Abstract
The roles of various core components, including α/β/γ-type small acid-soluble spore proteins (SASP), dipicolinic acid (DPA), core water content, and DNA repair by apurinic/apyrimidinic (AP) endonucleases or nonhomologous end joining (NHEJ), in Bacillus subtilis spore resistance to different types of ionizing radiation including X rays, protons, and high-energy charged iron ions have been studied. Spores deficient in DNA repair by NHEJ or AP endonucleases, the oxidative stress response, or protection by major α/β-type SASP, DPA, and decreased core water content were significantly more sensitive to ionizing radiation than wild-type spores, with highest sensitivity to high-energy-charged iron ions. DNA repair via NHEJ and AP endonucleases appears to be the most important mechanism for spore resistance to ionizing radiation, whereas oxygen radical detoxification via the MrgA-mediated oxidative stress response or KatX catalase activity plays only a very minor role. Synergistic radioprotective effects of α/β-type but not γ-type SASP were also identified, indicating that α/β-type SASP's binding to spore DNA is important in preventing DNA damage due to reactive oxygen species generated by ionizing radiation.
INTRODUCTION
Spores of Bacillus subtilis have been used extensively as biological indicators for industrial purposes such as sterilization or decontamination. Spores have also been shown to be suitable dosimeters for probing terrestrial and extraterrestrial ionizing radiation in environmental and astrobiological studies (1–3; reviewed in references 4, 5, 6, and 7). Ionizing radiation can damage cellular components through direct transfer of radiation energy into biomolecules (e.g., DNA, RNA, proteins) and indirectly by generating reactive oxygen species (ROS) from the radiolysis of intracellular H2O (8–11). The biological effects of ionizing radiation are thought to arise from the formation of single- and double-strand breaks (SSB and DSB) in cellular DNA and clustered DNA damage, e.g., two or more closely spaced lesions, including abasic sites, base lesions, SSB, and DSB. The biological effects of ionizing radiation depend on the quality and the dose of radiation and on the cell type (9–11). Linear energy transfer (LET) represents the energy lost per unit distance as an ionizing particle travels through a material, and it is used to quantify the effects of ionizing radiation on biological specimens (5, 10–13). High-LET radiation sources include protons, and high-energy-charged (HZE) particles give densely ionizing radiation, since they lose their energy in a small distance and thus cause dense ionization along their tracks and can give localized multiple DNA-damaging events. Low-LET radiation sources, such as X rays, give sparsely ionizing radiation, since they produce ionizations sparsely along their track and, hence, almost homogeneously within a cell. The biological effects of high-LET radiation are in general much higher than those of low-LET radiation of the same energy (10, 12, 13). This is because high-LET radiation deposits most of its energy within the volume of one cell and the damage to DNA is therefore larger (9–11). This is attributable to the formation of clusters of damage that result in two or more DNA lesions along the tracks of high-LET radiation sources, i.e., the physical path of the applied ionizing radiation source in a cell.
Previous studies have indicated that in both pro- and eukaryotic cells or under highly scavenging conditions mimicking those for ROS scavenging in the cell, one-fourth of the lesions induced in DNA by low-LET radiation can be ascribed to direct effects increasing up to 80% for high-LET particles (9, 11, 12). Hydrogen peroxide (H2O2) and hydroxyl radicals (HO·) are major oxidizing species produced by the radiolysis of water, and superoxide ions (O2·−) are formed in the presence of dissolved oxygen (12, 13). Lethal and mutagenic effects induced by ionizing radiation are thought to be the result of DNA damage caused during the course of irradiation (11, 14–16). Spore DNA resides in the innermost spore compartment, the core, and dormant spores of B. subtilis possess a complex arsenal of protective attributes in the core, in particular a low core water content as well as abundant novel core constituents such as (i) the calcium chelate of dipicolinic acid (Ca-DPA), which comprises ∼25% of core dry weight, and (ii) small, acid-soluble spore proteins (SASP) of the α/β type, which bind spore DNA and protect it from many types of damage, including UV photoproducts, apurinic/apyrimidinic (AP) sites, and oxidative lesions (17, 18, 47; reviewed in references 4, 6, and 19). Spore DNA is saturated with a group of unique proteins called α/β-type SASP, which are encoded by multiple genes and synthesized only during sporulation in the developing spore. These α/β-type SASP are nonspecific DNA-binding proteins that bind to random-sequence double-strand DNA and comprise approximately 5% of total spore protein (reviewed in references 6 and 18). The high levels of α/β-type SASP in spores are sufficient to saturate the spore DNA, and the DNA within this nucleoprotein complex is protected from a variety of environmental insults (4, 7).
