Abstract
We investigated the amplification and purification of phage preparations with respect to titer, contamination level, stability, and technical affordability. Using various production systems (wave bags, stirred-tank reactors, and Erlenmeyer flasks), we obtained peak titers of 109 to 1010 PFU/ml for T4-like coliphages. Phage lysates could be sterilized through 0.22-μm membrane filters without titer loss. Phages concentrated by differential centrifugation were not contaminated with cellular debris or bacterial proteins, as assessed by electron microscopy and mass spectrometry, respectively. Titer losses occurred by high-speed pelleting of phages but could be decreased by sedimentation through a sucrose cushion. Alternative phage concentration methods are prolonged medium-speed centrifugation, strong anion-exchange chromatography, and ultrafiltration, but the latter still allowed elevated lipopolysaccharide contamination. T4-like phages could not be pasteurized but maintained their infectivity titer in the cold chain. In the presence of 10 mM magnesium ions, phages showed no loss of titer over 1 month at 30°C.
INTRODUCTION
Antibiotic-resistant bacteria are observed with increasing prevalence. Alternatives to antibiotics have thus become a public health priority for the treatment of bacterial infections (1). Antibiotic resistance is not a concern just for intensive care units. Common infections such as Escherichia coli diarrhea in children (2–4) from developing countries are now highly resistant to many antibiotics. Some E. coli isolates, particularly from the Indian subcontinent, are now resistant to literally all known antibiotics (5), and even worse, these isolates are found in the environment outside hospitals (6).
In the past, we have explored alternatives to antibiotics for E. coli diarrhea in children from Bangladesh. While passive immunity via bovine milk immunoglobulins had therapeutic effects against rotavirus diarrhea (7), it demonstrated no effect against E. coli diarrhea (8). In contrast, oral application of probiotic lactobacilli showed treatment efficacy against bacterial but not rotavirus diarrhea (9). However, the production of high-titer probiotic lactobacilli is relatively costly. We therefore started to explore phage therapy for the treatment of E. coli diarrhea in children from Bangladesh (10). Since oral phages can amplify on the infecting E. coli strain in the guts of patients, low-titer, and thus cheap, phage preparations may be sufficient for treatment.
Phage therapy was developed in Eastern Europe (11, 12). The antibiotic crisis has rekindled the interest in phage therapy in the Western world. Commercial phage preparations, including preparations against E. coli diarrhea, are a registered medicine in Russia, where they are sold as over-the-counter products in pharmacies. So far, Russian companies have not documented the composition of their phage cocktails, their preparation and purity, or their safety and efficacy in humans with scientific reports. Previously, we characterized the composition of our T4-like phage cocktails (10) and examined the Russian E. coli-Proteus phage cocktail (13) for its phage composition and gene content. In the present study, we investigated the amplification and purification of T4-like coliphages for phage therapy purposes.
MATERIALS AND METHODS
Bacterial strains and phages.
From our collection of about 100 previously described T4-like E. coli phages (10), 11 T4-like phages, namely, NPC (Nestlé Phage Collection) 1000, 1001, 1002, 1003, 1004, 1005, 1006, 1007, 1008, 1009, and 1024, were selected for the experiments reported in the present report. These phages were isolated from the stools of children hospitalized with acute diarrhea at the International Centre for Diarrheal Diseases Research in Dhaka/Bangladesh (ICDDR,B) (14). Their selection was based on their suitability for composing a phage cocktail providing a good coverage of diarrhea-associated E. coli pathogens. Not all experiments were conducted with all 11 T4-like phages. The number and identity of phages used for a given experiment are given in the appropriate section of text. Phage NPC 1003 was propagated on E. coli strain WG5, obtained from J. Jofre, University of Barcelona (Spain), which is a common indicator strain used in phage ecology studies. All other NPC phages were propagated on E. coli strain K803, a K-12 derivative lacking prophage lambda, obtained from E. Kutter, Evergreen College, Olympia, WA. This strain lacks restriction-modification genes and contains only the core part of the lipopolysaccharide (LPS). NPC 1000 was also propagated on an E. coli B strain obtained from a culture collection (ATCC 11303). The bacterial strains were stored at −20°C on Protect Bacterial Preservers beads (Technical Service Consultants, Lancashire, United Kingdom). The phages were kept at 4°C in liquid bacterial growth medium, with the bacterial cells having been removed by centrifugation and filtration. For long-term storage, the phage solution was mixed with 15% glycerol, and aliquots were frozen in liquid nitrogen and stored at −20°C or −80°C.
Media and growth conditions.
The E. coli strains were grown at 37°C in brain heart infusion (BHI) broth and agar (Oxoid CM 225 and CM 375, respectively), Hershey broth, Luria broth (LB), and glycerol-Casamino Acids broth (GCA) (15). Hershey broth was supplemented with Casamino Acids (15 g/liter), and LB broth was supplemented with CaCl2 (0.03%) and glucose (0.2%). Cultures were kept at 4°C for a maximum of 30 days. Growth conditions are specified in Results.
Propagation and enumeration of phages.
