Abstract
In Bacillus subtilis, many membrane proteins localize to the sporulation septum, where they play key roles in spore morphogenesis and cell-specific gene expression, but the mechanism for septal targeting is not well understood. SpoIIQ, a forespore-expressed protein, is involved in engulfment and forespore-specific gene expression. We find that SpoIIQ dynamically localizes to the sporulation septum, tracks the engulfing mother cell membrane, assembles into helical arcs around the forespore and is finally degraded. Retention of SpoIIQ in the septum requires one or more mother cell-expressed proteins. We also observed that any forespore-expressed membrane protein initially localizes to the septum and later spreads throughout the forespore membrane, suggesting that membrane protein insertion occurs at the forespore septal region. This possibility provides an attractive mechanism for how activation of mother cell-specific gene expression is restricted to adjacent sister cells, since direct insertion of the signaling protein SpoIIR into the septum would spatially restrict its activity. In keeping with this hypothesis, we find that SpoIIR localizes to the septum and is transiently expressed.
Keywords: forespore septal membrane, membrane protein localization, sporulation, SpoIIQ, SpoIIR
Introduction
It is becoming increasingly clear that bacteria share with eucaryotes the ability to specifically and dynamically localize proteins within the cell. The best-studied example is FtsZ, a divergent tubulin homolog required for cell division, which assembles into rings at midcell during vegetative growth, moving to new division sites via spiral intermediates (Addinall and Holland, 2002; Ben-Yehuda and Losick, 2002; Lutkenhaus, 2002; Margolin, 2002). In fact, the number of bacterial proteins with dynamic localization is ever increasing: Escherichia coli MinCD oscillate at a rapid rate from pole to pole within the cell to promote correct placement of the division machinery (Marston and Errington, 1999; Justice et al, 2000; Pichoff and Lutkenhaus, 2001; Johnson et al, 2002); DivK of Caulobacter crescentus, a signal transduction protein required for polar morphogenesis and cell cycle progression, whose localization alternates between the cell pole and even dispersal throughout the membrane (Jacobs et al, 2001; Ausmees and Jacobs-Wagner, 2003); and Bacillus subtilis SpoIIIE, required for DNA translocation and membrane fusion during sporulation, relocalizes from the septal midpoint to the cell pole (Sharp and Pogliano, 1999, 2002; Bath et al, 2000). While the number of proteins with dynamic localization increases, the mechanism for the specific targeting of these proteins is still not well understood, particularly for membrane proteins. Most membrane proteins utilize the Sec translocation machinery for entry into the cytoplasmic membrane, but it has recently been shown that in E. coli, SecYEG are uniformly distributed throughout the membrane, further complicating the mechanism for membrane protein targeting (Brandon et al, 2003).
Sporulation in B. subtilis provides an excellent system to study dynamic protein localization, since it involves the dramatic rearrangement of the bacterial cell (Stragier and Losick, 1996), with specifically localized proteins mediating many key steps. The first morphological change of sporulation is the switch from medial to polar division (Figure 1A), resulting in the formation of two distinct cell types: a smaller forespore that eventually becomes the spore and a larger mother cell that lyses upon release of the spore. Relocalization of the division site is mediated by movement of the FtsZ ring from midcell to both poles (Levin and Losick, 1994; Ben-Yehuda and Losick, 2002), allowing recruitment of proteins required for cell-specific gene expression to the sporulation septum (Arigoni et al, 1999), which is ultimately synthesized at only one of the potential division sites. Following the onset of forespore and mother cell-specific gene expression, the mother cell membrane migrates around the forespore in a phagocytosis-like event known as engulfment (Figure 1A). During engulfment, the required mother cell-expressed membrane proteins localize to the leading edge of the engulfing membrane (Abanes-De Mello et al, 2002), while mother cell-expressed proteins required for coat assembly (Pogliano et al, 1995; Driks, 2002) and intracellular signal transduction (Rudner and Losick, 2002) localize to the mother cell-derived outer forespore membrane. With one exception, described below, little is known about the mechanism through which membrane proteins are localized during sporulation.
Figure 1.
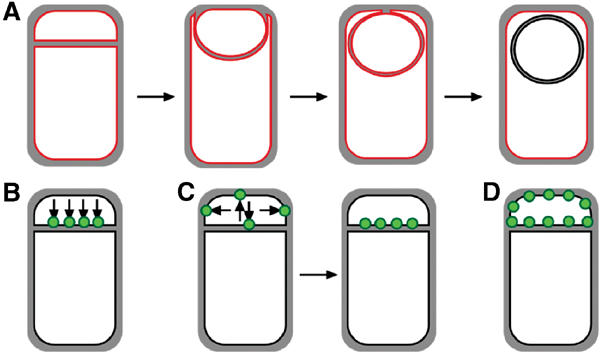
Engulfment pathway of sporulation in B. subtilis and models for targeting of proteins synthesized in the forespore. (A) Cartoon showing B. subtilis engulfment. Following polar septation, the membrane of the larger mother cell migrates around the forespore, until it is completely enclosed. Membranes stained by FM 4-64 are shown in red; following membrane fusion, FM 4-64 no longer stains the forespore membranes, providing an assay for the completion of engulfment. (B–D) Models for targeting forespore-expressed membrane proteins. Proteins (green) can either be directly inserted into the forespore septal region (B) or inserted elsewhere and migrate to the septal region (C). Then, they can either be retained during engulfment (C) or freely diffuse throughout the forespore membrane (D).
Two possible mechanisms can be envisioned for the specific localization of membrane proteins within the bacterial cell to the polar septum (Figure 1B and C). First, membrane proteins could be directly inserted into the membrane at the sporulation septum (Figure 1B, directed insertion). Alternatively, membrane protein insertion could occur elsewhere, with the proteins diffusing through the membrane until they are retained at the septum by a specific ligand (Figure 1C, lateral sorting or diffusion and capture). Recently, evidence has been provided that the mother cell-expressed SpoIVFB, required for activation of the late mother cell-specific σ factor, σK, inserts into the mother cell cytoplasmic membrane, diffusing to the septum where it is retained (Rudner et al, 2002).
Here we present evidence that in the forespore, membrane proteins can reach their correct destination by directed insertion and capture, as when non-native membrane proteins, such as those from E. coli, are expressed in the forespore, they initially localize to the sporulation septum and later diffuse throughout the forespore membrane (Figure 1D). In contrast, localized proteins can be selectively retained by interacting with a septum-specific ligand (Figure 1C), which in the case of SpoIIQ is a mother cell-expressed protein required to couple movement of SpoIIQ around the forespore to the engulfing mother cell membrane. Interestingly, SpoIIQ regulates the late forespore-specific transcription factor, σG (Sun et al, 2000), whose activity is tightly regulated both by the completion of engulfment and by an intracellular signal transduction cascade (Illing and Errington, 1991; Stragier and Losick, 1996; Serrano et al, 2003). Our cell biological studies thereby further support a role for SpoIIQ in this process, as it interacts with a mother cell-specific ligand, and displays a dynamic localization pattern coordinated with the completion of engulfment. We propose that the directed insertion of forespore-specific proteins into the septum delivers the SpoIIR signaling protein to the sporulation septum thereby restricting activity to its adjacent mother cell and preventing it from inappropriately activating σE in neighboring cells.
