation through sRNA-mediated fine-tuning of virulence gene expression.
For successful survival in the environment or colonization of a host, bacteria must adapt their phenotypes either through sensing and responding to changing conditions or through selection of beneficial mutations. The establishment of long-term infections and evasion of the immune system, in particular, require mechanisms to modulate gene expression, especially of genes encoding products that influence the interaction with the host. Phase variation represents a frequent and stochastic mechanism of genotype switching and facilitates phenotypic variation within bacterial populations (1). Besides a variety of mechanisms including site-specific recombination or epigenetic changes through DNA methylation, phase variation can occur due to highly mutable DNA sequences. These so called contingency loci are often associated with genes involved in LPS biosynthesis, bacterial surface structures, and DNA restriction or modification (2). In addition to deletions, gene conversions or point mutations, highly variable simple sequence repeats (SSRs) have been shown to be the major source of phase variation within these loci (2, 3).
Phase variation of SSRs is in most cases independent of recombination and occurs during replication through slipped-strand mispairing and polymerase slippage which leads to repeat length variation (3). SSRs have been described to affect virulence and host adaptation of several bacterial pathogens such as Bordetella pertussis (4), Neisseria meningitidis (5), Haemophilus influenzae (6), Campylobacter jejuni (7), or Helicobacter pylori (8, 9). Depending on their location, SSRs can either affect translation through the introduction of frame-shift mutations within coding regions leading to premature translation termination or altered C termini of proteins (intragenic SSRs) (5, 10–13) or influence transcription by changing the spacing of promoter elements or transcription factor binding sites (intergenic SSRs) (14–16). Whereas the mechanisms and roles of SSRs on gene regulation at the DNA level are established, effects on mRNA stability or posttranscriptional control are less understood (3).
Here, we show that length variation of a homopolymeric repeat can determine small RNA-mediated posttranscriptional regulation. Bacterial small regulatory RNAs (sRNAs) are posttranscriptional regulators of gene expression that have been implicated in stress response or virulence control (17, 18). Although some sRNAs can activate gene expression or directly modulate protein activity, most of the functionally characterized sRNAs act as antisense RNAs and repress translation and/or induce degradation of cis- or trans-encoded target mRNAs. So far, the majority of sRNAs has been investigated in enterobacteria, but genome-wide studies have been reporting an increasing number of sRNA candidates in various bacteria including several important human pathogens. Using RNA-seq, we recently identified more than 60 sRNAs in Helicobacter pylori, the causative agent of gastritis, ulcers, and gastric cancer (19, 20). Like 50% of all bacteria, Helicobacter lacks a homolog of the RNA chaperone Hfq, a key player in sRNA-mediated regulation in enterobacteria (21). Thus, sRNAs in these bacteria must act independently of Hfq and their study will provide new insights into mechanisms of sRNA-mediated regulation and virulence control (22).
Here we study a highly conserved and abundant sRNA, RepG (previously HPnc5490; ref. 19), and show that it targets a homopolymeric G-repeat in the mRNA leader of an acid-sensing chemotaxis receptor, TlpB (23, 24). The tlpB G-repeat length varies in diverse Helicobacter strains ranging from 6 to 16 guanines and was previously described as an intergenic promoter SSR (25). However, our global transcriptome map revealed that the G-repeat is located in a 5′ UTR (19) and, thus, is unlikely to affect tlpB promoter strength. We now demonstrate that variation of the G-repeat length influences posttranscriptional control of tlpB by RepG at the translational level and that there is strain-specific tlpB regulation by this sRNA. This length-dependent targeting of homopolymeric repeats by a trans-acting sRNA provides a twist in sRNA-mediated regulation and mechanisms of gene expression control. Considering the steadily increasing number of sRNAs in diverse bacteria, it is likely that also other phase-variable genes might be subject to posttranscriptional control and that SSRs in their mRNA leaders and also coding sequences might be sRNA target sites.
Results
The Abundant RepG sRNA Is Broadly Conserved in Helicobacter.
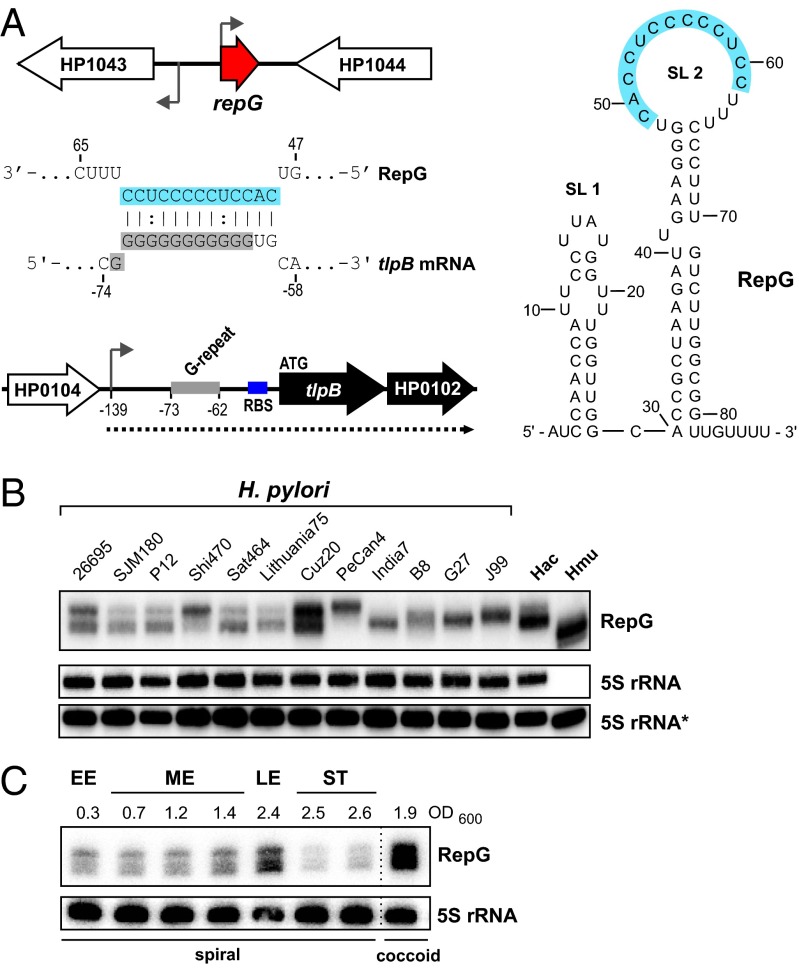
The 87-nt-long RepG sRNA (HPnc5490) was identified as one of the most abundant transcripts in our dRNA-seq study of H. pylori strain 26695 (19). It is transcribed from an intergenic region between genes encoding an orphan response regulator, HP1043, and a protein of unknown function, HP1044, and is predicted to fold into two stem-loop structures (Fig. 1A). Biocomputational searches for RepG homologs in 31 different H. pylori strains, Helicobacter acinonychis, and Helicobacter mustelae revealed that the RepG gene, its genomic context, and the predicted RepG secondary structure are highly conserved (SI Appendix, Fig. S1). Using Northern blot analysis, we confirmed RepG expression in diverse Helicobacter strains but observed variations in abundance and band patterns of RepG (Fig. 1B). Whereas multiple RepG bands were detected in H. pylori strain 26695, only one band was observed in G27, although both strains share an overall highly conserved sRNA sequence. Primer extension revealed that the different bands in strain 26695 correspond to RepG versions that vary slightly at their 5′ end (SI Appendix, Fig. S2A). In strain G27, the sRNA TSS is shifted to one nucleotide downstream compared with the main RepG species in 26695. Nevertheless, both strains showed a similar sRNA expression profile with an increase of RepG in late exponential growth, a decrease in stationary phase, and accumulation in coccoid forms (Fig. 1C and SI Appendix, Fig. S2B).
Fig. 1.
Conservation and expression of Helicobacter RepG sRNA. (A) The 87-nt-long RepG sRNA is transcribed from the intergenic region between an orphan response regulator, HP1043, and a hypothetical protein, HP1044. A C/U-rich single-stranded region (marked in blue) in the RepG terminator loop (SL 2) was predicted to base pair with a G-repeat (marked in gray) in the 5′ UTR of the dicistronic tlpB-HP0102 mRNA (dotted line) encoding a chemotaxis receptor and a hypothetical protein. Transcriptional start sites (19) are indicated by arrows and the tlpB G-repeat and ribosome binding site (RBS) by gray and blue bars, respectively. Numbers indicate the distance to the tlpB start codon. (B) Northern blot analysis of RepG expression at exponential growth phase in diverse H. pylori strains, H. acinonychis (Hac), and H. mustelae (Hmu) using 32P-labeled oligonucleotide CSO-0003. 5S rRNA served as loading control and was probed with two oligonucleotides: JVO-0485 (5S rRNA) for H. pylori and H. acinonychis and CSO-0053 (5S rRNA*) for H. mustelae, respectively. (C) Expression of RepG was analyzed during growth in H. pylori 26695 (EE, early exponential; ME, mid exponential; LE, late exponential; ST, stationary phase) at indicated optical densities (OD600). After ∼60 h, morphology changed from spiral to coccoid.