All 11 NPC test phages were propagated by inoculating the specified broth with 1% exponentially growing bacterial culture and 2% (final concentration) phage lysate (109 PFU/ml). If not indicated otherwise, broth was inoculated with 1% overnight bacterial culture and incubated at 37°C for 2 h to reach an approximate optical density at 600 nm (OD600) of 0.2 to 0.4 with exponentially growing cells. Phage lysate was then added at a multiplicity of infection (MOI) of 10, and the culture was incubated at 37°C for a further 5 h, with gentle agitation (100 rpm), until cell lysis occurred. The phage lysate was cleared of cellular debris by centrifugation (30 min, 5,000 × g, 4°C), and the supernatant was then filtered through a 0.2-μm-pore-size vacuum-driven disposable filtration system (Stericup; Millipore). Phage was enumerated using a plaque assay according to standard protocols (15). Phage dilutions were prepared in TS (1 g/liter tryptone, 8.5 g/liter NaCl): 100 μl of an appropriate dilution and 100 μl of an exponentially growing bacterial culture were mixed together, incubated at room temperature for 15 min, and used to inoculate 3 ml of top agar. Plates were incubated overnight at 37°C, and phage plaques were counted.
Phage amplification in fermenter.
T4-like phage NPC 1000 was amplified in a 16-liter stirred-tank L1523 bioengineering fermenter filled with 8 liters of LB medium (37°C; pH maintained at 7.0 with NaOH), with baffles and a sparger for the air supply. The medium was inoculated with 3% (vol/vol) of an overnight E. coli K-12 culture. At an OD600 of 0.5, the bacteria were infected with a single phage strain at an MOI of 1. The fermentation (stirrer speed, 400 rpm; aeration, 1 liter/min) was stopped after 7 h. Unlysed cells and cellular debris were removed by centrifugation (45 min, 5,000 × g, 4°C) and sterile filtration (0.22-μm Stericup filters; Millipore). Phage was titrated using the double-layer plaque assay as described previously (16).
Phage amplification in wave bag disposable system and purification using ultrafiltration.
T4-like phages NPC 1001, NPC 1002, and NPC 1005 were amplified separately in a wave bag disposable system (Biostat CultiBag RM system; Sartorius) with optical pH and pO2 probing. The bags were filled with LB medium (37°C; pH maintained at 7.0 with 10% NaOH) to half their total capacity. LB medium was inoculated with 3% of an overnight E. coli K-12 culture. At an OD600 of 0.5, 3% phage lysate was added to reach an MOI of 0.1. The rocker speed was increased gradually to the maximum speed over 2 h. Aeration at 1 liter/min to 6 liters/min was performed with pure oxygen. The fermentation trials were stopped after 4 h. Sixty-five liters of centrifugation-cleared (30 min, 5,000 × g, 4°C) phage lysates from 3 different fermentation experiments (25 liters of NPC 1001, 25 liters of NPC 1002, and 15 liters of NPC 1005) in the wave bag system was sterile filtered using a peristaltic pump and a cartridge filter (Durapore 0.22-μm hydrophilic cartridge filter; 10 in/25 cm; CVGL 01T P3). After sterile filtration, the phages from the three combined phage lysates were isolated using a Techsep 15-kDa ultrafiltration system (carbon M2, 0.32 m2; Novasep); the sterile-filtered phage lysates from the three phages were passed along the flow system. The retentate was washed with 50 liters of sterile phage buffer (150 mM NaCl, 10 mM MgSO4 in sterile water at pH 7.2) in a recirculation loop in order to change the buffer. Finally, the retentate was suspended in the phage buffer. Phage lysates obtained with all 11 test phages in 2-liter Erlenmeyer flasks (LB broth, 37°C, 16 h, 100 rpm, MOI = 0.5) were likewise tested for phage concentration by ultrafiltration.
Purification using centrifugation.
Phage lysates produced in 2-liter Erlenmeyer flasks were cleared of cellular debris by low-speed centrifugation (30 min, 4,500 × g, 4°C), and phage was then pelleted from the cleared lysate by ultracentrifugation (40 min, 35,000 × g, 4°C). The pellet was resuspended in either sterile water or phage buffer (150 mM NaCl and 10 mM MgSO4; pH adjusted to 7.2). Alternatively, the cleared lysate was layered on a 20% (wt/vol) sucrose cushion before ultracentrifugation.
Phage could also be concentrated from the cleared lysate by prolonged medium-speed centrifugation (4 h, 20,000 × g, 4°C).
Purification using chromatography.
T4-like phages NPC 1001, 1004, 1006, and 1008, propagated in 2-liter Erlenmeyer flasks, were also concentrated by strong anion-exchange chromatography on a CIM QA disk monolithic column (column volume = 8 ml; BiaSeparations, Slovenia). Cleared phage lysates were loaded onto the column by using a peristaltic pump and buffer A (125 mM NaHPO4, pH 7.0) as the mobile phase. The elution was done by a stepwise gradient with buffer A containing increasing concentrations of NaCl (250 mM = buffer B1, 500 mM = buffer B2, and 1 M = buffer B3). The flow rate was 14 ml/min.
PEG precipitation.
NPC 1000 phage lysate was produced in a 2-liter Erlenmeyer flask, and the cellular debris was removed using low-speed centrifugation (15 min, 1,500 × g, 4°C) (Sorvall RC 5C Plus centrifuge; SLA-1500 rotor). Bacteriophage was precipitated overnight at 4°C with polyethylene glycol (PEG) 6000 to 8000 (8 to 10% total [wt/vol]; Fluka) and NaCl (0.5 M; Fluka). The precipitated phage was collected by centrifugation (15 min, 11,000 × g, 4°C) (Sorvall RC 5C Plus centrifuge; SLA-1500 rotor) and suspended in phage buffer to obtain a 20- to 100-fold concentration. The phage was then extracted with 1 volume of chloroform (Fluka) for 20 min by gentle inversion; phases were then separated by centrifugation. The aqueous phase (upper phase), containing the phage, was centrifuged (15 min, 4,000 × g, 4°C) (Sorvall RC 5C Plus centrifuge; SLA-1500 rotor) and then filtered through 0.22-μm Stericup filters (Millipore).
