Abstract
Certain strains of Enterococcus faecium and Enterococcus faecalis contribute beneficially to animal health and food production, while others are associated with nosocomial infections. To determine whether there are structural and functional genomic features that are distinct between nonclinical (NC) and clinical (CL) strains of those species, we analyzed the genomes of 31 E. faecium and 38 E. faecalis strains. Hierarchical clustering of 7,017 orthologs found in the E. faecium pangenome revealed that NC strains clustered into two clades and are distinct from CL strains. NC E. faecium genomes are significantly smaller than CL genomes, and this difference was partly explained by significantly fewer mobile genetic elements (ME), virulence factors (VF), and antibiotic resistance (AR) genes. E. faecium ortholog comparisons identified 68 and 153 genes that are enriched for NC and CL strains, respectively. Proximity analysis showed that CL-enriched loci, and not NC-enriched loci, are more frequently colocalized on the genome with ME. In CL genomes, AR genes are also colocalized with ME, and VF are more frequently associated with CL-enriched loci. Genes in 23 functional groups are also differentially enriched between NC and CL E. faecium genomes. In contrast, differences were not observed between NC and CL E. faecalis genomes despite their having larger genomes than E. faecium. Our findings show that unlike E. faecalis, NC and CL E. faecium strains are equipped with distinct structural and functional genomic features indicative of adaptation to different environments.
INTRODUCTION
Enterococcus faecium and Enterococcus faecalis are Gram-positive bacteria in the Firmicutes phylum and are found on plants, in foods, and in the gastrointestinal tracts (GIT) of animals (1). These species are members of the polyphyletic group of bacteria known as lactic acid bacteria and have important roles in food and beverage fermentations. Certain strains of E. faecium confer beneficial or probiotic effects on animal and human health (2). Conversely, strains of E. faecium and E. faecalis are also associated with nosocomial infections resulting in endocarditis and bacteremia and represent a significant reservoir of antibiotic resistance genes (2).
The genetic features of E. faecium and E. faecalis were investigated previously to identify lineages specific to community and clinical environments (3–7). Virulence factors (VF), antibiotic resistance (AR) genes, mobile genetic elements (ME), and multilocus sequence typing (MLST) patterns are associated with the potential of E. faecium and E. faecalis to cause disease in humans (8, 9). Despite such studies, the structural and functional features of enterococcal genomes are not fully understood. We hypothesized that the lineage-specific differences observed previously were only a fraction of the greater, more significant distinctions between nonclinical (NC) and clinical (CL) strains, and the opposing environmental and health-specific associations are the result of broader niche-specific adaptations that can be observed in the genomes of those species. Thus, we compared structural and functional genomic features between NC and CL strains of E. faecium and E. faecalis.
MATERIALS AND METHODS
Identification of E. faecium and E. faecalis orthologous CDS.
All E. faecium and E. faecalis nucleotide sequences and annotations available at GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) were retrieved in the GenBank format in February 2012 and included complete and incomplete genomes, as well as nucleotide sequences individually deposited from other isolates (see Fig. S1 in the supplemental material). For identification of orthologs, the protein coding sequences (CDS) in the annotations were filtered to remove CDS containing premature stop codons (pseudogenes). Each CDS was then aligned to the entire CDS pool (which included pseudogenes) using GASSST (10) according to nucleotide sequence identity (−p 85, meaning ≥85% identity) and best sensitivity (−s 5). The aligned CDS were regarded as one ortholog, and the consensus sequence of each ortholog was determined using the CAP3 assembler with default options (11). The resulting E. faecium and E. faecalis orthologous CDS collections included all consensus CDS for the species.
Detection of CDS, VF, AR, and ME genes in each genome.
A total of 31 E. faecium and 38 E. faecalis genomes were retrieved from GenBank in May 2012 (Tables 1 and 2). Contigs for each genome sequence were fragmented sequentially into 50 bp at intervals of 7 bp, and each DNA fragment was aligned onto E. faecium and E. faecalis orthologous CDS using GASSST (10). Alignment coverage per gene was calculated, and genes highly covered by the fragments (≥90% of CDS length) were recognized to be present in the genome. The DNA fragments were also aligned onto nucleotide sequences of VF genes extensively studied for both species (8, 12), AR genes from the Antibiotic Resistance Genes Database (ARDB) (13), and ME genes for gene identification in the enterococcal genomes (see the supplemental materials for a list of AR and VF genes examined). Loci annotated as phage proteins, transposons, transposases, integrases, and insertion sequences (IS) were identified in the E. faecium genomes and collectively regarded as candidate ME genes.
TABLE 1.
E. faecium isolates and their genomes used in this study
| Group | Strain | Isolation origin(s) | MLSTa | Countryb (state) | Yrb | GenBank accession no. | Genome sizec (kb) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| NC | Com12 | Human, healthy volunteer, feces | ST107 | USA (MA) | 2006 | ACBC00000000 | 2,685 | 48 |
| NC | Com15 | Human, healthy volunteer, feces | ST583 | USA (MA) | 2006 | ACBD00000000 | 2,771 | 48 |
| NC | E1039 | Human, healthy volunteer, feces | ST42 | Netherlands | 1998 | ACOS00000000 | 2,503 | 39 |
| NC | E1071 | Human, hospital patient, not related to enterococcal infection outbreak, feces | ST32 | Netherlands | 2000 | ABQI00000000 | 2,701 | 39 |
| NC | E4452 | Dog, community, ampicillin-resistant enterococcus, rectal swabs | ST266 | Netherlands | 2008 | AEOU00000000 | 2,769 | 49 |
| NC | E4453 | Dog, community, ampicillin-resistant enterococcus, rectal swabs | ST192 | Netherlands | 2008 | AEDZ00000000 | 2,819 | 49 |
| NC | E980 | Human, healthy volunteer, feces | ST94 | Netherlands | 1998 | ABQA00000000 | 2,793 | 39 |
| NC | NRRL B-2354 | Food, milk | ST860d | USA | ≤1947 | CP004063 and CP004064 | 2,850 | 50 |
| NC | PC4.1 | Food, Mongolian yogurt (tarag) | ST720 | ≤2010 | ADMM00000000 | 2,811 | 51 | |
| NC | TX1330 | Human, healthy community volunteer, feces | ST107 | USA (TX) | 1994 | ACHL00000000 | 2,721 | 52 |
| CL | 1,141,733 | Human, clinical isolate, hospitalized patient, blood culture | ST327 | ACAZ00000000 | 2,865 | 48 | ||
| CL | 1,230,933 | Human, clinical isolate, hospitalized patient, wound swab | ST18 | ACAS00000000 | 2,952 | 48 | ||
| CL | 1,231,408 | Human, clinical isolate, hospitalized patient, blood culture | ST582 | ACBB00000000 | 2,889 | 48 | ||
| CL | 1,231,410 | Human, clinical isolate, skin and soft tissue infection | ST17 | ACBA00000000 | 2,944 | 48 | ||
| CL | 1,231,501 | Human, clinical isolate, hospitalized patient, blood culture | ST52 | ACAY00000000 | 2,799 | 48 | ||
| CL | 1,231,502 | Human, clinical isolate, hospitalized patient, blood culture | ST203 | ACAX00000000 | 2,926 | 48 | ||
| CL | Aus0004 | Human, liver transplant recipient, blood | ST17 | Australia | 1998 | CP003351 to CP003354 | 3,020 | 47 |
| CL | C68 | Human, clinical isolate, hospitalized patient, blood culture | ST16 | USA (OH) | 1998 | ACJQ00000000 | 2,941 | 53 |
| CL | D344SRF | Human, isolation site not specified | ST25 | France | 1985 | ACZZ00000000 | 2,745 | 3 |
| CL | DO | Human, endocarditis infection | ST18 | USA (TX) | 1994 | CP003583 to CP003586 | 3,053 | 4 |
| CL | E1162 | Human, clinical isolate, hospitalized patient, blood culture | ST17 | France | 1997 | ABQJ00000000 | 2,711 | 39 |
| CL | E1636 | Human, clinical isolate, hospitalized patient, blood culture | ST106 | Netherlands | 1961 | ABRY00000000 | 2,838 | 39 |
| CL | E1679 | Human, clinical isolate, vascular catheter tip | ST114 | Brazil | 1998 | ABSC00000000 | 2,928 | 39 |
| CL | TX0082 | Human, clinical isolate, endocarditis, blood | ST17 | USA (TX) | 1999 | AEBU00000000 | 2,691 | 52 |
| CL | TX0133A | Human, clinical isolate, blood | ST17 | USA (TX) | 2006 | AECH00000000 | 2,930 | 52 |
| CL | TX0133a01 | Human, clinical isolate, in the inhibition zone, blood | ST17 | USA (TX) | AECJ00000000 | 3,073 | ||
| CL | TX0133a04 | Human, clinical isolate, outside the inhibition zone, blood | ST17 | USA (TX) | AEBC00000000 | 2,922 | 52, 54 | |
| CL | TX0133B | Human, clinical isolate, blood | ST17 | USA (TX) | 2006 | AECI00000000 | 2,927 | 52, 54 |
| CL | TX0133C | Human, clinical isolate, blood | ST17 | USA (TX) | 2006 | AEBG00000000 | 2,907 | 52, 54 |
| CL | U0317 | Human, clinical isolate, urinary tract infection (urine) | ST78 | Netherlands | 2005 | ABSW00000000 | 2,893 | 39 |
| TC6 | Colonizing transconjugant between C68 and D344SRF | ST25 | USA (OH) | ACOB00000000 | 2,884 | 19 |
MLST were assigned according to each genome sequence used in this study. MLST of 24 isolates assigned by us were compared to the reported MLST. Twenty-three MLST were the same as the reported MLST, but that of D344SRF (ST25) was different from the reported MLST (ST21). MLST of 5 strains were assigned for the first time in this study.
