Abstract
Source waters sampled from Perpetual Spouter hot spring (pH 7.03, 86.4°C), Yellowstone National Park, WY, have low concentrations of total ammonia, nitrite, and nitrate, suggesting nitrogen (N) limitation and/or tight coupling of N cycling processes. Dominant small-subunit rRNA sequences in Perpetual Spouter source sediments are closely affiliated with the ammonia-oxidizing archaeon “Candidatus Nitrosocaldus yellowstonii” and the putatively nitrogen-fixing (diazotrophic) bacterium Thermocrinis albus, respectively, suggesting that these populations may interact at the level of the bioavailable N pool, specifically, ammonia. This hypothesis was evaluated by using a combination of geochemical, physiological, and transcriptomic analyses of sediment microcosms. Amendment of microcosms with allylthiourea, an inhibitor of ammonia oxidation, decreased rates of acetylene reduction (a proxy for N2 fixation) and nitrite production (a proxy for ammonia oxidation) and decreased transcript levels of structural genes involved in both nitrogen fixation (nifH) and ammonia oxidation (amoA). In contrast, amendment of microcosms with ammonia stimulated nitrite production and increased amoA transcript levels while it suppressed rates of acetylene reduction and decreased nifH transcript levels. Sequencing of amplified nifH and amoA transcripts from native sediments, as well as microcosms, at 2 and 4 h postamendment, indicates that the dominant and responsive populations involved in ammonia oxidation and N2 fixation are closely affiliated with Ca. Nitrosocaldus yellowstonii and T. albus, respectively. Collectively, these results suggest that ammonia-oxidizing archaea, such as Ca. Nitrosocaldus yellowstonii, have an apparent affinity for ammonia that is higher than that of the diazotrophs present in this ecosystem. Depletion of the bioavailable N pool through the activity of ammonia-oxidizing archaea likely represents a strong selective pressure for the inclusion of organisms capable of nitrogen fixation in geothermal communities. These observations help to explain the strong pattern in the codistribution of ammonia-oxidizing archaea and diazotrophs in circumneutral-to-alkaline geothermal springs.
INTRODUCTION
All life requires fixed sources of nitrogen (N), and its availability often limits productivity in natural ecosystems (1). Biological nitrogen fixation, or the reduction of dinitrogen (N2) to ammonia (NH3), is a keystone process in N-limited ecosystems, providing bioavailable fixed N for use in the synthesis of macromolecules (e.g., protein, nucleic acids) for both N2-fixing (diazotrophic) and non-N2-fixing members of the community (2). In addition to the use of total ammonia [i.e., NH4+(aq) and NH3o(aq), written as NH4(T)] in anabolic macromolecular synthesis, several lineages of bacteria and archaea have evolved to use NH4(T) as a source of reductant through the process of NH4(T) oxidation or the sequential oxidation of NH4(T) to nitrite (NO2−) (3). This process, documented to occur in a diversity of environments that span terrestrial and marine ecosystems (4–10), represents an additional sink on the bioavailable NH4(T) pool.
NH4(T) oxidation activity was recently demonstrated in a number of terrestrial hot springs (11–13) at temperatures that exceed the proposed ∼62°C upper limit of bacteria capable of NH4(T) oxidation (14). In these environments, NH4(T) oxidation was shown to be catalyzed by archaea affiliated with the phylum Thaumarchaeota (15). “Candidatus Nitrosocaldus yellowstonii,” the first cultivated thermophilic member of the phylum Thaumarchaeota (11), obligately uses NH4(T) as a source of reductant to drive inorganic carbon fixation under oxic conditions. Previous studies have shown Ca. Nitrosocaldus yellowstonii to be a widespread member of communities inhabiting circumneutral-to-alkaline hot springs in a number of globally distributed geothermal systems (11–13). More recently, screening of communities inhabiting geochemically heterogeneous springs in Yellowstone National Park (YNP) revealed a nearly universal distribution of archaeal ammonia monooxygenase genes (amoA), a proxy for NH4(T)-oxidizing archaea (16), in circumneutral and alkaline springs (17). All of the amoA sequences obtained from selected springs with temperatures of >60°C were found to be >97% identical at the nucleotide level to amoA from Ca. Nitrosocaldus yellowstonii (E. S. Boyd, unpublished data).