Results
Localization of heterologous GFP fusion proteins in the forespore
We were interested in identifying signals required for localization of integral membrane proteins to the forespore septal membrane, and therefore compared localization of membrane proteins not normally expressed during sporulation to that of sporulation-specific membrane proteins, such as SpoIIQ. Forespore-expressed proteins without localization signals should diffuse throughout the forespore membrane (Figure 1D), whereas localized proteins might first localize to the septal region of the forespore membrane, and later move around the forespore membrane together with the engulfing mother cell membrane (Figure 1C). The mechanism by which a forespore-expressed protein might track the engulfing mother cell membrane remains unclear, as the obvious morphological transformation of engulfment involves only the mother cell membrane (Sun et al, 2000; Abanes-De Mello et al, 2002).
We therefore constructed GFP fusions to sporulation-specific proteins, as well as to heterologous membrane proteins, and used the promoters and translational initiation sites from forespore-specific genes to mediate their expression. For the heterologous proteins, the first two membrane-spanning segments of five different membrane proteins (E. coli MalF, LacY, MotA and SecY, and B. subtilis AraP) were expressed from the promoter and translational initiation sites from one of four different forespore-specific promoters (spoIIQ, spoIIR, katX and csfB), with gfp fused to either the 5′ or 3′ end of the membrane protein-encoding region. For the native proteins, we constructed GFP fusions to the membrane proteins SpoIIQ and SpoIIR (at the N-terminus, since the C-termini are extracellular), to the soluble proteins CsfB and KatX and utilized native promoters for expression. Western blot analysis showed that these GFP fusion proteins were stable early in sporulation (data not shown), with the exception of GFP-SpoIIQ and GFP-SpoIIR (discussed further below).
Cells containing the GFP fusion proteins were induced to sporulate via resuspension (Materials and methods) and visualized using deconvolution fluorescence microscopy using fluorescent membrane stains to visualize the dramatic changes in membrane morphology associated with engulfment, providing a convenient method to discriminate between sporangia at different stages of sporulation since engulfment typically initiates 2 h after the onset of sporulation (t2) and completes within 60 min (t3) (Figure 1A) (Pogliano et al, 1999; Sharp and Pogliano, 1999). In early sporangia, with flat polar septa and faint GFP fluorescence, every heterologous membrane protein constructed localized first to the sporulation septum (MalF-GFP is shown in Figure 2A and B; MotA-GFP in Figure 2C; other proteins in Supplementary Figure 1; see arrows 1). We quantified this apparent septal enrichment for the most highly expressed protein (MalF-GFP, AR4) comparing fluorescence intensity of GFP and the membrane-specific stain FM 4-64 at the septum, forespore pole and mother cell cytoplasmic membrane (Materials and methods). As expected, the membrane stain was approximately twice as bright at the septum as at the cell pole or mother cell cytoplasmic membrane (79 700, 38 800 and 43 300 photons/μm2, respectively), since the septum is comprised of two parallel membranes (Figure 1A). In contrast, the GFP fluorescence was 5–30 times brighter at the septum than at the forespore pole, indicating that enrichment of the heterologous membrane fusion proteins is not due to a difference in the volume of membrane at the septum versus cell pole. In sporangia at early stages of engulfment (with curved sporulation septa), and slightly brighter GFP fluorescence, the membrane proteins remained enriched at the septum with faint fluorescence clearly visible at the cell pole (Figure 2A and C and Supplementary Figure 1, arrows 2), while late in engulfment each fusion protein was uniformly distributed throughout the forespore membrane (Figure 2B, arrows 3 and 4).
Figure 2.
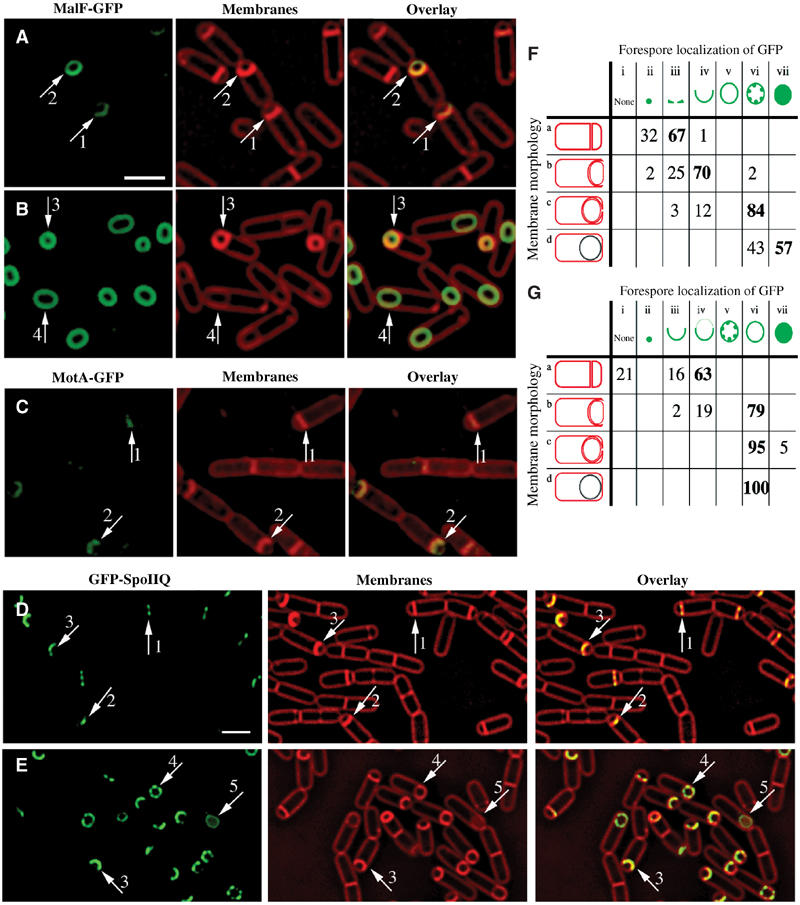
Localization patterns of forespore-expressed heterologous membrane proteins and the native forespore protein SpoIIQ. (A, B) MalF-GFP (green) 2 h (A, hereafter t2) and 3 h (B, hereafter t3) after the onset of sporulation stained with FM 4-64 (red). In most sporangia, engulfment is underway at t2 and complete by t3. (C) MotA-GFP (green) at t2 stained with Mitotracker Red (red). In panels A and C, arrows 1 and 2 represent sporangia with flat or slightly curving polar septa. In panel B, arrows 3 and 4 represent sporangia before and after membrane fusion, respectively. (D) GFP-SpoIIQ (green) at t2 and (E) t3 stained with FM 4-64 (red). In panels D and E, arrows 1, 2 and 3 represent early sporangia and arrows 4 and 5 represent sporangia late in engulfment. The scale bar in (A, D) is 2 μm. (F, G) Quantitation of GFP localization at various stages of engulfment (a–d). Numbers refer to the percentage of sporangia in each engulfment class with the indicated localization pattern; those in bold represent the predominant class. (F) GFP-SpoIIQ (AR126) localization. Seven subclasses of GFP localization were scored: (i) none; (ii) medial focus; (iii) ring; (iv) tracking mother cell engulfing membrane; (v) freely diffusible; (vi) punctate; (vii) soluble. A total of 504 sporangia were scored from t2 and t3. (G) MalF-GFP (AR4) localization. Seven subclasses for GFP localization were scored: (i) none; (ii) focus formation; (iii) tracking mother cell engulfing membrane; (iv) enriched at septum but ahead of engulfing membranes; (v) punctate; (vi) freely diffusible; (vii) soluble. A total of 296 sporangia were scored from t2 and t3.