Germinating spores also can detoxify ROS using enzymes such as catalase (KatX) and superoxide dismutase (SodA), although these enzymes play no role in dormant spore resistance to oxidizing agents, most likely because enzymes in the spore core are inactive due to the core's low water content (20–23). Mechanisms to prevent damage caused by ROS, in particular damage to DNA, include protective DNA-binding proteins as well as enzymes such as alkyl hydroperoxide reductases, catalases, and superoxide dismutases, which can destroy the oxidizing agents alkyl hydroperoxides, hydrogen peroxide, and superoxide, respectively. A study of the role of oxidative stress responsive proteins such as the DNA-binding protein MrgA, alkyl hydroperoxide reductase, catalases, and superoxide dismutases in the resistance of B. subtilis spores to oxidative stress caused by paraquat and hydrogen peroxide exposure showed that inactivation of genes coding for the protective enzymes or MrgA had no effect on the spores' heat or hydrogen peroxide resistance (21). Spore DNA damage can also be repaired during germination via a number of different pathways such as direct reversal, nonhomologous end joining (NHEJ, encoded by YkoV and YkoU), base and nucleotide excision repair (BER and NER, respectively), mismatch repair, recombination-mediated repair, and translesion synthesis (24–28; reviewed in references 6 and 29). Two AP endonucleases, ExoA and Nfo, have also been implicated in the repair of AP sites caused by oxidizing agents in germinating B. subtilis spores (24). However, there has been no systematic study of the relationship between the spore core protective features (i.e., α/β/γ-type SASP, DPA, and core hydration), the oxidative stress response (by MrgA), ROS detoxification (via SodA or KatX), and DNA repair (by NHEJ or BER) in spores' ionizing radiation resistance, and this is the subject of the present work.
MATERIALS AND METHODS
Bacterial strains, growth, sporulation, and spore purification.
The B. subtilis strains used in this work are listed in Table 1, and all are isogenic with the wild-type strain PS832. Spores were obtained by cultivation under vigorous aeration at 37°C for 7 days in double-strength liquid Schaeffer sporulation medium (30), and spores were purified and stored as described previously (31–33). When appropriate, chloramphenicol (5 μg/ml), neomycin (10 μg/ml), spectinomycin (100 μg/ml), erythromycin (1 μg/ml), or tetracycline (10 μg/ml) was added to the medium. Spore preparations consisted of single spores with no detectable clumps and were free (>99%) of growing cells, germinated spores, and cell debris, as seen in the phase-contrast microscope (32–34).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype and phenotype | Reference |
|---|---|---|
| PS832 | Trp+ revertant of strain 168 (wild type) | 40 |
| PS283a | ΔsspA, SASP-α−, Cmr | 45 |
| PS338a | ΔsspB, SASP-β−, Cmr | 45 |
| PS483a | ΔsspE, SASP-γ−, Cmr | 38 |
| PS356b | ΔsspA ΔsspB, α−β− | 22 |
| PS482a | ΔsspA ΔsspB ΔsspE, α−β−γ−, Cmr | 46 |
| PS1899a | ΔdacB::Cmr | 22 |
| PS2211a,b | ΔdacB::Cmr, α−β− | 22 |
| FB122c,d | ΔsleB::Spcr ΔspoVF::Tetr, DPA− | 39 |
| PS3664a,b,c,d | ΔsleB::Spcr ΔspoVF::Tetr, DPA−, α−β− | 47 |
| PS2496a | ΔmrgA::Cmr | 21 |
| PS2507a,b | ΔmrgA::Cmr, α−β− | 21 |
| PS2495a | ΔsodA::Cmr | 21 |
| PS2506a,b | ΔsodA::Cmr, α−β− | 21 |
| PS2558a | ΔkatX::Cmr | 21 |
| PS2559a,b | ΔkatX::Cmr, α−β− | 21 |
| PERM454d,e | ΔexoA::Tetr Δnfo::Neor | 24 |
| PERM450a,b,d,e | ΔexoA::Tetr Δnfo::Neor, α−β− | 24 |
| PS3722f | ΔykoVU::Ermr | 25 |
| PS3751a,b,f | ΔykoVU::Ermr, α−β− | 25 |
Cmr, resistant to chloramphenicol (5 μg/ml).
α−β−, spores lack SASP-α and -β and thus ∼80% of the spore's pool of α/β-type SASP (35).
Spcr, resistant to spectinomycin (100 μg/ml).
Tetr, resistant to tetracycline (10 μg/ml).
Neor, resistant to neomycin (10 μg/ml).
Ermr, resistant to erythromycin (1 μg/ml).
