Abstract
The abilities of enzymes to catalyze reactions in nonnatural environments of organic solvents have opened new opportunities for enzyme-based industrial processes. However, the main drawback of such processes is that most enzymes have a limited stability in polar organic solvents. In this study, we employed protein engineering methods to generate a lipase for enhanced stability in methanol, which is important for biodiesel production. Two protein engineering approaches, random mutagenesis (error-prone PCR) and structure-guided consensus, were applied in parallel on an unexplored lipase gene from Geobacillus stearothermophilus T6. A high-throughput colorimetric screening assay was used to evaluate lipase activity after an incubation period in high methanol concentrations. Both protein engineering approaches were successful in producing variants with elevated half-life values in 70% methanol. The best variant of the random mutagenesis library, Q185L, exhibited 23-fold-improved stability, yet its methanolysis activity was decreased by one-half compared to the wild type. The best variant from the consensus library, H86Y/A269T, exhibited 66-fold-improved stability in methanol along with elevated thermostability (+4.3°C) and a 2-fold-higher fatty acid methyl ester yield from soybean oil. Based on in silico modeling, we suggest that the Q185L substitution facilitates a closed lid conformation that limits access for both the methanol and substrate excess into the active site. The enhanced stability of H86Y/A269T was a result of formation of new hydrogen bonds. These improved characteristics make this variant a potential biocatalyst for biodiesel production.
INTRODUCTION
Lipases (triacylglycerol ester hydrolases, EC 3.1.1.3) are one of the most widely used groups of industrial biocatalysts, especially those of microbial origin (1–3). They are attractive biocatalysts due to several attributes, among them, their high catalytic yields, ease of genetic manipulation, relatively inexpensive production in bulk quantities, their capability to preserve their catalytic activity in nonaqueous media, and their high chemo-, regio-, and enantioselectivity (1–5). Under physiological aqueous conditions, they facilitate the hydrolysis of carboxyl ester bonds present in long-chain acylglycerols (4–6). However, under microaqueous conditions, lipases have the ability to perform the reverse reactions of esterification, transesterification, alcoholysis, acidolysis, or aminolysis on a wide range of natural and unnatural substrates (1, 4–6).
Generally, enzymatic reactions in organic solvent systems possess numerous advantages, such as increased solubility of water-insoluble substrates and products, changes in the thermodynamic equilibrium toward synthesis rather than hydrolysis reactions, avoidance of water-promoted side reactions, improved product recovery, and elimination of microbial contamination of the reaction system (7, 8). However, most enzymes have limited activity and stability in the presence of organic solvents, as they evolved through millions of years in natural aqueous environments. Enzymes' structural conformations in aqueous environments are maintained by the balance between electrostatic and hydrophobic interactions (7, 8). Inactivation by organic solvents is most likely due to conformational changes of the protein structure caused by changes in medium hydrophobicity and noncovalent interactions between the enzyme and the solvent molecules, which lead to protein unfolding and activity loss (7–10). Hydrophilic polar solvents are attractive solvents, as they create homogenous systems with water (7, 9). However, they can easily penetrate the enzyme surface or strip off essential water molecules from the enzyme, leading to denaturation and activity loss (7–9, 11). Therefore, biocatalysts with enhanced stability and activity in the presence of hostile hydrophilic polar solvents are of great interest for the chemical industry.
In recent years, enzymatic production of biodiesel by transesterification has drawn a lot of attention (12, 13). Biodiesel refers to monoalkyl esters of long-chain fatty acids from a renewable source (e.g., vegetable oil or animal fat) and short-chain alcohols (e.g., methanol or ethanol) for use in diesel engines as alternatives for the traditional fossil fuels (12–15). Biodiesel production is usually catalyzed by a chemical alkali catalyst. However, the use of an enzymatic catalyst allows the use low-grade oil feedstock containing a high percentage of free fatty acids, easy recovery of glycerol, mild reaction conditions, and reduced alkali or acidic wastewater (12–15). Enzymatic production of biodiesel by transesterification (methanolysis for the use of methanol as the acyl acceptor) involves lipases as catalysts in microaqueous reaction systems (12, 15). Methanol is frequently utilized as the acyl acceptor in the reaction, thanks to its high reactivity and low cost (14, 15). However, one of the main obstacles of enzymatic methanolysis is the limited stability of lipases in the presence of high methanol concentrations (14–16).
Different approaches have been used to enhance the stability of lipases and their activities in the presence of polar organic solvents. The most popular approaches have been chemical or physical immobilization onto support matrices (17–19) or physical entrapment in reversed micelles (20, 21), which improve the lipase stability and enable the reuse of the biocatalyst. Other approaches used have included chemical or physical modifications of the enzyme (22, 23), isolation of natural lipases with stability in organic solvents (24, 25), and protein engineering (26, 27). However, protein engineering of lipases for enhanced stability in methanol and, as a consequence, improved biodiesel synthesis ability had not been reported.
Protein engineering was used to create organic solvent-stable enzymes by amino acid alterations in the protein sequence. Enzyme stability in organic solvents can be successfully enhanced by a rational design approach (28–30). However, the main drawbacks of this approach are the lack of structural data, in many cases, together with a limited understanding of the enzyme-solvent interactions, and also the limited ability to predict how amino acid alternations influence the enzyme activity and stability. Therefore, directed evolution has been the main protein engineering approach used to produce enzymes tolerant to different denaturing organic solvents (27, 31–35). The main disadvantage of this approach is the time-consuming efforts involved in screening large numbers of clones (103 to 106) in the several rounds of mutagenesis cycles needed to improve the desired enzyme property (36–38).
Different studies have shown that many mutations that stabilize proteins at high temperatures also increase their stability in organic solvents (39–41). Protein engineering by introduction of ancestral/consensus residues to the enzyme protein sequence has been successfully used to evolve thermostable enzymes (39, 42–46). The underlying assumption of this approach is that conserved amino acids can contribute to the enzyme stability under extreme conditions (36, 43, 46–48). For example, Vazquez-Figueroa et al. used this approach to generate thermostable variants of glucose dehydrogenase (GDH) (39) that were also found to be more stable to organic solvents (40). Therefore, in this study we have chosen to use this approach to engineer a lipase for enhanced stability to methanol by screening directly for this property.
The goal of this study was to create a lipase with improved transesterification activity by enhancing its stability in high concentrations of methanol by using protein engineering methods. In this work, a lipase from a thermophilic bacterium, Geobacillus stearothermophilus T6 (lipase T6) (49), was used, since it was assumed that a lipase originating from a thermophilic organism would be more resistant to denaturing agents and stress conditions than other lipases (50). This lipase belongs to the I.5 lipase subfamily. Lipases of this group are usually highly active and relatively highly thermostable, making them suitable candidates for industrial processes (2). However, their poor stability in polar organic solvents limits their use in esterification and transesterification reactions. We applied random mutagenesis (error-prone PCR) and a structure-guided consensus approach to generate two libraries from which variants exhibiting improved stability in methanol were isolated via a high-throughput colorimetric assay. Both libraries generated variants with enhanced stability in 60% and 70% methanol. These variants were further evaluated for their ability to catalyze methanolysis of soybean oil. We found that the most stable variant in the hydrolysis of p-nitrophenyl laurate in 70% methanol also exhibited improved thermostability, as implied from differential scanning calorimetry measurements, and a 2-fold-improved methanolysis yield compared to the wild type. Elucidation of structure-function correlations was performed via in silico homology modeling.
MATERIALS AND METHODS
Chemicals.
Methanol and ethanol were purchased from Bio Labs (Jerusalem, Israel), and 2-propanol was obtained from J.T. Baker (Deventer, The Netherlands). Glycerol and ethyl acetate were purchased from Gadot (Haifa, Israel), and acetonitrile came from Spectrum Chemical MFG (Gardena, CA). Sodium taurocholate hydrate (≥97%), Trizma base, 4-nitrophenyl laurate (pNPL), and kanamycin were purchased from Sigma-Aldrich (Rehovot, Israel). p-Nitrophenol was purchased from Fluka (Switzerland). Refined soybean oil was purchased from a local grocery store. Methyl esters of palmitic acid, heptadecanoic acid, stearic acid, oleic acid, linoleic acid, and linolenic acid were purchased from Sigma-Aldrich (Rehovot, Israel). All materials used were of the highest purity available.
Bacterial strains, plasmids, and culture conditions.
The Geobacillus stearothermophilus T6 lip gene (EMBL accession number AF429311.1) (49) cloned into the vector pET9a (Novagen, Darmstadt, Germany) without the 90 bp encoding a 30-amino-acid leader peptide (MMKCCRRVALVLLGLWFVFCISVLGGRAEA) was kindly provided by Yuval Shoham (Faculty of Biotechnology and Food Engineering, Technion, Israel). The vector pET9a/lip T6 was transformed into Escherichia coli BL21(DE3) (Novagen, Darmstadt, Germany) for overexpression. E. coli transformants were routinely grown in 37°C in Luria-Bertani (LB) medium (51) containing 25 μg ml−1 kanamycin.
His6 tag introduction to lipase T6.
A histidine tag was introduced onto the C terminus of the recombinant lip T6 gene by using high-fidelity Phusion DNA polymerase (New England BioLabs, MA) and NdeI-F and BamHI-His-R primers (see Table S1 in the supplemental material). The resulting lip T6His gene PCR product was cloned into pET9a after double digestion with NdeI and BamHI (New England BioLabs, MA).
Lipase activity assay.
Lipase activity was determined by using a colorimetric assay based on the hydrolysis of pNPL in 96-well plates (Nunc A/S, Kamstrupvei, Denmark), and periodic measurement of the liberated p-nitrophenol (pNP) was based on the absorbance at 405 nm determined using a multiplate reader (OPTImax tunable microplate reader; Molecular Devices, Sunnyvale, CA). Ten microliters of purified lipase solution or soluble cell extract was added to 180 μl of assay mixture composed of 175 μl of reaction buffer (50 mM Tris-HCl [pH 8], 4% 2-propanol, and 1% acetonitrile) (52) and 5 μl of 50 mM CaCl2. The assay mixture containing the enzyme was equilibrated for 5 min at 40°C in the multiplate reader, and the reaction was started by the addition of 10 μl of 20 mM pNPL dissolved in 2-propanol. The reaction time was 3 min, and the absorbance was measured every 12 s. The activity assay following incubation in methanol or ethanol was conducted after the lipase solution was diluted with lipase buffer (1 mM CaCl2, 300 mM NaCl, 50 mM Tris-HCl [pH 7.2] buffer), so that the methanol or ethanol final concentration in the reaction mixture was <3.5 × 10−2%. The hydrolysis rate of pNPL was defined as the slope of the linear zone of the plot of absorbance versus time. A calibration curve of p-nitrophenol under the assay conditions was used for calculating the specific activity. All experimental results represent triplicates unless indicated otherwise.
Protein engineering by random mutagenesis.