Thus, at least five different membrane proteins not normally expressed in the forespore localize to the sporulation septum in early sporangia, but appear uniformly distributed throughout the forespore membrane in later sporangia. To reduce the possibility that either the untranslated sequences in the 5′ end of the mRNA or the first six amino acids of SpoIIQ directed septal localization, we expressed these fusion proteins from a total of four different promoters (including translational initiation signals and the first six codons), two of which encoded nonlocalized cytoplasmic proteins (csfB and katX; Supplementary Figure 1 and data not shown). In each case, the heterologous fusion proteins showed identical localization patterns regardless of the expression cassettes used, suggesting that any forespore-expressed membrane protein initially localizes to the sporulation septum, later diffusing throughout the forespore membrane. One possible explanation for these findings is that in the forespore, membrane proteins are directly inserted into the septal membrane region, but in the absence of specific localization signals, they later diffuse throughout the forespore membranes.
Dynamic localization of a native membrane protein, SpoIIQ
We compared the localization of the heterologous GFP fusions described above to two well-characterized σF-dependent membrane proteins, SpoIIQ and SpoIIR (described below). Transcriptional profiling experiments have identified only 66 genes under σF (and σF/σG) control (Fawcett et al, 2000) in contrast to the 253 genes controlled by σE (Eichenberger et al, 2003), with only a few membrane proteins of known function. We constructed a functional fusion of GFP to the cytoplasmic N-terminus of SpoIIQ, a protein essential for the synthesis and activation of the late forespore-specific transcription factor σG (Sun et al, 2000), and integrated this into a spoIIQ null strain (Materials and methods). The resulting strain (AR126) was similar to wild type for both engulfment and spore production. In early sporangia with very faint GFP fluorescence, GFP-SpoIIQ localized as a ring at the septum or a focus at the septal midpoint (Figure 2D, arrows 1 and 2, respectively). In sporangia that had initiated engulfment, GFP-SpoIIQ lined the septum, and closely followed the engulfing mother cell membrane as it moved around the forespore (Figure 2D and E, arrows 3). Subsequently, GFP-SpoIIQ displayed punctate membrane-localized fluorescence in sporangia near the completion of engulfment (Figure 2E, arrow 4). After the completion of membrane fusion, cytoplasmic GFP fluorescence was observed (Figure 2E, arrow 5), suggesting that GFP was released from the forespore membrane. A striking three-dimensional structure was observed at the transition between the last two stages. In the medial focal plane, we observed both soluble GFP fluorescence and six GFP-SpoIIQ foci, which appeared to mark the corners of a hexagonal forespore (Figure 3 and Supplementary Figure 2). In the uppermost focal plane, four of these foci resolved into two lines, which traversed the width of the forespore, suggesting that at the time that GFP-SpoIIQ is being degraded, it is arranged into two or more hoops that surround the forespore (Figure 3, +0.4 μm column). Indeed, three-dimensional modeling shows that SpoIIQ forms helical arcs around the forespore (Figure 3E; Supplementary movie) similar to those of the actin-like proteins MreB and Mbl (Jones et al, 2001; Carballido-Lopez and Errington, 2003; Daniel and Errington, 2003).
Figure 3.
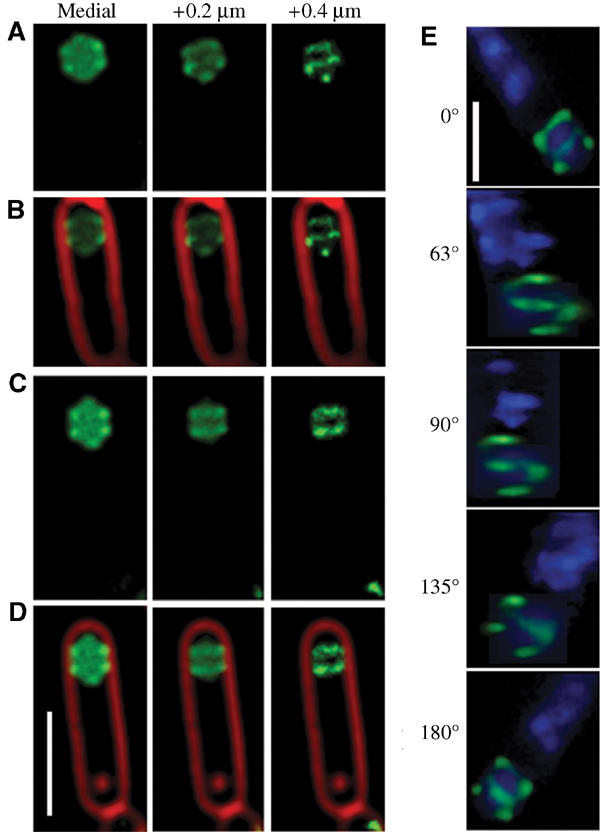
Three-dimensional visualization of GFP-SpoIIQ rings after membrane fusion. Optical sectioning deconvolution microscopy was used to obtain images from different focal planes within the specimen (A–D) and used to reconstruct the three-dimensional model of GFP-SpoIIQ (E). In all, 20 images were obtained from focal planes 0.1 μm (A–D) or 0.15 μm (E) apart. (A–D) Different focal planes of GFP-SpoIIQ (green) from two sporangia, starting with the medial GFP focal plane (far left) and +0.2 and +0.4 μm. For reference, the medial focal plane of FM 4-64-stained membrane (red) is included as an overlay (B, D). (E) Three-dimensional model of GFP-SpoIIQ. Z-stacks each separated by 0.15 μm were loaded into the volume builder function of the Applied Precision SoftWorx program (version 3.3) with modeled projections around the Y-axis shown and DAPI-stained chromosomes shown as a reference point (also shown in the Supplementary movie). Scale bar, 2 μm.
The localization of GFP-SpoIIQ correlated well with the various stages of engulfment (Figure 2F), and was dramatically different from that of MalF-GFP. First, in early sporangia, we never observed foci at the septal midpoint or ring formation with MalF-GFP or any other heterologous membrane protein. Second, MalF-GFP moved ahead of the engulfing mother cell membrane in 63% of sporangia (Figure 2A, arrow 2; Figure 2G, Class a-iv), whereas GFP-SpoIIQ never did. Finally, MalF-GFP remained associated with the forespore membrane even after the completion of engulfment (Figure 2B, arrow 4), in contrast to cytoplasmic GFP fluorescence observed for GFP-SpoIIQ (in 57% of sporangia; see Figure 2F, Class d-vii). Overall, most sporangia displayed uniform rings of MalF-GFP fluorescence (79% for sporangia with curving septa, 100% for sporangia that have completed engulfment; shown in Figure 2G, Class b-vi, c-vi and d-vi, respectively).
SpoIIQ is degraded during sporulation
The cytoplasmic GFP fluorescence visualized in late GFP-SpoIIQ sporangia suggested proteolytic release of GFP from the SpoIIQ membrane-spanning segment. To test this hypothesis, we performed immunofluorescence and Western blot analysis of GFP-SpoIIQ, SpoIIQ-myc, SpoIIQ-flag and GFP-SpoIIQ-flag (Figure 4 and data not shown) using GFP-, c-Myc- or Flag-specific antibodies, respectively. GFP-SpoIIQ was first detected at t2 and was markedly unstable, producing one major breakdown product approximately the size of soluble GFP (Figure 4A). This degradation product accumulated during sporulation, coincident with a decrease in the level of full-length protein (Figure 4A (*)). These results are in contrast to the more stable levels of MalF-GFP and its smaller GFP-sized band (data not shown). Similarly, SpoIIQ-myc was first detected at t1, with full-length protein peaking around t3 and decreasing with time, with no smaller degradation products detected (Figure 4B). Immunofluorescence data for GFP-SpoIIQ closely correlate with the localization observed in live cells (Figure 4C and D). In contrast, immunofluorescence analysis of SpoIIQ-myc revealed similar early localization patterns, but little soluble signal was detected within the forespore at later stages of engulfment (Figure 4E and F). Together, these results suggest that SpoIIQ is cleaved near the extracellular domain, producing a smaller GFP-sized product.