Information on isolation country and year (4).
Genome sizes were calculated as the total length of contigs for each genome.
New sequence type assigned during the course of this study (http://efaecium.mlst.net/).
TABLE 2.
E. faecalis isolates used in this study and their genomes
| Group | Strain | Isolation origin(s) | MLSTa | Countryb | Yrb | GenBank accession no. | Genome sizec (kb) | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| NC | 62 | Human, healthy infant, feces | ST66 | Norway | 2002–2003 | CP002491 to CP002495 | 3,131 | 55, 56 |
| NC | D6 | Pig | ST16 | Denmark | ACAT00000000 | 2,887 | 48, 57 | |
| NC | E1Sol | Human, commensal isolate, feces | ST93 | Solomon Islands | 1960s | ACAQ00000000 | 2,853 | 48, 57 |
| NC | Fly1 | Drosophila, commensal isolate | ST101 | USA | 2005 | ACAR00000000 | 2,791 | 48, 57 |
| NC | R712 | Human, feces | ST103 | USA | ADDQ00000000 | 3,037 | 52 | |
| NC | S613 | Human, feces | ST103 | USA | ADDP00000000 | 3,042 | 52 | |
| NC | X98 | Human, infant, feces | ST19 | 1934 | ACAW00000000 | 2,910 | 48, 57 | |
| CL | AR01/DG | Dog, wound isolate, wound | ST108 | New Zealand | 2001 | ACAK00000000 | 2,821 | 57 |
| CL | ATCC 29200 | Human, urogenital secretions | ST21 | Canada | ≤1974 | ACHK00000000 | 2,936 | 52 |
| CL | ATCC 29212 | Human, urine | ST30 | United Kingdom | ≤1903 | ALOD00000000 | 3,062 | 58 |
| CL | ATCC 4200 | Human, rheumatic fever isolate, blood | ST105 | 1926 | ACAG00000000 | 3,009 | 48, 57 | |
| CL | CH188 | Human, clinical isolate, liver | ST9 | USA | 1980s | ACAV00000000 | 3,159 | |
| CL | DAPTO 512 | Human, clinical isolate, blood | ST103 | AEBT00000000 | 3,054 | 52 | ||
| CL | DAPTO 516 | Human, clinical isolate, blood | ST103 | AEBS00000000 | 3,055 | 52 | ||
| CL | HH22 | Human, clinical isolate, urinary isolates | ST6 | USA | ≤1982 | ACIX00000000 | 3,050 | 59 |
| CL | HIP11704 | Human, clinical isolate | ST4 | USA | 2002 | ACAN00000000 | 3,130 | 48, 57 |
| CL | JH1 | Human, clinical isolate | ST40 | United Kingdom | ≤1974 | ACAP00000000 | 2,995 | 48, 57 |
| CL | Merz96 | Human, clinical isolate, blood | ST103 | USA | 2002 | ACAM00000000 | 3,038 | 48, 57 |
| CL | T11 | Human, clinical isolate, urine | ST65 | Japan | ≤1992 | ACAU00000000 | 2,729 | 48, 57 |
| CL | T2 | Human, clinical isolate, urine | ST11 | Japan | ≤1992 | ACAE00000000 | 3,205 | 48, 57 |
| CL | T3 | Human, clinical isolate, urine | ST67 | Japan | ≤1992 | ACAF00000000 | 2,784 | 48, 57 |
| CL | T8 | Human, clinical isolate, urine | ST8 | Japan | ≤1992 | ACOC00000000 | 2,985 | 48, 57 |
| CL | TUSoD Ef11 | Human, cause of urinary tract infections, bacteremia, and infective endocarditis | ST364 | USA | ACOX00000000 | 2,837 | 52 | |
| CL | TX0102 | Human, clinical isolate, blood | ST21 | USA | AEBD00000000 | 2,871 | 52 | |
| CL | TX0104 | Human, endocarditis isolate | ST2 | USA | ACGL00000000 | 3,107 | 6, 52 | |
| CL | TX0109 | Human, clinical isolate, endocarditis, blood | ST59 | USA | AEBY00000000 | 2,967 | 52 | |
| CL | TX0635 | Human, clinical isolate, urogenital tract | ST9 | USA | AEBZ00000000 | 3,168 | 52 | |
| CL | TX0855 | Human, clinical isolate, urogenital tract | ST4 | USA | AEBV00000000 | 2,986 | 52 | |
| CL | TX0860 | Human, clinical isolate, catheter tip, blood | ST11 | USA | AEBX00000000 | 3,062 | 52 | |
| CL | TX2134 | Human, clinical isolate, GIT | ST30 | USA | AEBW00000000 | 3,121 | 52 | |
| CL | TX4248 | Human, clinical isolate, lymph node | ST40 | USA | AEBR00000000 | 3,187 | 52 | |
| CL | V583 | Human, hospitalized patient, blood | ST6 | USA | 1989 | AE016830 to AE016833 | 3,360 | 60 |
| DS5 | ST55 | ≤1974 | ACAI00000000 | 3,128 | 48, 57 | |||
| OG1RF | Laboratory strain | ST1 | USA | ≤1975 | CP002621 | 2,740 | 61 | |
| T1 | Human | ST21 | ≤1950 | ACAD00000000 | 2,906 | 48, 57 | ||
| TX0411 | Human | ST90 | USA | 1954 | AECA00000000 | 3,124 | 52 | |
| TX0470 | Human | ST110 | USA | 1963 | AECC00000000 | 2,877 | 52 | |
| TX1322 | Human | ST64 | USA | 1994 | ACGM00000000 | 2,930 | 52 |
MLST were reassigned using each genome sequence used in this study. MLST of 26 isolates assigned by us were compared to reported MLST. All of them were the same as the reported MLST. MLST of 12 strains were assigned for the first time in this study.
Information on isolation country and year (6).
Genome sizes were calculated as the total length of contigs for each genome.
Analysis of functional categories of genes.
Genome sequences (FASTA files) were uploaded to the RAST server (14) with default options to obtain information on gene functional categories, called subsystems. The gene amounts were counted for each subsystem. Differentially overrepresented gene numbers in each subsystem were examined between NC and CL genomes using Student's t test.
Hierarchical isolate clustering.
The presence or absence of each E. faecium and E. faecalis orthologous CDS in a genome was used for hierarchical isolate clustering using the Euclidean distance method implemented in the R package (15). Existence of NC1, NC2, and CL clades was statistically examined by 10,000 bootstrap resamplings using an R package, Pvclust (16).