The lip T6His gene (1,188 bp) was amplified using error-prone PCR. A 50-μl reaction mixture contained 5 mM MgCl2, 0.1 mM MnCl2, 1M betaine, 0.2 mM dATP and dGTP each, 1 mM dCTP and dTTP each, 5 U Taq DNA polymerase (MyTaq; Bioline, London, United Kingdom), 2.4 ng μl−1 of each primer (NdeI-F and T7-R [see Table S1 in the supplemental material]) and 20 ng of template DNA. The PCR program comprised 5 min at 95°C, followed by 30 cycles of 95°C for 1 min, 60°C for 45 s, and 72°C for 1 min, with a final extension of 72°C for 10 min in a thermocycler (Labcycler; SensoQuest GmbH, Germany). The resulting randomized PCR product was incubated for 3 h at 37°C with 0.8 U μl−1 of DpnI endonuclease (New England BioLabs, MA) in order to digest the template DNA. The PCR product was cloned into pET9a after double digestion with NdeI and BamHI. The resulting library was transformed into E. coli BL21(DE3) competent cells via electroporation. Single colonies were picked into Nunclon 96-well plates (Nunc A/S, Kamstrupvei, Denmark) containing 70 μl LB with 25 μg ml−1 kanamycin. Two colonies of E. coli BL21(DE3) containing pET9a/lip T6His wild type were also picked into every plate as controls. After 12 h of incubation at 37°C, 70 μl of sterile 50% glycerol was added to each well, and the plates were stored at −80°C. These plates served as master plates for the library screening.
Structure-guided consensus.
The structure-guided consensus approach was used to generate a small-sized mutant library, based on the methods described by Vázquez-Figueroa et al. with a few modifications (39). A total of 205 homologous sequences of lipase T6 of bacterial origin were collected using the ConSurf server (http://consurf.tau.ac.il) from the Clean_Uniprot database (a modified version of the UniProt database aimed to screen the more reliable sequences) (53). A total of 59 homologous sequences were chosen after limiting the search to 30 to 95% sequence identity in order to avoid similarities that were too high or too low. Sequences were narrowed to 59 by considering only complete protein sequences of active lipases and putative genes. Sequences were further narrowed to 9 sequences that represented the main branches of a phylogenetic tree constructed by using ClustalW (http://www.genome.jp/tools/clustalw/) (see Fig. S1 in the supplemental material), in order to reduce bias toward closely related homologous sequences. The additional 9 lipases sequences ranged in amino acid identity from 35 to 56% relative to the wild type (see Table S2 in the supplemental material). Final alignment was carried out using PRALINE (using the default settings) (see Fig. S2 in the supplemental material), a multiple sequence alignment tool that integrates homology-extended and secondary structure information (54, 55). From the PRALINE alignment, 42 of the 389 positions (not including the His tag sequence) were found to have at least a 50% frequency in the consensus sequence. Of these optional substitutions for mutagenesis, we excluded positions with distances below 6 Å from the active site residues or ones that were situated on the α-helix lid, in order to minimize impairment to activity (39). In addition, no substitutions on the calcium- or zinc-binding sites were chosen, to minimize loss of stability, and furthermore, the substitution had to be represented in both of the two main braches of the phylogenetic tree (see Fig. S1 in the supplemental material). For the application of these criteria, a three-dimensional (3D) homology model of wild-type lipase T6 was created by using the SWISS-MODEL server and the crystal structure of the lipase from G. stearothermophilus P1 (PDB code 1JI3 [56]), which is 95% identical to lipase T6. The visualization of the 3D model was generated by using PyMOL (http://www.pymol.org/). After the application of these criteria, 22 positions were chosen for mutagenesis. Twenty-five specific mutations were designed, as in three positions two substitutions were chosen. The library was constructed using oligonucleotides of these specific mutations (see Table S3 in the supplemental material) obtained from Sigma-Aldrich (Rehovot, Israel). We generated a DNA library of lip T6His with randomized specific mutations by applying the Herman and Tawfic ISOR protocol (incorporating synthetic Oligonucleotides via gene reassembly [57]) with a few modifications. The lip T6His gene (1,188 bp) was amplified from the pET9a/lip T6His plasmid by PCR using high-fidelity Phusion DNA polymerase (New England BioLabs, MA) and 2.4 ng μl−1 of each primer (T7-F and T7-R [see Table S1 in the supplemental material]). The PCR product was purified with a PCR purification kit (Qiagen, CA). Eight micrograms of the purified PCR product in 50 μl digestion buffer (DNase I buffer, 10 mM MnCl2) was equilibrated at 20°C in a thermocycler. The DNA digestion was started by transferring the 50 μl of PCR product to a new PCR tube containing 2 μl of 1 U μl−1 DNase I (New England BioLabs, MA). The DNA digestion proceeded for 2, 4, and 6 min at 20°C and was terminated by transferring aliquots of 15 μl to a new tube containing 5 μl of EDTA (0.25 M solution) and heating to 90°C for 10 min. The digested gene products were loaded onto a 2% low-melt agarose gel (Amresco, OH) for separation. Fragments of 70 to 150 bp in size were cut out and purified using a DNA isolation kit (Biological Industries, Beit Haemek, Israel). The gene was reassembled, and the oligonucleotides were hybridized using the PCR program described in the ISOR protocol (57) in a thermocycler (SensoQuest GmbH, Germany). The assembly PCR included a 25-μl reaction mixture composed of 120 ng of purified DNA fragments, 1 μl spiking oligonucleotides mixture (0.05 pmol μl−1 of each oligonucleotide in the mix solution), 0.5 U Phusion DNA polymerase (New England BioLabs, MA), 5 μl 5× Phusion buffer, and 1 mM each deoxynucleoside triphosphate. The assembly product was further amplified in a nested PCR. One microliter of assembly product was used as a template in a 50-μl reaction mixture with high-fidelity Phusion DNA polymerase and 2.4 ng μl−1 of each primer (NdeI-F and BamHI-R [see Table S1]) under standard conditions. The purified nested PCR product was cloned into pET9a after double digestion with NdeI and BamHI. The resulting library was transformed into E. coli BL21(DE3) competent cells via electroporation. Plasmids from 15 colonies were extracted using a minikit (Qiagen, CA) and sequenced in order to evaluate the reassembly and hybridization success. Master plates were created as described above for the library screening.
Site-specific saturation mutagenesis.
Three gene libraries encoding all possible amino acids at positions H86, A269, and Q185 of lip T6His were constructed by replacing the target codon with an NNK degenerate codon (N represents A, T, G, or C, and K represents G or T) by using the QuikChange II site-directed mutagenesis kit (Stratagene). Library T6His-H86 was created using primers H86-F and H86-R (see Table S1), library T6His-A269 was created using primers A269-F and A269-R (see Table S1), and library T6His-Q185 was created using primers Q185-F and Q185-R (see Table S1). The resulting libraries were transformed into E. coli BL21(DE3) competent cells via electroporation. Master plates were created as described above with addition of two positive control wells containing E. coli BL21(DE3) cells expressing lipase T6 variant H86Y in the T6His-H86 library, A269T in the T6His-A269 library, and variant Q185L in the T6His-Q185 library.
Site-directed mutagenesis.
The double mutant Q185L/A269T was created by joining mutations Q185L and A269T using the QuikChange II site-directed mutagenesis kit (Stratagene). Mutation Q185L was added to pET9a/lip T6His-A269T by using primers Q185L-F and Q185L-R (see Table S1). The plasmid pET9a/lip T6His-Q185L/A269T was transformed into E. coli BL21(DE3) competent cells via electroporation.
Screening for enhanced methanol stability.
A high-throughput assay for isolation of enhanced methanol stability variants was developed based on the protocol described by Bottcher and Bornscheuer for measuring the activities of lipases and esterases (58) with some modifications. The colonies were transferred into 96-deep-well polypropylene plates with a plastic lid (ABgene, Thermo Fisher Scientific, Epson, United Kingdom) containing 1.2 ml of LB with 25 μg ml−1 kanamycin by using a library copier (VP 381; V&P Scientific, Inc., San Diego, CA) and grown overnight for 14 h at 37°C with shaking at 200 rpm in an incubator shaker (TU-400 orbital shaker incubator; MRC, Holon, Israel) to an optical density at 600 nm (OD600) of 0.4 to 0.6. After the growth stage, cells were harvested by centrifugation at 2,500 × g for 10 min at 4°C in a 4K15 centrifuge (Sigma, Osterode, Germany). The cell pellets were broken by resuspension in 300 μl lysis buffer (50 mM sodium phosphate buffer [pH 8], 300 mM NaCl, 0.15% lysozyme [Amresco], and 1 unit ml−1 DNase I [Sigma]) in an epMotion 5070 robotic system (Eppendorf). The suspended cells were incubated for 30 min at 4°C, followed by freezing at −80°C for 1 h and then thawing for 1 h at 37°C with shaking at 250 rpm in an incubator shaker. The cell lysate was diluted by addition of 300 μl 50 mM sodium phosphate buffer (pH 8) in an epMotion 5070 robotic system, followed by centrifugation at 3,000 × g for 30 min at 4°C. The supernatant was used as the enzyme solution for the activity assay before and after incubation in 60% methanol. Twenty microliters of supernatant was diluted 100-fold with lipase buffer, and 10 μl of the diluted enzyme solution was used for the lipase activity assay with pNPL (without incubation in methanol solution). Aliquots of 150 μl of supernatant were transferred into 96-deep-well polypropylene plates together with 225 μl methanol (final concentration of methanol, 60%), incubated at room temperature for 30 min, and then centrifuged at 3,000 × g for 10 min at 4°C. The supernatant was diluted 10-fold with lipase buffer and used for the lipase activity assay with pNPL (after incubation in 60% methanol). The relative residual activity was obtained by calculating the residual activity of each variant (the ratio between the activity after and before incubation in 60% methanol) and compared to the residual activity of the wild type. Variants that showed a relative activity of >1.5 compared to the wild type were isolated, sequenced, and taken for further validation.
Validation of positive variants.
The validation of positive variants was performed as described before (59) with a few modifications. Single colonies of transformants were grown in 5 ml LB with 25 μg ml−1 kanamycin for 6 h at 37°C with shaking at 250 rpm. A 1-ml aliquot of the starter broth was inoculated into 20 ml TB medium with 25 μg ml−1 kanamycin for overnight growth. Cells were harvested by centrifugation (8,000 × g, 10 min, 10°C) and resuspended with 10 ml lipase buffer. Cell breakage was carried out by sonication (VibraCell VCX750 with a CV33 transducer and SM0401 tip; Sonics & Materials Inc., Newtown, CT) in a 3-min treatment at a relative output power of 0.35 with 0.5 duty periods (60). The cell lysate was centrifuged for 20 min at 16,000 × g at 15°C, and the supernatant was used as a crude cellular extract for lipase activity assays before and after incubation in 60% methanol. A 200-μl aliquot of the supernatant was mixed with 300 μl methanol (final concentration of 60% methanol) in a 1.7-ml tube (Eppendorf) and incubated for 30 min at room temperature followed by a 2-min centrifugation at 12,100 × g (MiniSpin; Eppendorf) for the removal of denatured proteins from the solution. The supernatant was used for lipase activity assays after incubation in 60% methanol. The residual activity (E/E0) was calculated and normalized to the total protein concentration, which was determined by the method of Bradford. Protein samples of every colony were analyzed on standard 12% Lammeli discontinuous sodium dodecyl sulfate (SDS)-polyacrylamide gels (51) in order to ensure protein expression levels were similar.