RepG Represses tlpB via a G-Repeat.
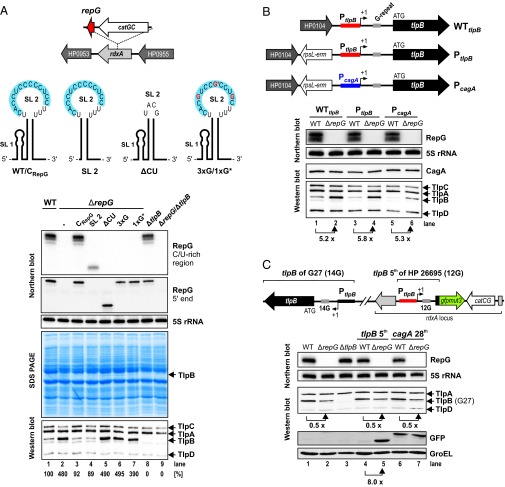
Sequence alignment of RepG homologs showed that the predicted C/U-rich terminator loop of the sRNA is highly conserved, even in more distant species such as the ferret colonizing H. mustelae (SI Appendix, Fig. S1B). Predictions for potential target mRNAs using the TargetRNA program (26) indicated that RepG might base pair with its C/U-rich sequence to a homopolymeric G-repeat in the 5′ UTR of the mRNA encoding the chemotaxis receptor TlpB (HP0103) (Fig. 1A), and preliminary analysis of a repG deletion mutant indicated that the sRNA represses tlpB expression at the mRNA and protein level (19). To study the potential regulation of tlpB by RepG, we complemented the ΔrepG mutant of H. pylori 26695 with wild-type (CRepG) or mutant RepG sRNAs expressed from the PrepG promoter at the unrelated rdxA locus. Mutants we tested were SL 2, which expresses only the second stem-loop; ΔCU, in which the C/U-rich region of the RepG terminator loop was replaced by an extrastable tetraloop; and 3xG and 1xG*, in which three or one C residue(s) in the predicted interaction site were exchanged to G(s) (Fig. 2A).
Fig. 2.
In vivo validation of tlpB regulation by RepG sRNA. (A, Upper) The ΔrepG mutant was complemented with wild-type RepG (CRepG) or several mutant sRNAs in the rdxA locus. SL 2 consists of only the second stem-loop (nucleotides 30–87) of RepG. In ΔCU the tlpB binding site (marked in blue) was replaced by an extrastable tetraloop (UACG). Triple or single* C to G point mutations at position 52, 56, and 60 (3xG) or at position 56 (1xG*) are indicated in red. (A, Lower) H. pylori 26695 wild type (WT), ΔrepG, and complementation strains (CRepG, SL 2, ΔCU, 3xG, and 1xG*), as well as ΔtlpB, and ΔrepG/ΔtlpB double deletion mutants were grown to exponential growth phase and RNA and protein samples were analyzed by Northern blot and SDS-PAGE or Western blot, respectively. RepG was detected with CSO-0003 (binds to the C/U-rich loop) and JVO-2134 (binds to RepG 5′ end). 5S rRNA served as loading control (JVO-0485). Whole cell protein fractions (OD600 of 0.1 for Coomassie gel or 0.01 for Western blot) were directly stained with Coomassie or chemotaxis receptors TlpA, B, C, and D were detected by a polyclonal rabbit anti-TlpA22 antiserum. (B, Upper) Schematic illustration of the tlpB locus including its promoter region (WTtlpB). The cagA promoter region (blue bar) together with a rpsL-erm resistance cassette (PcagA) or rpsL-erm alone (PtlpB) were inserted upstream of the tlpB promoter (red bar). The tlpB G-repeat is marked by a gray box. (B, Lower) H. pylori 26695 strains with either the WT tlpB locus (WTtlpB) or mutants that carry the rpsL-erm cassette insertion (PtlpB, lanes 3 and 4) or express tlpB from the cagA promoter (PcagA, lanes 5 and 6) in the wild-type (WT) or repG deletion (ΔrepG) background were grown to exponential growth phase and RNA and protein samples were analyzed on Northern and Western blot, respectively. 5S rRNA and CagA protein served as controls. (C, Upper) The H. pylori 26695 tlpB promoter region, its 5′ UTR and the first five amino acids of the tlpB coding region were fused to gfpmut3 and inserted into the rdxA locus of H. pylori G27. (C, Lower) H. pylori G27 WT, ΔrepG, and ΔtlpB mutant strains, as well as WT and ΔrepG strains which carry either the tlpB-5th::gfpmut3 (lanes 4 and 5) or cagA-28th::gfpmut3 (lanes 6 and 7) fusions were grown to exponential phase and RNA and protein samples were analyzed by Northern and Western blot, respectively. Note that H. pylori strain G27 expresses only three chemotaxis receptors, TlpA, B, and D.
Analysis of whole cell protein fractions and RNA samples of these strains by SDS/PAGE and Northern blot, respectively, showed that complementation with the wild-type sRNA and the SL 2 mutant which harbors the predicted C/U-rich tlpB interaction site both restore tlpB repression (Fig. 2A). In contrast, deletion of the C/U-rich binding site and introduction of triple or single point mutations abolished tlpB regulation, confirming that the sRNA terminator loop is important for tlpB repression. Western blot analysis using an antiserum against all four chemotaxis receptors of H. pylori gave results consistent with the SDS/PAGE. RepG specifically represses tlpB ∼fivefold, whereas the levels of the other chemotaxis receptors TlpA, TlpC, and TlpD remained unaltered upon deletion of the sRNA. Analysis of ΔtlpB and ΔtlpB/ΔrepG double deletion mutants confirmed that the up-regulated band is indeed TlpB (Fig. 2A). Despite threefold lower levels, the SL 2 mutant represses tlpB expression to the same extent as the wild-type sRNA, indicating that RepG levels are not limiting for tlpB regulation under this condition. Moreover, this simple stem-loop structure of 58 nt, corresponding to the RepG terminator, is sufficient to act as a regulatory RNA and, thus, could be used for the optimized design of synthetic RNA regulators, which commonly consist of multiple domains (27).
RepG Acts Posttranscriptionally to Repress tlpB.
To investigate whether RepG regulates tlpB expression at the posttranscriptional level, we exchanged the tlpB promoter with the unrelated cagA promoter of the major effector protein CagA. To replace the endogenous tlpB promoter, we inserted the cagA promoter in the chromosome upstream of the tlpB transcriptional start site (19, 28) together with an rpsL-erm resistance cassette (Fig. 2B). Western blot analysis for endogenous CagA confirmed that cagA expression itself is not affected by RepG. As a control, the resistance cassette alone was inserted upstream of the tlpB promoter (PtlpB), and Western blot analysis confirmed that this did not interfere with tlpB expression and its regulation by RepG (Fig. 2B, lanes 1–2 and 3–4). When tlpB was expressed from the cagA promoter (PcagA), we observed a two- to threefold reduced TlpB protein level compared with the wild-type (WTtlpB) and control (PtlpB) strains. Nevertheless, a ∼fivefold increase of the TlpB protein level was observed upon deletion of RepG similar as in WTtlpB, indicating that regulation occurs at the posttranscriptional level (Fig. 2B, lanes 5–6).
Translational reporter systems based on gfp or lacZ have been successfully used to study sRNA-mediated regulation and to define mRNA and sRNA interaction sites (29, 30). Therefore, we adapted the use of a GFP-variant, gfpmut3, which was previously used in transcriptional reporter plasmids (31), as a translational reporter in the chromosome of H. pylori (Fig. 2C). To confirm that the tlpB 5′ UTR is sufficient for RepG-mediated repression, we fused the coding region for the first five amino acids, the promoter region, and 5′ UTR of tlpB from H. pylori strain 26695 to gfpmut3. Transformation of gfpmut3 fusions seemed to be toxic for strain 26695 but not for strain G27. Western blot analysis of the TlpB::GFP fusion protein in H. pylori G27 showed a ∼eightfold up-regulation upon repG deletion (Fig. 2C, lanes 4 and 5). A cagA::gfpmut3 (cagA-28th::GFP) control fusion was not affected by repG deletion (Fig. 2C, lanes 6–7), which is in line with what we observed for endogenous CagA (Fig. 2B). Together, our in vivo results indicate that the terminator loop of RepG contains the tlpB binding site and that RepG represses tlpB at the posttranscriptional level by interacting with its 5′ UTR.