Figure 4.
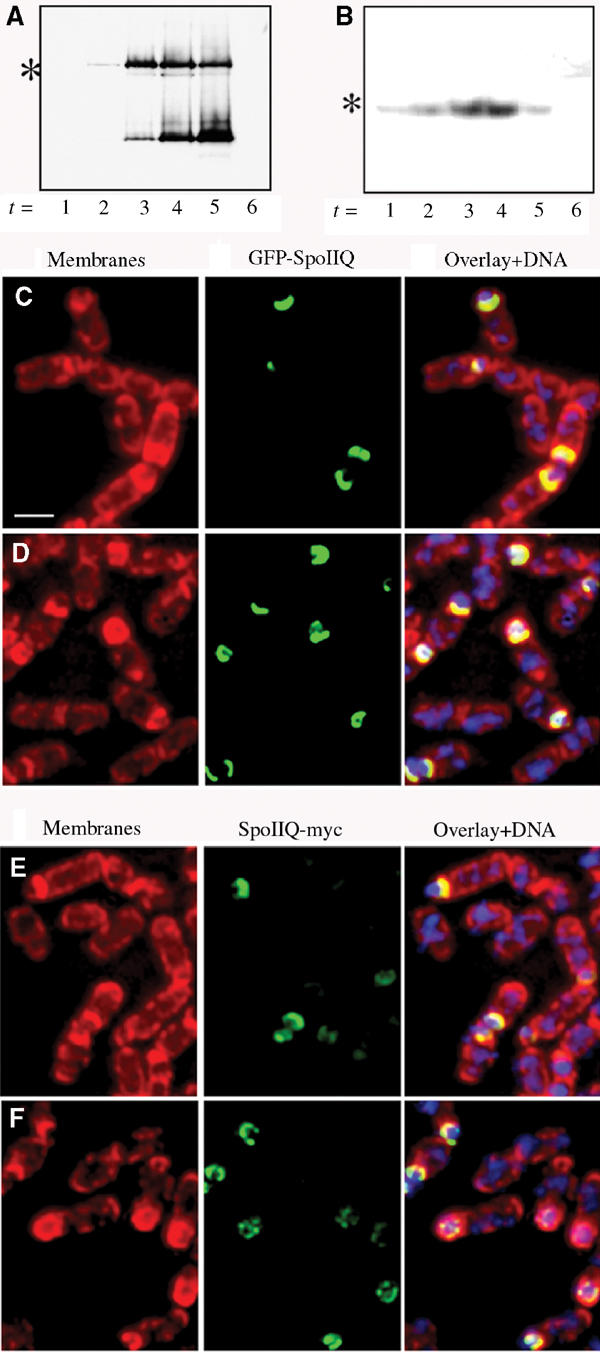
Western blot analysis and immunofluorescence of GFP-SpoIIQ and SpoIIQ-myc. (A) Immunoblot of GFP-SpoIIQ depicting specific proteolytic cleavage during engulfment. Full-length GFP-SpoIIQ is marked with an asterisk (*). (B) Western blot of SpoIIQ-myc shows no smaller degradation product. Full-length SpoIIQ-myc is marked with an asterisk (*). Immunofluorescence samples were taken at (C, E) t2 and (D, F) t3 and stained with Mitotracker Red (membranes in red), DAPI (DNA in blue) and SpoIIQ (GFP or Myc in green). (C, D) Localization patterns for GFP-SpoIIQ observed for fixed cells are the same as with live cells. (E, F) Localization of SpoIIQ-myc suggests that degradation occurs at the C-terminus of the protein since early patterns are similar to GFP-SpoIIQ, but little signal is observed in late sporangia. Scale bar, 2 μm.
Movement of GFP-SpoIIQ around the forespore depends on engulfment
GFP-SpoIIQ appeared to track the engulfing mother cell membrane as it moved around the forespore; however, it might move around the forespore by an engulfment-independent mechanism and only coincidentally track the engulfing membrane. To determine more precisely if GFP-SpoIIQ tracked the engulfing membranes, we tested if its movement around the forespore depended on the mother cell-expressed engulfment proteins SpoIIM, SpoIIP and SpoIID. Mutants lacking these proteins fail to initiate engulfment, and ultimately the forespore breaks through the septum into the mother cell (Lopez-Diaz et al, 1986; Smith et al, 1993; Frandsen and Stragier, 1995), forming bulges at the septum that are readily visualized by fluorescence microscopy (Pogliano et al, 1999; Perez et al, 2000; Abanes-De Mello et al, 2002). We constructed strains expressing either GFP-SpoIIQ or MalF-GFP in spoIIP (AR139 or AR154), spoIID (AR137 or AR152) or spoIIM (AR140 or AR153), and also in a strain lacking all three engulfment proteins. In strains lacking any one or all three of the engulfment proteins, GFP-SpoIIQ localized to the septum as in wild type, forming a focus and lining the septum (Figure 5C; quantitation shown in Supplementary Figure 3). However, movement around the forespore was abolished, and GFP-SpoIIQ stayed in the septum without moving into the bulge (Figure 5D; 69% in spoIIP, 58% in spoIID and 79% in spoIIM). In contrast, MalF-GFP was initially enriched in the septum (Figure 5I, arrow), but then moved around the forespore and into the bulge (Figure 5J; 72% in spoIIP, 63% in spoIID and 83% in spoIIM). Thus, movement of SpoIIQ, but not MalF, throughout the forespore membrane is coupled to migration of the mother cell membrane during engulfment.
Figure 5.
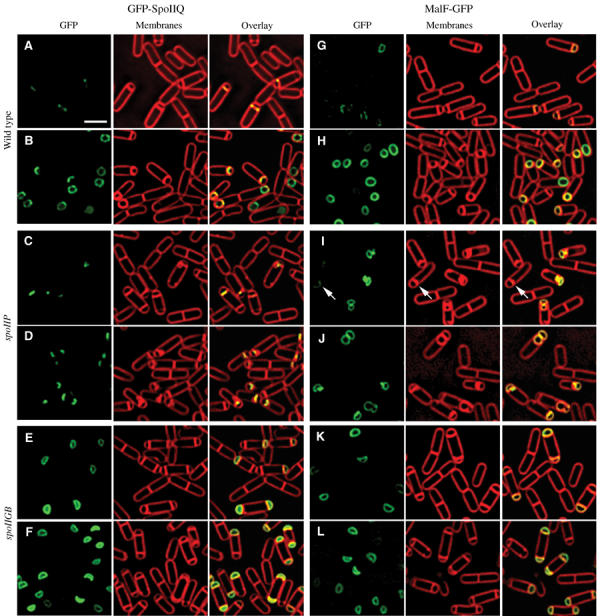
Localization of GFP-SpoIIQ and MalF-GFP in different spo backgrounds. Samples were taken at t2 (A, C, E, G, I, K) and t3 (B, D, F, H, J, L) after resuspension and stained with FM 4-64 (red, middle column). GFP images (green) are on the left with overlaid images on the right. (A) AR126 (spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat) at t2 and (B) t3. (C) AR139 (spoIIQ∷spc spoIIP∷tet amyE∷PspoIIQgfp-spoIIQΩcat) at t2 and (D) t3. GFP-SpoIIQ remains at the septum. (E) AR156 (spoIIQ∷spc spoIIGB∷erm amyE∷PspoIIQgfp-spoIIQΩcat) at t2 and (F) t3. GFP-SpoIIQ diffuses throughout the forespore membrane. (G) AR4 (amyE∷PspoIIQmalF12-gfpΩcat) at t2 and (H) t3. (I) AR154 (spoIIP∷tet amyE∷PspoIIQmalF12-gfpΩcat) at t2 and (J) t3. The arrow in (I) shows enrichment at the septum. (K) AR16 (spoIIGB∷erm amyE∷PspoIIQmalF12-gfpΩcat) at t2 and (L) t3. Scale bar, 2 μm.