Purification of His6-tagged lipase T6.
His6-tagged lipase T6 and its improved variants were purified in one step by using an Ni(II)-bound affinity column (HisTrap HP; Amersham Biosciences, Giles, United Kingdom). Single colonies of E. coli BL21(DE3) cells harboring pET9a/lip T6His were grown in 20 ml LB with 25 μg ml−1 kanamycin for 6 h at 37°C with shaking at 250 rpm. A 1-ml aliquot of the starter broth was inoculated into 0.5 liter of 1 mM CaCl2 TB medium with 25 μg ml−1 kanamycin for overnight growth at 37°C with shaking at 250 rpm. The cells were harvested by centrifugation (8,000 × g for 10 min at 4°C) and resuspended in a binding buffer (20 mM Tris-HCl buffer [pH 7.2], 500 mM NaCl, and 20 mM imidazole). The cells were broken by using a pressure cell press (French press; Spectronic Instruments Inc., Rochester, NY). Immediately after cell breakage, sodium taurochlate hydrate (Sigma) was added to the cell extract (1%, vol/wt) in order to improve the lipase recovery. The cell debris was removed by centrifugation (16,000 × g for 20 min at 15°C), and the supernatant was subjected to heat treatment of 50°C for 15 min in order to denature many of the E. coli BL21(DE3) host proteins. After another centrifugation step (16,000 × g for 20 min at 15°C), the supernatant was applied to the Ni(II)-bound affinity column, which was equilibrated with the binding buffer before use. The nonbinding proteins were washed with the binding buffer, and the lipase was subsequently eluted from the column with elution buffer (20 mM Tris-HCl buffer [pH 7.2], 500 mM NaCl, and 500 mM imidazole). The lipase-containing fractions were collected and dialyzed against lipase buffer with 5% glycerol at 4°C for 24 h.
Kinetic analysis.
Km and Vmax values for lipase T6 and its variants were determined using the colorimetric assay of pNPL hydrolysis in a 200-μl reaction volume in 96-well plates as described above. A 0.002-μg portion of purified enzyme was employed with increasing substrate (pNPL) concentrations ranging from 0.01 to 1 mM. The Michaelis-Menten curves for determination of the kinetic constants were obtained using GraFit version 5 (Erithacus Software Ltd., United Kingdom).
Stability of wild-type lipase T6 and its variants in alcohol.
The stability of wild-type lipase T6 and its variants in high methanol and ethanol concentrations was determined by measuring the residual activity of the lipase after 1 h of incubation in different alcohol solutions. A 100-μl volume of 0.2 mg ml−1 lipase solution was mixed with an alcohol solution ranging from 50 to 80% in a final volume of 500 μl (60% solution; 100 μl lipase solution, 100 μl lipase buffer, and 300 μl methanol or ethanol) and incubated in closed 1.7-ml Eppendorf tubes at room temperature. The incubation was stopped by taking 20-μl aliquots from the mixture and transferring them into 180 μl lipase buffer. The residual activity was determined in a lipase activity assay as described above.
Half-life determinations in methanol or ethanol.
Half-life (t[1/2]) values of lipase T6 in 50%, 60%, and 70% methanol and 70% ethanol were calculated by measuring the residual activities after different incubation periods. A 300-μl volume of a 0.2-mg ml−1 lipase solution was mixed with alcohol solution ranging from 50 to 70% to a final volume of 1 ml (for example, to make a 50% methanol solution we combined 300 μl lipase solution, 200 μl lipase buffer, and 500 μl methanol). The incubation was stopped periodically by removing 20-μl aliquots from the mixture and transferring them into 180 μl lipase buffer, and the residual activity was determined as described above.
Calorimetric studies of lipase T6 and its variants.
The thermal-induced unfolding of wild-type lipase T6 and variants A269T, Q185L, Q185L/A269T, and H86Y/A269T was measured using differential scanning calorimetry (DSC) on a MicroCalVP-DSC instrument (GE Healthcare Bio-Sciences AB, Sweden). The reaction cell contained 1 ml (1 to 1.5 mg ml−1) of purified lipase in lipase buffer with 5% glycerol, and the reference cell contained 1 ml of the same buffer without the enzyme. The measurements were performed by scanning from 35°C to 90°C at 1°C min−1. DSC measurements were analyzed using Origin software (MicroCal Inc.), and JMP 7 software was used for the statistical analysis of the results.
Lipase-catalyzed methanolysis of soybean oil.
Methanolysis reactions catalyzed by lipase T6 were carried out in 4.5-ml closed glass vials filled with 1 g soybean oil. Generally, in the full hydrolysis of 1 mol of triglyceride, 3 mol of free fatty acid is liberated (15). Therefore, 3 mol of methanol is needed for the full conversion of oil to free fatty acid methyl esters (FAME). In this research, we used 1.5:1, 2:1,and 3:1 methanol-to-soybean oil ratios. A 50-μl aliquot of a 1-mg ml−1lipase T6 solution (0.5 mM CaCl2, 2.5% glycerol, 100 mM NaCl, 50 mM Tris-HCl [pH 7.5]) was added to the reaction mixture, resulting in 0.005% lipase and 5% water content based on oil weight. The addition of the methanol (which does not dissolve in the oil but rather in the buffer phase) resulted in 58, 65, and 73% methanol solutions, providing methanol:oil ratios of 1.5:1, 2:1, and 3:1, respectively. Before the lipase addition, the methanol was mixed with the oil in order to avoid direct contact of the lipase with high methanol concentrations. The reaction mixture was maintained at 45°C in a VibraMax 100 orbital shaker (Heidolph Instruments, Germany) at 1,350 rpm. After 4 h, 200-μl aliquots were taken from the reaction mixture and centrifuged for 3 min at 20,000 × g. Ten microlayers of the upper layer were precisely weighed and mixed with 490 μl of 1 mg ml−1 heptadecanoic acid methyl ester (as internal standard) in ethyl acetate for gas chromatographic analysis.
Gas chromatography analysis of FAME.
The FAME content in the reaction mixture was analyzed and quantified using a 6890N GC instrument (Agilent Technologies, CA) equipped with a capillary DB-23 column (60 m by 250 μm by 0.25 μm; Agilent Technologies) and a flame ionization detector. One-microliter samples were injected in a split mode (1/10). The initial column temperature was 170°C and was kept for 1 min, raised to 230°C at 5°C/min, and maintained at this temperature for 4 min. The temperatures of the injector and detector were set at 270°C and 320°C, respectively. Nitrogen was used as a carrier gas at a column flow rate of 2.2 ml min−1. FAME were identified based on the retention times of FAME standards (Sigma-Aldrich, Rehovot, Israel) and quantified using proper calibration curves based on the peak area ratio between each FAME and the internal standard (heptadecanoic acid methyl ester). FAME yield was defined as the percentage of the sample weight.
RESULTS
Screen for enhanced stability in methanol.
Two protein engineering approaches, random mutagenesis and the data-driven approach of structure-guided consensus, were chosen for creating lipase T6 variants which are more stable in methanol. These approaches were chosen based on our limited ability to predict a mutation's effects on enzyme stability and activity in organic solvents. Thus, two libraries were created and screened for enhanced stability in methanol.
The structure-guided consensus approach refers to the introduction of specific mutations to the protein sequence predicted by utilizing sequence-based alignment, phylogenetic analysis, structural information, and bioinformatics (39, 61). About 1,000 colonies of the consensus library were initially screened for improved activity following incubation for 30 min in 50% methanol. The mutation rate of the library (calculated based on the sequencing of 15 randomly picked colonies) was 2.5 ± 1.7 (mean ± standard error of the mean) consensus mutations (amino acid substitutions) and 1.1 ± 1 random mutations (bp) per lip T6His gene. The library was highly functional, as about 90% of the transformants were active. Eighteen colonies were found with improved stability in the screening assay (relative activity, >1.5-fold above wild-type activity). Seventeen variants had the substitution A9Y in their protein sequence. However, after testing their stability in methanol via the validation assay (incubation in 60% methanol), none of them showed improved stability. Furthermore, the SDS-PAGE gel revealed that all of the variants possessing A9Y had overexpression of the protein compared to wild type (see Fig. S3 in the supplemental material). Therefore, the library was screened again, although this time the residual activity of each variant was calculated by measuring the activity before and after incubation in 60% methanol in order to avoid false-positive results caused by lipase overexpression. Using this method, 6 variants were found with improved stability in the validation assay (Fig. 1). Two single mutations were found, H86Y and A269T, with improved residual activities of 3.7- and 21-fold, respectively. However, their combined double mutant, H86Y/A269T, which was also found in the screen, did not show an additive effect in the screening validation assay. The best four variants selected possessed the substitution A269T and showed improved relative activity, in the range of 11- to 21-fold (Fig. 1). The highest methanol-stable variant had the substitution A269T as a single mutation.
FIG 1.
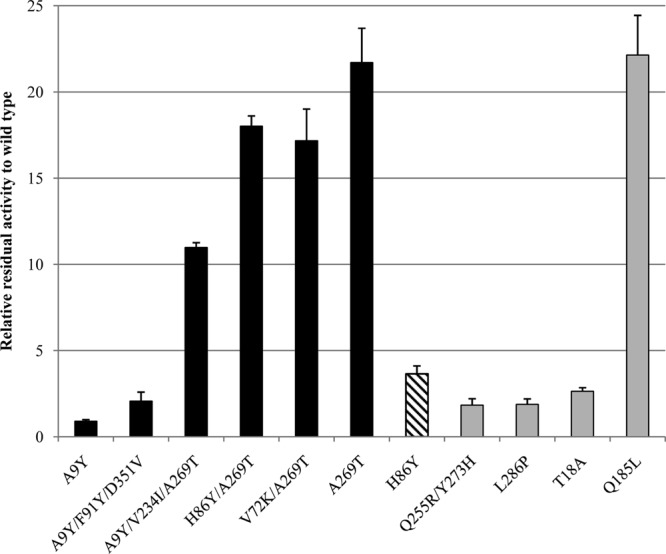
Screening results of random mutagenesis and structure-guided consensus libraries. The columns represent residual activities of soluble cell extracts of lipase T6 variants after 30 min of incubation in 60% methanol compared to the wild type. Black bars, variants obtained from structure-guided consensus; striped bar, variant obtained from random mutagenesis by error-prone PCR; gray bars, variants found in both libraries. The wild-type activity before incubation was 330 U/mg (total protein), and it lost 70% of its activity after 30 min of incubation in 60% methanol. U, the amount of enzyme releasing 1 μmol pNP/min.