Measurement of spore resistance to ionizing radiation.
Preparation of spore samples for radiation exposure has been described in detail previously (33, 34). In brief, suspensions of spores of the different B. subtilis strains (Table 1) were prepared in sterile distilled water to a final concentration of 1 × 108 spores per ml. Triplicate samples of spores in water (100 μl) were individually exposed to three different types of ionizing radiation: X rays (200 keV/15 mA), protons (with energy of 218 MeV, LET of 0.4 keV/μm, and a range of 301 mm in water), and high-energy-charged iron ions (Fe; with energy of 500 MeV/nucleon, LET of 200 keV/μm, and a range of 99 mm in water). X ray, proton, and Fe irradiations were conducted at the German Aerospace Center, Institute of Aerospace Medicine (DLR-ME), in Cologne, Germany, at the University of Florida Proton Therapy Institute (UF-PTI), Jacksonville, Florida, USA, and at the heavy ion medical accelerator (HIMAC) facility at the National Institute for Radiological Sciences (NIRS) in Chiba, Japan, respectively. Details on the irradiation geometry of the NIRS-HIMAC, DLR-ME, and UF-PTI facilities, beam monitoring, dosimetry, and dose calculations have been described (34–37).
Numerical and statistical analysis.
Spore survival was determined from appropriate dilutions in distilled water as colony-forming ability after incubation overnight at 37°C on nutrient broth (NB) agar plates (Difco, Detroit, MI, USA) (33, 34, 36). The surviving fraction of B. subtilis spores was determined from the ratio N/N0, where N is the number of CFU of the irradiated sample and N0 that of the nonirradiated controls. Spore inactivation curves were obtained as described previously (34). Data are reported as D10-values, the dose of ionizing radiation killing 90% of the initial spore population (34). All data are expressed as averages ± standard deviations (n = 3). The significance of the differences in the survival rates and relative sensitivities were determined by analysis of variance (ANOVA), using SigmaPlot software Version 12.0 (Systat Software GmbH, Erkrath, Germany). Values were evaluated in multigroup pairwise combinations, and differences with P values of <0.05 were considered statistically significant (33, 34, 36).
RESULTS
The importance of the two types of SASP, the DNA-binding α/β type and non-DNA-binding γ type, alone and in combination with various other protective spore core components, including core water content and DPA, has been assessed to establish their roles as radioprotectants. B. subtilis spores have several additional potential mechanisms for minimizing damage to spore DNA including BER via AP endonucleases (by ExoA and Nfo), DNA double-strand break repair (via NHEJ by YkoU and V), and radical detoxification (superoxide dismutase SodA and major spore catalase KatX), as well as via the oxidative stress response (DNA-binding protein MrgA); spores of strains lacking these various gene products have been also included in this study. Wild-type and mutant spores (Table 1) were subjected to three different types of ionizing radiation, X rays, protons, and high-energy Fe ions.
Role of spore core components in ionizing radiation resistance.
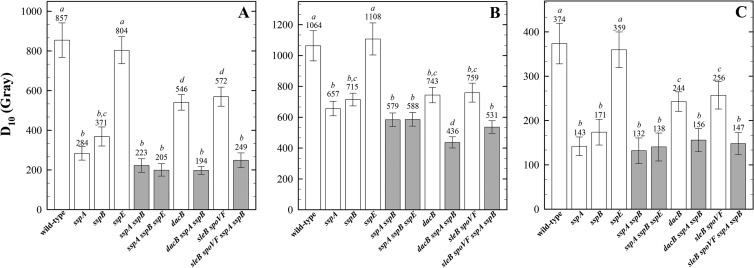
Spore structures (e.g., the spore coat and cortex) and components in the spore core (e.g., Ca-DPA, α/β-type SASP) have previously been demonstrated to protect spores from inactivation after exposure to various sporicidal treatments such as exposure to UV radiation, hydrogen peroxide, or heat (reviewed in references 4, 6, and 7). Spores lacking various spore core components were irradiated with X rays (Fig. 1A), protons (Fig. 1B), or Fe ions (Fig. 1C). In order, from most to least sensitive, were spores of mutants in sspA (encoding one of two major α/β-type SASP [38]) > sspB (encoding the less abundant major α/β-type SASP [38]) > dacB, previously shown to exhibit a 1.6-fold-increased core water content (52 to 65%) compared to that of wild-type spores (34 to 41%) (22, 23) > sleB plus spoVF (which allow isolation of stable spores that lack Ca-DPA [39]) > sspE (encoding the only γ-type SASP; termed γ− spores), which is equivalent to the wild type (Fig. 1).