MLST of E. faecium and E. faecalis.
Multilocus sequence typing (MLST) assignment was based on partial sequences of the 7 E. faecium housekeeping genes, atpA, adk, ddl, gdh, purK, gyd, and pstS (http://efaecium.mlst.net/) (17), or 7 E. faecalis housekeeping genes, aroE, gdh, gki, gyd, pstS, xpt, and yqiL (http://efaecalis.mlst.net/). The sequences were retrieved from each genome and compared to the MLST database to identify allele types for the 7 genes and assign each strain to a specific MLST group. Concatenated MLST sequences from 31 strains were also aligned to each other and further analyzed for phylogenetic relationships by using the neighbor joining method with 1,000 resamplings.
Phylogenetic analysis of pbp5 sequences.
Full-length pbp5 CDS were retrieved from each E. faecium genome and analyzed by using the neighbor joining method with 1,000 resamplings and the maximum likelihood method with 500 resamplings. MEGA5 was used for the phylogenetic analysis (18).
Detection of NC- and CL-enriched genes.
E. faecium and E. faecalis isolates were divided into NC and CL groups according to isolation origin (Tables 1 and 2). The frequency of each orthologous CDS was enumerated for the NC and CL groups. A significant difference in CDS count between the groups was determined using Fisher's exact test.
PCR detection of NC- and CL-enriched genes.
E. faecium NRRL B-2354 was obtained from the USDA Agricultural Research Service Culture Collection (Peoria, IL; receiving date, 22 July 2011). E. faecium 1,231,502 was provided by Michael Gilmore (Harvard University, Boston, MA), and Lactobacillus plantarum WCFS1 was provided by Michiel Kleerebezem, NIZO Food Research, The Netherlands. E. faecium strains were cultivated in brain heart infusion (BHI) broth (Becton, Dickinson and Company, USA) at 37°C for 8 h without aeration. L. plantarum was cultivated in de Man, Rogosa, and Sharpe (MRS) (Oxoid, England) broth under the same conditions as E. faecium. Cells were harvested by centrifugation at 21,000 × g for 1 min, and genomic DNA was then extracted using the DNeasy blood and tissue kit (Qiagen). The PCR was performed for 3 NC-enriched and 2 CL-enriched genes using GoTaq DNA polymerase (Promega) and 200 nM each gene-specific primer (see Table S1 in the supplemental material) with the following steps: 95°C for 5 min; 35 cycles of 95°C for 30 s, 55°C for 35 s, and 72°C for 45 s; and 72°C for 7 min.
Analysis of gene proximity.
Different types of genes (ME/VF/AR genes and NC-/CL-enriched genes) were located on E. faecium genome sequences using GASSST with ≥90% sequence similarity and best sensitivity. Two genes localized together within 1,000 bp were regarded as colocalized. The colocalized rate (%) was calculated for each genome.
RESULTS
General features of E. faecium and E. faecalis pangenomes.
Orthologous CDS collections were constructed using all publicly available nucleotide sequences for the two species (see Table S2 and Fig. S1 in the supplemental material), including 31 E. faecium and 38 E. faecalis genomes (Tables 1 and 2). According to ortholog clustering based on sequence similarity, a total of 7,017 and 8,032 orthologous CDS were identified for E. faecium and E. faecalis species, respectively (see Table S2). The orthologs were identified using existing genome annotations available in GenBank. CDS collections constructed when all genomes were examined using a common annotation pipeline (RAST) (14) provided similar results (data not shown). E. faecalis CDS contained a greater number of genes with gene designations (1,724 CDS; 21.46%) than E. faecium (1,133 genes; 16.15%). However, the majority of E. faecalis CDS (6,308; 78.54%) and E. faecium CDS (5,884; 83.85%) were unassigned to gene designations. Among the 7,017 E. faecium orthologs, 1,755 core genes (25.0%) were shared by all 31 E. faecium genomes analyzed. The pangenome contained 1,327 accessory genes (18.9%) unique to specific strains and another 240 genes (3.4%) in the public databases not yet associated with published genome sequences (see Fig. S2A in the supplemental material). E. faecalis orthologous CDS comprised a greater number of core genes (2,184 genes; 27.2%) than those of E. faecium. The pangenome contained 1,538 accessory genes (19.1%) unique to specific strains and another 828 genes (10.3%) with single representatives in GenBank (see Fig. S2B).
The average genome size of E. faecium (2,847.2 ± 122.1 kb) was significantly smaller than that of E. faecalis (3,000.8 ± 143.0 kb) (P < 0.001) (Table 3). At 3,122.5 ± 132.1 CDS per genome, E. faecium genomes contained significantly fewer genes than E. faecalis (3,247.6 ± 180.9 CDS per genome; P < 0.01) (Table 3). Although E. faecium genomes harbor fewer AR genes (Table 3), this difference was not sufficient to account for the smaller genome size. Moreover, the number of ME was not significantly different between the two species, and while a greater number of VF genes were identified in E. faecalis genomes, this distinction might be due to gaps in knowledge of enterococcus pathogenesis (8).
TABLE 3.
Genome comparisons between two species and between NC and CL isolatesa
| Parameter | E. faecium (n = 31) | E. faecalis (n = 38) | Significanceb (t test) |
E. faecium |
E. faecalis |
||||
|---|---|---|---|---|---|---|---|---|---|
| NC (n = 10) | CL (n = 20) | Significance | NC (n = 7) | CL (n = 25) | Significance | ||||
| Genome size (kb) | 2,847.2 ± 122.1 | 3,000.8 ± 143.0 | P < 0.001 | 2,742.4 ± 99.4 | 2,897.8 ± 101.5 | P < 0.001 | 2,950.0 ± 121.9 | 3,027.0 ± 144.8 | NS |
| No. of CDS | 3,122.5 ± 132.1 | 3,247.6 ± 180.9 | P < 0.01 | 2,999.7 ± 107.5 | 3,182.0 ± 101.8 | P < 0.001 | 3,187.4 ± 168.6 | 3,276.9 ± 178.2 | NS |
| G+C content (%) | 37.9 ± 0.2 | 37.3 ± 0.2 | P < 0.001 | 38.0 ± 0.1 | 37.8 ± 0.2 | P < 0.05 | 37.4 ± 0.1 | 37.3 ± 0.2 | NS |
| No. of VF genesc | 14.0 ± 3.3 | 44.7 ± 4.1 | NA | 11.4 ± 2.4 | 15.4 ± 2.9 | P < 0.001 | 43.1 ± 3.0 | 45.3 ± 4.4 | NS |
| No. of AR genesd | 2.9 ± 2.3 | 5.7 ± 2.2 | P < 0.001 | 1.3 ± 2.4 | 3.8 ± 1.8 | P < 0.05 | 5.4 ± 2.8 | 6.2 ± 2.0 | NS |
| No. of ME genese | 97.1 ± 35.8 | 107.3 ± 27.8 | NS | 62.0 ± 24.0 | 111.8 ± 26.5 | P < 0.001 | 103.9 ± 30.5 | 110.6 ± 25.7 | NS |
Values are shown as means ± standard deviations.
P values were calculated by Student's t test. NA and NS indicate not analyzed and no significance, respectively.
Comparisons between virulence factor gene numbers were not applicable, because the genes were selected using different criteria depending on the species. Extensively studied VF genes were used for these comparisons (for more detail, see the supplemental material).
Known AR genes from the antibiotic resistance gene database (ARDB) (for more detail, see the supplemental material).
ME genes were regarded to be all phage, transposon, transposase, integrase, or insertion sequences (IS) designated according to the genome annotations.
Hierarchical clustering of E. faecium and E. faecalis based on genome contents.