Nearly 1,200 colonies of the random mutagenesis library were screened for enhanced stability in methanol by using the modified screen (incubation in 60% methanol). The mutation rate of the library (calculated based on the sequencing of 15 randomly picked colonies) was 3.5 ± 1.7 random mutations (bp) per lip T6His gene, which resulted in 2 ± 1.5 amino acid substitutions in the protein sequence. However, this library was less functional than the consensus library, as only 50% of the transformants were active. Six colonies were found with improved stability in the screen, and 5 of them showed improved stability in the validation assay. These variants had improved residual activity in the range of 1.8- to 22-fold compared to the wild type (Fig. 1). The most stable variant, Q185L, had 22-fold-enhanced residual activity (Fig. 1). Variant H86Y, which was previously found in the consensus library, was also found in the random mutagenesis library, where it had the second best improved stability compared to the wild type.
The best variants, A269T from the consensus library and Q185L from the random mutagenesis library were combined using site directed mutagenesis to create the double mutant Q185L/A269T. These mutants together with variant H86Y/A269T from the consensus library were purified for further characterization.
Site-specific saturation mutagenesis at positions A269, Q185, and H86.
Substitutions A269T, Q185L, and H86Y were found beneficial for enhancing lipase T6 stability in methanol. In order to find the most beneficial amino acid substitution at these positions and to improve our understanding of how these substitutions contribute to the enzyme stability in methanol, three saturation mutagenesis libraries encoding all possible amino acids at positions A269, Q185, and H86 of the lip T6His gene were constructed by using NNK codon degeneracy (32 codons/20 amino acids). Roughly 300 colonies of each library were screened for enhanced stability in methanol in order to ensure that all possible amino acids were evaluated. It was previously demonstrated by Reetz et al. that for mutagenesis of one position using NNK codon degeneracy, screening of 94 colonies is sufficient to ensure that all 32 possible outcomes are evaluated with a probability of 95% (38).
Twelve colonies were found to have improved methanol stability in the screening of the A269 library. Half of them had serine residue at position 269 (A269S), and the other half had threonine (A269T), which was the mutation discovered in the consensus library. Both amino acids had a relatively short side chain with a hydroxyl functional group. Variant A269S had 11-fold-improved relative residual activity above the wild type, while A269T was 21-fold improved.
In the screening of library for Q185, eight colonies were found to have improved stability in methanol. All of them were residues with a hydrophobic side chain: methionine, valine, alanine, phenylalanine, and leucine. In the validation assay, substitutions Q185-to-Met, -Val, -Ala, or -Phe had an improved residual activity in the range of 8- to 12-fold above the wild type but lower than that of L185 (22-fold).
In the screening library for H86, only two colonies, besides H86Y, showed improved stability in methanol. In the screening validation assay, these improved variants, A3T/H86M and H86L, showed only slightly improved stability, 1.5- to 2-fold above the wild type, but that was lower than that of Y86 (3.7-fold).
Stability in methanol and ethanol at various concentrations.
Wild-type lipase T6 and the improved variants Q185L, A269T, H86Y/A269T, and the combined variant Q185L/A269T were purified using an Ni(II)-bound affinity column. Their stabilities in high concentrations of hydrophilic solvents, methanol and ethanol, was evaluated by measuring their hydrolytic activities on pNPL after 1 h of incubation in methanol or ethanol solutions. The hydrolytic activity of all enzymes decreased as the solvent concentrations increased (Fig. 2). In addition, ethanol was more denaturing than methanol. However, the decline in activity of the wild type in both solvents was dramatic compared to the improved variants, as after incubation in 60% methanol or 50% ethanol it lost 70% and 89% of its original activity, respectively (Fig. 2). Variant A269T had the highest activity in buffer, followed by variants Q185L/A269T and H86Y/A269T. Variant Q185L exhibited a significant activity decrease of 15% in buffer compared to A269T (P < 0.05). Nonetheless, after incubation in alcohol, Q185L showed elevated stability compared to the wild type and A269T. The most destructive effect on the wild type was measured after incubation in 70% alcohol; more than 99% of its activity was lost. However, under the same conditions, A269T and Q185L lost 81% and 71% of their activity in methanol, respectively, and 89% and 80% losses in ethanol. Moreover, the double mutant Q185L/A269T exhibited an additive stabilizing effect, as it lost 53% and 73% of its activity after incubation in methanol and ethanol, respectively. However, the best improvement was obtained with double mutant H86Y/A269T, which lost only 25% and 47% of its activity after incubation in 70% methanol and ethanol, respectively. Furthermore, this variant showed improved stability in 80% alcohol solutions and showed higher activity than all other variants after incubation in 70% methanol or ethanol (Fig. 2).
FIG 2.
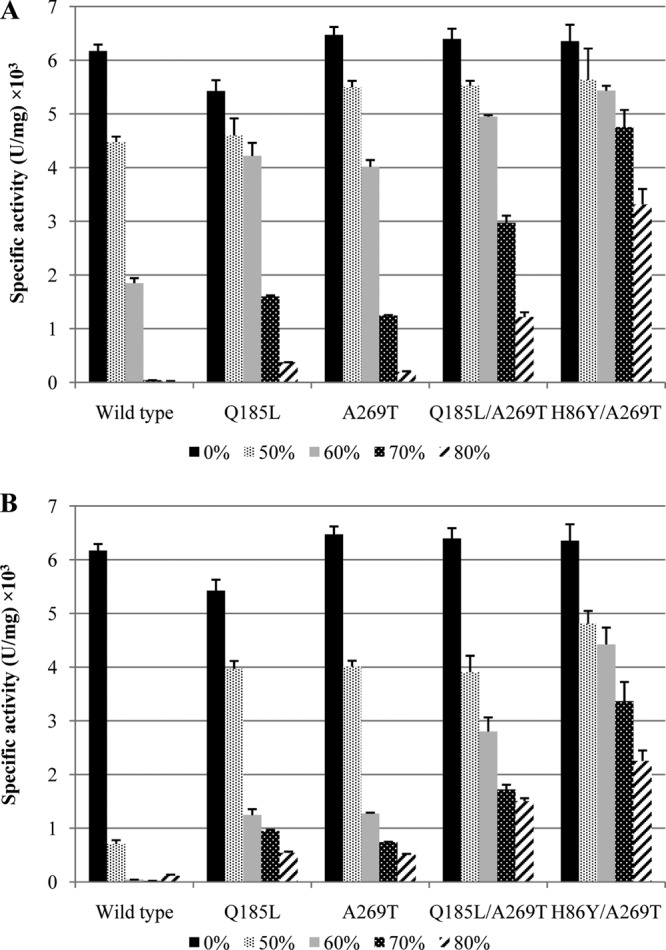
Specific activity of purified lipase T6 and variants A269T, Q185L, Q185L/A269T, and H86Y/A269T after incubation in methanol (A) and ethanol (B) solutions. The activity was measured under standard reaction assay conditions (0.002 μg enzyme in 200 μl of 50 mM Tris-HCl [pH 8] buffer containing 1 mM pNPL as substrate) after 1 h of incubation in different alcohol concentrations. The results represent triplicate measurements. U, the amount of enzyme releasing 1 μmol pNP/min.
Half-life determinations in alcohol.
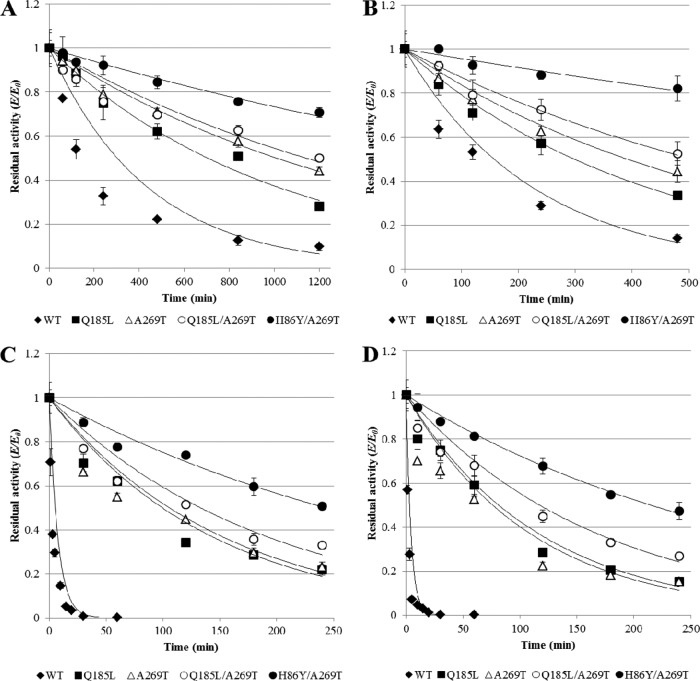
The stabilities of wild-type lipase T6 and variants Q185L, A269T, H86Y/A269T, and Q185L/A269T in 50 to 70% alcohol were determined by calculating the half-life values from residual activity (E/E0) plots, which represent the deactivation profiles of the enzyme (Fig. 3). E0 is the lipase activity under the standard activity assay conditions without an incubation period in alcohol, and E is the lipase activity after incubation in methanol or ethanol solutions. The t1/2 is the time required for the enzyme to lose half of its original activity under the conditions tested. The half-life values were calculated from the exponential model E/E0 = exp(−Kdt). Kd represents the enzyme inactivation constant, and t represents the treatment time. This model was previously shown to agree with other enzyme denaturation profiles in aqueous-organic media (62). Wild-type stability in 50 to 70% methanol and 70% ethanol was very poor, and the results showed a steep decline in the residual activity as the alcohol concentration increased (Fig. 3). The most destructive effect was shown in 70% methanol and ethanol, in which the wild-type residual activity dropped to less than 1% after 1 h of incubation (Fig. 3). However, variants A269T, Q185L, Q185L/A269T, and H86Y/A269T had elevated stability compared to the wild type in 70% alcohol, and after the maximal incubation time (240 min) they maintained 20, 25, 33, and 52% of their activity after incubation in methanol and 15, 15, 27, and 47% after incubation in ethanol, respectively (Fig. 3).
FIG 3.
Deactivation profiles of wild-type lipase T6 and improved variants after different incubation times in 50% methanol (A), 60% methanol (B), 70% methanol (C), and 70% ethanol (D). The activity of purified lipase T6 variants under the standard activity assay conditions (0.002 μg enzyme in 200 μl of 50 mM Tris-HCl [pH 8] buffer containing 1 mM pNPL as substrate) was measured after different incubation times in different alcohol concentrations. The residual activity (E/E0) was calculated by comparing the activity of each variant before (E) and after (E0) incubation in alcohol. The results represent triplicates.