Concentrations of NH4(T) in alkaline geothermal systems, including those documented to contain abundant populations of putative NH4(T)-oxidizing archaea affiliated with Ca. Nitrosocaldus yellowstonii (11, 17), tend to be low (<100 μM) relative to those in acidic geothermal springs (18) (Fig. 1A). The low concentrations of NH4(T) in alkaline geothermal springs is due, in part, to geological, chemical, and hydrological factors, most notably, the equilibration of aqueous NH4+(aq) with NH3(g) (pK = 7.6 at 90°C) and the subsequent volatilization of NH3(g) out of the system (18). The success of NH4(T)-oxidizing archaea in other NH4(T)-limited ecosystems, such as the marine water column, has been attributed to their high affinity for reduced nitrogen (19), which enables these populations to obtain NH4(T) at ambient concentrations much lower (>10 nM) than those of bacterial NH4(T) oxidizers and other members of the community. The thermophilic NH4(T)-oxidizing archaea in NH4(T)-limited alkaline geothermal ecosystems may have similar adaptations to acquire this nutrient at low concentrations and/or may play an active role in maintaining the low concentrations of NH4(T) in these systems.
FIG 1.
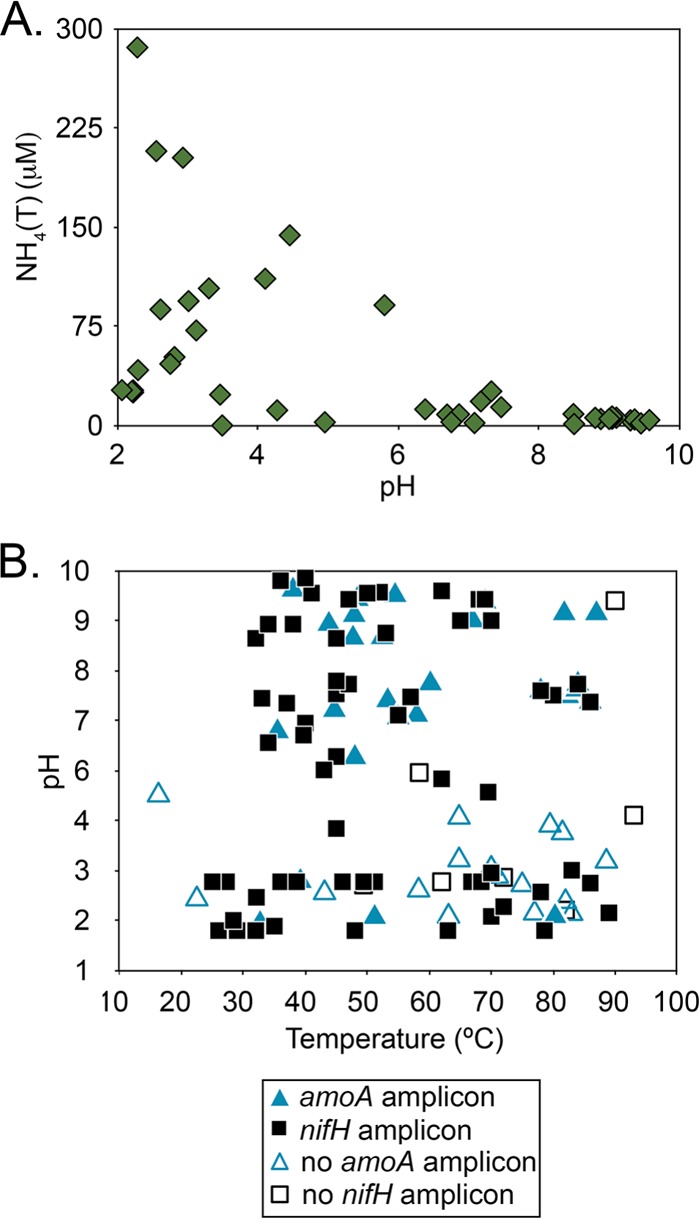
(A) NH4(T) concentrations in YNP geothermal springs plotted as a function of pH (data extrapolated from reference 17). (B) Distribution of nifH and amoA in mat and sediment samples from YNP geothermal springs plotted as a function of spring water temperature and pH, as compiled from previous reports (17, 20). Closed triangles (amoA) and closed squares (nifH) indicate sites where PCR amplicons were detected, and open triangles (amoA) and open squares (nifH) indicate sites where PCR amplicons were not detected.