Chloroform extraction.
Phage NPC 1000 particles were also recovered from imperfectly lysed E. coli K-12 and B cells at the end of the infection process. The fermentation broth was centrifuged (80 min, 5,500 × g, 4°C) (Sorvall RC 5C Plus centrifuge; SLA-1500 rotor), and the cell pellet was suspended and homogenized in phage buffer and agitated with a magnetic stirrer and 10% chloroform (Fluka). After sedimentation of the chloroform, the aqueous phase (upper phase) was centrifuged (15 min, 4,000 × g, 4°C) and the supernatant filtered through 0.22-μm filters (Millipore).
Phage stability experiments.
Using E. coli strain K-12, phage NPC 1024 was amplified in Hershey broth and LB. Half of each lysate was filtered, enumerated, and diluted in Hershey broth or LB to obtain a volume containing approximately 109 PFU/ml. The other half of each lysate was filtered, centrifuged at 16,000 × g for 30 min, and resuspended in 0.9% NaCl to obtain concentrated aliquots containing approximately 1011 PFU/ml. Aliquots were stored at 4°C for up to 96 days. After 7, 14, 21, 28, 56, and 96 days, aliquots were enumerated in triplicate by using a plaque assay. For short-term stability experiments, phage NPC 1000 was amplified as described in “Propagation and enumeration of phages.”
Mass spectrometry.
Mass spectrometry has become a reference method to distinguish closely related phages and bacterial strains. This method also allows assessment of the degree of contamination of phage preparations with compounds of the propagating bacterial strain. Therefore, mass spectra for all 11 test phages were recorded on a Bruker Autoflex matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Bremen, Germany) operating in delayed-extraction linear positive-ion mode. Ions formed upon irradiation by a pulsed UV laser beam (nitrogen laser, 337 nm) were accelerated to 20 kV. In parallel, the propagating bacterial host strains were investigated by this technology. For this purpose, 10 μl of sterile water was added to the cell pellet and homogenized. Twenty microliters of 70% formic acid was added to the pellet, and the sample contents were mixed vigorously. After centrifugation (2 min at 4,500 × g), the supernatant was used as a lysed bacterial sample.
A saturated solution of α-cyano-4-hydroxycinnamic acid (HCCA) in acetonitrile (30%) and 5% trifluoroacetic acid (TFA) was used as a matrix for the bacteriophages and the lysed bacteria. Typically, 1 μl of the saturated matrix was mixed with 1 μl of purified and concentrated bacteriophage sample or 1 μl of lysed bacteria. The resulting mixture was then deposited on a ground-steel target and allowed to dry at room temperature. Each mass spectrum was produced by averaging 250 laser shots spread over the entire sample spot surface. Several replicate spectra were taken for each sample under the same experimental conditions. The six replicate spectra with the highest overall signal intensity were selected to represent each sample. Peak lists were extracted with the Bruker Autoflex instrument software, admitting only peaks with a signal-to-noise (S/N) ratio of >15. External mass calibration was performed with a protein mixture containing bovine insulin, ubiquitin, cytochrome c, and myoglobin.
LPS test.
For all 11 test phages, the LPS (endotoxin) contamination level in the phage preparations was measured with the Limulus amoebocyte lysate test (Charles River, Wilmington, MA).
RESULTS
Phage cultivation. (i) Phage amplification in wave bags.
In our pilot plant, single-use bags made of a multilayer film of pharmaceutical-grade polyethylene were used to amplify the T4-like phages NPC 1001, 1002, and 1005. The bags were subjected to a rocking motion resulting in a low shear stress ideal for mass cell culture, including the propagation of bacterial cells. We inoculated 10-liter bags with LB and the laboratory E. coli strain K-12 and 60 min later added a T4-like phage (one phage strain per bag) at an MOI of 0.1. Figure 1 shows the infection course with the T4-like phage NPC 1005. The cell density, measured as the OD600, increased for an hour after infection, in parallel with a gradual increase in phage titer. However, when the phage titer reached 2 × 108 PFU/ml, the OD value decreased dramatically, although not completely. A phage titer of 1 × 109 PFU/ml was obtained in wave bags. Phages NPC 1001 and NPC 1002 behaved in the same way (data not shown).
FIG 1.
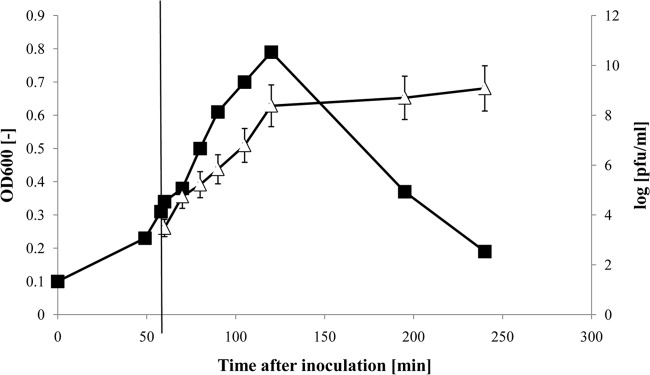
Amplification of T4-like phage NPC 1005 on E. coli strain K-12 in a 10-liter wave bag system (Biostat Cultibag RM; Sartorius). The time course after inoculation of the broth with K-12 bacteria is given on the abscissa. The infection time with phage NPC 1005 is indicated by the vertical line at 60 min. The growth of the bacterial culture was followed by reading the OD (filled squares), and the growth of phage was determined by infectivity titration. The phage titers are given as mean PFU/ml, with standard errors of the means, on a logarithmic scale (open triangles).
(ii) Phage amplification in stirred fermentation tank and in Erlenmeyer flasks.