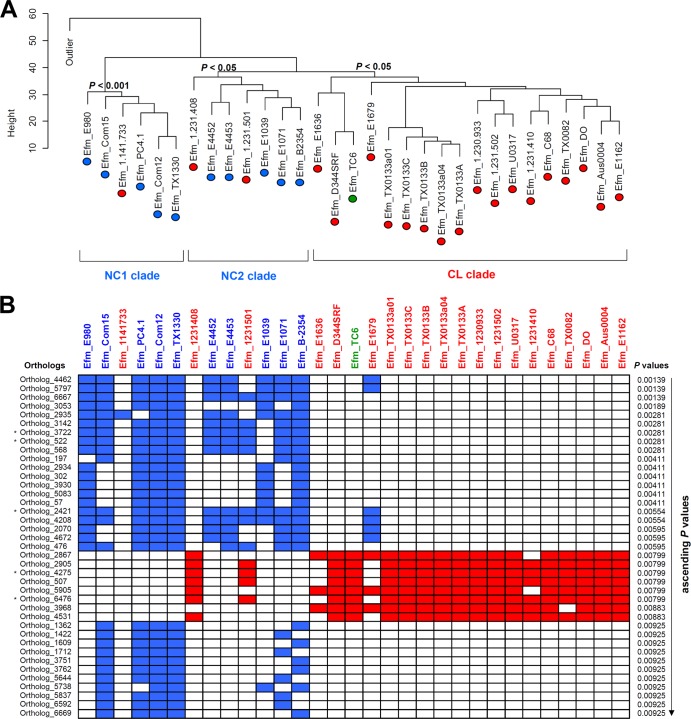
CDS presence/absence was measured for E. faecium and E. faecalis to determine whether community-associated strains form a lineage distinct from those isolated from nosocomial infections. Among the 31 E. faecium genomes available for comparison (Table 1) were 10 NC strains from community origins, including human/animal feces and dairy products, 20 CL strains isolated from human blood and tissues, and 1 strain from an experimental transconjugation between two clinical strains (19). Hierarchical clustering of the 7,017 E. faecium CDS orthologs showed that NC strains clustered into two clades (NC1 and NC2) and were distinct from a CL clade consisting of 17 CL strains and a transconjugant (Fig. 1A). The NC1 clade was also identified by MLST (see Fig. S3 in the supplemental material) and is identical to the NC-enriched clade that was previously found using a concatenated sequence alignment of 299 core orthologous proteins (20). The NC2 clade was not distinguishable by MLST (see Fig. S3) and represents a newly described clade. Despite the good separation between NC and CL strains, three CL strains (1,141,733, 1,231,501, and 1,231,408) were clustered together with NC strains, as shown in other studies (4, 20). The same result was found for 1,141,733 by MLST, and because the other 2 strains were in NC2, they were not distinguishable from CL strains by MLST. Notably, strains from similar origins were not necessarily closely related. For example, two dairy strains, PC4.1 and NRRL B-2354, were located in different NC clades, and one of the five strains from healthy human feces (TX1330) was associated with a different NC clade from the others. Hierarchical clustering was also performed using the 8,032 E. faecalis orthologs identified for 38 E. faecalis genomes representing 7 NC and 25 CL strains. In contrast to E. faecium, no evidence of distinct lineages was found (see Fig. S4).
FIG 1.
Hierarchical clustering of E. faecium isolates and NC-/CL-enriched genes. (A) In the hierarchical clustering of E. faecium isolates, NC isolates (blue ovals) were enriched in clades NC1 and NC2, and CL isolates (red ovals) were enriched in the CL clade. A transconjugant, TC6 (a green oval), was associated with the CL clade. One artificially generated outlier was included to distinguish between E. faecium strains. Height represents a relative distance between strains. A bootstrap analysis was performed to confirm the reliability of the clustering. The resampling size was 10,000, and clusters with P values less than 0.05 were considered present. (B) The abundances of 39 enriched genes in NC or CL isolates were visualized in the heat map. The left side of the box array shows E. faecium orthologous CDS numbers. The P values from Fisher's exact test are shown in an ascending order at the right side of the box array. The upper side of the box array indicates isolate names with origin information: 10 NC isolates (blue), 20 CL isolates (red), and 1 transconjugant (green). The isolate names are listed in the same order as that shown in the hierarchical clustering. Filled blue or red boxes indicate the presence of each NC- or CL-enriched gene, respectively, in a given genome, and white blank boxes indicate gene absence. Asterisks indicate genes that were examined by PCR (see Fig. S5 in the supplemental material).
Structural and functional genomic differences between NC and CL E. faecium strains.
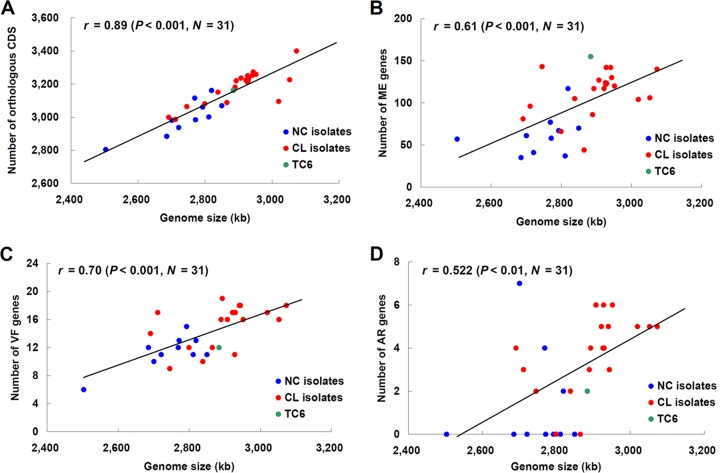
The genomes of E. faecium CL strains were, on average, 155 kb larger than strains isolated from community sources (P < 0.001). CL genomes contained, on average, approximately 182 more genes, and the number of predicted genes was positively correlated with genome size (r = 0.89, P < 0.001) (Fig. 2A). Contributing to the larger genome size of the CL isolates was the higher number of genes coding for VF, AR, and ME (Table 3 and Fig. 2B, C, and D). Approximately 4-, 3-, and 50-more VF, AR, and ME genes were found in the CL genomes, respectively (Table 3). Similar to the lack of evidence for different lineages of E. faecalis based on hierarchical gene clustering, genome-level differences between 7 NC and 25 CL E. faecalis isolates were not found (Table 3).
FIG 2.
Correlation of genome size with numbers of orthologous genes and clinically relevant genes in E. faecium. The numbers of orthologous CDS (A), ME genes (B), VF genes (C), and AR genes (D) in an individual strain are plotted against the genome size of that strain. Pearson's correlation coefficients and P values were calculated and are shown in each scatter plot. The color of each circle indicates NC isolates (blue), CL isolates (red), and a transconjugant (green).
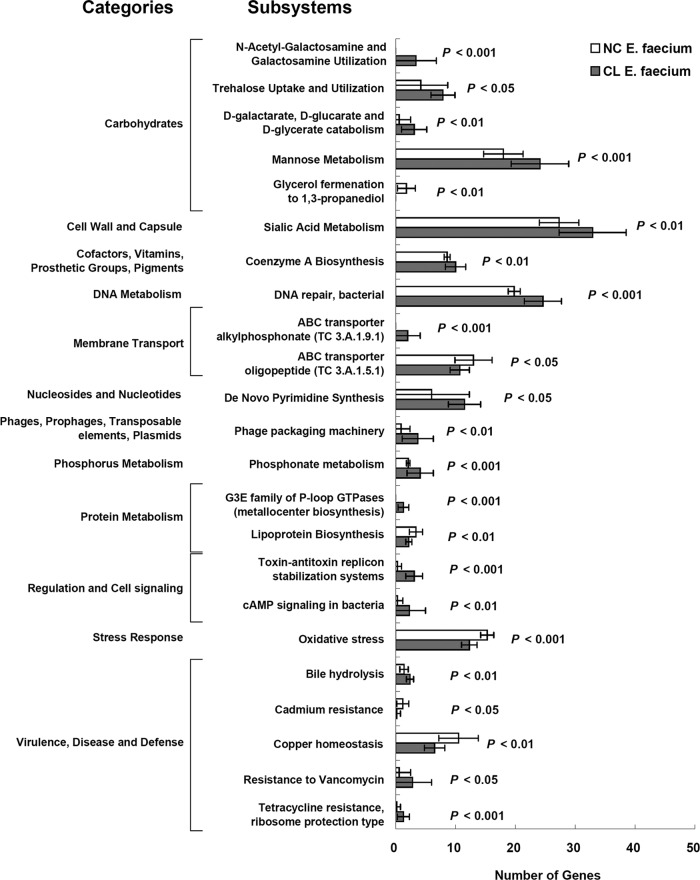
Further characterization of NC and CL genomes showed that E. faecium CL strains have genes overrepresented in 17 RAST subsystems (14). CL strains were found to be enriched in functional gene groups encompassing antibiotic resistance and lysogenic phages (Fig. 3). Genes belonging to N-acetyl-galactosamine and galactosamine utilization, ABC transporter alkylphosphonate (TC 3.A.1.9.1), and the G3E family of P-loop GTPase (metallocenter biosynthesis) subsystems were exclusively found in CL strains. E. faecium NC genomes contained genes related to the glycerol-1,3-propanediol fermentation pathway as well as an overrepresentation of genes in 6 subsystems predominantly involved in metabolism, oxidative stress, and metal homeostasis.