Half-life values and inactivation constants of the wild type and its variants in methanol and ethanol were determined (Table 1). All variants showed improved stability in methanol and ethanol, and the relative stability of the variants compared to the wild type was increased in higher alcohol concentrations, as reflected from the half-life values and inactivation constants (Table 1). The wild-type half-life values at 70% alcohol were very low, 3.9 and 2.9 min in methanol and ethanol, respectively, whereas the mutants' half-lives were elevated. Under these conditions A269T and Q185L showed 25- and 23-fold improved stability above the wild type for incubation in methanol (Table 1). For incubation in ethanol, both Q185L and A269T expressed 27-fold-improved stability. Q185L/A269T showed an additive effect of the single mutations of 36- and 40-fold-improved stability above the wild type for methanol and ethanol, respectively (Table 1). Moreover, the most improved stability in alcohol was shown by the double mutant, H86Y/A269T, which displayed elevated half-life values of 258 and 231 min for methanol and ethanol, respectively. Theses prominent half-life values were even higher than the wild-type half-life value in 60% methanol and reflected 66- and 80-fold-improved stability above that of the wild type for methanol and ethanol, respectively (Table 1).
TABLE 1.
Half-lives and first-order inactivation constants for wild-type lipase T6 and the mutants in methanol and ethanol
| Variant | 50% methanol |
60% methanol |
70% methanol |
70% ethanol |
||||
|---|---|---|---|---|---|---|---|---|
| t1/2 (min)a | Kd (10−4 min−1) | t1/2(min)a | Kd (10−4 min−1) | t1/2 (min)a,b | Kd (10−4 min−1) | t1/2 (min)a,b | Kd (10−4 min−1) | |
| Wild type | 347 | 20 | 173 | 40 | 3.9 | 1,760 | 2.9 | 2,350 |
| Q185L | 693 | 10 | 277 | 25 | 89 | 78 | 77 | 90 |
| A269T | 990 | 7 | 395 | 18 | 96 | 72 | 77 | 90 |
| Q185L/A269T | 1,265 | 5.5 | 577 | 12 | 139 | 50 | 116 | 60 |
| H86Y/A269T | 2,429 | 2.9 | 1,777 | 3.9 | 258 | 27 | 231 | 30 |
t1/2 values were calculated from the exponential model E/E0 = exp(−Kdt). The correlation coefficient of the model was generally >0.85 for all variants.
t1/2 values of the wild type for incubation in 70% methanol and ethanol solutions were calculated by considering time points 0, 1, 3, 5, 10, 15, 20, and 30 min, because for longer times its activity was very low and did not match the model.
Kinetic analysis of the wild type and variants.
Kinetic constants of the wild type and variants were determined for the hydrolysis reaction of pNPL (lipase activity assay without incubation in alcohol). It was evident that substitutions Q185L and A269T did not cause a drastic effect on the wild type's hydrolysis activity of pNPL. Generally, most mutants' catalytic parameters were unchanged from the wild-type enzyme and their affinities for the substrate were at the same level, with Km values ranging between 7.0 × 10−2 and 8.1 mM × 10−2 (Table 2). However, variant Q185L showed a 10% decrease in the kcat value compared to the wild type.
TABLE 2.
Kinetic parameters of wild-type lipase T6 and improved variants on pNPL
| Variant | Km (10−2 mM)a | kcat (103 s−1) | kcat/Km (103 s−1 mM−1) |
|---|---|---|---|
| Wild type | 7.9 ± 0.6 | 4.7 | 59 |
| Q185L | 7.5 ± 0.7 | 4.2 | 56 |
| A269T | 8.1 ± 0.7 | 4.8 | 59 |
| Q185L/A269T | 7.0 ± 0.5 | 4.5 | 64 |
| H86Y/A269T | 7.3 ± 0.7 | 4.6 | 63 |
Values are means ± standard errors of the means.
Calorimetric studies of lipase T6.
The positive correlation between enzyme thermostability and stability in organic solvents was previously demonstrated in numerous cases (40, 41, 63, 64). Therefore, thermal-induced unfolding was measured using DSC. The thermal-induced unfolding of wild-type lipase T6 occurred as a single transition, with a melting temperature (Tm) of 66.8 ± 0.2°C (Fig. 4). The single substitution Q185L did not affect the lipase unfolding temperature (Tm, 66.8 ± 0.6°C), but the substitution A269T resulted in a significant increase in the Tm, to 70.2 ± 0.4°C and 69.4 ± 0.4°C for A269T and Q185L/A269T, respectively (P < 0.05). Furthermore, the combination of substitution H86Y together with A269T resulted in an addition of 4.3°C to the wild-type Tm, to 71.1 ± 0.1°C, which was also significantly higher than all other variants (P < 0.05).
FIG 4.
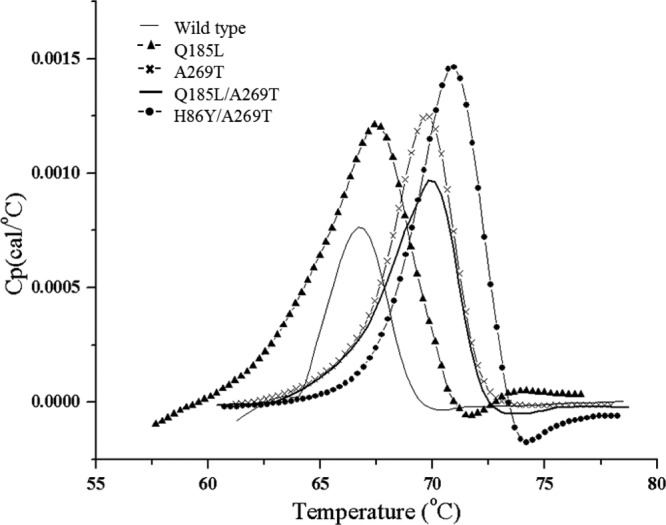
Typical DSC thermograms of wild-type lipase T6 and variants Q185L, A269T, Q185L/A269T, and H86Y/A269T. The unfolding temperatures of purified wild-type lipase T6 and its improved variants were measured by DSC (1 mM CaCl2, 5% glycerol, 300 mM NaCl, 50 mM Tris-HCl [pH 7.2]). The thermal-induced unfolding of lipase T6 occurs as a single transition. The thermograms were generated by using Origin software (MicroCal Inc.).
Synthesis of biodiesel from soybean oil.
Methanol-induced unfolding of lipases is one of the major drawbacks of lipase-catalyzed biodiesel production (13, 15, 65). Therefore, methanolysis of soybean oil catalyzed by lipase T6 and its variants was conducted in order to evaluate the stability of the enzymes in the nonaqueous environment. Solvent-free systems were used with lipase dissolved in buffer and utilizing three different methanol:oil molar ratios (1.5:1, 2:1, and 3:1).
The methanolysis results showed a clear decrease in the FAME yield by all variants as the methanol concentration increased (Fig. 5). The highest FAME conversion for all methanol:oil molar ratios was achieved by variant H86Y/A269T, which also had the highest half-life values in alcohol. This variant enabled FAME yields of 36.8, 25.1, and 15.5% in methanol:oil molar ratios of 1.5:1, 2:1, and 3:1, respectively. These conversion values reflected, in general, 2-fold-improved activity above the wild type (Fig. 5). The second-best variant was A269T. Not surprisingly, the lowest conversion results were obtained with variant Q185L, with lower performance than the wild type. The activity exhibited by double mutant Q185L/A269T was a combination of those of the single mutants and was slightly lower than the wild type (Fig. 5). These overall results imply that, despite the improvement in stability in methanol by variant Q185L, this substitution leads to a poor activity in methanolysis.
FIG 5.
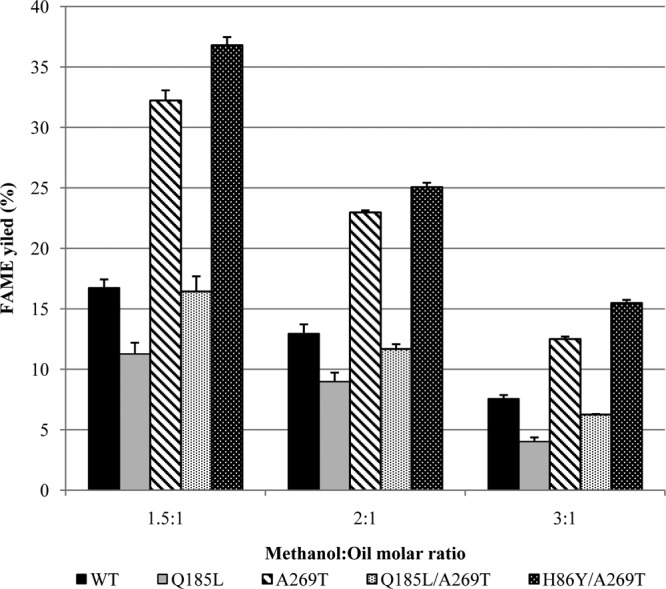
Effect of the methanol:oil molar ratio on biodiesel production from soybean oil by wild-type lipase T6 and mutants. Reaction conditions were the following: 1 g soybean oil, 5% water, and 0.005% lipase content (based on oil weight), 1,350 rpm, 4 h, 45°C. The results represent triplicates.
DISCUSSION
Lipase's ability to catalyze chemical reactions in hydrophilic polar organic solvents, such as methanol, holds great opportunities for the chemical industry and biodiesel production. Therefore, organic solvent-stable lipases have great potential for use in commercial processes. The goal of this work was to enhance the stability in methanol of a lipase from the thermophilic bacterium G. stearothermophilus T6, which naturally has relatively poor performance in this medium. Random mutagenesis (error-prone PCR) and a structure-guided consensus approach were used to create a methanol-stable lipase with improved performance in transesterification of soybean oil.
Wild-type lipase T6 exhibited high activity and thermostabiltiy in water yet poor stability in high methanol and ethanol concentrations, with the latter having a more destructive effect (Fig. 2 and 3; Table 1). This denaturing influence of alcohols on the structure of a close homologue, G. stearothermophilus lipase P1,which shares 95% sequence identity with lipase T6, was predicted by Chakravorty and coworkers, who used bioinformatic and structural data for the analysis of organic solvent effects on lipases (10). It was suggested that the lipase structure will be unstable in the presence of hydrophilic solvents (logP < 2) as they interact with the active site pocket residues and may hinder hydrogen bonds between these residues and water molecules (10). Nonetheless, in this study, both protein engineering approaches were successfully used to generate variants with improved methanol stability (Fig. 1).
The data-driven consensus library was found to be very functional, with 90% of active transformants compared to about 50% in the random mutagenesis library. The high functionality of the consensus library can be attributed to the choice of specific point mutations and the application of eliminating criteria to reduce destructive mutations. It was previously shown that the data-driven consensus approach is useful in creating mutant libraries of high functionality, which enables the screening effort to be minimized (39, 42, 47, 61). Low functionality of a random mutagenesis library may be a result of a high mutation rate and multiple alterations that can lead to destabilization and unfolding of the enzyme (36, 66). Moderate functionality (61% and 33%) of random mutagenesis libraries (generated by error-prone PCR) with similar mutation frequencies as those determined in this work was previously reported by Bloom et al. for the mutagenesis of cytochrome P450 (66).