Not all bacteriology laboratories are equipped with the wave bag fermentation system, which is more typical of pilot laboratories doing eukaryotic cell culture. Phage amplification can also be achieved in stirred fermentation tanks, which are more common in bacteriology laboratories. In a test with phage NPC 1000 on strain K-12 in LB medium, a high phage titer was also achieved in the lysate produced in a stirred-tank fermenter (see Fig. S1 in the supplemental material). Since a different phage strain, incubation time, and MOI were used, the titers cannot be compared directly with the phage titers produced in the wave bag system. Sophisticated technical installations are not needed for phage amplification. All 11 test phages could also be propagated to about 109 PFU/ml in large Erlenmeyer flasks fixed to a rocking support. For a given stirred phage batch culture (NPC 1000), final phage titers increased 10-fold, at most, when volumes changed over a thousandfold range. The choice of broth (LB, Hershey broth, or BHI) had only a weak effect on the final phage titer (Fig. 2). The infection of rapidly dividing cells in the early exponential growth phase gave much higher titers than those obtained by infecting cells which were still in the lag phase or in the late exponential growth phase (Fig. 2). Supplementing the LB with extra calcium or glucose led to a phage titer increase of <0.2 log. The phage strain and the time between bacterial and phage inoculation had the greatest impacts on final phage titers.
FIG 2.
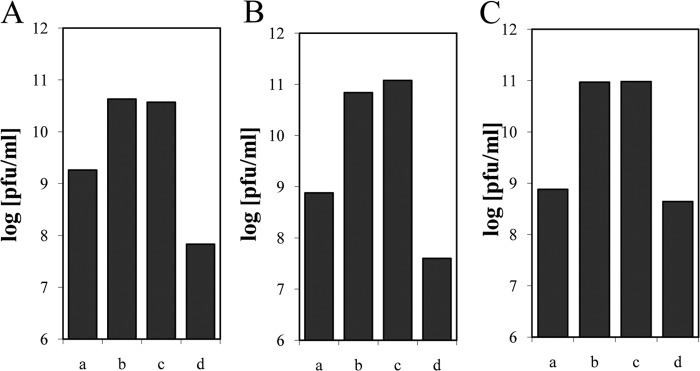
Effects of medium and growth phase on phage titer. T4-like phage NPC 1024 was propagated on E. coli strain K-12 in Hershey broth (A), BHI (B), and LB supplemented with calcium and glucose (C). The overnight culture was infected either directly (a) or after 2 (b), 3 (c), or 4 (d) h of bacterial growth. Typical values for one of three experiments are shown.
Cell association of phage and PEG precipitation.
When T4-infected E. coli cells are superinfected with T4 phage, a situation commonly encountered during phage propagation in bulk culture, lysis inhibition is observed and prevents the release of the intracellular T4 phage into the lysate. With K-12 cells infected with NPC 1000, half of the phage infectivity remained cell associated, as determined by chloroform extraction of the cell pellet. With E. coli strain B infected with NPC 1000, 10 times more phage infectivity was recovered by chloroform extraction from the cell pellet as was found in the cell-free supernatant (see Fig. S2A in the supplemental material).
A standard procedure for the purification of T4 phage consists of PEG precipitation followed by chloroform extraction of the pellet to remove PEG (17). For NPC 1000 produced on E. coli B, 90% of the phage infectivity from the supernatant could be recovered in the PEG pellet. However, for NPC 1000 produced on E. coli K-12, only 30% of the infectivity in the cell-free lysate was precipitated with PEG (see Fig. S2B in the supplemental material). When data for both properties were combined (cell association and PEG precipitation of phage), it was seen that comparable amounts of phage NPC 1000 were achieved on both E. coli propagation strains.
Ultracentrifugation and membrane filtration.
Cell-associated phage was not used for phage isolation. Phage was prepared only from the cell-free lysate, by a combination of low-speed centrifugation and sterile filtration across a 0.22-μm membrane. Since T4-like phages measure 220 nm along their long axis, the phage size was close to the pore size of the membrane filters, such that filtration could have led to phage loss (Fig. 3A). However, in comparing the infectivity titers of the 11 T4-like test phages before and after sterile filtration of fresh lysates, a <0.5-log (NPC 1003 and 1006) or no titer loss was observed (see Fig. S3 in the supplemental material).
FIG 3.
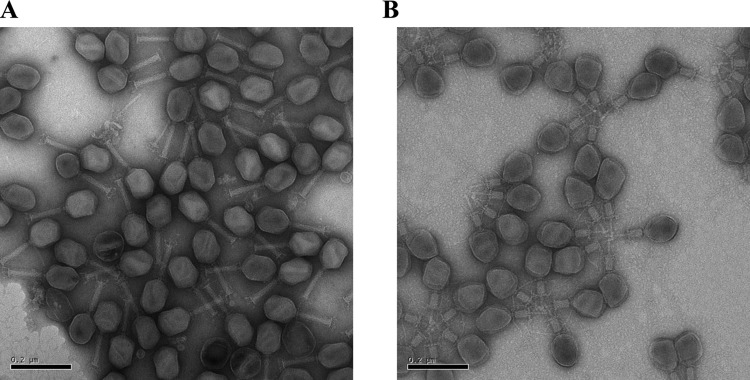
Negative-staining electron microscopic pictures of T4-like phages after ultracentrifugation. (A) Phage NPC 1000, showing T4-like phage particles with noncontracted tails. (B) Phage NPC 1003, showing T4-like phage particles with contracted tails. Phages form rosettes by aggregation via their exposed contracted terminal tail structures.