FIG 1.
Spore resistance to irradiation with X rays (A), protons (B), and high-energy charged Fe ions (C). Spores differing in various core components, including major α/β-type SASP, the single γ-type SASP, and Ca-DPA (due to loss of DPA synthetase encoded by spoVF), or core water content (due to the dacB mutation), with (white bars) and without (gray bars) protection by α/β-type SASP. Spores were irradiated in water, and D10 values are expressed as averages ± standard deviations (n = 3) as described in the text. Lowercase letters above the bars denote groups significantly different by ANOVA (P < 0.05). Note the differences in the y axes of panels A, B, and C.
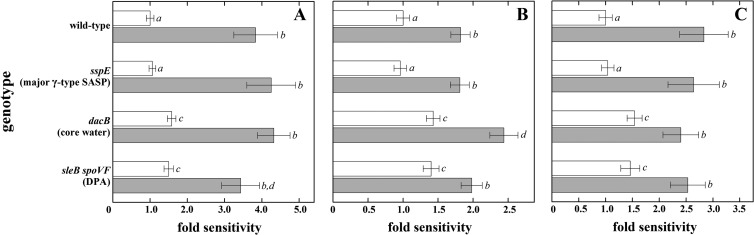
Radiation resistance of spores carrying mutations in sspE, dacB, or both sleB and spoVF was tested either in a wild-type background or in an sspA sspB background lacking both major α/β-type SASP (termed α−β− spores) (Fig. 2). The combined effects on spore sensitivity, from most sensitive to least, were as follows: α−β− dacB > α−β− sleB spoVF > α−β− ≫ wild-type ≈ γ− (Fig. 2). The results show that spore core dehydration and DPA, but not γ-type SASP, function in addition to α/β-type SASP as radioprotectants in the spore core.
FIG 2.
Impact of spore core-specific components on spore resistance to different types of ionizing radiation: X rays (A), protons (B), and high-energy charged Fe ions (C) with (white bars) and without (gray bars) protection by α/β-type SASP. Relative spore sensitivity was expressed as the ratio of the D10 value of spores of each mutant strain to the D10 value of spores of the corresponding reference strain (wild-type or α−β−) from each irradiation, using data from Fig. 1. Data are averages and standard deviations (n = 3). Actual data values are given above the corresponding columns in Fig. 1. Lowercase letters next to the bars denote groups significantly different by ANOVA (P < 0.05).
Role of oxidative stress responses and DNA repair in spore ionizing radiation resistance.
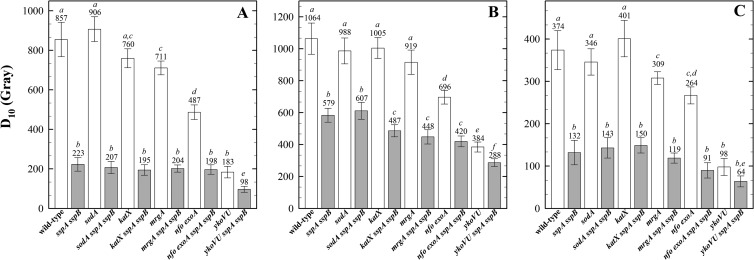
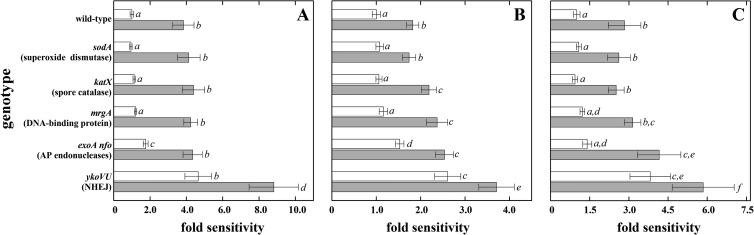
Spores of strains defective in ROS detoxification, the oxidative stress response DNA-binding protein MrgA, or DNA repair were exposed to X rays, protons, and Fe ions, and their sensitivities were compared (Fig. 3). Spores deficient in radical detoxification (sodA or katX spores) were no more sensitive than wild-type spores to any type of ionizing radiation tested (Fig. 3), as expected given the minimal activity of enzymes in the spore core. Spores lacking a major oxidative-stress-protective protein (mrgA spores) were significantly more sensitive than wild-type spores to X rays and Fe ions, but not to protons, which could be due to the different spectra of lesions induced by the three types of radiation (9, 11, 15) (Fig. 3). Spores lacking BER (exoA nfo spores) and double-strand break repair (ykoVU spores) were both significantly more sensitive to all types of radiation than wild-type spores (Fig. 3). Comparison of the spore radiation resistance in wild-type and α−β− backgrounds showed that in response to X rays only the ykoVU spores showed an additive effect with the loss of α/β-type SASP (Fig. 4A). In response to proton exposure, the effects of the katX, mrgA, exoA nfo, and ykoVU mutations (but not sodA) were all potentiated in the α−β− background (Fig. 4B). In response to Fe ions, only the exoA nfo (BER-deficient) and ykoVU (NHEJ-deficient) mutant spores demonstrated a significantly enhanced sensitivity in the α−β− background (Fig. 4C).