Three beneficial mutations were found in the consensus library. Substitutions H86Y and A269T improved the lipase stability in methanol, and A9Y increased the recombinant lipase expression in the E. coli host cells. Various factors can influence the expression levels of recombinant protein genes in E. coli cells, among them, codon usage, tRNA concentrations, mRNA primary and secondary structures, and the interactions between the sequence following the initiation codon and 5′-end of the 16S rRNA (67, 68). This mutation can be introduced at a later stage to the improved lipase variants to enhance their expression levels in order to achieve larger enzyme quantities in the purification process.
In this study, random mutagenesis (error-prone PCR) was also found to be useful for creating organic solvent-stable lipases. Random mutagenesis is beneficial, especially in cases of limited structural data and structure-stability correlations, as shown in other studies (27, 31). Q185L showed the highest stability in methanol, and the second best was H86Y, which was also found in the consensus library. It was interesting to see that both approaches resulted in the same mutation. However, the mutations at the nucleotide level were different. In the consensus library, the point mutations were designed to best fit the E. coli codon frequencies (CAC → TAT, two transition mutations), and in the random mutagenesis the mutation was the result of a single transition mutation (CAC → CAT). Based on this result, we assume that substitution A269T could have been found as well in the random mutagenesis library, as only a single transition difference between the codons (GCC → ACC). However, this would have required the exploitation of more resources and screening efforts.
Variants Q185L and A269T contributed the most to the lipase stability in methanol (Fig. 1), and their combination, Q185L/A269T, resulted in an additive stabilizing effect. Nevertheless, the double mutant from the consensus library, H86Y/A269T, showed the highest thermostability and most improved stability in alcohol (Fig. 2 and 3; Table 1).
The deactivation profiles of the wild type and the improved variants in the presence of methanol and ethanol followed the first-order inactivation model (Fig. 3 and Table 1). Similar observations on the inactivation effect of organic solvents have been reported for other enzymes, such as Pseudomonas aeruginosa LST-03 lipase (27) and horseradish peroxidase (62).
As our research goal was to create a lipase for enhanced stability in methanol with improved performance in esterification and transesterification reactions, we evaluated the wild type and improved variants for biodiesel synthesis. The methanolysis activity of all variants decreased as the methanol concentrations in the reaction system increased (Fig. 5). This decreased activity at high methanol concentrations was probably a result of the methanol-induced unfolding of lipase, which led to inactivation. It was previously reported by Shimada et al. that a decline in methanolysis performance was observed for immobilized Candida antarctica lipase in reaction systems with methanol:oil ratios higher than 1.5:1 (16).
Mutants H86Y/A269T and A269T, which showed the best methanolysis activities in all reaction systems, also possessed improved half-life values in methanol (Fig. 3 and Table 1) and elevated thermostability (Fig. 4). However, mutants Q185L and Q185L/A269T, which showed improved stability in methanol, had inferior methanolysis activity compared to the wild type in nonaqueous media (Fig. 5). Therefore, we suggest that the improved methanolysis activities of variants A269T and H86Y/A269T were a result of their improved stabilities in methanol, as there were no significant differences in their kinetic parameters compared to the wild type (Table 2). Subsequently, we assume that the decreased methanolysis activities of Q185L and Q185L/A269T were a result of the Q185L mutation, which hinders the lipase activity.
This work demonstrated the effectiveness of protein engineering to design a lipase for enhanced stability in methanol and as a result to improve its performance in biodiesel synthesis. Our research results also emphasize the power of the structure-guided consensus approach for generating active and stable enzymes by using smaller and smarter libraries, while minimizing the screening efforts and shortening the time to achieve the desired biocatalyst. The random mutagenesis approach was also useful in creating methanol-stable variants. However, this approach requires larger libraries and a few mutagenesis cycles to find improved variants (directed evolution), which may expand the screening efforts and the time for the target (36, 61, 69).
A 3D homology model of lipase T6 and the methanol-stable variants was created based on the crystal structure of G. stearothermophilus lipase P1 (PDB code 1JI3) (56) in order to improve our understanding of the mutagenesis effect on lipase stability and activity. According to the model, all mutations, H86Y, Q185L, and A269T, are located on the enzyme surface and could have direct interactions with the solvent (Fig. 6). It was previously reported that the interactions between surface residues and the solvent molecules have a key role in enzyme stability in organic solvents (7, 8). Ogino and coworkers found that substitutions of residues located on the enzyme surface resulted in a shift of their stability in various organic solvents. P. aeruginosa PST-01 protease stability in organic solvents was reduced (70), and P. aeruginosa LST-03 lipase stability in organic solvents was enhanced as a result of surface residue substitutions (27). In both cases, it was suggested that the surface residues have an important role in preventing solvent molecule penetration through the enzyme surface, and therefore mutagenesis of such residues caused a change in the enzyme's organic solvent stability (27, 70).
FIG 6.
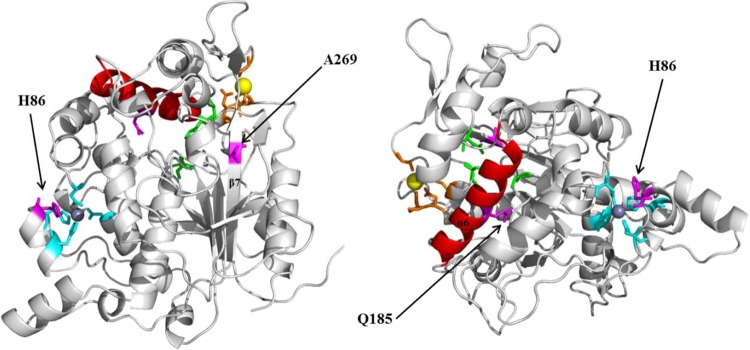
Structure models of wild-type lipase T6 with mutation location sites. The active site catalytic triad residues (Ser114, Asp318, and His359) are marked in green. The α-helix lid (α6; Asp176-Ala192) is marked in red. The calcium-binding residues (Glu361, Gly287, Pro367, and Asp366) are marked in orange, with calcium in yellow. The zinc-binding residues (Asp62, His88, Asp239, and His82) are marked in cyan, with zinc in dark gray. Residues subjected to mutagenesis are marked in magenta. The wild-type lipase T6 model was visualized using PyMOL, based on the crystal structure of G. stearothermophilus P1 lipase (PDB code 1JI3).
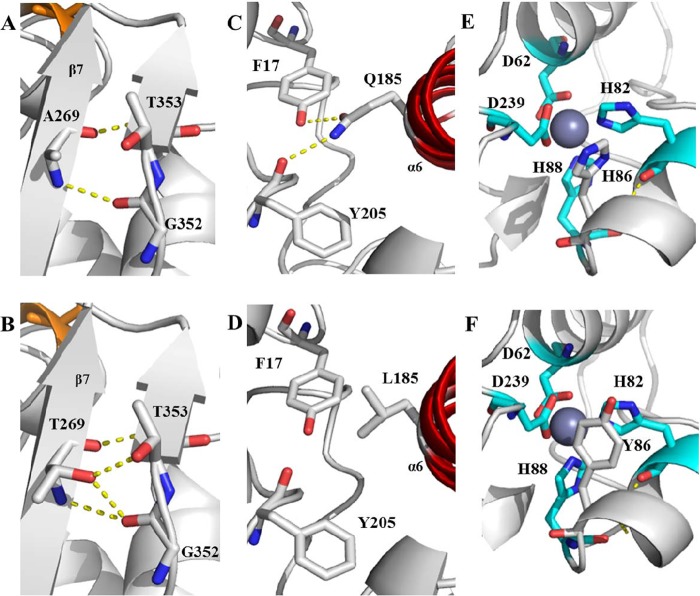
Alanine 269 is located on a β-sheet and is exposed to the protein surface (Fig. 6). The replacement with threonine resulted in improved stability in the presence of methanol and elevated the thermal-induced unfolding Tm by 3.4, 2.6, and 4.3°C for A269T, Q185L/A269T, and H86Y/A269T, respectively (Fig. 4). Based on the model, T269 can form two new hydrogen bonds with the hydroxyl group of T353 and with the G352 hydroxyl backbone (Fig. 7A and B). This assumption is supported by the saturation mutagenesis results, in which the only residues found to improve the enzyme stability were threonine and serine. Both have a hydroxyl that can create new hydrogen bonds with other residues. However, A269S showed lower stability than A269T. This observation can be explained with consideration of the relatively high conformational flexibility of the serine residue compared to the threonine, which has a methyl group that can limit the conformational possibilities at this position. Introduction of hydrogen bonds and electrostatic interactions for improving the interactions between amino acids on the enzyme surface were previously reported by Reetz et al. as contributing to the thermostability (71) and organic solvent stability (41) of lipase from Bacillus subtilis. Similarly, organic solvent stability was achieved by the introduction of new hydrogen bonds and facilitating new salt bridges, as reported by Kawata and Ogino for Pseudomonas aeruginosa LST-03 lipase (27).
FIG 7.
Structure models with hydrogen bonds for wild-type lipase T6 and the improved mutants. (A) Wild-type A269; (B) A269T; (C) wild-type Q185; (D) Q185L; (E) wild-type H86; (F) H86Y. The α-helix lid (α6) is marked in red (C and D), and the zinc-binding residues are marked in cyan, with zinc in dark gray. Possible hydrogen bonds are marked by yellow dashed lines. The models were visualized using PyMOL based on the crystal sturucture of G. stearothermophilus P1 lipase (PDB code 1JI3).
Glutamine 185 is situated on the lipase α-helix lid (α6), which has an important role in the lipase “interfacial activation” (Fig. 6 and 7). Depending on the medium hydrophobicity, it can undergo a conformational change from “closed” to “open” and enable the substrate access to the active site (5, 6). Tyndall and coworkers pointed out the role of Gln184 in G. stearothermophilus lipase P1, homologues to Gln185 in lipase T6, as a possible lid anchor that maintains the “closed” lid conformation by creating hydrogen bonds with the backbone carbonyl oxygen of Phe16 (56). Based on our model, another hydrogen bond can possibly be formed between the Gln185 side chain and the Tyr205 hydroxyl side chain (Fig. 7C and D). The Q185L substitution resulted in improved organic solvent stability. However, as opposed to A269T, it did not elevate the thermal-induced unfolding Tm (Fig. 4). Such observations were previously reported by Kawata and Ogino on enhanced stability in organic solvent by single mutations that did not affect the P. aeruginosa LST-03 lipase thermostability (27). The contribution of a hydrophobic residue at this position was also supported by the saturation mutagenesis results, in which only hydrophobic amino acids (Ala, Val, Leu, Met, and Phe) were found to improve the lipase stability in methanol. In addition, this substitution resulted in slightly lower activity and catalytic efficiency compared to the wild type and the other mutants in the standard activity assay without incubation in alcohol (Fig. 2 and Table 2) and impaired methanolysis (Fig. 5). We suggest that the replacement of the polar residue glutamine by the hydrophobic residue leucine improved the structural stability of the enzyme by facilitating the interaction between the solvent molecules and the lid surface. Martinez and Arnold have demonstrated the stabilizing effect of surface charges in substitutions by more hydrophobic ones on the stability of α-lytic protease in the presence of 84% dimethylformamide (32). Furthermore, we assume that the lower activity of Q185L is a result of a more favorable “closed” lid conformation than that of the wild-type lid as a result of hydrophobic interactions between the leucine residue and the hydrophobic residues of the active site pocket. This can result in limited substrate access to the active site and lower activity. Thus, it is suggested that the differences between the minor activity decrease in the pNPL hydrolysis and the major decrease in methanolysis activity can be explained by the differences in the substrate sizes. When we tested the consensus alignment at position 185, we realized that all the organic solvent-stabilizing residues found in the saturation mutagenesis were also found in the alignment (see Fig. S2 in the supplemental material). However, this position was eliminated from the final candidates in order to reduce the mutagenesis effect on activity as it was placed on the helix lid.