A standard procedure for the concentration of T4 phage is ultracentrifugation of the cell-free lysate supernatant at 35,000 × g for 20 min (17). With respect to the recovery of phage infectivity from the lysate, ultracentrifugation yielded mixed results for the 11 test phages. The phage pellet from NPC 1000 showed mainly morphologically intact T4-like phage, together with a few tailless phage heads, empty phage capsids, and isolated phage tails or tail tubes from the T4-like phage, while cellular debris was not seen (Fig. 3A). After centrifugation, the phage infectivity of the filtered lysate from NPC 1000 was nearly quantitatively recovered in the phage pellet (see Fig. S3 in the supplemental material). However, phages NPC 1003, 1006, and 1009 showed high-titer losses (2 log or greater) after ultracentrifugation (see Fig. S3A). After ultracentrifugation, the phage pellet of NPC 1003 showed a large percentage of T4-like phage particles with a contracted tail (Fig. 3B). NPC 1003 particles with contracted tails also showed a tendency to aggregate across the exposed tail tubes. Filtering these preparations across a 0.22-μm membrane caused a further substantial loss of phage infectivity (see Fig. S3B). However, in general, ejection of the phage genome was not seen in the NPC 1003 phage particles with contracted tails, and great differences with respect to phage titer loss by ultracentrifugation were observed for different preparations of phages NPC 1003, 1006, and 1009 (see Fig. S3A).
We suspected that the T4-like phages suffered damage from being pelleted under high gravitational force. This problem was solved by collecting the T4-like phages by high-speed centrifugation through a density cushion of 20% sucrose: the recovery rates of infectious T4-like phages increased to 50 to 90% for NPC 1003, 1006, and 1009.
Medium-speed centrifugation.
To decrease the mechanical stress to the phage particles, we reduced the gravitational force to 20,000 × g and prolonged the centrifugation time to 4 h in a refrigerated medium-speed centrifuge. T4-like phage particles are large enough to sediment under these conditions (4). However, the average recovery rate was only 60%, and frequently as much phage infectivity remained in the supernatant as was recovered with the phage pellet (see Fig. S4A in the supplemental material). While tails from the collected T4-like phage were not contracted, a higher degree of debris (only tails, only empty capsids, and membrane fragments) was recovered with medium-speed centrifugation (see Fig. S4B) than with high-speed centrifugation through a sucrose cushion. Apparently, the sucrose cushion presented a density barrier preventing the sedimentation of less-dense material such as proteins and lipids.
Chromatography.
A classical procedure for phage purification practiced in Eastern Europe is adsorption to DEAE-cellulose and recovery by low-speed centrifugation. Ninety percent of the phage infectivity was adsorbed when DEAE-cellulose was added at a concentration of 10% to the phage lysate (see Fig. S5 in the supplemental material). However, the adsorption capacity of DEAE-cellulose was not high for T4-like phages. When the concentration of DEAE-cellulose was decreased to 5 or 1% (wt/vol), only 38 or 12%, respectively, of the phage infectivity was adsorbed from the lysate (see Fig. S5).
More recently, a biotech company developed a commercial strong anion-exchange chromatography column (quaternary amine disk monolithic column; BiaSeparations) particularly adapted for the purification of T4 phages.
This system has the advantage that with 8-liter columns, hundreds of liters of cleared phage lysate can be treated in a reasonable time, and without the need for pressure application to the column. Phages NPC 1001, 1004, 1006, and 1008 were purified on this type of column. About 70% of the applied phage infectivity could be recovered in a single fraction (Fig. 4). However, up to 10,000 endotoxin units (EU) of LPS/ml, which represents the totality of the LPS present in the initial sample, coeluted with the phage fraction in all four purification assays.
FIG 4.
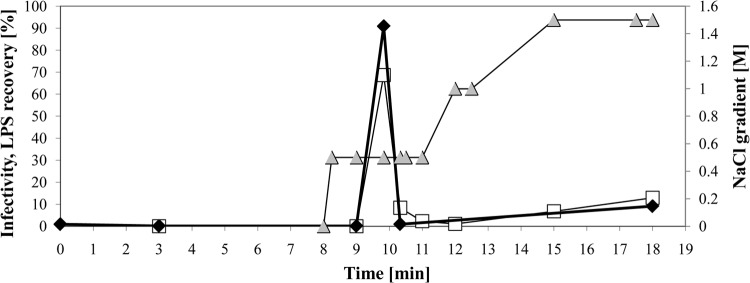
Chromatographic purification of T4-like phage NPC 1008 on BiaSeparations column. The elution time of the indicated fraction from the column is given in minutes on the abscissa. The phage infectivity recovered in the indicated fraction is given as a percentage of the phage infectivity (left ordinate) applied to the column (open squares). The LPS contents of the fractions are likewise given as percentages of the LPS concentration applied to the column (filled diamonds). Phage was eluted from the column by a gradient of NaCl (filled triangles), whose molarity is indicated on the right ordinate.
Ultrafiltration.
Volumes of up to 65 liters of sterile-filtered phage lysate were processed with a Techsep ultrafiltration system that has a 15-kDa cutoff. T4-like phages could be concentrated in less than 1/100 of the initial volume, with 100% recovery.
Ten T4-like test phages were concentrated individually from smaller volumes (800 μl) by ultrafiltration using a 30-kDa cutoff. Three phages (NPC 1000, 1001, and 1005) showed only 1 to 10% recovery with respect to the initial lysate, while good recoveries were achieved by ultrafiltration for the other 7 test phages (see Fig. S6 in the supplemental material). High levels of LPS were recovered by ultrafiltration for phages NPC 1002, 1003, 1005, and 1007. The six remaining phages displayed only low LPS contamination levels (see Fig. S7). We then used Endotrap affinity columns (Hyglos) that were specifically designed to remove lipopolysaccharides from biological samples targeted for medical application. However, with a single passage, we failed to reduce the LPS concentration in the T4-like phage preparations (data not shown).