FIG 3.
Spore resistance to irradiation with X rays (A), protons (B), and high-energy charged Fe ions (C). Spores deficient in radical detoxification by SodA (superoxide dismutase), KatX (major spore catalase), DNA protection by MrgA (oxidative stress resistance DNA-binding protein), DNA repair by ExoA and Nfo, or YkoV and YkoU (with [white bars] and without [gray bars] protection by α/β-type SASP), were exposed in water, and D10 values are expressed as averages ± standard deviations (n = 3) as described in the text. Lowercase letters above the bars denote groups significantly different by ANOVA (P < 0.05). Note the differences in the y axes of panels A, B, and C.
FIG 4.
Relative ionizing radiation sensitivities of spores lacking radical detoxification by SodA or KatX, DNA protection by MrgA, or DNA repair by ExoA plus Nfo or YkoV plus YkoU to X rays (A), protons (B), and high-energy charged Fe ions (C) with (white bars) and without (gray bars) protection by α/β-type SASP. Relative spore sensitivity is expressed as the ratio of the D10 value of spores of each mutant strain to the D10 value of spores of the corresponding reference strain (wild-type or α−β−) from each irradiation, using data from Fig. 3. Data are averages and standard deviations (n = 3). Actual data values are given above the corresponding columns. Lowercase letters next to the bars denote groups significantly different by ANOVA (P < 0.05).
DISCUSSION
In regard to the characteristic resistance to ionizing radiation, spores of the Gram-positive bacterium B. subtilis have often been used to evaluate sterilization efforts in areas of food preservation, medical sterilization, and decontamination of potential biohazardous materials (3, 4, 6). Ionizing radiation is known to cause damage to numerous targets within the cell, including DNA, due both to (i) direct interaction of the radiation with the target molecule and to (ii) indirect effects due to the production of reactive species such as oxygen radicals (9, 10, 12, 13, 15). Ionizing radiation induces a large variety of damages to DNA bases and through interaction with the sugar moiety can cause formation of DNA strand breaks, either SSB or DSB. DNA strand breaks were reported to be the major lesions in the genome of spores exposed to ionizing radiation (reviewed in references 4 and 7), caused either directly by the highly energetic photons or accelerated particles or indirectly via interaction of radiation-induced free radicals, e.g., ROS, with the DNA (9, 13, 15, 16). The results of several radiobiological studies at accelerators and in space experiments lead to the assumption that at least two qualitatively different lesions are formed depending on the type and energy of the applied ionizing radiation. The spore's structure and chemical composition play major roles in spore resistance (6, 7, 18). The spore's general structure is very different from that of a growing cell, with a number of features and constituents unique to spores (4). Several of these features have been shown to be involved in spore resistance to some chemicals and physical treatments, but little is known of their role in spore resistance to ionizing radiation. Spore DNA is saturated with a group of SASP that bind to DNA largely on the outside of the DNA helix and straighten and stiffen the DNA while changing the DNA to an A-like helix (reviewed in reference 6 and references therein). As a consequence, DNA properties in spores are dramatically different from those in vegetative cells, e.g., spore DNA complexed with SASP is much more resistant to chemical attack, thermal degradation, and UV radiation (6, 7, 17, 22, 40), strongly suggesting that SASP may also be important for spore resistance to ionizing radiation, as indicated in earlier studies testing mutants lacking a single SASP (17, 33).