Retention of GFP-SpoIIQ in the septum requires a mother cell-expressed protein
One simple mechanism for a forespore-specific protein to track the engulfing mother cell membrane would be for its extracellular domain to interact with that of a mother cell-specific membrane protein. This zipper-like interaction would allow both proteins to localize to the boundary between the two cells, the septum. We therefore tested if retention of SpoIIQ in the septum required mother cell-specific gene expression, by localizing GFP-SpoIIQ in a strain lacking σE. Such mutants fail to initiate engulfment and divide at a second potential division site within the mother cell, producing sporangia with two forespores and a single anucleate mother cell (Kenney and Moran, 1987). Strikingly, in the absence of σE-directed gene expression, GFP-SpoIIQ freely diffused throughout the forespore membrane, identical to MalF-GFP (Figure 5E, F, K and L; quantitation in Supplementary Figure 3). These results suggest that a σE-dependent protein is required to tether GFP-SpoIIQ to the sporulation septum, as well as to mediate its assembly into foci and hoops.
SpoIIR
The initial localization of integral membrane proteins expressed in the forespore to the septum suggests that membrane protein insertion is limited to this site, and that there is a barrier to diffusion away from the septal membrane domain. This possibility could explain a puzzling aspect of cell-specific gene expression: mother cell-specific gene expression requires activation of a sigma factor produced before polar septation, pro-σE, as well as a secreted protein produced in the forespore, SpoIIR (Karow et al, 1995), and yet only adjacent daughter cells activate σE (Fujita and Losick, 2002). While purified SpoIIR can be added to cultures to allow premature activation of σE in cells that have not yet synthesized the polar septum, this normally never occurs (Hofmeister et al, 1995). It has been proposed, but never demonstrated, that SpoIIR secretion is restricted to the sporulation septum, thereby preventing release of SpoIIR into the culture supernatant and the consequent aberrant activation of σE in neighboring cells.
We therefore localized SpoIIR, using a functional N-terminal GFP fusion to SpoIIR (Materials and methods), which fully complemented a spoIIR null mutation. Sporulation was induced by resuspension and GFP-SpoIIR localized at t2 and t3 (Figure 6A and B), respectively. In early sporangia, GFP-SpoIIR first localized as a focus at the septal midpoint (Figure 6A, arrow 1), similar to GFP-SpoIIQ. Later, GFP-SpoIIR lined the septum, moving along with the mother cell membranes (Figure 6A, arrows 2 and 3). GFP-SpoIIR was only transiently present during sporulation, and as engulfment neared completion no GFP fluorescence was observed, suggesting that GFP-SpoIIR is completely degraded (Figure 6B, arrow 4). We were unable to detect GFP-SpoIIR by Western blot analysis, likely because of its low expression relative to SpoIIQ (Karow et al, 1995).
Figure 6.
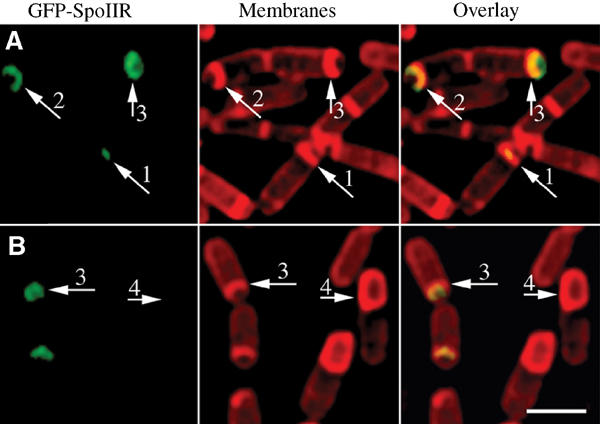
Localization of GFP-SpoIIR. Samples were taken at t2 (A) and t3 (B) and stained with Mitotracker Red (red). Arrows 1 and 2 represent early sporangia, whereas arrows 3 and 4 represent late sporangia. GFP-SpoIIR (green) first localizes as a focus in the middle of the sporulation septum, and tracks with the engulfing mother cell membrane, but fluorescence is rapidly lost. Scale bar, 2 μm.
Discussion
The results presented here provide evidence that localization of proteins to the bacterial septum can be mediated by an extremely simple mechanism involving an interaction between extracellular domains of proteins located on either side of the septum (Figure 7). During sporulation, these zipper-like interactions provide an effective means for forespore proteins to track the engulfing mother cell membrane, and for mother cell proteins to be retained in the septum (Figure 7A–D). If not, as is the case with GFP-SpoIIQ in the absence of its mother cell-specific ligand or a non-native protein (MalF-GFP), the protein is freely diffusible in the forespore membrane (Figure 7E). The case we describe here likely involves at least three different proteins, since septal localization of SpoIIQ requires an unidentified mother cell-expressed protein, and we have recently found that localization of the mother cell-expressed protein SpoIIIAH to the septum requires SpoIIQ. Although the extracellular domains of SpoIIIAH and SpoIIQ directly interact, this interaction is not essential for the SpoIIQ septal retention, suggesting that the primary tether for SpoIIQ is another mother cell-expressed protein (B Blaylock, X Jiang, A Rubio, C Moran and K Pogliano, in preparation). We note that a similar mechanism could mediate septal localization in nondifferentiating bacterial cells, since interactions between the extracellular domains of cytoplasmic membrane proteins would be restricted to membrane invaginations, such as the septum.
Figure 7.
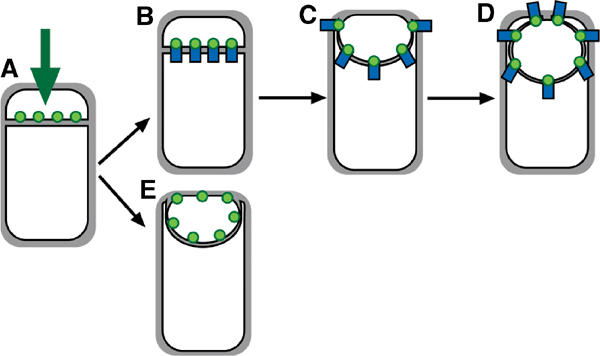
Model for localization of forespore membrane proteins. (A) Any membrane protein (green) synthesized by a σF-dependent promoter inserts directly into the forespore septal domain. (B–D) Localized proteins (such as SpoIIQ) are retained through an interaction with a partner protein (blue) made in the mother cell or the septal peptidoglycan. (E) Nonlocalized proteins (such as MalF-GFP) freely diffuse throughout the forespore membrane.