In contrast to the repression of the translational tlpB-5th::gfpmut3 fusion from strain 26695 by RepG, we observed that the level of the native TlpB protein of H. pylori G27 was decreased about twofold upon repG deletion (Fig. 2C, lanes 1–2). Because RepG is highly conserved and the tlpB 5′ UTRs are overall very similar in both strains but carry different G-repeat lengths, namely 12 and 14 Gs, we reasoned that the G-repeat length could determine strain-specific RepG-mediated tlpB regulation, which we examined in more detail below.
RepG and the tlpB mRNA Directly Base Pair.
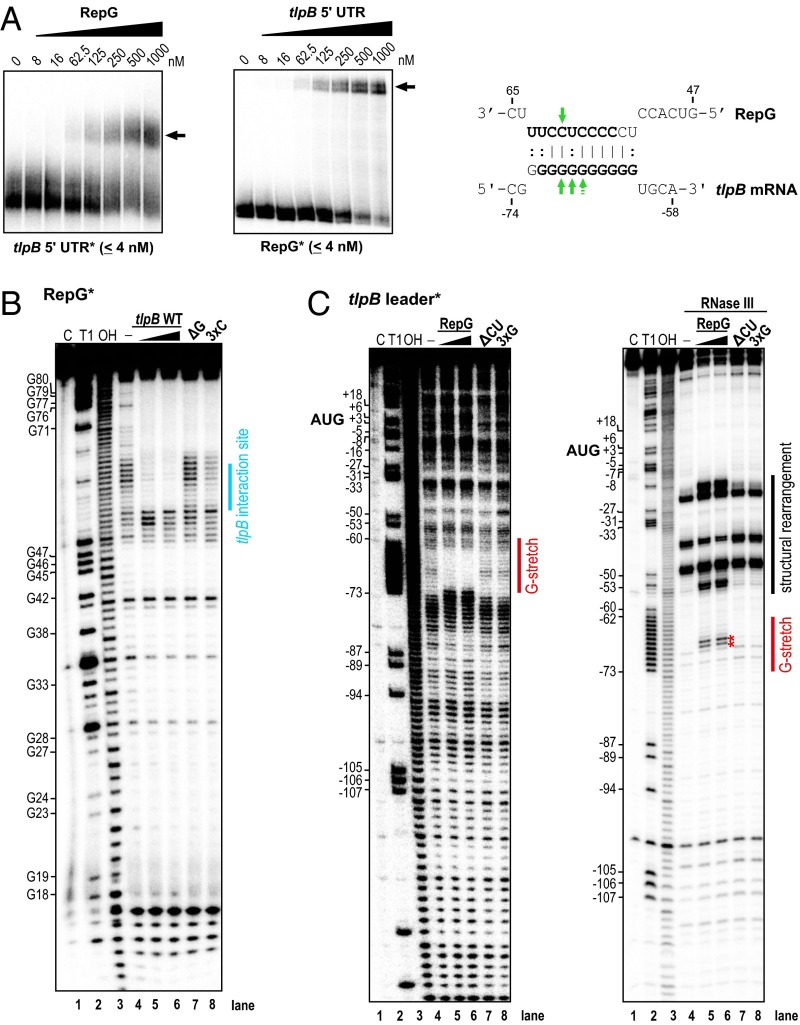
To further test for a direct interaction between the C/U-rich region of RepG and the tlpB G-repeat, we performed binding studies using in vitro transcribed RNAs. Gel-shift assays with 5′ end-labeled tlpB mRNA leader from strain 26695 and increasing amounts of RepG and vice versa revealed a RNA–RNA complex formation at a comparable molecular ratio of 1:15.6 (4 nM:62.5 nM) (Fig. 3A). Modifications of either the tlpB binding site in RepG (ΔCU, 3xG, 1xG*) or the G-repeat in tlpB leader variants, which either lack the G-repeat (ΔG) or contain triple and single nucleotide exchanges in the G-stretch (3xC, 1xC*), abolished RepG–tlpB mRNA interaction (SI Appendix, Fig. S3). Only gel-shifts with RNA pairs with compensatory base pair exchanges (i.e., RepG 3xG with tlpB 3xC or RepG 1xG* with tlpB 1xC*) restored the interaction between RepG and tlpB mRNA, albeit with lower affinities compared with the respective wild-type versions. Overall, our gel-shifts confirmed a direct interaction between the G-repeat in the tlpB 5′ UTR and the C/U-rich terminator loop of RepG in vitro and that mutations in the interaction sites abolish duplex formation.
Fig. 3.
In vitro gel-shift and structure probing assays show a direct interaction between RepG sRNA and the tlpB leader. (A, Left) Gel-shift experiments with in vitro synthesized RepG and 26695 tlpB mRNA leader (-139 to +78 relative to the annotated start codon). About 0.04 pmol 32P-labeled RNA (RepG* or tlpB 5′ UTR*) was incubated with increasing concentrations of unlabeled RNA. Arrows indicate RNA-RNA complex formation. (A, Right) Predicted 11 bp-long duplex between the C/U-rich loop of RepG and the G-repeat in the tlpB 5′ UTR based on structure probing assays. Identified RNase III cleavage sites in the RepG-tlpB mRNA duplex are indicated by arrows and nucleotides that are protected from cleavage in in-line probing assays are marked in bold. Note that the interacting nucleotides based on the structure probing results are slightly shifted compared with the predicted interaction in Fig. 1A. (B) In-line probing of ∼0.2 pmol 32P-labeled RepG in the absence (lane 4) or presence of either 20 nM (lane 5) or 200 nM tlpB mRNA leader (lane 6), or 200 nM of tlpB leader mutants, ΔG (lacks the G-repeat, lane 7) or 3xC (triple G to C substitutions in the G-stretch, lane 8). Spontaneous cleavages of single-stranded regions were analyzed on a 10% PAA gel under denaturing conditions. Untreated RNA (lane C), partially alkali (lane OH) or RNase T1 (lane T1) digested RepG served as ladders. (C, Left) In-line probing experiment with ∼0.2 pmol 32P-labeled tlpB mRNA leader in the absence or presence of either 20 nM (lane 5) or 200 nM RepG (lane 6), or 200 nM of the RepG mutants, ΔCU (lane 7) or 3xG (lane 8). (C, Right) In vitro structure mapping of the RepG-tlpB mRNA duplex using RNase III cleavage. About 0.1 pmol 32P-labeled tlpB mRNA leader was treated with RNase III in the absence or presence of either 100 nM (lane 5) or 1,000 nM (lane 6) RepG or 1,000 nM RepG ΔCU or 3xG mutant RNAs (lanes 7 and 8). RepG-mediated RNase III cleavage sites in the G-repeat and structural rearrangements in the tlpB 5′ UTR are indicated by red stars and black bars, respectively.
The C/U-Rich RepG Terminator Loop Region Interacts with the G-Repeat of tlpB.
To map the sRNA and mRNA interaction in vitro, we performed footprinting assays of in vitro transcribed RepG in the absence or presence of unlabeled tlpB leader using in-line probing (32) or enzymatic and chemical cleavages (Fig. 3B and SI Appendix, Fig. S4A). The in-line probing method takes advantage of the fact that single-stranded or unstructured RNA regions undergo spontaneous cleavage of phosphoester linkages faster than structured regions. Thus, besides secondary structure probing, it can be used to monitor ligand binding or RNA–RNA interactions as a reduction in spontaneous RNA degradation events. Cleavage patterns observed in the in-line as well as RNase T1 (cleaves single-stranded G-residues), lead (II)-acetate (cleaves single-stranded nucleotides), and RNase III (cleaves double-stranded RNAs) probing assays agreed with single- and double-stranded regions according to the two biocomputationally predicted stem-loops in RepG (Figs. 1A and 3B, SI Appendix, Figs. S4A and S5A). The predicted single-stranded 17-nt terminator loop harboring the C/U-rich tlpB interaction site showed slight protection against lead (II)-cleavage as well as some RNase III cleavages (SI Appendix, Fig. S4A), indicating that some of the nucleotides within the C/U-rich loop region might be involved in a tertiary structure. However, upon addition of unlabeled tlpB leader, we observed a clear footprint in the RepG terminator loop region in the in-line probing assay, suggesting that the C/U-rich site is indeed involved in the sRNA–mRNA interaction (Fig. 3B). In line with the predicted interaction, this protection from spontaneous cleavages within the C/U-rich region was not observed with a tlpB mutant RNA that lacks the G-repeat (ΔG) and was only slightly visible upon addition of the 3xC tlpB mutant RNA.