Histidine 86 is located on an α-helix relatively close to the zinc-binding residues H82 and H88 (Fig. 7E and F). The zinc-binding domain is a common characteristic of the I.5 lipase subfamily and may have an important contribution to the lipase thermostability (2, 56). The H86Y substitution did not seem to affect the zinc binding, as the distance between the tyrosine and the zinc was >6 Å. We assume that the contribution of this mutation for the improved stability in methanol is via the introduction of new hydrogen bonds between the tyrosine hydroxyl group directly to other amino acid residues or to structurally important water molecules, as its effect was similar to that of A269T.
The double mutants, Q185L/A269T and H86Y/A269T, expressed an additive stabilizing effect compared to the two single mutations (Fig. 2 and 3; Table 1). We assume that the additive stabilizing effect of these mutations was due to the relatively distant location on the enzyme structure without direct interaction between them, and therefore the individual contribution of each mutation was reflected in the combined mutant. Such an additive effect of several mutations on enzyme stability was previously demonstrated for thermostability and organic solvent stability (27, 31, 32, 39, 40).
In conclusion, we have successfully engineered lipase T6 for enhanced stability in the presence of methanol and ethanol by applying the protein engineering approaches of random mutagenesis and structure-guided consensus. In this study, we have demonstrated for the first time the power of the data-driven approach of structure-guided consensus for creating a lipase with improved methanolysis activity by improving its stability in methanol. The best mutant created by this approach exhibited enhanced stability in polar organic solvents together with improved thermostability while maintaining the catalytic activity. These characteristics could make it a powerful biocatalyst for biodiesel production. The lipase performance in organic solvents can be further improved by using additional protein engineering methods, such as the B-FIT and ISM approaches (38, 41) and by immobilization to a support matrix for recycling. The enzymes will be further investigated by determining their X-ray crystal structures.
Supplementary Material
ACKNOWLEDGMENT
This project was partly funded by the Israel-Mexico Energy Research Fund.
Footnotes
Published ahead of print 20 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03371-13.
REFERENCES
- 1.Gupta R, Gupta N, Rathi P. 2004. Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64:763–781. 10.1007/s00253-004-1568-8 [DOI] [PubMed] [Google Scholar]
- 2.Guncheva M, Zhiryakova D. 2011. Catalytic properties and potential applications of Bacillus lipases. J. Mol. Catal. B Enzym. 68:1–21. 10.1016/j.molcatb.2010.09.002 [DOI] [Google Scholar]
- 3.Hasan F, Shah AA, Hameed A. 2006. Industrial applications of microbial lipases. Enzyme Microb. Technol. 39:235–251. 10.1016/j.enzmictec.2005.10.016 [DOI] [Google Scholar]
- 4.Villeneuve P, Muderhwa JM, Graille J, Haas MJ. 2000. Customizing lipases for biocatalysis: a survey of chemical, physical and molecular biological approaches. J. Mol. Catal. B Enzym. 9:113–148. 10.1016/S1381-1177(99)00107-1 [DOI] [Google Scholar]
- 5.Jaeger KE, Dijkstra BW, Reetz MT. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53:315–351. 10.1146/annurev.micro.53.1.315 [DOI] [PubMed] [Google Scholar]
- 6.Jaeger KE, Ransac S, Dijkstra BW, Colson C, van Heuvel M, Misset O. 1994. Bacterial lipases. FEMS Microbiol. Rev. 15:29–63 [DOI] [PubMed] [Google Scholar]
- 7.Arnold FH. 1990. Engineering enzymes for non-aqueous solvents. Trends Biotechnol. 8:244–249. 10.1016/0167-7799(90)90186-2 [DOI] [PubMed] [Google Scholar]
- 8.Doukyu N, Ogino H. 2010. Organic solvent-tolerant enzymes. Biochem. Eng. J. 48:270–282. 10.1016/j.bej.2009.09.009 [DOI] [Google Scholar]
- 9.Ogino H, Ishikawa H. 2001. Enzymes which are stable in the presence of organic solvents. J. Biosci. Bioeng. 91:109–116. 10.1016/S1389-1723(01) [DOI] [PubMed] [Google Scholar]
- 10.Chakravorty D, Parameswaran S, Dubey V, Patra S. 2012. Unraveling the rationale behind organic solvent stability of lipases. Appl. Biochem. Biotechnol. 167:439–461. 10.1007/s12010-012-9669-9 [DOI] [PubMed] [Google Scholar]
- 11.Schulze B, Klibanov AM. 1991. Inactivation and stabilization of stabilisins in neat organic solvents. Biotechnol. Bioeng. 38:1001–1006. 10.1002/bit.260380907 [DOI] [PubMed] [Google Scholar]
- 12.Bajaj A, Lohan P, Jha PN, Mehrotra R. 2010. Biodiesel production through lipase catalyzed transesterification: an overview. J. Mol. Catal. B Enzym. 62:9–14. 10.1016/j.molcatb.2009.09.018 [DOI] [Google Scholar]
- 13.Du W, Li W, Sun T, Chen X, Liu D. 2008. Perspectives for biotechnological production of biodiesel and impacts. Appl. Microbiol. Biotechnol. 79:331–337. 10.1007/s00253-008-1448-8 [DOI] [PubMed] [Google Scholar]
- 14.Al-Zuhair S. 2007. Production of biodiesel: possibilities and challenges. Biofuels Bioprod. Bioref. 1:57–66. 10.1002/bbb.2 [DOI] [Google Scholar]
- 15.Xiao M, Mathew S, Obbard JP. 2009. Biodiesel fuel production via transesterification of oils using lipase biocatalyst. GCB Bioenergy 1:115–125. 10.1111/j.1757-1707.2009.01009.x [DOI] [Google Scholar]
- 16.Shimada Y, Watanabe Y, Samukawa T, Sugihara A, Noda H, Fukuda H, Tominaga Y. 1999. Conversion of vegetable oil to biodiesel using immobilized Candida antarctica lipase. J. Am. Oil Chem. Soc. 76:789–793 [Google Scholar]
- 17.Kumar S, Ola RP, Pahujani S, Kaushal R, Kanwar SS, Gupta R. 2006. Thermostability and esterification of a polyethylene-immobilized lipase from Bacillus coagulans BTS-3. J. Appl. Polym. Sci. 102:3986–3993. 10.1556/AMicr.53.2006.2.8 [DOI] [Google Scholar]
- 18.Bagi K, Simon LM, Szajáni B. 1997. Immobilization and characterization of porcine pancreas lipase. Enzyme Microb. Technol. 20:531–535 [Google Scholar]
- 19.Kawakami K, Oda Y, Takahashi R. 2011. Application of a Burkholderia cepacia lipase-immobilized silica monolith to batch and continuous biodiesel production with a stoichiometric mixture of methanol and crude Jatropha oil. Biotechnol. Biofuels 4:42. 10.1186/1754-6834-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho CML, Cabral JMS. 2000. Reverse micelles as reaction media for lipases. Biochimie 82:1063–1085. 10.1016/S0300-9084(00)01187-1 [DOI] [PubMed] [Google Scholar]
- 21.Yang C-L, Gulari E. 1994. Enzymic esterification of diols in reverse micellar media. Biotechnol. Prog. 10:269–276 [Google Scholar]
- 22.Mine Y, Fukunaga K, Yoshimoto M, Nakao K, Sugimura Y. 2001. Modification of lipases with poly(ethylene glycol) and poly(oxyethylene) detergents and their catalytic activities in organic solvents. J. Biosci. Bioeng. 92:539–543. 10.1016/S1389-1723(01)80312-1 [DOI] [PubMed] [Google Scholar]
- 23.Cabrera Z, Fernandez-Lorente G, Fernandez-Lafuente R, Palomo JM, Guisan JM. 2009. Enhancement of Novozym-435 catalytic properties by physical or chemical modification. Process Biochem. 44:226–231. 10.1016/j.procbio.2008.10.005 [DOI] [Google Scholar]
- 24.Uttatree S, Winayanuwattikun P, Charoenpanich J. 2010. Isolation and characterization of a novel thermophilic-organic solvent stable lipase from Acinetobacter baylyi. Appl. Biochem. Biotechnol. 162:1362–1376. 10.1007/s12010-010-8928-x [DOI] [PubMed] [Google Scholar]
- 25.Khan M, Jithesh K. 2012. Expression and purification of organic solvent stable lipase from soil metagenomic library. World J. Microbiol. Biotechnol. 28:2417–2424. 10.1007/s1274-012-1051-0 [DOI] [PubMed] [Google Scholar]
- 26.Di Lorenzo M, Hidalgo A, Molina R, Hermoso JA, Pirozzi D, Bornscheuer UT. 2007. Enhancement of the stability of a prolipase from Rhizopus oryzae toward aldehydes by saturation mutagenesis. Appl. Environ. Microbiol. 73:7291–7299. 10.1128/AEM.01176-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawata T, Ogino H. 2009. Enhancement of the organic solvent-stability of the LST-03 lipase by directed evolution. Biotechnol. Prog. 25:1605–1611. 10.1002/btpr.264 [DOI] [PubMed] [Google Scholar]
- 28.Park H, Joo J, Park K, Yoo Y. 2012. Stabilization of Candida antarctica lipase B in hydrophilic organic solvent by rational design of hydrogen bond. Biotechnol. Bioprocess Eng. 17:722–728. 10.1007/s12257-012-0092-4 [DOI] [Google Scholar]
- 29.Takagi H, Hirai K, Maeda Y, Matsuzawa H, Nakamori S. 2000. Engineering subtilisin E for enhanced stability and activity in polar organic solvents. J. Biochem. 127:617–625. 10.1093/oxfordjournals.jbchem.a022649 [DOI] [PubMed] [Google Scholar]
- 30.Badoei-Dalfard A, Khajeh K, Asghari SM, Ranjbar B, Karbalaei-Heidari HR. 2010. Enhanced activity and stability in the presence of organic solvents by increased active site polarity and stabilization of a surface loop in a metalloprotease. J. Biochem. 148:231–238. 10.1093/jb/mvq057 [DOI] [PubMed] [Google Scholar]
- 31.Song JK, Rhee JS. 2001. Enhancement of stability and activity of phospholipase A1 in organic solvents by directed evolution. Biochim. Biophys. Acta 1547:370–378. 10.1016/S0167-4838(01)00204-7 [DOI] [PubMed] [Google Scholar]