FIG 3.
Comparisons of subsystems between NC and CL E. faecium genomes. Gene abundance in 23 subsystems is shown. P values were calculated according to Student's t test. cAMP, cyclic AMP.
NC- and CL-enriched genes.
Differences among individual orthologous CDS were compared to identify genes that contributed to the genetic separation between the E. faecium NC and CL strains. A total of 68 and 153 genes were significantly enriched among NC and CL E. faecium strains, respectively, at P < 0.05 (Fisher's exact test) (see Table S3 in the supplemental material). At P < 0.01 (Fisher's exact test), a subset of 31 NC- and 8 CL-enriched genes was detected (Fig. 1B and Table 4; also see Table S3). In contrast, CL- and NC-enriched genes were not found in the E. faecalis genomes at P < 0.01 (Fisher's exact test).
TABLE 4.
NC- and CL-enriched genes in E. faecium
| Genea | P valuesb | Product(s) | Strain group enriched |
|---|---|---|---|
| Ortholog_4462 | 0.00139 | Hypothetical protein | NC |
| Ortholog_5797 (fnr-1) | 0.00139 | Crp/Fnr family transcriptional regulator | NC |
| Ortholog_6667 | 0.00139 | Hypothetical protein | NC |
| Ortholog_3053 | 0.00189 | Hypothetical protein | NC |
| Ortholog_2935 | 0.00281 | Hypothetical protein | NC |
| Ortholog_3142 | 0.00281 | Hypothetical membrane protein | NC |
| Ortholog_3722 | 0.00281 | NAD-dependent epimerase/dehydratase | NC |
| Ortholog_522 | 0.00281 | Hypothetical 3-demethylubiquinone-9 3-methyltransferase | NC |
| Ortholog_568 (thiJ) | 0.00281 | ThiJ/PfpI, C56 or DJ-1/PfpI family intracellular protease/amidase, hypothetical chaperone protein HSP31 | NC |
| Ortholog_197 (epsB) | 0.00411 | Undecaprenyl-phosphate galactosephosphotransferase, glucosyltransferase EpsB, galactosyltransferase | NC |
| Ortholog_2934 (cobO) | 0.00411 | Cob(I)alamin adenosyltransferase | NC |
| Ortholog_302 | 0.00411 | Cobalamin ECF transporter, lipoprotein | NC |
| Ortholog_3930 (cbrT) | 0.00411 | Substrate-specific component CbrT of cobalamin ECF transporter | NC |
| Ortholog_5083 | 0.00411 | Hypothetical protein | NC |
| Ortholog_57 | 0.00411 | Adenosylcobalamin (coenzyme B12)-dependent ribonucleoside-diphosphate reductase | NC |
| Ortholog_2421 (dps) | 0.00554 | Non-specific DNA-binding protein Dps, iron-binding ferritin-like antioxidant protein/ferroxidase | NC |
| Ortholog_4208 (copZ) | 0.00554 | Copper chaperone copZ, heavy metal-associated domain, MerTP family mercury (Hg2+) permease | NC |
| Ortholog_2070 | 0.00595 | Hypothetical protein | NC |
| Ortholog_4672 | 0.00595 | Hypothetical protein | NC |
| Ortholog_476 | 0.00595 | HTH ArsR-type DNA-binding transcriptional regulator | NC |
| Ortholog_2867 | 0.00799 | Transposase IS3/IS911 | CL |
| Ortholog_2905 | 0.00799 | Hypothetical protein | CL |
| Ortholog_4275 | 0.00799 | Hypothetical protein | CL |
| Ortholog_507 | 0.00799 | Hypothetical protein | CL |
| Ortholog_5905 | 0.00799 | IS3-family transposase, integrase, transposase InsK, IS150-like transposase | CL |
| Ortholog_6476 | 0.00799 | Hypothetical protein | CL |
| Ortholog_3968 | 0.00883 | Hypothetical protein | CL |
| Ortholog_4531 | 0.00883 | Hypothetical protein | CL |
| Ortholog_1362 | 0.00925 | D12 class N6 adenine-specific DNA methyltransferase | NC |
| Ortholog_1422 (cps4F) | 0.00925 | Capsular polysaccharide biosynthesis protein Cps4F, glycosyl transferase group 1 | NC |
| Ortholog_1609 | 0.00925 | Hypothetical protein | NC |
| Ortholog_1712 | 0.00925 | UDP-glucose 4-epimerase, NAD-dependent epimerase/dehydratase | NC |
| Ortholog_3751 | 0.00925 | Hypothetical protein | NC |
| Ortholog_3762 (mga) | 0.00925 | M protein trans-acting positive regulator (MGA) | NC |
| Ortholog_5644 (capD) | 0.00925 | Polysaccharide biosynthesis protein CapD, NAD-binding protein, UDP-glucose 4-epimerase | NC |
| Ortholog_5738 | 0.00925 | Hypothetical protein | NC |
| Ortholog_5837 (wecB) | 0.00925 | UDP-N-acetylglucosamine 2-epimerase | NC |
| Ortholog_6592 (cap5F) | 0.00925 | NAD-dependent epimerase/dehydratase, capsular polysaccharide biosynthesis protein Cap5F | NC |
| Ortholog_6669 | 0.00925 | Hypothetical protein | NC |
Full sets of NC- or CL-enriched genes are listed in Table S3 in the supplemental material.
P values were obtained by Fisher's exact test.
Several different functional categories were represented among the 31 NC-enriched genes at P < 0.01, including hypothetical (membrane) proteins, capsule and vitamin biosynthesis, and sugar metabolism (Table 4). Several transcriptional regulators were also among those NC-enriched genes as well as loci associated with stress responses, including dps, a gene coding for iron-binding ferroxidase (ortholog_2421). The 8 CL-enriched genes (P < 0.01) consisted of two insertion sequence elements and 6 genes encoding hypothetical proteins (Table 4; also see Table S3 in the supplemental material).
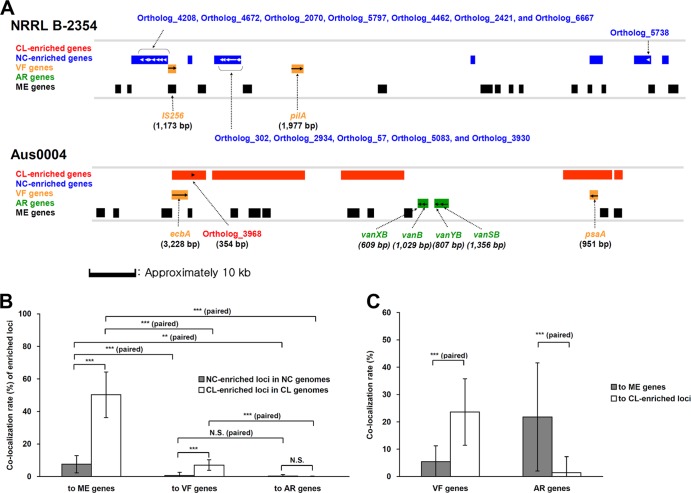
Proximity analysis showed that the E. faecium NC-enriched genes were often localized within 1,000 bp of each other in the NC E. faecium genomes, and the same result was found for CL-enriched genes in CL E. faecium (Fig. 4A; also see Fig. S5 in the supplemental material). Among the NC genomes, the 68 enriched genes (P < 0.05) were segregated into 20 loci, with an average of 2.2 genes per locus per NC genome. For the 153 CL-enriched genes, an average of 49 loci were identified with 2 genes per locus per CL genome.