The current work has shown that protective components in the spore core are important determinants of spore resistance to X ray, proton, and heavy Fe ion bombardment with the order of importance being as follows: α/β-type SASP ≫ core dehydration > DPA. In contrast, the single major γ-type SASP does not bind to DNA and plays no protective role in spore ionizing radiation resistance, as has also been found when spore resistance to many other agents has been tested (23, 37; reviewed in reference 6) (Fig. 1 and 2). In the dormant state, spores remain metabolically inactive, and thus substantial DNA, protein, tRNA, and ribosome damage may accumulate, and the spores are incapable of repairing and/or degrading such damaged DNA and proteins (6, 7, 18). Consequently, damage to DNA, proteins, or other essential components of bacterial spores poses a unique problem, since such repair of such damage does not occur until the processes of germination and outgrowth, when spores reactivate and prepare to return to vegetative growth (6, 7, 28). Repair of DSB and base modification/loss via NHEJ and BER appear to be important mechanisms for spore resistance to ionizing radiation, and the MrgA-mediated oxidative stress response (with effects in wild-type spores for X-ray stress and in α−β− spores for proton radiation resistance) plays a lesser role. In contrast, ROS detoxification by catalase or superoxide dismutase appears to play a very minor role, if any, in spore resistance to ionizing radiation, as is also the case in spore resistance to oxidizing agents such as hydrogen peroxide and superoxide (20, 21) (Fig. 3 and 4). Clearly the data indicate that spore DNA is a major target of lethal ionizing radiation damage, based on the observation that the major factors contributing significantly to spore survival (Fig. 3 and 4) in response to different types of ionizing radiation are known to be important in either DNA protection in spores or DNA repair in spore germination and outgrowth. The findings in this communication thus complement the results from microarray experiments demonstrating upregulation of stress-related regulons responding to DNA damage (SOS response) and oxidative stress (PerR regulon) during germination of spores exposed to space conditions, including the effects of galactic cosmic radiation (41).
Spores appear to have two possible ways to minimize deleterious effects of ionizing radiation: (i) by protecting dormant spore DNA from damage in the first place (Fig. 1 and 2) and (ii) by ensuring detoxification and repair of direct and indirect damage during spore germination (Fig. 3 and 4). Because ionizing radiation causes such a wide spectrum of direct and indirect DNA damage, it will be interesting to determine the contributions of additional error-free and error-prone DNA repair pathways (e.g., mismatch repair, translesion synthesis, and homologous recombination [29, 42–44]), to gain further detailed insights into spore resistance to ionizing radiation.
ACKNOWLEDGMENTS
We thank Andrea Schröder and Petra Schwendner for their excellent technical assistance during parts of this work and Mario Pedraza-Reyes for his generous donation of B. subtilis strains. We are very grateful to all UF-PTI and HIMAC operators for their excellent support during proton and Fe ion irradiations. We express gratitude to Thomas Berger and Daniel Matthiä for their consulting, providing technical data and discussion.
R.M., M.R., and G.R. were supported by DLR grant DLR-FuE-Projekt ISS Nutzung in der Biodiagnostik, Programm RF-FuW, Teilprogramm 475, and DLR Forschungssemester-fellowship (to R.M.). Work in the Setlow lab was supported by a U.S. Department of Defense Multi-University Research Initiative through the U.S. Army Research Laboratory and the U.S. Army Research Office under contract number W911NF-09-1-0286.
These results will be included in the research reports of the HIMAC project 13J302 (hiiSPORES project).
Footnotes
Published ahead of print 11 October 2013
REFERENCES
- 1.Blatchley ER, Meeusen A, Aronson AI, Brewster L. 2005. Inactivation of Bacillus spores by ultraviolet or gamma radiation. J. Environ. Eng. 131:1245–1252. 10.1061/(ASCE)0733-9372(2005)131:9(1245) [DOI] [Google Scholar]
- 2.Horneck G. 1994. HZE particle effects in space. Acta Astronaut. 11:749–755 [DOI] [PubMed] [Google Scholar]
- 3.Niebuhr SE, Dickson JS. 2003. Destruction of Bacillus anthracis strain Sterne 34F2 spores in postal envelopes by exposure to electron beam irradiation. Lett. Appl. Microbiol. 37:17–20. 10.1046/j.1472-765X.2003.01337.x [DOI] [PubMed] [Google Scholar]