In a reciprocal experiment, we mapped the structure of 5′ end-labeled tlpB leader in the absence or presence of unlabeled RepG sRNA (Fig. 3C and SI Appendix, Fig. S4B). In combination with secondary structure predictions using RNAstructure (33), our structure probing results indicated a stem-loop structure upstream of the ribosome binding site (RBS) of the 139-nt-long tlpB mRNA leader (SI Appendix, Fig. S5B). Moreover, we observed only minor cleavage events within the G-repeat in the in-line probing assay and a protection against RNase T1 and lead (II)-cleavages as well as two RNase III cleavage sites, indicating a potential intra- or intermolecular structure, which could not be resolved with the applied methods (Fig. 3C and SI Appendix, Fig. S4B). Apart from this potentially structured region, the cleavage patterns in the in-line and lead (II) probing assays indicate a rather flexible structure or multiple conformations of the tlpB leader. Upon addition of increasing amounts of RepG, a footprint in the in-line reactions (Fig. 3C, Left) and RNase III cleavage sites (Fig. 3C, Right, red asterisks) were observed in the tlpB G-repeat, indicating the formation of a RepG–tlpB leader duplex. The footprint as well as RNase III cleavages and several other structural rearrangements in the tlpB leader, especially in the stem-loop upstream of the ribosome binding site, were not observed upon addition of ΔCU or 3xG RepG mutant RNAs. In summary, the structure probing results support an interaction between the C/U-rich terminator loop of RepG and the G-repeat in the tlpB 5′ UTR (Fig. 3A).
Posttranscriptional Regulation of tlpB Varies in Different H. pylori Strains.
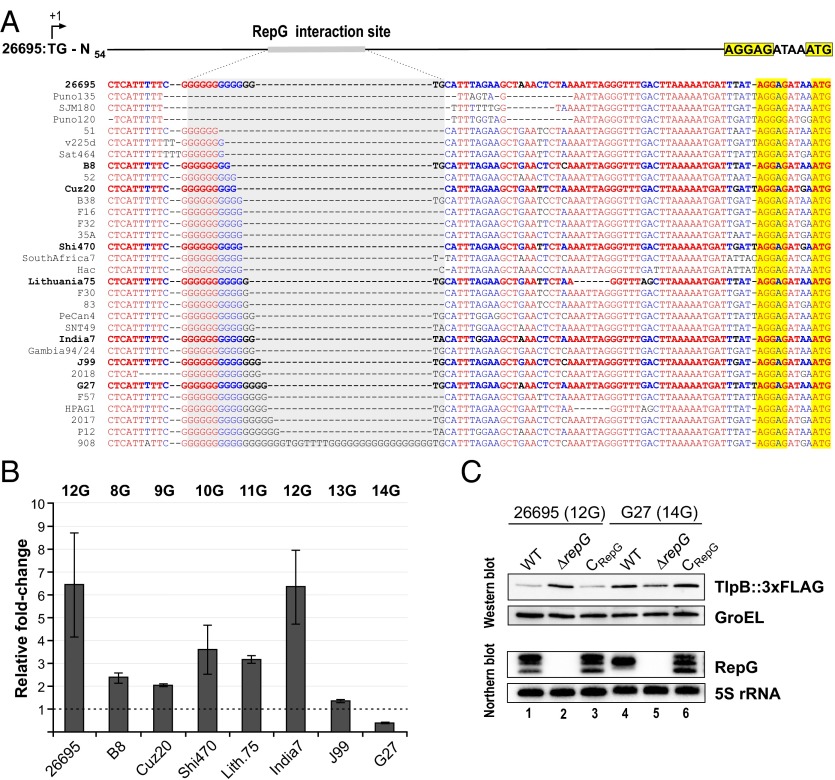
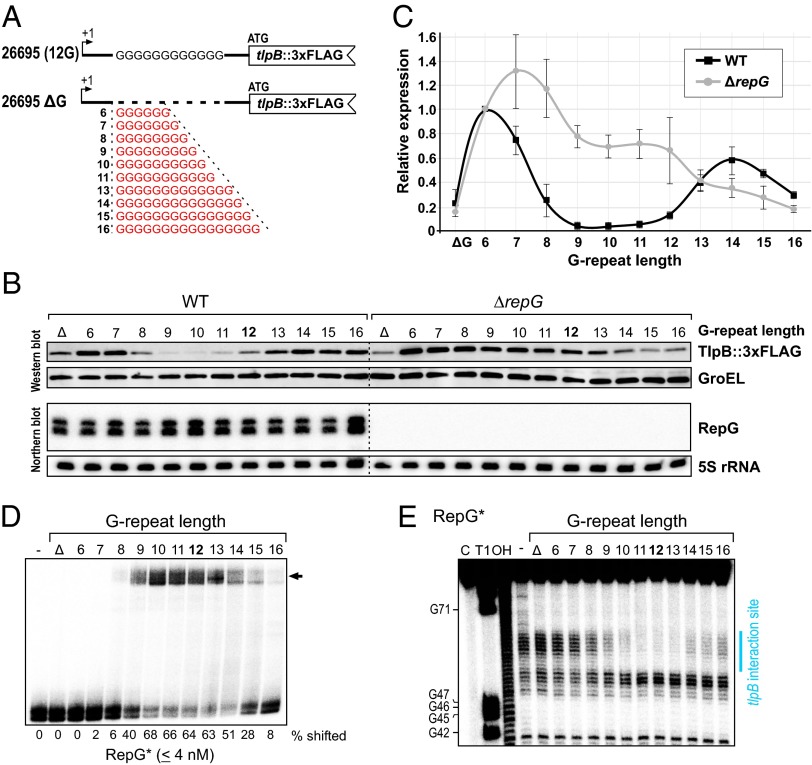
Our conservation analysis revealed that RepG and especially its C/U-rich loop are highly conserved among different H. pylori strains (Fig. 1 and SI Appendix, Fig. S1). In contrast, despite high conservation of the tlpB promoter, RBS, and start codon (SI Appendix, Fig. S6A), the G-repeat in the tlpB leader varies among diverse H. pylori strains ranging from 6 to 16 guanines (Fig. 4A). In strain 26695, in which tlpB is repressed by RepG, the G-repeat comprises 12Gs. In contrast, the G-repeat is completely absent in strains SJM180, Puno120, and Puno135, or contains a duplication of two sets of 17Gs separated by TGGTTTT in strain 908. Because the G-repeat is located in a 5′ UTR, its variation could neither lead to frame-shift mutations nor affect transcription of tlpB.
Fig. 4.
Variations in the G-repeat length and tlpB regulation in diverse Helicobacter strains. (A) Sequence alignment of the tlpB 5′ UTR (+57 to +139 relative to the transcriptional start site) including the RepG binding site (gray), RBS (yellow), and annotated ATG start codon (yellow) from different H. pylori strains. Strains that are marked in bold were used for the analysis of tlpB regulation shown in B. (B) Relative fold-changes of tlpB expression upon repG deletion in diverse H. pylori strains were determined by Western blot analysis for TlpB in comparison with the respective wild-type backgrounds (see SI Appendix, Fig. S6B) and are represented in the bar diagram. Error bars indicate SDs among two or three biological replicates. (C) The 26695 and G27 strains comprise tlpB mRNA leaders with different G-repeat lengths of 12 and 14Gs, respectively. Protein and RNA samples from strains 26695 and G27 wild-type (WT), repG deletion (ΔrepG), and RepG complementation with RepG from strain 26695 (CRepG), which carried a chromosomally tagged tlpB::3xFLAG gene, were harvested at exponential growth phase (OD600 of ∼0.9) and analyzed by Western and Northern blot. TlpB::3xFLAG was detected with α-FLAG antibody and GroEL served as loading control. RepG was probed with 32P-labeled CSO-0003 and 5S rRNA with JVO-0485, respectively.
Based on the observed RepG-mediated regulation of tlpB expression, we predicted that variations in the G-repeat length could influence posttranscriptional regulation of tlpB by RepG. Therefore, we deleted repG in strains B8, Cuz20, Shi470, Lithuania75, India7, J99, and G27, which comprise G-repeat lengths from 8 to 14Gs. Western blot analysis showed that, although the basal protein levels of the four chemotaxis receptors vary slightly among the different H. pylori strains, only the TlpB level was affected by repG deletion (Fig. 4B and SI Appendix, Fig. S6B). We detected a ∼two- to tenfold increase in TlpB protein level in the ΔrepG mutants of B8, Cuz20, Shi470, Lithuania75, and India7, which harbor homopolymeric repeats of 8–12Gs. However, the TlpB protein level was not significantly affected in a ΔrepG mutant of strain J99 with a 13G-long repeat.