Comparison of methods.
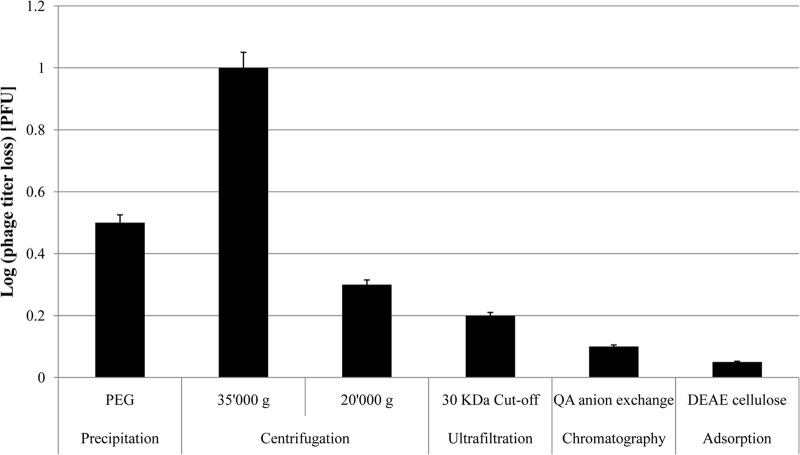
To allow a direct comparison of the different purification methods, we prepared lysates of K-12 cells infected with the same seed of phage NPC 1001, which were then purified by the above-mentioned methods. The phage titer in the purified fraction was compared to the phage titer in the lysate. Ultracentrifugation and PEG precipitation led to 1-log and 0.5-log phage losses, respectively, while medium-speed centrifugation, ultrafiltration, and the two chromatographic methods were associated with only minimal titer losses (Fig. 5). Ultracentrifugation at 35,000 × g led to 90% LPS removal, while neither ultrafiltration nor chromatography diminished the LPS loads of the phage preparations.
FIG 5.
Phage infectivity titer losses for NPC 1001 purified by six different preparation methods, as specified under the bars. Loss is expressed as the log PFU titer decrease with respect to the infectivity present in the lysate of the infected bacterial culture. The error bars represent the standard errors of the means for three experiments.
Mass spectrometry for phage identification.
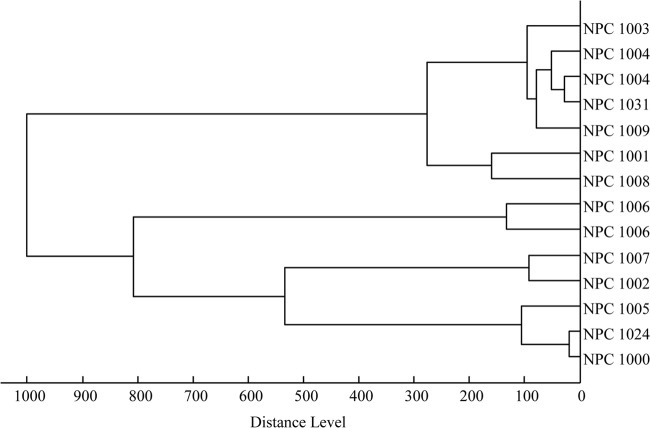
In a laboratory handling many T4-like phages, cross-contamination is a constant risk. It is thus important to have a quick method to distinguish T4-like phages to allow correct identification and tracking of phages from the seed stock to the phage cocktail. Due to chemical modification of T4 phage DNA, restriction analysis is not a useful analytical method. Sequencing of g23 PCR products is a possibility, but it tests only one, highly conserved phage gene, and this analysis is time-consuming. We therefore looked for quicker analytical tools to characterize T4-like phage isolates. T4-like phages purified by a combination of low-speed centrifugation, sterile filtration, and high-speed ultracentrifugation (without a sucrose cushion) yielded a simple mass spectrum with few peaks by MALDI-TOF analysis. Different T4-like phage strains showed distinct mass spectra (data not shown). The peak patterns of the mass spectra were automatically analyzed, and a dendrogram was constructed (Fig. 6). Already with the small set of analyzed T4-like phage preparations, the distinction of three branches could be visualized, which concurred with the definition of T4-like phage subgroups based on the much larger number of gene sequences of the major head gene, g23 (18). Independent isolations from the same T4-like phage strain yielded highly reproducible mass spectra (see branches for NPC 1004 and NPC 1006 in Fig. 6). The mass spectra of T4-like phages did not reveal molecule fragments shared with artificially lysed noninfected E. coli cells (see Fig. S8 in the supplemental material), thus excluding major contamination of the phage pellet with cellular compounds.
FIG 6.
Dendrogram calculated from the peak patterns of the mass spectra and separating the investigated T4-like phages into three groups concurring with the g23 DNA sequence classification (10). Identical NPC numbers refer to independent preparations of the same phage strain.
Conservation and storage.