- 4.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of bacterial endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548–572. 10.1128/MMBR.64.3.548-572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horneck G, Klaus DM, Mancinelli RL. 2010. Space microbiology. Microbiol. Mol. Biol. Rev. 74:121–156. 10.1128/MMBR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setlow P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514–525. 10.1111/j.1365-2672.2005.02736.x [DOI] [PubMed] [Google Scholar]
- 7.Setlow P. 2007. I will survive: DNA protection in bacterial spores. Trends Microbiol. 15:172–180. 10.1016/j.tim.2007.02.004 [DOI] [PubMed] [Google Scholar]
- 8.Cadet J, Delatour T, Douki T, Gasparutto D, Pouget JP, Ravanat JL, Sauvaigo S. 1999. Hydroxyl radicals and DNA base damage. Mutat. Res. 424:9–21 [DOI] [PubMed] [Google Scholar]
- 9.Goodhead DT. 1994. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol. 65:7–17 [DOI] [PubMed] [Google Scholar]
- 10.Hutchinson F. 1985. Chemical changes induced in DNA by ionizing radiation. Prog. Nucleic Acid Res. Mol. Biol. 32:115–154 [DOI] [PubMed] [Google Scholar]
- 11.Hada M, Sutherland BM. 2006. Spectrum of complex DNA damages depends on the incident radiation. Radiat. Res. 165:223–230. 10.1667/RR3498.1 [DOI] [PubMed] [Google Scholar]
- 12.Sutherland BM, Bennett PV, Sidorkina O, Laval J. 2000. Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry 39:8026–8031. 10.1021/bi9927989 [DOI] [PubMed] [Google Scholar]
- 13.Lomax ME, Gulston MK, O'Neill P. 2002. Chemical aspects of clustered DNA damage induction by ionising radiation. Radiat. Prot. Dosimetry 99:63–68. 10.1093/oxfordjournals.rpd.a006840 [DOI] [PubMed] [Google Scholar]
- 14.Dianov GL, O'Neill P, Goodhead DT. 2001. Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. Bioessays 23:745–749. 10.1002/bies.1104 [DOI] [PubMed] [Google Scholar]
- 15.Yokoya A, Shikazono N, Fujii K, Urushibara A, Akamatsu K, Watanabe R. 2008. DNA damage induced by the direct effect of radiation. Radiat. Phys. Chem. 77:1280–1285. 10.1016/j.radphyschem.2008.05.021 [DOI] [Google Scholar]
- 16.Asaithamby A, Chen DJ. 2011. Mechanism of cluster DNA damage repair in response to high-atomic number and energy particles radiation. Mutat. Res. 711:87–99. 10.1016/j.mrfmmm.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granger AC, Gaidamakova EK, Matrosova VY, Daly MJ, Setlow P. 2011. Effects of Mn and Fe levels on Bacillus subtilis spore resistance and effects of Mn2+, other divalent cations, orthophosphate, and dipicolinic acid on protein resistance to ionizing radiation. Appl. Environ. Microbiol. 77:32–40. 10.1128/AEM.01965-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leggett MJ, McDonnell G, Denyer SP, Setlow P, Maillard JY. 2012. Bacterial spore structures and their protective role in biocide resistance. J. Appl. Microbiol. 113:485–498. 10.1111/j.1365-2672.2012.05336.x [DOI] [PubMed] [Google Scholar]
- 19.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 11:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagyan I, Casillas-Martinez L, Setlow P. 1998. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by SigmaF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 180:2057–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casillas-Martinez L, Setlow P. 1997. Alkyl hydroperoxide reductase, catalase, MrgA, and superoxide dismutase are not involved in resistance of Bacillus subtilis spores to heat or oxidizing agents. J. Bacteriol. 179:7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popham DL, Sengupta S, Setlow P. 1995. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl. Environ. Microbiol. 61:3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moeller R, Setlow P, Reitz G, Nicholson WL. 2009. Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation. Appl. Environ. Microbiol. 75:5202–5208. 10.1128/AEM.00789-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibarra JR, Orozco AD, Rojas JA, López K, Setlow P, Yasbin RE, Pedraza-Reyes M. 2008. Role of the Nfo and ExoA apurinic/apyrimidinic endonucleases in repair of DNA damage during outgrowth of Bacillus subtilis spores. J. Bacteriol. 190:2031–2038. 10.1128/JB.01625-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16–37. 10.1016/j.jmb.2006.01.059 [DOI] [PubMed] [Google Scholar]
- 26.Pitcher RS, Brissett NC, Doherty AJ. 2007. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 61:259–282. 10.1146/annurev.micro.61.080706.093354 [DOI] [PubMed] [Google Scholar]
- 27.Salas-Pacheco JM, Setlow B, Setlow P, Pedraza-Reyes M. 2005. Role of the Nfo (YqfS) and ExoA apurinic/apyrimidinic endonucleases in protecting Bacillus subtilis spores from DNA damage. J. Bacteriol. 187:7374–7381. 10.1128/JB.187.21.7374-7381.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setlow B, Setlow P. 1996. Role of DNA repair in Bacillus subtilis spore resistance. J. Bacteriol. 178:3486–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. 2012. DNA repair and genome maintenance in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 76:530–564. 10.1128/MMBR.05020-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 45:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nicholson WL, Setlow P. 1990. Sporulation, germination, and outgrowth, p 391–450 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. J. Wiley and Sons, Sussex, England [Google Scholar]