In contrast to tlpB repression for strains with 8–12Gs, we observed that longer G-repeat lengths lead to activation of tlpB expression by RepG. For example, in strain G27 with a 14G repeat, tlpB was about twofold down-regulated upon deletion of repG, which we had also noticed for the endogenous TlpB levels in G27 during our GFP–reporter fusion experiments (Fig. 2C). Despite the small differences at their 5′ ends, RepG sRNAs from strains 26695 and G27 are very similar (SI Appendix, Figs. S1B and S2). Complementation of the sRNA deletion mutant in G27 with RepG from strain 26695 restored the TlpB protein level to the wild-type level of G27, indicating that the difference in regulation is not due to sRNA variations but rather due to differences within the G-repeat length (Fig. 4C). In line with this finding, RepG from strain G27 represses the tlpB::gfpmut3 fusion with the 12G tlpB 5′ UTR from strain 26695 (Fig. 2C). Overall, the differences in RepG regulation in diverse H. pylori strains indicate that variations in the G-repeat length could influence sRNA-mediated regulation of tlpB.
The Length of the Homopolymeric G-Repeat Determines Posttranscriptional Regulation of tlpB.
To test whether different G-repeat lengths can lead to the observed strain-specific tlpB regulation, we varied the length of the G-repeat in the tlpB 5′ UTR of strain 26695, which normally contains 12Gs. We either completely deleted the G-repeat (ΔG) or modulated the G-repeat length of the tlpB leader from 6 to 16 guanines (6G to 16G) (Fig. 5A). Western blot analysis of TlpB::3xFLAG of these G-repeat variants in the wild-type and the ΔrepG deletion background revealed that RepG-mediated tlpB regulation is dependent on the length of the G-repeat (Fig. 5 B and C). Whereas a lack of the G-repeat had only a minor influence on TlpB protein levels compared with 26695 wild type (12G), the TlpB protein level was increased in the variant carrying 6Gs. A gradual RepG-dependent decrease in TlpB protein level was observed with an increased number of guanines in the tlpB leader in the wild-type background that reaches its minimum in mutants with a 9- to 11-nt-long G-repeat. Further extension of the G-repeat from 12 to 16Gs resulted in an increase in TlpB protein levels. Although deletion of repG did not affect tlpB expression in the ΔG, 6G, and 13G mutants, an increased TlpB protein level was observed for G-repeat lengths of 7–12Gs, indicating these lengths as an optimal window for RepG-mediated repression. In line with our observations for strain G27, a homopolymeric repeat of 14–16Gs resulted in a slight down-regulation of TlpB levels upon repG deletion. Because RepG is expressed at similar levels in all G-repeat mutants, the differences in TlpB expression are likely a result of the variation in the G-repeat length rather than sRNA levels.
Fig. 5.
Variation of the G-repeat length in H. pylori 26695 determines tlpB regulation by RepG. (A) Scheme of tlpB mRNA leader mutants, which either lack the homopolymeric G-repeat (ΔG) or comprise diverse G-repeat lengths ranging from 6 to 16 guanine residues (6G to 16G). All mutants were constructed in a H. pylori 26695 tlpB::3xFLAG strain. (B) Western and Northern blot analyses of tlpB leader mutants (ΔG, 6G to 16G) in wild-type (WT) or ΔrepG backgrounds at exponential growth phase. TlpB protein was detected using α-FLAG antibody and GroEL served as loading control. RepG was detected using 32P-labeled CSO-0003 and the loading control 5S rRNA with JVO-0485, respectively. (C) Quantification of the relative TlpB protein levels in the different tlpB mRNA leader mutants determined by Western blot (B) in the WT (black) and ΔrepG (gray) background. The TlpB protein level in the tlpB 6G leader was used as reference and set to 1. (D) Gel-shift assay with ∼0.04 pmol 32P-labeled RepG* in the absence or presence of 1,000 nM unlabeled tlpB leader variants that either lack the homopolymeric G-repeat (ΔG) or comprise different G-repeat lengths (6 to 16G). The arrow indicates RNA-RNA duplex formation and the amount of shifted RepG* for each variant is given in percent. The result of a representative experiment (out of three) is shown. (E) In-line probing of ∼0.2 pmol 32P- labeled RepG in the absence or presence of 20 nM tlpB mRNA leader variants with indicated G-repeat lengths. The footprint in the RepG terminator loop which is observed upon addition of several tlpB variants is marked by a blue bar and corresponds to the tlpB interaction site.
To investigate whether the different G-repeat lengths influence the interaction between RepG and the tlpB leader, we performed gel-shift assays and in-line probing experiments with RepG and different tlpB mRNA leader variants (Fig. 5 D and E). Gel-shift assays showed that RepG efficiently base pairs with tlpB leaders with a repeat of 9–14Gs, with the strongest affinity for variants with 10–13Gs. In contrast, shorter (ΔG, 6–8Gs) or longer G-repeats (>14Gs) abolished or reduced the interaction between RepG and the tlpB leader. Reciprocal experiments with selected labeled tlpB G-repeats variants (10, 12, 13, and 14G) and increasing concentrations of RepG confirmed that the different tlpB leaders bind RepG with different affinities (SI Appendix, Figs. S3 and S7). Differences in the footprint strength observed in the terminator loop of RepG upon addition of different G-repeat variants in in-line probing experiments confirmed that the strength of the interaction between both RNAs is influenced by the tlpB G-repeat length (Fig. 5E). Overall, the pattern of different binding affinities for different G-repeat variants closely correlates with the observed pattern of RepG-mediated tlpB regulation in vivo, indicating that the G-repeat length influences the interaction with RepG and in turn posttranscriptional tlpB regulation (Fig. 5 B and C).
RepG Regulates tlpB mRNA Translation Depending on the G-Repeat Length.
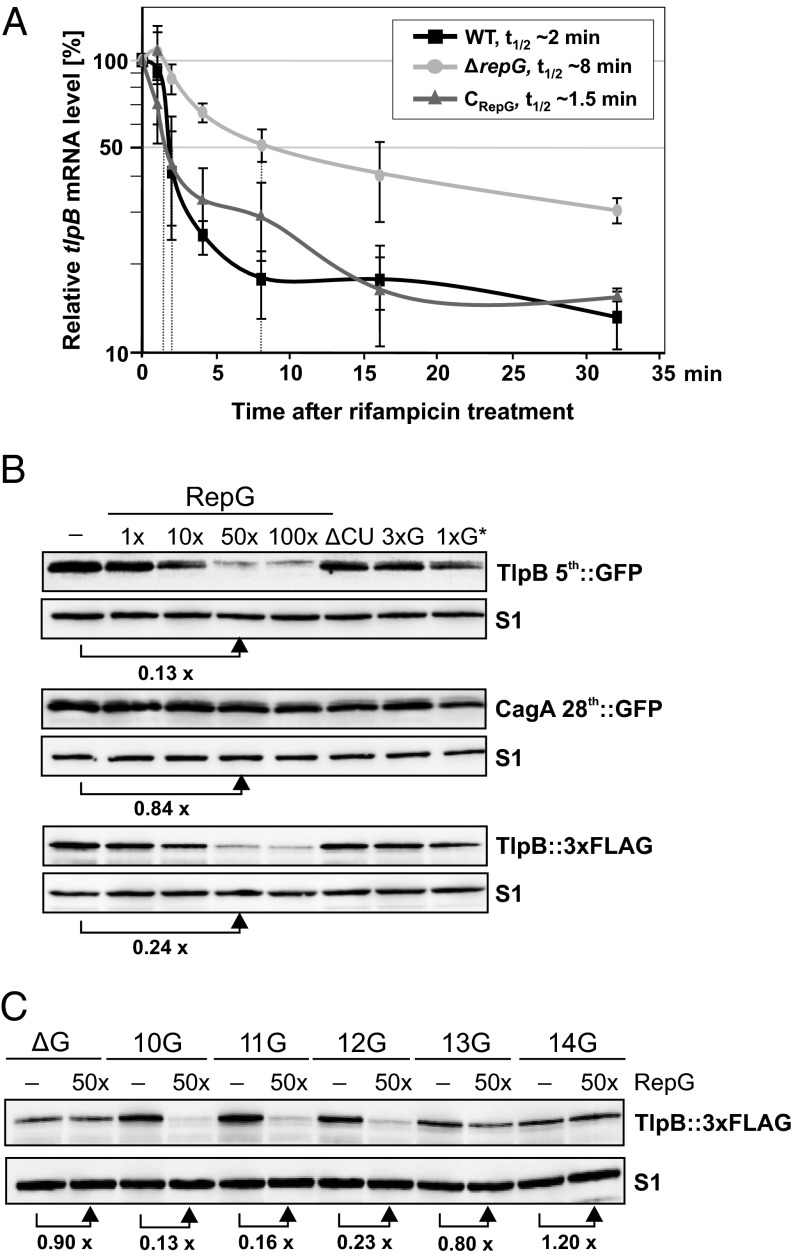
To further study the influence of the G-repeat length on RepG-mediated posttranscriptional control of tlpB, we investigated the underlying molecular mechanism. Our preliminary quantitative RT-PCR data indicated that the tlpB mRNA is fivefold up-regulated in a ΔrepG mutant (19). To examine a potential effect of RepG on tlpB mRNA stability, we determined the half-life (t1/2) of tlpB mRNA in H. pylori 26695 wild-type, ΔrepG deletion, and complementation (CRepG) strains (Fig. 6A). Rifampicin stability assays showed that tlpB mRNA was less stable in the wild-type (t1/2 WT ∼2 min) and complementation strains (t1/2 CRepG ∼1.5 min) than in the repG deletion strain (t1/2 ΔrepG ∼8 min), indicating that RepG reduces tlpB mRNA stability.