Some phage strains are relatively heat resistant and are able to survive pasteurization temperatures. However, purified T4-like phages showed rapid titer losses at 70°C. After 30 min, all infectivity was lost, precluding the possibility of postprocessing pasteurization of T4-like phage preparations (see Fig. S9 in the supplemental material). At refrigeration temperature, T4-like phages in broth maintained infectivity titers over extended periods (Fig. 7A). A cocktail of nine NPC phages maintained its titer (<0.5-log titer decrease) for 2 years under refrigeration in the phage buffer described in Materials and Methods. However, in developing countries, a cold chain cannot always be maintained, and the high ambient temperatures might compromise the viability of phages. Therefore, we tested whether purified phages diluted in a buffer could be maintained over extended periods at an elevated ambient temperature of 30°C. Addition of a cryoprotective agent (trehalose) did not prevent a substantial phage titer loss at 30°C. However, addition of Mg2+ led to a phage preparation that maintained a good residual titer even after storage for more than a month at 30°C (Fig. 7B).
FIG 7.
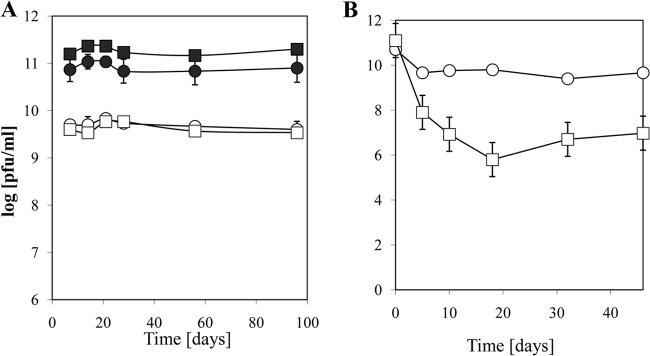
Long-term stability of T4-like phages under refrigeration and elevated ambient temperature. (A) Mean residual log10 infectivity titers and standard errors of the means for T4-like phage NPC 1024 stored in broth (open symbols) (circles, Hershey broth; squares, LB) or concentrated phage stored in 0.9% NaCl (filled symbols) for the indicated numbers of days at 4°C. (B) Mean residual log10 infectivity titers and standard errors of the means for T4-like phage NPC 1024 stored at 30°C in 0.9% NaCl (squares) and 0.9% NaCl containing 10 mM MgSO4 (circles).
DISCUSSION
To represent a potential practical solution for E. coli diarrhea in children from developing countries, large-scale phage amplification must be achieved in an inexpensive way, while ensuring good laboratory and manufacturing practice. In a prior study (6), we established that bacteriophages targeting pathogenic E. coli can be propagated on nonpathogenic laboratory E. coli strains, such as K-12, B, and WG-5, allowing propagation of phages outside biohazard safety laboratories, which substantially facilitates the production task. Nonpathogenic E. coli strains differ in their production of progeny phage and the release or retention of phage from the infected cell, which should be considered in choosing the optimal host for phage infection. Here we demonstrated that T4-like coliphage titers of ≥109 PFU/ml lysate can be achieved in pilot plant-sized volumes by use of wave bags, stirred-tank fermenters, or simply large Erlenmeyer flasks as a production system. Inexpensive microbiological media containing only food-grade supplements can be used for growing bacteria and phages.
T4-like phages have a suitable thermal stability: with refrigeration, they are stable for at least 1 year. When bivalent ions are added, even interruptions of the cold chain for at least 1 month are survived without substantial titer loss. With temperatures in excess of 70°C, T4-like phages are quickly killed, allowing their elimination from the production unit by steam sterilization. All T4-like phages from our phage strain collection had previously been characterized for their gene content, and no undesired genes were found; in addition, previous biological safety tests in healthy adults did not reveal adverse effects (10). A health risk for the personal handling of very large amounts of these bacterial viruses is thus unlikely.
Costs are incurred not only by the phage production system but also—and perhaps to an even greater extent—by the selected purification scheme. To remain affordable for populations in the developing world who have the most need for alternative approaches for the treatment of E. coli diarrhea, the purification technique must remain simple and inexpensive. We therefore did not consider up-scaling standard laboratory techniques, such as CsCl density equilibrium centrifugation, which are known to deliver highly purified phage preparations. Such preparations are necessary for considering intravenous injection but dispensable for oral phage application. Since the gut is physiologically exposed to decaying bacteria, including E. coli, humans have a relatively high tolerance level to bacterial lipopolysaccharides. While endotoxin limits for parenteral drugs have been established (19), there is no defined oral endotoxin limit dose. Oral administration of E. coli endotoxin to mice at a dose of up to 106 EU/mouse produced no evidence of toxicity (20). A different situation is encountered when oral phages infect the intestinal pathogen in the guts of diarrhea patients and release endo- and enterotoxins from the lysed pathogen in patients with a compromised gut barrier. Under these conditions, adverse reactions could occur and need to be investigated in future safety trials.
Ultrafiltration is probably the most cost-efficient phage concentration method and is a routine industrial procedure. However, this procedure is essentially a concentration method, not a purification method, for any molecules which are larger than the cutoff of the membrane. In the present study, LPS was concentrated with the phages, resulting in very high endotoxin levels in some phage preparations. It might still be possible to decrease LPS contamination by working with a 100-kDa cutoff. Much lower LPS concentrations were obtained with methods involving differential centrifugation or chromatography. Ultrafiltration, like ultracentrifugation and QA chromatography, requires costly equipment. We have not investigated what is needed in a country such as Bangladesh to produce phage, starting with a phage stock and ending with a phage cocktail ready to be used for treatment. Medium-speed centrifugation, PEG precipitation, and DEAE-cellulose chromatography in batch processes are probably the technologies most adapted to the possibilities of developing countries. Anticipating a staff trained in microbiology and basic biotechnology, as found in many food fermentation environments and instructed by experienced phage microbiologists, such techniques can be established within months. Once installed, the technology is very flexible, as demonstrated by the “personalized medicine” approach in Eastern Europe, where the pathogen is isolated from the patient, sent to a phage collection center for selection and amplification of an appropriate phage, and sent back to the hospital for a tailored phage therapy approach.