- 32.Martinez P, Arnold FH. 1991. Surface charge substitutions increase the stability of α-lytic protease in organic solvents. J. Am. Chem. Soc. 113:6336–6337 [Google Scholar]
- 33.Chen K, Robinson AC, Van Dam ME, Martinez P, Economou C, Arnold FH. 1991. Enzyme engineering for nonaqueous solvents. II. Additive effects of mutations on the stability and activity of subtilisin E in polar organic media. Biotechnol. Prog. 7:125–129 [DOI] [PubMed] [Google Scholar]
- 34.Zumárraga M, Bulter T, Shleev S, Polaina J, Martínez-Arias A, Plou FJ, Ballesteros A, Alcalde M. 2007. In vitro evolution of a fungal laccase in high concentrations of organic cosolvents. Chem. Biol. 14:1052–1064. 10.1016/j.chembiol.2007.08.010 [DOI] [PubMed] [Google Scholar]
- 35.Takahashi T, Ng KK-S, Oyama H, Oda K. 2005. Molecular cloning of the gene encoding vibrio metalloproteinase vimelysin and isolation of a mutant with high stability in organic solvents. J. Biochem. 138:701–710. 10.1093/jb/mvi173 [DOI] [PubMed] [Google Scholar]
- 36.Kazlauskas RJ, Bornscheuer UT. 2009. Finding better protein engineering strategies. Nat. Chem. Biol. 5:526–529. 10.1038/nchembio0809-526 [DOI] [PubMed] [Google Scholar]
- 37.Turner NJ. 2009. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5:567–573. 10.1038/nchembio.203 [DOI] [PubMed] [Google Scholar]
- 38.Reetz MT, Kahakeaw D, Lohmer R. 2008. Addressing the numbers problem in directed evolution. Chembiochem 9:1797–1804. 10.1002/cbic.200800298 [DOI] [PubMed] [Google Scholar]
- 39.Vázquez-Figueroa E, Chaparro-Riggers J, Bommarius AS. 2007. Development of a thermostable glucose dehydrogenase by a structure-guided consensus concept. Chembiochem 8:2295–2301. 10.1002/cbic.200700500 [DOI] [PubMed] [Google Scholar]
- 40.Vazquez-Figueroa E, Yeh V, Broering JM, Chaparro-Riggers JF, Bommarius AS. 2008. Thermostable variants constructed via the structure-guided consensus method also show increased stability in salts solutions and homogeneous aqueous-organic media. Protein Eng. Des. Sel. 21:673–680. 10.1093/protein/gzn048 [DOI] [PubMed] [Google Scholar]
- 41.Reetz MT, Soni P, Fernandez L, Gumulya Y, Carballeira JD. 2010. Increasing the stability of an enzyme toward hostile organic solvents by directed evolution based on iterative saturation mutagenesis using the B-FIT method. Chem. Commun. (Camb.) 46:8657–8658. 10.1039/c0cc02657c [DOI] [PubMed] [Google Scholar]
- 42.Polizzi KM, Chaparro-Riggers JF, Vazquez-Figueroa E, Bommarius AS. 2006. Structure-guided consensus approach to create a more thermostable penicillin G acylase. Biotechnol. J. 1:531–536. 10.1002/biot.200600029 [DOI] [PubMed] [Google Scholar]
- 43.Miyazaki J, Nakaya S, Suzuki T, Tamakoshi M, Oshima T, Yamagishi A. 2001. Ancestral residues stabilizing 3-isopropylmalate dehydrogenase of an extreme thermophile: experimental evidence supporting the thermophilic common ancestor hypothesis. J. Biochem. 129:777–782. 10.1093/oxfordjournals.jbchem.a002919 [DOI] [PubMed] [Google Scholar]
- 44.Watanabe K, Ohkuri T, Yokobori S-I, Yamagishi A. 2006. Designing thermostable proteins: ancestral mutants of 3-isopropylmalate dehydrogenase designed by using a phylogenetic tree. J. Mol. Biol. 355:664–674. 10.1016/j.jmb.2005.10.011 [DOI] [PubMed] [Google Scholar]
- 45.Lehmann M, Loch C, Middendorf A, Studer D, Lassen SF, Pasamontes L, van Loon APGM, Wyss M. 2002. The consensus concept for thermostability engineering of proteins: further proof of concept. Protein Eng. 15:403–411. 10.1093/protein/15.5.403 [DOI] [PubMed] [Google Scholar]
- 46.Steipe B, Schiller B, Plückthun A, Steinbacher S. 1994. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 240:188–192 [DOI] [PubMed] [Google Scholar]
- 47.Amin N, Liu AD, Ramer S, Aehle W, Meijer D, Metin M, Wong S, Gualfetti P, Schellenberger V. 2004. Construction of stabilized proteins by combinatorial consensus mutagenesis. Protein Eng. Des. Sel. 17:787–793. 10.1093/protein/gzh091 [DOI] [PubMed] [Google Scholar]
- 48.Khersonsky O, Rosenblat M, Toker L, Yacobson S, Hugenmatter A, Silman I, Sussman JL, Aviram M, Tawfik DS. 2009. Directed evolution of serum paraoxonase PON3 by family shuffling and ancestor/consensus mutagenesis, and its biochemical characterization. Biochemistry (Wash.) 48:6644–6654. 10.1021/bi900583y [DOI] [PubMed] [Google Scholar]
- 49.Meshulam-Simon G. 2001. Isolation and characterization of lipases from thermophilic bacteria for the preparation of optically active compounds. Ph.D. thesis. Technion—Israel Institute of Technology, Haifa, Israel [Google Scholar]
- 50.Kristjansson JK. 1989. Thermophilic organisms as sources of thermostable enzymes. Trends Biotechnol. 7:349–353. 10.1016/0167-7799(89)90035-8 [DOI] [Google Scholar]
- 51.Sambrook J, Russel DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 52.Rashid N, Shimada Y, Ezaki S, Atomi H, Imanaka T. 2001. Low-temperature lipase from psychrotrophic Pseudomonas sp. strain KB700A. Appl. Environ. Microbiol. 67:4064–4069. 10.1128/AEM.67.9.4064-4069.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. 2010. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38:W529–W533. 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simossis VA, Heringa J. 2005. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 33:289–294. 10.1093/nar/gki170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simossis VA, Kleinjung J, Heringa J. 2005. Homology-extended sequence alignment. Nucleic Acids Res. 33:816–824. 10.1093/nar/gki233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tyndall JDA, Sinchaikul S, Fothergill-Gilmore LA, Taylor P, Walkinshaw MD. 2002. Crystal structure of a thermostable lipase from Bacillus stearothermophilus P1. J. Mol. Biol. 323:859–869. 10.1016/S0022-283.6(02)01004-5 [DOI] [PubMed] [Google Scholar]
- 57.Herman A, Tawfik DS. 2007. Incorporating synthetic oligonucleotides via gene reassembly (ISOR): a versatile tool for generating targeted libraries. Protein Eng. Des. Sel. 20:219–226. 10.1093/protein/gzm014 [DOI] [PubMed] [Google Scholar]
- 58.Bottcher D, Bornscheuer UT. 2006. High-throughput screening of activity and enantioselectivity of esterases. Nat. Protoc. 1:2340–2343. 10.1038/nprot.2006.391 [DOI] [PubMed] [Google Scholar]
- 59.Ben-Yosef VS, Sendovski M, Fishman A. 2010. Directed evolution of tyrosinase for enhanced monophenolase/diphenolase activity ratio. Enzyme Microb. Technol. 47:372–376. 10.1016/j.enzmictec.2010.08.008 [DOI] [Google Scholar]
- 60.Solano F, Garcia E, Perez D, Sanchez-Amat A. 1997. Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium. Appl. Environ. Microbiol. 63:3499–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bommarius AS, Blum JK, Abrahamson MJ. 2011. Status of protein engineering for biocatalysts: how to design an industrially useful biocatalyst. Curr. Opin. Chem. Biol. 15:194–200. 10.1016/j.cbpa.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 62.Fishman A, Levy I, Cogan U, Shoseyov O. 2002. Stabilization of horseradish peroxidase in aqueous-organic media by immobilization onto cellulose using a cellulose-binding-domain. J. Mol. Catal. B Enzym. 18:121–131. 10.1016/S1381-1177(02)00075-9 [DOI] [Google Scholar]
- 63.Sellek GA, Chaudhuri JB. 1999. Biocatalysis in organic media using enzymes from extremophiles. Enzyme Microb. Technol. 25:471–482 [Google Scholar]
- 64.Yao C, Cao Y, Wu S, Li S, He B. 2013. An organic solvent and thermally stable lipase from Burkholderia ambifaria YCJ01: purification, characteristics and application for chiral resolution of mandelic acid. J. Mol. Catal. B Enzym. 85–86:105–110. 10.1016/j.molcatb.2012.08.016 [DOI] [Google Scholar]
- 65.Gog A, Roman M, Toşa M, Paizs C, Irimie FD. 2012. Biodiesel production using enzymatic transesterification: current state and perspectives. Renewable Energy 39:10–16. 10.1016/j.renene.2011.08.007 [DOI] [Google Scholar]
- 66.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. 2006. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. U. S. A. 103:5869–5874. 10.1073/pnas.0510098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cèbe R, Geiser M. 2006. Rapid and easy thermodynamic optimization of the 5′-end of mRNA dramatically increases the level of wild type protein expression in Escherichia coli. Protein Expr. Purif. 45:374–380. 10.1016/j.pep.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 68.Degryse E. 1990. Influence of the second and third codon on the expression of recombinant hirudin in E. coli. FEBS Lett. 269:244–246. 10.1016/0014-5793(90)81164-J [DOI] [PubMed] [Google Scholar]
- 69.Eijsink VGH, Gåseidnes S, Borchert TV, van den Burg B. 2005. Directed evolution of enzyme stability. Biomol. Eng. 22:21–30. 10.1016/j.bioeng.2004.12.003 [DOI] [PubMed] [Google Scholar]
- 70.Ogino H, Uchiho T, Doukyu N, Yasuda M, Ishimi K, Ishikawa H. 2007. Effect of exchange of amino acid residues of the surface region of the PST-01 protease on its organic solvent-stability. Biochem. Biophys. Res. Commun. 358:1028–1033. 10.1016/j.bbrc.2007.05.047 [DOI] [PubMed] [Google Scholar]
- 71.Reetz MT, Soni P, Acevedo JP, Sanchis J. 2009. Creation of an amino acid network of structurally coupled residues in the directed evolution of a thermostable enzyme. Angew. Chem. Int. Ed. Engl. 121:8418–8422. 10.1002/anie.200904209 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.