FIG 4.
Proximity of NC-/CL-enriched loci to ME, VF, and AR genes. (A) Representative loci are shown from E. faecium NRRL B-2354 (plasmid) and Aus0004 (chromosome) containing NC- and CL-enriched genes. NC-enriched (blue) and CL-enriched (red) genes are indicated, as well as VF, AR, and ME loci. A scale bar is provided. Gene clusters identified in other strains are shown in Fig. 5S in the supplemental material. (B) Colocalization rates of E. faecium NC-/CL-enriched loci to ME, VF, and AR genes was compared in NC (n = 10) and CL (n = 20) genomes. (C) Colocalization rates of VF and AR genes to ME genes and CL-enriched loci were also compared among CL genomes (n = 20). Colocalization rates are shown as means ± standard deviations. N.S., no significance; P < 0.01 (**) and P < 0.001 (***) according to Student's t test using unpaired or paired data.
To confirm the presence of the NC- and CL-enriched genes in E. faecium, several genes were selected for PCR amplification in representative NC (NRRL B-2354) and CL (1,231,502) strains. The three NC-specific genes tested (ortholog_522, ortholog_3722, and ortholog_2421) were detected in E. faecium strain NRRL B-2354 but not E. faecium strain 1,231,502 (see Fig. S6 in the supplemental material). The two CL-enriched genes (ortholog_4275 and ortholog_6476) were found only in strain 1,231,502 (see Fig. S6).
Structural proximity of NC-/CL-enriched loci to ME, VF, and AR genes.
As ME genes have contributed to the emergence of VF and AR genes and are often colocalized in E. faecium (21–23), proximity analysis was performed for ME, VF, and AR genes on the E. faecium genomes. The majority of ME genes were located within 1,000 bp from niche-specific loci (Fig. 4A), and CL-enriched genes (50.3% ± 14.0% loci/CL genome) were more frequently colocalized to ME than NC-enriched genes (7.6% ± 5.3% loci/NC genome) (Fig. 4B). CL-enriched loci were also more frequently found in close proximity to VF genes (7.1% ± 3.3% loci/CL genome) (Fig. 4B). In each CL genome, VF genes were also located in close proximity to ME, although colocalization of VF genes to CL-enriched genes was even higher (Fig. 4C). In contrast, AR genes were more often associated with ME (Fig. 4C).
DISCUSSION
This study revealed novel structural (genome size and colocalization of NC-/CL-enriched genes to ME, VF, and AR genes) and functional (overrepresented functional categories and NC-/CL-enriched genes) features between E. faecium NC and CL genomes. E. faecalis genomes were found to contain more core genes than E. faecium but do not show clade separation between NC and CL isolates. The results indicate that NC and CL E. faecium are each equipped with different genes for adaptation to the GIT and extraintestinal sites.
A set of 68 genes (P < 0.05) and a subset of 31 genes (P < 0.01) enriched among community (NC) E. faecium strains were identified. These genes constitute novel NC adaptive loci and may be used to distinguish community/commensal E. faecium from strains that are more likely to cause infection. Among the NC-enriched genes are 7 polysaccharide biosynthesis genes (ortholog_1422 [cps4F], ortholog_5644 [capD], ortholog_6592 [cap5F], ortholog_197 [epsB], ortholog_3722, ortholog_1712, and ortholog_5837 [wecB]). Those genes are associated with formation of cell wall and capsule, and 6 out of the 7 (cps4F, cap5F, capD, epsB, wecB, and ortholog_1712) are identical to the capsule genes that are located in the variable regions of NC strains (5). UDP-glucose 4-epimerases (ortholog_1712 and capD) are known to be involved in glycogen and sucrose synthesis in plants (24), suggesting that they have a role in Enterococcus adaptation to plant environments. The capD gene is also known to confer degradation from H-capsule to lower-molecular-weight L-capsule, an essential molecule to escape from host defenses (25). epsB is involved in exopolysaccharide (EPS) I biosynthesis (26). Although capD and epsB are regarded as virulence factors, the clinical relevance of extracellular carbohydrates and enterococcal virulence is currently unclear (27).
NC-enriched genes also included 4 cobalamin-related genes (ortholog_2934 [cobO], ortholog_302, ortholog_3930 [cbrT], and ortholog_57) and an iron-binding ferritin-like ferroxidase gene (ortholog_2421 [dps]). While synthesis or uptake of cobalamin (vitamin B12) is not essential for enteric pathogens, including Escherichia coli and Salmonella (28), iron is well documented to be required for pathogen survival and virulence in the host (29, 30). Hence, the dps gene may have a role in E. faecium iron acquisition in the gut or from other low-iron food or plant environments. This distinction indicates that dps constitutes a niche factor rather than a virulence factor (31).
E. faecium NC genomes also have greater numbers of genes involved in glycerol fermentation to 1,3-propanediol. Biosynthesis of 1,3-propanediol was reported for several lactic acid bacteria, including Lactobacillus reuteri (32), Lactobacillus brevis (33), and Lactobacillus buchnerii (33), but not E. faecium. The pathway from glycerol to 1,3-propanediol is associated with restoring redox balance in L. reuteri during anaerobic growth on glucose (32). The 1,3-propanediol pathway is dependent on adenosylcobalamin, the synthesis of which is mediated by cobO (ortholog_2934), an NC-enriched gene. Although 6 out of the 10 E. faecium NC isolates have two or three genes responsible for glycerol transport and adenosylcobalamin synthesis, key converting enzymes are missing from those strains. Thus, this pathway might have a function other than restoring redox balance in E. faecium.
Genes associated with responses to oxidative stress were also overrepresented in NC E. faecium. A recent study showed that AsrR-mediated sensing of oxidative stress by Enterococcus influenced levels of antibiotic resistance, interactions with host cells, and virulence (34). Because the mucus layer is exposed to oxygen diffused from the bloodstream and intestinal epithelial cells (35), E. faecium cells might need mechanisms to tolerate oxygen in the intestine. These genes are not as commonly shared among NC strains as the NC-enriched genes; rather, this indicates adaptations of E. faecium to different environmental sites.
CL E. faecium strains contained 153 genes (P < 0.05) and a subset of 8 genes (P < 0.01) distinct from the NC isolates. CL-enriched genes included IS16 (ortholog_6478), a sequence that has been extensively used to screen for CL E. faecium (36–39). However, according to the genome comparisons performed here, two other ME (ortholog_2867 and ortholog_5905) were more highly associated with CL strains and might be more useful predictors of CL genotypes among E. faecium strains.
E. faecium CL strains also contained greater numbers of genes for N-acetyl-galactosamine and galactosamine metabolism, alkylphosphonate transport, and bile metabolism. N-acetyl-galactosamine is a terminal carbohydrate in the human blood group A antigen, and it has been shown that the carbohydrate is associated with blood group A-specific ear infections by Pseudomonas aeruginosa (40). Hence, the genes for N-acetyl-galactosamine and galactosamine metabolism may have a role in human blood infection by E. faecium CL strains. Alternatively, N-acetyl-galactosamine is also a primary monosaccharide of intestinal mucins, and some vancomycin-resistant E. faecium strains were reported to ferment monosaccharides released from mucins as energy sources, which would support colonization and survival in the gut (41). Because alkylphosphonate is known to be utilized as a phosphorus source in Enterobacter aerogenes (42), the overrepresentation of genes for alkylphosphonate transport in CL E. faecium indicates enhanced levels of phosphorus utilization in the human body. Lastly, E. faecium CL strains also have more genes for bile hydrolysis than NC strains. This corresponds to the variation in bile salt hydrolysis (BSH) activity found for strains of E. faecium (43). Although the role of BSH activity of E. faecium CL strains in human health is still largely unknown, bile hydrolysis is associated with the survival of probiotic microorganisms in the intestine and might contribute to CL survival in the gut (44).