Fig. 6.
RepG reduces tlpB mRNA stability and regulates translation of tlpB mRNA. (A) The tlpB mRNA half-life at exponential growth phase was determined in H. pylori 26695 WT, ΔrepG, and RepG complementation (CRepG) strains using rifampicin assays and quantitative RT-PCR. The tlpB mRNA abundance at 0 min was set to 100% and percentage of tlpB mRNA remaining at indicated time points after rifampicin treatment was plotted. The time points at which 50% of tlpB mRNA remained (dotted lines) were used to determine the half-lives (t1/2) of tlpB mRNA in the three strains based on three biological replicates. (B) Western blot of TlpB::GFP, TlpB::3xFLAG or CagA::GFP proteins synthesized during in vitro translation assays with 0.1 µM in vitro transcribed tlpB-5th::gfpmut3, tlpB::3xFLAG or cagA-28th::gfpmut3 mRNA in the absence or presence of 0.1, 1, 5, and 10 µM RepG (1- to 100-fold excess). As control, the effect of 10 µM of RepG mutants ΔCU, 3xG or 1xG* on tlpB translation was examined in parallel. TlpB::GFP and CagA::GFP were detected using α-GFP antibody and TlpB::3xFLAG with monoclonal α-FLAG antibody, respectively. The ribosomal protein S1 served as loading control. (C) In vitro translation assay with 0.1 µM in vitro synthesized mRNAs of FLAG-tagged tlpB mRNAs with different leader variants that either lack the homopolymeric G-repeat (ΔG) or comprise a G-repeat length of 10–14Gs in the absence (−) or presence of 50-fold excess (50x) of RepG. For B and C, a representative Western blot (out of two or three experiments) is shown.
Because changes in mRNA stability could be due to inhibition of translation by sRNAs, which is often coupled to mRNA degradation in vivo, we examined the influence of RepG on tlpB translation in an in vitro translation system. In vitro transcribed mRNAs of tlpB::3xFLAG and the translational reporter fusion tlpB-5th::gfpmut3 were translated using reconstituted ribosomes, and protein synthesis was monitored on Western blots (Fig. 6B). We observed reduced TlpB::3xFLAG or TlpB::GFP protein levels upon addition of increasing concentrations of wild-type RepG, whereas mutant RNAs had no effect on protein synthesis. Translation of the control mRNA of the cagA-28th::gfpmut3 fusion was not affected by RepG, confirming a specific effect on tlpB translation by RepG. Overall, the in vitro translation assays fully recapitulated the observed regulation in vivo (Fig. 2 A and C) and indicate that RepG regulates tlpB expression at the translational level.
Next, we performed in vitro translation reactions with different tlpB leader variants (ΔG, 10–14G) in the absence or presence of RepG (Fig. 6C). In line with our in vivo results (Fig. 5 B and C), RepG reduced translation for tlpB variants with a G-repeat of 10–12Gs, had no effect on tlpB mRNAs that lack the G-stretch or contain 13Gs, and slightly increased translation of the 14G-long tlpB mRNA. Because the RepG binding site is still present in the tlpB leader and binding still occurs (although to a lesser extent, Fig. 5 D and E), longer G-repeats (13–14G) might fold into a structure that affects translation of tlpB and thereby lead to the reversal of RepG regulation. Overall, our data indicate that the G-repeat length determines the outcome of RepG-mediated posttranscriptional regulation of the chemotaxis receptor gene tlpB and that its activation or repression occurs at the translational level.
Discussion
In our study, we showed that a variable SSR contributes to sRNA-mediated posttranscriptional regulation. We demonstrated that the length of a G-repeat in the leader of tlpB mRNA encoding a chemotaxis receptor in H. pylori determines tlpB repression or activation through the abundant sRNA RepG. This modulation of tlpB expression through length variation of a SSR represents an unexpected twist in sRNA-mediated regulation and connects it with gene expression control and phenotypic variation through variable repeats. Such gradual posttranscriptional regulation through a SSR in a 5′ UTR allows for a fine-tuning of gene expression, whereas intragenic SSRs mediate rather digital ON–OFF switches through frame-shift mutations. Promoter associated SSRs (intergenic SSRs) mainly result in strong, moderate or low expression due to changing the spacing of promoter elements or transcription factor binding sites (3). An increase in the number of 7-bp tandem repeats between two promoters has been shown to result in a step-wise decrease in mRNA and protein production of adhesins in Haemophilus influenzae and occurs during natural infection in humans (34). Because phase variation by SSRs facilitates host adaptation for a variety of bacterial pathogens (2), the gradual control through SSRs at the transcriptional or posttranscriptional level could be important for the fine-tuning of virulence gene expression. Our work demonstrates the functional characterization of a trans-acting sRNA in Helicobacter and shows that studying sRNAs in bacteria that lack the common RNA chaperone Hfq can reveal unexpected mechanisms of sRNA-mediated gene regulation.
RepG Binds to a G-Repeat in the tlpB 5′ UTR and Affects Translation of tlpB mRNA.
The majority of sRNAs regulate gene expression by base-pairing close to the RBS or the start codon of their target mRNAs (17). Our in vitro translation assays indicate that RepG influences tlpB expression at the translational level (Fig. 6). This translational control could either occur at the level of translation initiation or translation elongation. Because the tlpB G-repeat, which is the RepG interaction site, is far upstream of the RBS (Fig. 3), we assume that repression of tlpB expression is based on structural changes or binding to a ribosome stand-by site rather than on a direct masking of the RBS. Several sRNAs bind far upstream of the RBS and affect gene expression through the sequestration of translational enhancers or ribosome stand-by sites (35, 36). Moreover, structural rearrangements can lead to inhibition of translation, transcript destabilization, transcription attenuation, or termination (17, 37, 38). Following translational inhibition, interacting RNAs often become substrates for endoribonucleases such as RNase E and RNase III (38). Also the observed repression of tlpB translation might be coupled to an increased transcript degradation and thereby lead to the RepG-mediated reduction in tlpB mRNA stability observed in vivo.
The Homopolymeric G-Repeat Forms an Inter- or Intramolecular Structure.
Our in vitro structure probing indicated that the tlpB G-repeat might form an inter- or intramolecular structure. Repetitive guanine-rich DNA and RNA sequences have the ability to form so called G-quadruplex structures (39, 40). In eukaryotes, such G-quadruplexes have been implicated in, for example, DNA maintenance, telomere homeostasis, recombination, or gene expression regulation. Introduction of G-quadruplex-forming sequences close to the RBS in bacterial mRNAs has been shown to affect gene expression in E. coli (41). A DNA G-quadruplex in the promoter of the pilin locus in Neisseria gonorrheae also has been shown to be required for antigenic variation (42). Interestingly, transcription of a cis-encoded antisense RNA that originated within the guanine-rich sequence has been suggested to be crucial for formation of this DNA G-quadruplex and antigenic variation (43). Future studies will be required to determine the exact structure of the G-repeat in the tlpB 5′ UTR, its interaction with RepG, and to resolve RepG-mediated structural rearrangements that could be the underlying mechanism of RepG-mediated tlpB translational regulation.
RepG and Its Genomic Context Are Highly Conserved in Diverse H. pylori Strains.
RepG is one of the most conserved small RNAs in Helicobacter, particularly its C/U-rich terminator loop, indicating that RepG uses this loop to interact with other mRNAs. Our own unpublished whole transcriptome analyses indicate that multiple genes are affected upon repG deletion and potentially targeted at G-rich sequences. Consequently, a mutation in the highly conserved C/U-rich region would abolish the global regulatory function of RepG. In contrast, variation of the tlpB G-repeat length and its influence on RepG regulation facilitates uncoupling of a single target from a sRNA regulon through modification of its targeting site.
During our search for RepG homologs, we observed that repG in H. pylori is always encoded upstream of homologs of the orphan response regulator HP1043 (SI Appendix, Fig. S1). Because sRNAs are often encoded next to their transcriptional regulators (44, 45), HP1043 might control repG expression. In addition, a positive control of tlpB by this regulator was suggested based on in vitro promoter binding studies (28), indicating a potential feed-forward loop of tlpB regulation involving HP1043 and RepG. Examination of repG expression under various stress conditions or in transcriptional regulator mutants will provide further insights into its own regulation.