It does not appear that this zipper-like interaction directs the initial localization of forespore-expressed membrane proteins to the septum, since we have found that any membrane protein expressed in the forespore is initially localized to the septum (Figure 7A), prior to diffusing throughout the rest of the forespore membrane. Indeed, there seem to be only two requirements for the initial localization of proteins to the forespore septum: a σF-dependent promoter and one or more membrane-spanning segments. These results seem most consistent with directed insertion of membrane proteins into the forespore septum followed by their slow diffusion throughout the forespore membrane (Figure 7A and E) since we found that the heterologous GFP fusion proteins (i.e. MalF-GFP), after their direct insertion into the septum, freely diffuse in the forespore membrane. However, it remains possible that membrane protein insertion occurs throughout the forespore membrane and newly synthesized proteins either quickly diffuse to the septum and are degraded everywhere except the septum, or that the folding of GFP occurs most rapidly at the septal membrane domain. While further experiments are necessary to discriminate among these models, we favor the directed insertion hypothesis, because the forespore is a unique and tiny bacterial cell that likely consists of one cell pole and a newly synthesized septum. Prior studies have demonstrated that bacterial cell poles are static structures, with little turnover of surface proteins, membrane or cell wall material compared to the longitudinal envelope (Anderson et al, 1978; Mobley et al, 1984; Koch and Burdett, 1986; Clarke-Sturman et al, 1989; Merad et al, 1989). Thus, if the forespore has only a small volume of longitudinal envelope, the septum might comprise the bulk of its metabolically active cell envelope, thereby becoming the preferred site of membrane protein insertion. Regardless of the precise mechanism, our results suggest that in the forespore newly synthesized membrane proteins are initially restricted to the sporulation septum and later either captured at a specific location or released.
One clear mechanism by which membrane proteins could be restricted to the septum would be if the membrane protein biogenesis machinery were localized to the septum, or if there were a sporulation-specific protein insertion complex. We tested if the known or likely components of the membrane protein insertion machinery are localized to the septum by constructing a functional GFP fusion to SecDF however, this protein was uniformly distributed throughout the cell membrane during both vegetative growth and sporulation, with no quantifiable enrichment at either vegetative or sporulation septa (A Rubio, X Jiang and K Pogliano, in preparation). Recently, it has been shown by Brandon et al (2003) that in E. coli, SecYEG is uniformly distributed throughout the membrane. We found the same to be true of two candidates for a sporulation-specific protein insertion complex, SpoIIIJ and YqjG, which share homology with YidC from E. coli, a protein required for membrane protein insertion (Samuelson et al, 2000; Scotti et al, 2000). However, while SpoIIIJ is essential for sporulation, the available evidence suggests that SpoIIIJ and YqjG are involved in membrane protein folding rather than insertion (Murakami et al, 2002; Serrano et al, 2003).
We do not believe it is likely that newly synthesized membrane proteins are restricted to the sporulation septum in the much larger mother cell, which has a large area of longitudinal cell envelope. Our attempts to test directly the localization of mother cell-expressed membrane proteins, such as MalF, failed, because the hybrid proteins failed to reach high steady-state levels. However, Rudner and Losick have presented evidence that in the mother cell, septal localization is mediated by the diffusion of randomly inserted membrane proteins followed by their capture at the septum (Rudner et al, 2002). In addition, we have also obtained evidence that SpoIIIAH is randomly inserted into the mother cell membrane, and only later retained in the septum by its interaction with SpoIIQ (B Blaylock, X Jiang, A Rubio, C Moran and K Pogliano, in preparation), in support of the diffusion and capture model.
We propose that initial localization of forespore-expressed exported proteins to the septum plays two crucial roles in the establishment of polarity. First, the initial restriction of newly synthesized membrane proteins to the septum would increase the local concentrations of proteins (such as SpoIIQ) required for septal localization of mother cell-expressed proteins (such as SpoIIIAH), thereby increasing the affinity of the mother cell proteins for the septum. Second, restriction of forespore membrane proteins to the septum could also limit the forespore to mother cell signaling event necessary for the onset of mother cell-specific gene expression. This crucial event requires the forespore-expressed and secreted protein SpoIIR, which can be added to the culture medium to mediate the premature activation of mother cell transcription in sporangia lacking a sporulation septum (Hofmeister et al, 1995). Based on our localization studies of GFP-SpoIIR, we propose that the restriction of SpoIIR secretion to the sporulation septum prevents its release into the culture supernatant, which would have devastating effects on the timing of mother cell-specific gene expression.
Our results also demonstrate that SpoIIQ displays a remarkably dynamic and complex localization pattern consistent with its multiple roles during sporulation. It is interesting that during the punctate and soluble stages of GFP-SpoIIQ localization, the forespore is hexagonal, with GFP-SpoIIQ assembling helical arcs at the vertices. This structure is reminiscent of the actin-like proteins MreB and Mbl, and the helical incorporation of peptidoglycan dictated by Mbl (Jones et al, 2001; Carballido-Lopez and Errington, 2003; Daniel and Errington, 2003). Therefore, it is possible that the assembly of SpoIIQ into arcs is mediated either by MreB or Mbl, or the peptidoglycan itself, along which the engulfing membrane has been proposed to track (Illing and Errington, 1991; Kellner et al, 1996; Pogliano et al, 1999; Abanes-De Mello et al, 2002).
Previous studies demonstrated that under certain growth conditions SpoIIQ is required for the phagocytosis-like process of engulfment, with mutants unable to allow membrane migration across the cell pole. However, SpoIIQ is essential for forespore-specific gene expression under all conditions, as it is required for both the synthesis and activation of the second forespore-specific transcription factor σG (Londono-Vallejo et al, 1997; Sun et al, 2000). Interestingly, both of these latter functions require a signal from the mother cell to the forespore: expression of the gene encoding σG requires an unidentified mother cell-expressed protein (Evans et al, 2003), while the activation of σG requires both the completion of engulfment, a process that depends on several mother cell-expressed proteins (SpoIID, M, P), and the eight mother cell-expressed proteins encoded by the spoIIIA operon (Illing and Errington, 1991; Kellner et al, 1996; Pogliano et al, 1999; Abanes-De Mello et al, 2002). Clearly, SpoIIQ is poised to play a key role in both of these functions, as it interacts with at least two mother cell-expressed proteins: its unidentified tether and SpoIIIAH (tethered by SpoIIQ). We postulate that the striking SpoIIQ assemblage described here represents an intracellular signal transduction system comprised of proteins made in both the mother cell and the forespore, coupling the completion of engulfment to cell-specific gene expression.
Materials and methods
Bacterial strains, genetic manipulations and growth conditions
B. subtilis strains listed in Table I are derivatives of wild-type strain PY79 (Youngman et al, 1984) and were constructed by standard methods (Dubnau and Davidoff-Abelson, 1971). Construction of GFP fusions is described below with plasmids listed in Table I; all plasmids were sequenced by the Shared Resource UCSD Cancer Center (funded in part by NCI Cancer Center Support Grant #2 P30 CA23100-18). The GFP fusion protein encoding plasmids were introduced into the B. subtilis chromosome by marker replacement at the amyE locus, selecting chloramphenicol resistance (5 μg/ml) and confirmed by spectinomycin sensitivity (100 μg/ml). Strains carrying these fusions produced wild-type levels of heat-resistant spores (∼108 spores/ml). Sporulation was induced by resuspension with t representing the hours after onset of sporulation (Sterlini and Mandelstam, 1969). Sporulation efficiency was determined after 24 h of sporulation at 37oC, by heating samples to 80oC for 20 min, plating on rich media and quantifying the number of heat-resistant spores per milliliter.
Table 1.