ME genes are more abundant in E. faecium CL genomes (4), and transposases and transposon genes were predominant among the 153 CL-enriched genes identified here. ME might influence E. faecium CL genomes by introducing novel ME-associated, CL-enriched VR and AR loci. These additions would also result in an increase in genome size, as was found for the CL isolates. This is supported by the finding that CL-enriched genes are colocalized more frequently to ME than NC-enriched genes (Fig. 4B). ME associations with VF genes (hyl and esp) (21, 22) and AR genes (erm, tetM, tetS, and vanB) (23) are known and confirmed here for a greater number of genes using genome proximity analysis. VF were also frequently in close proximity to CL-enriched genes in E. faecium CL genomes, indicating that CL-enriched genes are associated with virulence and potentially novel virulence-associated loci or pathogenicity islands.
Using whole-genome CDS, we identified a novel E. faecium NC clade (NC2) (Fig. 1A) that has not been found or specifically designated in previous studies (3–5, 7, 20). Interestingly, the NC2 clade clustered with the majority of CL isolates. The finding of the new NC clade is supported by a hypothesis that the hospital-associated (HA) clade includes community-based ampicillin-resistant E. faecium (ARE) with the pbp5-R genotype (4). Two community-based ARE strains (E4452 and E4453) are included in the NC2 clade. A phylogenetic tree of pbp5 confirmed that all of the NC2 strains except 1,231,501 clustered together with CL strains (see Fig. S7 in the supplemental material). Unlike the separation between hospital-associated and community-associated (CA) E. faecium clades at least 300,000 years ago (3), NC2 and CL strains might have diverged from the common ancestral HA lineage due to the relatively recent development of hospital environments in which antibiotics have been frequently used (8). An alternative hypothesis is that NC2-clade strains represent hybrid genomes of NC1 and CL strains (5). Although NC2-clade strains do have a few CL-enriched genes, both hypotheses should be tested in additional studies using more NC genomes.
Although E. faecalis and E. faecium are highly related, the genomes of E. faecalis are larger and contain more core genes, indicating that E. faecalis strains have more complex gene networks than E. faecium (45). E. faecalis genes were also more frequently assigned gene designations than E. faecium. This difference might be related to the fact that the first E. faecalis complete genome sequence was published 9 years earlier than genomes of E. faecium (46, 47), and E. faecalis has been extensively studied because of its high levels of antibiotic resistance (8). Also unlike E. faecium, the genomes of NC and CL E. faecalis lacked specific structural and functional features, and clade separation based on ortholog presence/absence between NC and CL strains was not applicable to E. faecalis. These differences indicate that E. faecalis strains examined thus far constitute a single lineage specifically adapted to the intestinal tract which has been subjected to genome expansion. Such distinctions may be the cause of the earlier appearance of antibiotic-resistant strains of E. faecalis than of E. faecium (8).
Our findings show genome-wide species and origin-specific differences of two closely related opportunistic pathogens with commensal lifestyles. The NC- and CL-enriched genes and other genomic features identified here can be employed to elucidate the mechanisms of E. faecium pathogenesis and distinguish strains adapted for foods and the GIT.
Supplementary Material
ACKNOWLEDGMENTS
We thank Linda J. Harris and Lauren Kopit for their review of the manuscript.
This study was supported by a grant from the Almond Board of California.
We also kindly thank Michael Gilmore for the E. faecium strain 1,231,502, Michiel Kleerebezem for the L. plantarum strain WCFS1, and Janetta Top and Rob Willems for their support on the MLST assignment of strain NRRL B-2354.
Footnotes
Published ahead of print 18 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03108-13.
REFERENCES
- 1.McGowan LL, Jackson CR, Barrett JB, Hiott LM, Fedorka-Cray PJ. 2006. Prevalence and antimicrobial resistance of enterococci isolated from retail fruits, vegetables, and meats. J. Food Prot. 69:2976–2982 [DOI] [PubMed] [Google Scholar]
- 2.Willems RJL, van Schaik W. 2009. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 4:1125–1135. 10.2217/fmb.09.82 [DOI] [PubMed] [Google Scholar]
- 3.Galloway-Peña J, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7:e30187. 10.1371/journal.pone.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin X, Galloway-Pena J, Sillanpaa J, Roh J, Nallapareddy S, Chowdhury S, Bourgogne A, Choudhury T, Muzny D, Buhay C, Ding Y, Dugan-Rocha S, Liu W, Kovar C, Sodergren E, Highlander S, Petrosino J, Worley K, Gibbs R, Weinstock G, Murray B. 2012. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 12:135. 10.1186/1471-2180-12-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer KL, Godfrey P, Griggs A, Kos VN, Zucker J, Desjardins C, Cerqueira G, Gevers D, Walker S, Wortman J, Feldgarden M, Haas B, Birren B, Gilmore MS. 2012. Comparative genomics of enterococci: variation in Enterococcus faecalis, clade structure in E. faecium, and defining characteristics of E. gallinarum and E. casseliflavus. mBio 3:e00112–12. 10.1128/mBio.00112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solheim M, Brekke M, Snipen L, Willems R, Nes I, Brede D. 2011. Comparative genomic analysis reveals significant enrichment of mobile genetic elements and genes encoding surface structure-proteins in hospital-associated clonal complex 2 Enterococcus faecalis. BMC Microbiol. 11:3. 10.1186/1471-2180-11-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebreton F, van Schaik W, Manson McGuire A, Godfrey P, Griggs A, Mazumdar V, Corander J, Cheng L, Saif S, Young S, Zeng Q, Wortman J, Birren B, Willems RJL, Earl AM, Gilmore MS. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4:e00534–13. 10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat. Rev. Microbiol. 10:266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227–10. 10.1128/mBio.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizk G, Lavenier D. 2010. GASSST: global alignment short sequence search tool. Bioinformatics 26:2534–2540. 10.1093/bioinformatics/btq485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Madan A. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Xiong Z, Sun L, Yang J, Jin Q. 2012. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 40:D641–D645. 10.1093/nar/gkr989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Pop M. 2009. ARDB–antibiotic resistance genes database. Nucleic Acids Res. 37:D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team 2012. R: a language and environment for statistical computing, 2.15.2. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 16.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. 10.1093/bioinformatics/btl117 [DOI] [PubMed] [Google Scholar]
- 17.Homan WL, Tribe D, Poznanski S, Li M, Hogg G, Spalburg E, van Embden JDA, Willems RJL. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963–1971. 10.1128/JCM.40.6.1963-1971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice LB, Laktičova V, Carias LL, Rudin S, Hutton R, Marshall SH. 2009. Transferable capacity for gastrointestinal colonization in Enterococcus faecium in a mouse model. J. Infect. Dis. 199:342–349. 10.1086/595986 [DOI] [PubMed] [Google Scholar]
- 20.Willems RJL, Top J, van Schaik W, Leavis H, Bonten M, Sirén J, Hanage WP, Corander J. 2012. Restricted gene flow among hospital subpopulations of Enterococcus faecium. mBio 3:e00151–12. 10.1128/mBio.00151-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arias CA, Panesso D, Singh KV, Rice LB, Murray BE. 2009. Cotransfer of antibiotic resistance genes and a hylEfm-containing virulence plasmid in Enterococcus faecium. Antimicrob. Agents Chemother. 53:4240–4246. 10.1128/AAC.00242-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leavis H, Top J, Shankar N, Borgen K, Bonten M, van Embden J, Willems RJ. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 186:672–682. 10.1128/JB.186.3.672-682.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 16:541–554. 10.1111/j.1469-0691.2010.03226.x [DOI] [PubMed] [Google Scholar]