Possible Role for Phase Variation of the Helicobacter Chemotaxis Receptor TlpB in Virulence.
Motility and chemotaxis are important for virulence and efficient colonization by H. pylori (20, 46–48). H. pylori strain 26695 carries four methyl-accepting chemotaxis receptors, TlpA, TlpB, TlpC, and TlpD. Our study shows that RepG sRNA specifically regulates tlpB expression whereas the other chemotaxis receptors are not affected (Fig. 2). TlpB has been shown to sense quorum sensing molecules and pH, whereby acid acts as a repellant (49). Moreover, TlpB has been implicated in colonization and inflammation during mice and gerbil infections (23, 47, 48). We observed that the length of the G-repeat in the tlpB 5′ UTR determines the outcome of sRNA-mediated tlpB regulation in different H. pylori strains (Fig. 4). Analysis of tlpB sequences of sequential H. pylori isolates from human patients (50–52) and from strains reisolated from animal colonization experiments (46, 53) indicates that the G-repeat not only varies between strains from different patients but also between isolates from the same host, suggesting that tlpB can undergo phase variation during infection (SI Appendix, Table S1). How differential tlpB expression is connected to host adaptation remains to be shown. Because pH-taxis is crucial for the spatial orientation of H. pylori along the mucus pH gradient (54), sRNA-mediated regulation and fine-tuning of tlpB expression could be important for colonization of different niches within the stomach. It is also possible that the gene downstream of tlpB, HP0102, which encodes for a putative glycosyltransferase and is coregulated with tlpB in the dicistronic tlpB-HP0102 mRNA by RepG (19), is important for host adaptation. In a global transposon screen both genes were identified as candidate loci that contribute to stomach colonization of mice (55). Thus, RepG could have an impact on virulence of H. pylori, which needs to be addressed in future studies.
H. pylori Exploits Phase Variation for Host Adaptation and Persistent Colonization.
It has been suggested that persistent colonization of the human host by H. pylori is facilitated through its extensive genetic diversity due to an elevated mutation rate, impaired DNA repair system, horizontal gene transfer, frequent recombination events, and phase variation (56, 57). Based on the presence of simple sequence or tandem repeats, around 50 candidate phase-variable genes have been identified in H. pylori (25, 53, 58). The products of these phase-variable genes are often involved in surface structures and, in turn, host recognition or adhesion (8, 9, 59–61), motility (13), or in DNA restriction and modification (62, 63). Although the influence of phase variation on transcription and translation has mainly been attributed to length variation of SSRs within promoters or coding regions, we now showed that the length of a G-repeat in an mRNA leader affects expression of a chemotaxis receptor through posttranscriptional regulation by a sRNA. The only other example of a 5′ UTR-associated G-repeat so far has been described for the UspA1 adhesin in Moraxella catarrhali and has been shown to influence uspA1 mRNA levels (64). Moreover, the length of a heteropolymeric tetranucleotide repeat in the leader of uspA2 mRNA was shown to affect mRNA stability and protein level of this adhesin and thereby contributes to serum resistance in M. catarrhali (65). However, in both cases the underlying mechanism remained unclear and it is possible that also these SSRs might be targeted by sRNAs. Apart from posttranscriptional control through SSRs in 5′ UTRs, a phase-variable invertible element in the cwpV leader of Clostridium difficile has been shown to determine transcription elongation through formation of an intrinsic transcription terminator depending on the orientation of the DNA element (66).
Besides in Helicobacter, length variations of poly-G tracts have been observed under selective environmental conditions and passage through animals also in other Epsilonproteobacteria such as the food-borne pathogen Campylobacter jejuni (7, 67). These G-repeats could also be potential target sites of sRNAs which have recently been identified in this pathogen (68). Comparisons of homopolymeric SSR locations with our global transcriptional start site maps of H. pylori and C. jejuni (19, 68) showed that the majority of SSRs are found in promoter or coding regions, but revealed about 10 genes that carry a SSR in their 5′ UTR which might act by influencing posttranscriptional regulation (SI Appendix, Table S2). Besides base-pairing with translation initiation regions, bacterial sRNAs can also regulate gene expression by targeting coding sequences (69). Therefore, several of the intragenic SSRs could also be targeted by trans-encoded sRNAs and our previous transcriptome study also identified several cis-encoded antisense RNAs to SSRs in H. pylori (19). Overall, this posttranscriptional mode of gene regulation through homopolymeric repeats is likely to be more widespread and SSRs not only in 5′ UTRs but also within the coding sequence could be targeting sites of sRNAs.
Materials and Methods
Bacterial Strains, Oligonucleotides, and Plasmids.
Helicobacter and Escherichia coli strains used in this study are listed in SI Appendix, Table S3. DNA oligodesoxynucleotides used for cloning, T7 transcription template generation, and Northern blot probes are listed in SI Appendix, Table S4. Plasmids are summarized in SI Appendix, Table S5, and sequences of all RepG and tlpB variants in SI Appendix, Tables S6 and S8.
Bacterial Growth and Construction of Helicobacter Mutants.
E. coli strains were grown in Luria Bertani (LB) medium supplemented with 100 µg/mL ampicillin and/or 20 µg/mL chloramphenicol if applicable. H. pylori media used for growth on plates or in liquid cultures as well as culture conditions are described in SI Appendix. Details about the generation of H. pylori mutant strains are also listed in SI Appendix.
RNA Preparation, Northern Blot Analysis, and Stability Assays.
If not mentioned otherwise, H. pylori was grown in liquid culture to midexponential growth phase (OD600 nm 0.5–0.9) and cells corresponding to an OD600 of 4 were harvested, mixed with 0.2 volumes stop mix [95% (vol/vol) EtOH/5% (vol/vol) phenol], and immediately shock-frozen in liquid nitrogen. Frozen cell pellets were thawed on ice, centrifuged for 10 min at 3,250 × g at 4 °C, and resuspended in TE buffer (pH 8.0) containing 0.5 mg/mL lysozyme and 1% SDS. Cell lysis was completed by incubation at 65 °C for 2 min. RNA was extracted using the hot phenol method as described (19). For Northern blot analysis, 5–10 µg of total RNA was separated on 6% (vol/vol) polyacrylamide (PAA) gels containing 7 M urea and subsequently blotted to Hybond-XL membranes (GE-Healthcare). After blotting, total RNA was UV cross-linked to the membrane and hybridized with 5′ end-labeled (γ32P) DNA oligonucelotides as described (29). Details about rifampicin stability assays and quantitative RT-PCR are listed in SI Appendix.
SDS/PAGE and Immunoblotting.
For protein analysis, cells corresponding to an OD600 of 1 from H. pylori cells grown to midexponential growth phase were collected by centrifugation at 16,100 × g at 4 °C for 2 min. Cell pellets were dissolved in 100 µL of 1× protein loading buffer [62.5 mM Tris⋅HCl pH 6.8, 100 mM DTT, 10% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.01% bromophenol blue]. After boiling at 95 °C for 8 min, protein samples corresponding to 0.1 OD600 were separated by 12% (vol/vol) one-dimensional SDS-PAA gels and stained by Coomassie (Fermentas, #R0571). For Western blot analysis, protein samples corresponding to an OD600 of 0.01 or 0.005 were separated by 12% or 10% (vol/vol) SDS/PAGE and transferred to a PVDF membrane by semidry blotting. Membranes were blocked for 1 h with 10% (wt/vol) milk powder/TBS-T and incubated over night with primary antibody at 4 °C. Afterward, membranes were washed with TBS-T, followed by 1 h incubation with secondary antibody linked to horseradish peroxidase. After additional washing steps, chemiluminescence was detected using ECL-reagent. Details about the used antisera and antibodies are listed in SI Appendix.
In Vitro Structure Probing and Gel Mobility Shift Assays.
DNA templates that contain the T7 promoter sequence for in vitro transcription using the MEGAscript T7 Kit (Ambion) were generated by PCR. Oligos and DNA templates used to generate the individual T7 templates are listed in SI Appendix, Tables S7 and S8. Details about in vitro T7 transcription, structure probing, and footprinting assays, as well as gel-shift experiments and in vitro translation reactions, are listed in SI Appendix.