B. subtilis strains and plasmids used in this study
| Strain | Genotype | Source |
|---|---|---|
| PY79 | Youngman et al (1984) | |
| KP8 | spoIID∷Tn917ΩHU298 | Lopez-Diaz et al (1986) |
| KP161 | spoIIGB∷erm | Kenney and Moran (1987) |
| KP298 | amyE∷sspE(2G)-lacZΩcat spoIIIG∷neo | Oke and Losick (1993) |
| KP513 | spoIIP∷tet | Frandsen and Stragier (1995) |
| KP519 | spoIIM∷Tn917ΩHU287 | Smith et al (1993) |
| KP575 | spoIIQ∷spc | Londono-Vallejo et al (1997) |
| AR4 | amyE∷PspoIIQmalF12-gfpΩcat | This work |
| AR6 | amyE∷PspoIIQgfpΩcat | This work |
| AR16 | spoIIGB∷erm amyE∷PspoIIQmalF12-gfpΩcat | This work |
| AR29 | amyE∷PspoIIQlacY12-gfpΩcat | This work |
| AR31 | amyE∷PspoIIQaraP12-gfpΩcat | This work |
| AR77 | amyE∷PspoIIRgfpΩcat | This work |
| AR78 | amyE∷PspoIIRmalF12-gfpΩcat | This work |
| AR90 | amyE∷PcsfBmalF12-gfpΩcat | This work |
| AR92 | amyE∷PcsfBgfpΩcat | This work |
| AR104 | amyE∷PspoIIQsecY12-gfpΩcat | This work |
| AR111 | amyE∷PspoIIQmotA12-gfpΩcat | This work |
| AR126 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat | This work |
| AR130 | spoIIR∷kan | Karow et al (1995) |
| AR132 | spoIIR∷kan amyE∷PspoIIRgfp-spoIIRΩcat | This work |
| AR137 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat | This work |
| spoIID∷ Tn917ΩHU298 | ||
| AR139 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat spoIIP∷tet | This work |
| AR140 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat | This work |
| spoIIM∷Tn917ΩHU287 | ||
| AR151 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat spoIIIG∷neo | This work |
| AR152 | amyE∷PspoIIQmalF12-gfpΩcat spoIID∷ Tn917ΩHU298 | This work |
| AR153 | amyE∷PspoIIQmalF12-gfpΩcat spoIIM∷Tn917ΩHU287 | This work |
| AR154 | amyE∷PspoIIQmalF12-gfpΩcat spoIIP∷tet | This work |
| AR156 | spoIIQ∷spc amyE∷PspoIIQgfp-spoIIQΩcat spoIIGB∷erm | This work |
| AR161 | amyE∷PspoIIQmalF12-gfpΩcat spoIIIG∷neo | This work |
| Plasmid |
Insert |
Source |
| pMDS12 | amyE∷gfpΩcat | Sharp and Pogliano (2002) |
| pMDS13 | amyE∷PspoIIQgfpΩcat | Sharp and Pogliano (2002) |
| pMDS78 | amyE∷PspoIIRgfpΩcat | Sharp and Pogliano (2002) |
| pAR1 | amyE∷PspoIIQmalF12-gfpΩcat | This work |
| pAR5 | amyE∷PspoIIQlacY12-gfpΩcat | This work |
| pAR7 | amyE∷PspoIIQaraP12-gfpΩcat | This work |
| pAR22 | amyE∷PspoIIRmalF12-gfpΩcat | This work |
| pAR23 | amyE∷PcsfBgfpΩcat | This work |
| pAR28 | amyE∷PcsfBmalF12-gfpΩcat | This work |
| pAR34 | amyE∷PspoIIQsecY12-gfpΩcat | This work |
| pAR39 | amyE∷PspoIIQmotA12-gfpΩcat | This work |
| pAR46 | amyE∷PspoIIRgfp-spoIIRΩcat | This work |
| pAR47 | amyE∷PspoIIQgfp-spoIIQΩcat | This work |
GFP fusion constructions
Heterologous GFP fusions were constructed by amplification of membrane-spanning segments 1 and 2 from E. coli membrane proteins MalF, LacY, SecY and MotA and B. subtilis membrane protein AraP, as predicted by HMMTOP and TMpred programs (Tusnady and Simon, 2001). All PCR products contained flanking BamHI restriction sites for ease of cloning into existing GFP-containing plasmids (Sharp and Pogliano, 2002) and resulted in plasmids with membrane-spanning segments upstream and in frame with gfp (see Table I). GFP-SpoIIQ was constructed by PCR amplification from PY79 using primers 5′-CAGAATGTTGCGGCCGCGGGGTGCAACAATGAGAGAG-3′ and 5′-GATAGACGTGCGGCCGCAATTAAGACTGTTCAGT-3′ (NotI restriction site shown in bold). This fragment was then digested with NotI and ligated to an existing GFP-containing plasmid (pMDS13) digested with EagI (Sharp and Pogliano, 2002) to yield pAR47. This plasmid, which has the spoIIQ promoter, translational initiation site and first six codons driving expression of gfp fused in frame with the rest of spoIIQ, was transformed into KP575, producing AR126. GFP-SpoIIR was constructed by PCR amplification using primers 5′-CACGGACGGCCGCGGTGGGGAGCGATGAAAAAAACA-3′ and 5′-TTTTAGACCCGGCCGTTAGGAAAAAAG-3′ (EagI restriction site shown in bold). This fragment and pMDS78 were digested with EagI and ligated to yield pAR46, which has the spoIIR promoter driving expression of gfp fused in frame with spoIIR. pAR46 was transformed into AR130, producing AR132.
Construction of myc-tagged SpoIIQ
Primers 5′-AAGGAATTCACTTAGCTGAGAAAGACG-3′ (EcoRI restriction site shown in bold) and 5′-TTTCTCGAGAGACTGTTCAGTGTCTTC-3′ (AvaI restriction site shown in bold) were used to amplify a 486 bp fragment from PY79 corresponding to the 3′ end of spoIIQ. The vector, pKL94 (Perez et al, 2000) and this PCR fragment were digested with EcoRI and AvaI and ligated to yield pAR50, with the 3′ end of spoIIQ in frame with the c-myc epitope. Transformation of pAR50 into PY79 via a single homologous recombination event integrating the spoIIQ-myc-containing plasmid at the chromosomal spoIIQ locus, resulted in strain AR190.
Microscopy and image analysis
For GFP visualization, live cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.2 μg/ml; Molecular Probes) and Mitotracker Red (0.1 μg/ml; Molecular Probes) or with DAPI (0.2 μg/ml) and FM 4-64 (5 μg/ml; Molecular Probes) as described (Sharp and Pogliano, 1999), and images were collected with an Applied Precision optical sectioning microscope. GFP enrichment at the forespore septal membrane was quantified using the ‘edit polygon' function of Delta Vision software. Briefly, GFP fluorescence at the septum, pole and mother cell cytoplasmic membrane was compared to the FM 4-64- or Mitotracker Red-stained membranes (Pogliano et al, 1999, 2002), after manually drawing polygons. The integrated intensity of each region from 15 cells was determined using the ‘polygon statistics' function. Immunofluorescence microscopy was performed as described (Perez et al, 2000).
Western blot analysis
Samples were prepared as described (Pogliano et al, 1997), heated at 50oC for 10 min, loaded on a 12.5% SDS–polyacrylamide gel and transferred to nitrocellulose (Perez et al, 2000). A 1:500 dilution of mouse monoclonal anti-GFP antibodies (Roche) was used, followed by a 1:500 dilution of horseradish peroxidase-labeled anti-mouse antibodies, and enhanced chemiluminescence (ECL, Amersham) was used for detection.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplemental movie. Three dimentional modeling of GFP-SpoIIQ. A 180° rotation of GFP-SpoIIQ sporangia imaged as in Supplemental figure 2, but showing DAPI-stained chromosomes rather than membrane staining. The Quick Projection function of the SoftWoRx program (Applied Precision) was used to generate the rotation, which was imported into QuickTimePro.