- 24.Li C, Wang Y, Liu L, Hu Y, Zhang F, Mergen S, Wang G, Schläppi MR, Chu C. 2011. A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genet. 7:e1002196. 10.1371/journal.pgen.1002196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino S-I, Watarai M, Cheun H-I, Shirahata T, Uchida I. 2002. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J. Infect. Dis. 186:227–233. 10.1086/341299 [DOI] [PubMed] [Google Scholar]
- 26.Huang J, Schell M. 1995. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol. Microbiol. 16:977–989. 10.1111/j.1365-2958.1995.tb02323.x [DOI] [PubMed] [Google Scholar]
- 27.Mundy LM, Sahm DF, Gilmore M. 2000. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13:513–522. 10.1128/CMR.13.4.513-522.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampson BA, Gotschlich EC. 1992. Elimination of the vitamin B12 uptake or synthesis pathway does not diminish the virulence of Escherichia coli K1 or Salmonella typhimurium in three model systems. Infect. Immun. 60:3518–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sritharan M. 2006. Iron and bacterial virulence. Indian J. Med. Microbiol. 24:163–164 [PubMed] [Google Scholar]
- 30.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. 10.1371/journal.ppat.1000949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill C. 2012. Virulence or niche factors: what's in a name? J. Bacteriol. 194:5725–5727. 10.1128/JB.00980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens M, Vollenweider S, Meile L, Lacroix C. 2011. 1,3-Propanediol dehydrogenases in Lactobacillus reuteri: impact on central metabolism and 3-hydroxypropionaldehyde production. Microb. Cell Fact. 10:61. 10.1186/1475-2859-10-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schütz H, Radler F. 1984. Anaerobic reduction of glycerol to propanediol-1.3 by Lactobacillus brevis and Lactobacillus buchneri. Syst. Appl. Microbiol. 5:169–178. 10.1016/S0723-2020(84)80018-1 [DOI] [Google Scholar]
- 34.Lebreton F, van Schaik W, Sanguinetti M, Posteraro B, Torelli R, Le Bras F, Verneuil N, Zhang X, Giard J-C, Dhalluin A, Willems RJL, Leclercq R, Cattoir V. 2012. AsrR is an oxidative stress sensing regulator modulating Enterococcus faecium opportunistic traits, antimicrobial resistance, and pathogenicity. PLoS Pathog. 8:e1002834. 10.1371/journal.ppat.1002834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 9:577–589. 10.1038/nrgastro.2012.156 [DOI] [PubMed] [Google Scholar]
- 36.European Food Safety Authority 2012. Guidance on the safety assessment of Enterococcus faecium in animal nutrition. EFSA J. 10:2682–2692. 10.2903/j.efsa.2012.2682 [DOI] [Google Scholar]
- 37.Werner G, Fleige C, Geringer U, van Schaik W, Klare I, Witte W. 2011. IS element IS16 as a molecular screening tool to identify hospital-associated strains of Enterococcus faecium. BMC Infect. Dis. 11:80. 10.1186/1471-2334-11-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice LB, Carias L, Rudin S, Vael C, Goossens H, Konstabel C, Klare I, Nallapareddy SR, Huang W, Murray BE. 2003. A potential virulence gene, hylEfm, predominates in Enterococcus faecium of clinical origin. J. Infect. Dis. 187:508–512. 10.1086/367711 [DOI] [PubMed] [Google Scholar]
- 39.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. 10.1186/1471-2164-11-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steuer MK, Hofstadter F, Probster L, Beuth J, Strutz J. 1995. Are ABH antigenic determinants on human outer ear canal epithelium responsible for Pseudomonas aeruginosa infections? ORL J. Otorhinolaryngol. Relat. Spec. 57:148–152. 10.1159/000276728 [DOI] [PubMed] [Google Scholar]
- 41.Pultz NJ, Hoskins LC, Donskey CJ. 2006. Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb. Drug Resist. 12:63–67. 10.1089/mdr.2006.12.63 [DOI] [PubMed] [Google Scholar]
- 42.Murata K, Higaki N, Kimura A. 1988. Detection of carbon-phosphorus lyase activity in cell free extracts of Enterobacter aerogenes. Biochem. Biophys. Res. Commun. 157:190–195. 10.1016/S0006-291X(88)80031-7 [DOI] [PubMed] [Google Scholar]
- 43.Franz CM, Specht I, Haberer P, Holzapfel WH. 2001. Bile salt hydrolase activity of Enterococci isolated from food: screening and quantitative determination. J. Food Prot. 64:725–729 [DOI] [PubMed] [Google Scholar]
- 44.Patel AK, Singhania RR, Pandey A, Chincholkar SB. 2010. Probiotic bile salt hydrolase: current developments and perspectives. Appl. Biochem. Biotechnol. 162:166–180. 10.1007/s12010-009-8738-1 [DOI] [PubMed] [Google Scholar]
- 45.Szathmáry E, Jordán F, Pál C. 2001. Can genes explain biological complexity? Science 292:1315–1316. 10.1126/science.1060852 [DOI] [PubMed] [Google Scholar]
- 46.Paulsen IT, Banerjei L, Myers GSA, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071–2074. 10.1126/science.1080613 [DOI] [PubMed] [Google Scholar]
- 47.Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PDR, Howden BP, Stinear TP. 2012. Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194:2334–2341. 10.1128/JB.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palmer KL, Carniol K, Manson JM, Heiman D, Shea T, Young S, Zeng Q, Gevers D, Feldgarden M, Birren B, Gilmore MS. 2010. High-quality draft genome sequences of 28 Enterococcus sp. isolates. J. Bacteriol. 192:2469–2470. 10.1128/JB.00153-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Regt MJ, van Schaik W, van Luit-Asbroek M, Dekker HA, van Duijkeren E, Koning CJ, Bonten MJ, Willems RJ. 2012. Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One 7:e30319. 10.1371/journal.pone.0030319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speck ML. 1947. The resistance of Micrococcus freudenreichii in laboratory high-temperature-short-time pasteurization of milk and ice cream mix. J. Dairy Sci. 30:975–981. 10.3168/jds.S0022-0302(47)92426-0 [DOI] [Google Scholar]
- 51.Hadji-Sfaxi I, El-Ghaish S, Ahmadova A, Batdorj B, Le Blay-Laliberté G, Barbier G, Haertlé T, Chobert J-M. 2011. Antimicrobial activity and safety of use of Enterococcus faecium PC4.1 isolated from Mongol yogurt. Food Control 22:2020–2027. 10.1016/j.foodcont.2011.05.023 [DOI] [Google Scholar]
- 52.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH human microbiome project. Genome Res. 19:2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carias LL, Rudin SD, Donskey CJ, Rice LB. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias CA, Torres HA, Singh KV, Panesso D, Moore J, Wanger A, Murray BE. 2007. Failure of daptomycin monotherapy for endocarditis caused by an Enterococcus faecium strain with vancomycin-resistant and vancomycin-susceptible subpopulations and evidence of in vivo loss of the vanA gene cluster. Clin. Infect. Dis. 45:1343–1346. 10.1086/522656 [DOI] [PubMed] [Google Scholar]
- 55.Brede DA, Snipen LG, Ussery DW, Nederbragt AJ, Nes IF. 2011. Complete genome sequence of the commensal Enterococcus faecalis 62, isolated from a healthy Norwegian infant. J. Bacteriol. 193:2377–2378. 10.1128/JB.00183-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solheim M, Aakra A, Snipen L, Brede D, Nes I. 2009. Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genomics 10:194. 10.1186/1471-2164-10-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBride SM, Fischetti VA, LeBlanc DJ, Moellering RC, Jr, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. PLoS One 2:e582. 10.1371/journal.pone.0000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim EB, Kopit LM, Harris LJ, Marco ML. 2012. Draft genome sequence of the quality control strain Enterococcus faecalis ATCC 29212. J. Bacteriol. 194:6006–6007. 10.1128/JB.01423-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murray BE, Mederski-Samaroj B. 1983. Transferable beta-lactamase. A new mechanism for in vitro penicillin resistance in Streptococcus faecalis. J. Clin. Investig. 72:1168–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahm DF, Kissinger J, Gilmore MS, Murray PR, Mulder R, Solliday J, Clarke B. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588–1591. 10.1128/AAC.33.9.1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murray BE, Singh KV, Ross RP, Heath JD, Dunny GM, Weinstock GM. 1993. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J. Bacteriol. 175:5216–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.