Supplementary Material
Acknowledgments
We thank G. Storz, J. Vogel, A. Westermann, F. Darfeuille, D. Lopez, S. Gorski, and N. Machuy for critical comments on our manuscript, B. Aul, A. Lins, S. Bauer, N. Kapica, and E. Schönwetter for excellent technical assistance, and M. Alzheimer and P. Tan for help with the construction of GFP reporter fusions. Moreover, we thank F. Darfeuille for providing genomic DNA of a RepG complementation strain, D. S. Merrell for gfpmut3 plasmids, and K. Ottemann and R. Haas for antisera against H. pylori chemotaxis receptors and CagA, respectively. In addition, we thank S. Suerbaum and C. Josenhans for sharing genomic DNA of sequential H. pylori isolates, and D. E. Berg, R. Haas, and T. F. Meyer for providing H. pylori wild-type strains. The C.M.S. laboratory received financial support from the ZINF Young Investigator program at the Research Center for Infectious Diseases (ZINF, Würzburg, Germany), DFG project Sh580/1-1, and the Young Academy program of the Bavarian Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315152111/-/DCSupplemental.
References
- 1.van der Woude MW. Phase variation: How to create and coordinate population diversity. Curr Opin Microbiol. 2011;14(2):205–211. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Moxon R, Bayliss C, Hood D. Bacterial contingency loci: The role of simple sequence DNA repeats in bacterial adaptation. Annu Rev Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 3.Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol Rev. 2013 doi: 10.1111/1574-6976.12036. 10.1111/1574-6976.12036. [DOI] [PubMed] [Google Scholar]
- 4.Stibitz S, Aaronson W, Monack D, Falkow S. Phase variation in Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989;338(6212):266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- 5.Stern A, Brown M, Nickel P, Meyer TF. Opacity genes in Neisseria gonorrhoeae: Control of phase and antigenic variation. Cell. 1986;47(1):61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 6.Hood DW, et al. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc Natl Acad Sci USA. 1996;93(20):11121–11125. doi: 10.1073/pnas.93.20.11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerome JP, et al. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS ONE. 2011;6(1):e16399. doi: 10.1371/journal.pone.0016399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appelmelk BJ, et al. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in alpha3-fucosyltransferase genes. Infect Immun. 1999;67(10):5361–5366. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solnick JV, Hansen LM, Salama NR, Boonjakuakul JK, Syvanen M. Modification of Helicobacter pylori outer membrane protein expression during experimental infection of rhesus macaques. Proc Natl Acad Sci USA. 2004;101(7):2106–2111. doi: 10.1073/pnas.0308573100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59(4):657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 11.Yang QL, Gotschlich EC. Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med. 1996;183(1):323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder LA, et al. Simple sequence repeats in Helicobacter canadensis and their role in phase variable expression and C-terminal sequence switching. BMC Genomics. 2010;11:67. doi: 10.1186/1471-2164-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josenhans C, Eaton KA, Thevenot T, Suerbaum S. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect Immun. 2000;68(8):4598–4603. doi: 10.1128/iai.68.8.4598-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willems R, Paul A, van der Heide HG, ter Avest AR, Mooi FR. Fimbrial phase variation in Bordetella pertussis: A novel mechanism for transcriptional regulation. EMBO J. 1990;9(9):2803–2809. doi: 10.1002/j.1460-2075.1990.tb07468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Ham SM, van Alphen L, Mooi FR, van Putten JP. Phase variation of H. influenzae fimbriae: Transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73(6):1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- 16.Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER. Microsatellite instability regulates transcription factor binding and gene expression. Proc Natl Acad Sci USA. 2005;102(10):3800–3804. doi: 10.1073/pnas.0406805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8(1):116–127. doi: 10.1016/j.chom.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 19.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 20.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr Opin Microbiol. 2010;13(1):24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Pernitzsch SR, Sharma CM. Transcriptome complexity and riboregulation in the human pathogen Helicobacter pylori. Front Cell Infect Microbiol. 2012;2:14. doi: 10.3389/fcimb.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol. 2006;188(7):2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goers Sweeney E, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20(7):1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol Microbiol. 1998;27(6):1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 26.Tjaden B, et al. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34(9):2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Na D, et al. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31(2):170–174. doi: 10.1038/nbt.2461. [DOI] [PubMed] [Google Scholar]
- 28.Delany I, Spohn G, Rappuoli R, Scarlato V. Growth phase-dependent regulation of target gene promoters for binding of the essential orphan response regulator HP1043 of Helicobacter pylori. J Bacteriol. 2002;184(17):4800–4810. doi: 10.1128/JB.184.17.4800-4810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35(3):1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72(3):551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter BM, et al. Expanding the Helicobacter pylori genetic toolbox: Modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl Environ Microbiol. 2007;73(23):7506–7514. doi: 10.1128/AEM.01084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 33.Mathews DH. RNA secondary structure analysis using RNAstructure. Curr Protoc Bioinformatics. 2006;12 doi: 10.1002/0471250953.bi1206s13. Unit 12.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawid S, Barenkamp SJ, St Geme JW., 3rd Variation in expression of the Haemophilus influenzae HMW adhesins: A prokaryotic system reminiscent of eukaryotes. Proc Natl Acad Sci USA. 1999;96(3):1077–1082. doi: 10.1073/pnas.96.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21(21):2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26(3):381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3(12):a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caron MP, Lafontaine DA, Massé E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol. 2010;7(2):140–144. doi: 10.4161/rna.7.2.11056. [DOI] [PubMed] [Google Scholar]
- 39.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3(4):495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 40.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: Stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13(11):770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieland M, Hartig JS. Investigation of mRNA quadruplex formation in Escherichia coli. Nat Protoc. 2009;4(11):1632–1640. doi: 10.1038/nprot.2009.111. [DOI] [PubMed] [Google Scholar]
- 42.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325(5941):764–767. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cahoon LA, Seifert HS. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013;9(1):e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: Role as a pleiotropic regulator and antimutator. Cell. 1997;90(1):43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 45.Vanderpool CK, Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol. 2004;54(4):1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 46.Behrens W, et al. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect Immun. 2013;81(10):3534–3551. doi: 10.1128/IAI.00750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams SM, et al. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun. 2007;75(8):3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGee DJ, et al. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun. 2005;73(3):1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rader BA, et al. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology. 2011;157(Pt 9):2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devi SH, et al. Genome of Helicobacter pylori strain 908. J Bacteriol. 2010;192(24):6488–6489. doi: 10.1128/JB.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avasthi TS, et al. Genomes of two chronological isolates (Helicobacter pylori 2017 and 2018) of the West African Helicobacter pylori strain 908 obtained from a single patient. J Bacteriol. 2011;193(13):3385–3386. doi: 10.1128/JB.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kennemann L, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108(12):5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salaün L, Ayraud S, Saunders NJ. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiology. 2005;151(Pt 3):917–923. doi: 10.1099/mic.0.27379-0. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber S, et al. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci USA. 2004;101(14):5024–5029. doi: 10.1073/pnas.0308386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldwin DN, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect Immun. 2007;75(2):1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5(6):441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 57.Salama NR, Hartung ML, Müller A. Life in the human stomach: Persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11(6):385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 59.Yamaoka Y, et al. Helicobacter pylori infection in mice: Role of outer membrane proteins in colonization and inflammation. Gastroenterology. 2002;123(6):1992–2004. doi: 10.1053/gast.2002.37074. [DOI] [PubMed] [Google Scholar]
- 60.Kennemann L, et al. In vivo sequence variation in HopZ, a phase-variable outer membrane protein of Helicobacter pylori. Infect Immun. 2012;80(12):4364–4373. doi: 10.1128/IAI.00977-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodwin AC, et al. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology. 2008;154(Pt 8):2231–2240. doi: 10.1099/mic.0.2007/016055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Vries N, et al. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J Bacteriol. 2002;184(23):6615–6623. doi: 10.1128/JB.184.23.6615-6623.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srikhanta YN, Fox KL, Jennings MP. The phasevarion: Phase variation of type III DNA methyltransferases controls coordinated switching in multiple genes. Nat Rev Microbiol. 2010;8(3):196–206. doi: 10.1038/nrmicro2283. [DOI] [PubMed] [Google Scholar]
- 64.Lafontaine ER, Wagner NJ, Hansen EJ. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J Bacteriol. 2001;183(5):1540–1551. doi: 10.1128/JB.183.5.1540-1551.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Attia AS, Hansen EJ. A conserved tetranucleotide repeat is necessary for wild-type expression of the Moraxella catarrhalis UspA2 protein. J Bacteriol. 2006;188(22):7840–7852. doi: 10.1128/JB.01204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emerson JE, et al. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol Microbiol. 2009;74(3):541–556. doi: 10.1111/j.1365-2958.2009.06812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bayliss CD, et al. Phase variable genes of Campylobacter jejuni exhibit high mutation rates and specific mutational patterns but mutability is not the major determinant of population structure during host colonization. Nucleic Acids Res. 2012;40(13):5876–5889. doi: 10.1093/nar/gks246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dugar G, et al. High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates. PLoS Genet. 2013;9(5):e1003495. doi: 10.1371/journal.pgen.1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16(8):840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.