In conclusion, large volumes containing high phage titers can be produced for several T4-like phages under standard conditions of the biotechnology industry. A sterile preparation can be achieved by filtration through 0.2-μm membrane filters. The phage cocktail is stable over time under refrigeration and survives interruptions in the cold chain. The approach is flexible: when a phage cocktail is needed for a different geographical area, one can isolate the prevalent E. coli pathogens and choose the corresponding phages from a sequenced T4-like phage collection (10) for optimal host coverage.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laurent Crosset-Perrotin for advice in the setup of the pilot-scale ultrafiltration trial, Florence Charton for help in bacteriophage characterization, and Wolfram Brück and Enea Rezzonico for critical readings of the manuscript.
Footnotes
Published ahead of print 20 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03357-13.
REFERENCES
- 1.World Health Organization 2012. The evolving threat of antimicrobial resistance. Options for action. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/publications/2012/9789241503181_eng.pdf Accessed 1 February 2013 [Google Scholar]
- 2.Djie-Maletz A, Reither K, Danour S, Anyidoho L, Saad E, Danikuu F, Ziniel P, Weitzel T, Wagner J, Bienzle U, Stark K, Seidu-Korkor A, Mockenhaupt FP, Ignatius R. 2008. High rate of resistance to locally used antibiotics among enteric bacteria from children in Northern Ghana. J. Antimicrob. Chemother. 61:1315–1318. 10.1093/jac/dkn108 [DOI] [PubMed] [Google Scholar]
- 3.Ochoa TJ, Ruiz J, Molina M, Del Valle LJ, Vargas M, Gil AI, Ecker L, Barletta F, Hall E, Cleary TG, Lanata CF. 2009. High frequency of antimicrobial drug resistance of diarrheagenic Escherichia coli in infants in Peru. Am. J. Trop. Med. Hyg. 81:296–301 [PMC free article] [PubMed] [Google Scholar]
- 4.Qadri F, Svennerholm AM, Faruque AS, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin. Microbiol. Rev. 18:465–483. 10.1128/CMR.18.3.465-483.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10:597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- 7.Sarker SA, Casswall TH, Mahalanabis D, Alam NH, Albert MJ, Brüssow H, Fuchs GJ, Hammerström L. 1998. Successful treatment of rotavirus diarrhea in children with immunoglobulin from immunized bovine colostrum. Pediatr. Infect. Dis. J. 17:1149–1154. 10.1097/00006454-199812000-00010 [DOI] [PubMed] [Google Scholar]
- 8.Casswall TH, Sarker SA, Faruque SM, Weintraub A, Albert MJ, Fuchs GJ, Alam NH, Dahlstrom AK, Link H, Brüssow H, Hammarström L. 2000. Treatment of enterotoxigenic and enteropathogenic Escherichia coli-induced diarrhoea in children with bovine immunoglobulin milk concentrate from hyperimmunized cows: a double-blind, placebo-controlled, clinical trial. Scand. J. Gastroenterol. 35:711–718. 10.1080/003655200750023372 [DOI] [PubMed] [Google Scholar]
- 9.Sarker SA, Sultana S, Fuchs GJ, Alam NH, Azim T, Brüssow H, Hammarström L. 2005. Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics 116:e221–e228. 10.1542/peds.2004-2334 [DOI] [PubMed] [Google Scholar]
- 10.Sarker SA, McCallin S, Barretto C, Berger B, Pittet AC, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A, Reuteler G, Brüssow H. 2012. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434:222–232. 10.1016/j.virol.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 11.Sulakvelidze A, Alavidze Z, Morris JG., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sulakvelidze A, Kutter E. 2005. Bacteriophage therapy in humans, p 381–436 In Kutter E, Sulakvelidze A. (ed), Bacteriophages—biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 13.McCallin S, Sarker SA, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, Brüssow H. 2013. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology 443:187–196. 10.1016/j.virol.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 14.Chibani-Chennoufi S, Sidoti J, Bruttin A, Dillmann ML, Kutter E, Qadri F, Sarker SA, Brüssow H. 2004. Isolation of Escherichia coli bacteriophages from the stool of pediatric diarrhea patients in Bangladesh. J. Bacteriol. 186:8287–8294. 10.1128/JB.186.24.8287-8294.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karam JD. (ed). 1994. Molecular biology of bacteriophage T4 American Society for Microbiology, Washington, DC [Google Scholar]
- 16.Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brüssow H. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob. Agents Chemother. 48:2558–2569. 10.1128/AAC.48.7.2558-2569.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson K. 2005. Working with bacteriophages: common techniques and methodological approaches, p 437–494 In Kutter E, Sulakvelidze A. (ed), Bacteriophages—biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 18.Zuber S, Ngom-Bru C, Barretto C, Bruttin A, Brüssow H, Denou E. 2007. Genome analysis of phage JS98 defines a fourth major subgroup of T4-like phages in Escherichia coli. J. Bacteriol. 189:8206–8214. 10.1128/JB.00838-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Food and Drug Administration Center for Drug Evaluation and Research 1987. Guideline on validation of the Limulus amebocyte lysate test as an end-product endotoxin test for human and animal parenteral drugs, biological products, and medical devices. FDA Center for Drug Evaluation and Research, Rockville, MD [Google Scholar]
- 20.Harper MS, Carpenter C, Klocke DJ, Carlson G, Davis T, Delaney B. 2011. E. coli lipopolysaccharide: acute oral toxicity study in mice. Food Chem. Toxicol. 49:1770–1772. 10.1016/j.fct.2011.04.025 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.