- 32.Moeller R, Stackebrandt E, Reitz G, Berger T, Rettberg P, Doherty AJ, Horneck G, Nicholson WL. 2007. Role of DNA repair by non-homologous end joining (NHEJ) in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV and ionizing radiation. J. Bacteriol. 189:3306–3311. 10.1128/JB.00018-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeller R, Setlow P, Horneck G, Berger T, Reitz G, Rettberg P, Doherty AJ, Okayasu R, Nicholson WL. 2008. Roles of the major, small, acid-soluble spore proteins and spore-specific and universal DNA repair mechanisms in resistance of Bacillus subtilis spores to ionizing radiation from X-rays and high-energy charged-particle bombardment. J. Bacteriol. 190:1134–1140. 10.1128/JB.01644-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okayasu R, Okada M, Okabe A, Noguchi M, Takakura K, Takahashi S. 2006. Repair of DNA damage induced by accelerated heavy ions in mammalian cells proficient and deficient in the non-homologous end-joining pathway. Radiat. Res. 165:59–67. 10.1667/RR3489.1 [DOI] [PubMed] [Google Scholar]
- 35.Moeller R, Reitz G, Li Z, Klein S, Nicholson WL. 2012. Multifactorial resistance of Bacillus subtilis spores to high-energy proton radiation: role of spore structural components and the homologous recombination and non-homologous end joining DNA repair pathways. Astrobiology 12:1069–1077. 10.1089/ast.2012.0890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Z, Lopatiuk-Tirpak O, Zeidan O, Sruprisan SI, Meeks SL, Slopsema R, Flampouri S, Li Z. 2012. An experimental investigation into the effect of periodic motion on proton dosimetry using polymer gel dosimeters and a programmable motion platform. Phys. Med. Biol. 57:649–663. 10.1088/0031-9155/57/3/649 [DOI] [PubMed] [Google Scholar]
- 37.Setlow B, Setlow P. 1993. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl. Environ. Microbiol. 59:3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hackett RH, Setlow P. 1988. Properties of spores of Bacillus subtilis strains which lack the major small, acid-soluble protein. J. Bacteriol. 170:1403–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magge A, Granger AC, Wahome PG, Setlow B, Vepachedu VR, Loshon CA, Peng L, Chen D, Li YQ, Setlow P. 2008. Role of dipicolinic acid in the germination, stability, and viability of spores of Bacillus subtilis. J. Bacteriol. 190:4798–4807. 10.1128/JB.00477-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loshon CA, Genest PC, Setlow B, Setlow P. 1999. Formaldehyde kills spores of Bacillus subtilis by DNA damage and small, acid-soluble spore proteins of the alpha/beta-type protect spores against this DNA damage. J. Appl. Microbiol. 87:8–14 [DOI] [PubMed] [Google Scholar]
- 41.Nicholson WL, Moeller R, Team PROTECT. Horneck G. 2012. Transcriptomic responses of germinating Bacillus subtilis spores exposed to 1.5 years of space parameters and simulated martian conditions on the EXPOSE-E experiment PROTECT. Astrobiology 12:469–486. 10.1089/ast.2011.0748 [DOI] [PubMed] [Google Scholar]
- 42.Ayora S, Carrasco B, Cárdenas PP, César CE, Cañas C, Yadav T, Marchisone C, Alonso JC. 2011. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 35:1055–1081. 10.1111/j.1574-6976.2011.00272.x [DOI] [PubMed] [Google Scholar]
- 43.Bolz NJ, Lenhart JS, Weindorf SC, Simmons LA. 2012. Residues in the N-terminal domain of MutL required for mismatch repair in Bacillus subtilis. J. Bacteriol. 194:5361–5367. 10.1128/JB.01142-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivas-Castillo AM, Yasbin RE, Robleto E, Nicholson WL, Pedraza-Reyes M. 2010. Role of the Y-family DNA polymerases YqjH and YqjW in protecting sporulating Bacillus subtilis cells from DNA damage. Curr. Microbiol. 60:263–267. 10.1007/s00284-009-9535-3 [DOI] [PubMed] [Google Scholar]
- 45.Mason JM, Setlow P. 1986. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J. Bacteriol. 167:174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wuytack EY, Boven S, Michiels CW. 1998. Comparative study of pressure-induced germination of Bacillus subtilis spores at low and high pressures. Appl. Environ. Microbiol. 64:3220–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Setlow B, Atluri S, Kitchel R, Koziol-Dube K, Setlow P. 2006. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective alpha/beta-type small acid-soluble proteins. J. Bacteriol. 188:3740–3747. 10.1128/JB.00212-06 [DOI] [PMC free article] [PubMed] [Google Scholar]