Acknowledgments
We thank Xin Jiang for many helpful discussions and Joe Pogliano for critical reading of the manuscript. This work was supported by the National Science Foundation (NSF 0135955) and the National Institutes of Health (GM57045). AR was supported by a National Science Foundation Minority Postdoctoral Fellowship (DBI-0109229).
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K (2002) A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev 16: 3253–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall SG, Holland B (2002) The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J Mol Biol 318: 219–236 [DOI] [PubMed] [Google Scholar]
- Anderson AJ, Green RS, Sturman AJ, Archibald AR (1978) Cell wall assembly in Bacillus subtilis: location of wall material incorporated during pulsed release of phosphate limitation, its accessibility to bacteriophages and concanavalin A, and its susceptibility to turnover. J Bacteriol 136: 886–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F, Guerout-Fleury AM, Barak I, Stragier P (1999) The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol Microbiol 31: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Ausmees N, Jacobs-Wagner C (2003) Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol 57: 225–247 [DOI] [PubMed] [Google Scholar]
- Bath J, Wu LJ, Errington J, Wang JC (2000) Role of Bacillus subtilis SpoIIIE in DNA transport across the mother cell-prespore division septum. Science 290: 995–997 [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Losick R (2002) Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109: 257–266 [DOI] [PubMed] [Google Scholar]
- Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB (2003) IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol 50: 45–60 [DOI] [PubMed] [Google Scholar]
- Carballido-Lopez R, Errington J (2003) The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell 4: 19–28 [DOI] [PubMed] [Google Scholar]
- Clarke-Sturman AJ, Archibald AR, Hancock IC, Harwood CR, Merad T, Hobot JA (1989) Cell wall assembly in Bacillus subtilis: partial conservation of polar wall material and the effect of growth conditions on the pattern of incorporation of new material at the polar caps. J Gen Microbiol 135 (Part 3): 657–665 [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- Driks A (2002) Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol 10: 251–254 [DOI] [PubMed] [Google Scholar]
- Dubnau D, Davidoff-Abelson R (1971) Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor–recipient complex. J Mol Biol 56: 209–221 [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R (2003) The sigmaE regulon and the identification of additional sporulation genes in Bacillus subtilis. J Mol Biol 327: 945–972 [DOI] [PubMed] [Google Scholar]
- Evans L, Clarkson J, Yudkin MD, Errington J, Feucht A (2003) Analysis of the interaction between the transcription factor sigmaG and the anti-sigma factor SpoIIAB of Bacillus subtilis. J Bacteriol 185: 4615–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett P, Eichenberger P, Losick R, Youngman P (2000) The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 97: 8063–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frandsen N, Stragier P (1995) Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol 177: 716–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R (2002) An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol 43: 27–38 [DOI] [PubMed] [Google Scholar]
- Hofmeister AE, Londono-Vallejo A, Harry E, Stragier P, Losick R (1995) Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83: 219–226 [DOI] [PubMed] [Google Scholar]
- Illing N, Errington J (1991) The spoIIIA operon of Bacillus subtilis defines a new temporal class of mother-cell-specific sporulation genes under the control of the sigma E form of RNA polymerase. Mol Microbiol 5: 1927–1940 [DOI] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L (2001) Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA 98: 4095–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, Lackner LL, de Boer PA (2002) Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J Bacteriol 184: 2951–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Justice SS, Garcia-Lara J, Rothfield LI (2000) Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol Microbiol 37: 410–423 [DOI] [PubMed] [Google Scholar]
- Karow ML, Glaser P, Piggot PJ (1995) Identification of a gene, spoIIR, that links the activation of sigma E to the transcriptional activity of sigma F during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 92: 2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner EM, Decatur A, Moran CP Jr (1996) Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol 21: 913–924 [DOI] [PubMed] [Google Scholar]
- Kenney TJ, Moran CP Jr (1987) Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol 169: 3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch AL, Burdett ID (1986) Biophysics of pole formation of Gram-positive rods. J Gen Microbiol 132 (Part 12): 3451–3457 [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R (1994) Characterization of a cell division gene from Bacillus subtilis that is required for vegetative and sporulation septum formation. J Bacteriol 176: 1451–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C, Stragier P (1997) SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol 24: 29–39 [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J (1986) spoIID operon of Bacillus subtilis: cloning and sequence. J Gen Microbiol 132 (Part 2): 341–354 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J (2002) Unexpected twist to the Z ring. Dev Cell 2: 519–521 [DOI] [PubMed] [Google Scholar]
- Margolin W (2002) Bacterial sporulation: FtsZ rings do the twist. Curr Biol 12: R391–R392 [DOI] [PubMed] [Google Scholar]
- Marston AL, Errington J (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol 33: 84–96 [DOI] [PubMed] [Google Scholar]
- Merad T, Archibald AR, Hancock IC, Harwood CR, Hobot JA (1989) Cell wall assembly in Bacillus subtilis: visualization of old and new wall material by electron microscopic examination of samples stained selectively for teichoic acid and teichuronic acid. J Gen Microbiol 135 (Part 3): 645–655 [DOI] [PubMed] [Google Scholar]
- Mobley HL, Koch AL, Doyle RJ, Streips UN (1984) Insertion and fate of the cell wall in Bacillus subtilis. J Bacteriol 158: 169–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Haga K, Takeuchi M, Sato T (2002) Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J Bacteriol 184: 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oke V, Losick R (1993) Multilevel regulation of the sporulation transcription factor sigma K in Bacillus subtilis. J Bacteriol 175: 7341–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K (2000) SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J Bacteriol 182: 1096–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J (2001) Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol 183: 6630–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K (1999) A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol Microbiol 31: 1149–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Sharp MD, Pogliano K (2002) Partitioning of chromosomal DNA during establishment of cellular asymmetry in Bacillus subtilis. J Bacteriol 184: 1743–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K, Harry E, Losick R (1995) Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol 18: 459–470 [DOI] [PubMed] [Google Scholar]
- Pogliano K, Hofmeister AE, Losick R (1997) Disappearance of the sigma E transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol 179: 3331–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R (2002) A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization. Genes Dev 16: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM (2002) Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci USA 99: 8701–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE (2000) YidC mediates membrane protein insertion in bacteria [see comments]. Nature 406: 637–641 [DOI] [PubMed] [Google Scholar]
- Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J (2000) YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J 19: 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Corte L, Opdyke J, Moran CP Jr, Henriques AO (2003) Expression of spoIIIJ in the prespore is sufficient for activation of sigma G and for sporulation in Bacillus subtilis. J Bacteriol 185: 3905–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc Natl Acad Sci USA 96: 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K (2002) Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295: 137–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Bayer ME, Youngman P (1993) Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol 175: 3607–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J (1969) Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R (1996) Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet 30: 241–297 [DOI] [PubMed] [Google Scholar]
- Sun YL, Sharp MD, Pogliano K (2000) A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J Bacteriol 182: 2919–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17: 849–850 [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R (1984) A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet 195: 424–433 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplemental movie. Three dimentional modeling of GFP-SpoIIQ. A 180° rotation of GFP-SpoIIQ sporangia imaged as in Supplemental figure 2, but showing DAPI-stained chromosomes rather than membrane staining. The Quick Projection function of the SoftWoRx program (Applied Precision) was used to generate the rotation, which was imported into QuickTimePro.