The N-limited nature of most alkaline geothermal springs in YNP is expected to represent a strong selective pressure for inclusion of microbial populations with the capacity to fix N2 via the enzyme nitrogenase in resident communities. Indeed, a recent analysis of the distribution of the gene encoding the nitrogenase iron protein (nifH, a proxy for nitrogen fixation potential) in a range of YNP environments revealed a nearly universal distribution in circumneutral-to-alkaline ecosystems (20), similar to the distributional pattern of archaeal amoA (17) (Fig. 1B). More recently, nifH genes were shown to be actively transcribed in a number of high-temperature circumneutral-to-alkaline YNP springs (21), consistent with a role in N2 fixation in these systems. In the present study, we hypothesized that the strong pattern in the codistribution of putative NH4(T)-oxidizing archaea and putative N2-fixing microorganisms across YNP circumneutral-to-alkaline hot spring environments (Fig. 1B) is the result of the activity of NH4(T)-oxidizing archaea, which function to maintain low bioavailable NH4(T) concentrations, which in turn select for biological N2 fixation (diazotrophy). We evaluated this hypothesis by using a combination of inhibitor- and suppressor-based microcosm manipulations at Perpetual Spouter (PS) hot spring, a circumneutral geothermal spring in YNP where evidence of the presence of populations putatively involved in NH4(T) oxidation (17) and N2 fixation (20) has been documented.
MATERIALS AND METHODS
Description of sample sites and experimental design.
Microcosm-based biological activity assays, geochemical assays, and genetic/transcriptional assays (described in detail below) were conducted at PS hot spring (YNP thermal inventory ID NBB113), Norris Geyser Basin, YNP, WY, in May 2012.
Chemical analyses.
Spring temperature and pH were determined with a Cole-Parmer temperature-compensated model 59002-00 electrode. An alcohol thermometer was used to confirm the temperature. Conductivity was measured with a YSI model 33 S-C-T meter (Yellow Springs Instrument Company, Inc., Yellow Springs, OH). Conductivity values were standardized to a common temperature of 25.0°C as previously described (20). Early attempts to quantify dissolved NH4+ and NO2− in spring source waters and microcosm fluid via ion chromatography were confounded by the elevated Na+ and Cl− concentrations in PS source water (∼22 mM each [22]). Thus, dissolved NO2− and NH4(T) were determined by colorimetric approaches with a Hach DR/890 spectrophotometer and AccuVac ampuls for NO2−, and an AmVer ammonia reagent set for NH4(T) (Hach Company, Loveland, CO). Water for analysis was filtered through a 0.2-μm Supor filter prior to analysis. NH4(T) refers to the sum of the dissolved species of aqueous NH3 and NH4+ as measured by colorimetry (18).
DNA extraction and qPCR of 16S rRNA genes.
Sediments for molecular analyses (DNA or RNA) were collected aseptically with a flame-sterilized spatula, placed in 1.5-ml centrifuge tubes containing 500 μl RNAlater (Qiagen, Valencia, CA), and immediately flash frozen in a dry ice-ethanol slurry for transport to Montana State University (MSU), where they were kept at −80°C for use in molecular analyses. Genomic DNA was extracted from ∼250 mg of PS sediments as previously described (24) and was quantified with the Qubit DNA Assay kit (Life Technologies, Grand Island, NY) and a Qubit 2.0 Fluorometer (Life Technologies). Quantitative PCR (qPCR) was used to estimate the number of archaeal and bacterial 16S rRNA gene sequences in genomic DNA as described previously (10). Assays were performed in triplicate in a Rotor-Gene 300 real-time qPCR machine (Qiagen) with the SsoFast EvaGreen Supermix qPCR kit (Bio-Rad Laboratories, Hercules, CA). The qPCR cycling conditions used are described in more detail in the supplemental material, and primer sequences and annealing temperatures are provided in Table S1 in the supplemental material. Standard curves relating the template copy number to the threshold qPCR amplification signal (see Table S2 in the supplemental material) were generated with plasmid DNA generated from the cloning of bacterial and archaeal 16S rRNA genes generated from PS as previously described (10).
RNA extraction, cDNA synthesis, amplicon cloning, and sequencing.
RNA was extracted from native sediments (represents time zero in microcosm studies) and from sediments that were incubated for 2 or 4 h (microcosm studies) with the FastRNA Pro Soil-Direct kit (MP Biomedical, Solon, OH). RNA was extracted in triplicate from ∼400 mg of sediment from each microcosm. Following extraction and purification, equal volumes of RNA from each replicate extraction were pooled and subjected to DNase I digestion (Roche) for 1 h at room temperature (∼22°C). Following digestion, RNA was further purified with a High Pure RNA Isolation kit (Roche, Indianapolis, IN) and stored at −80°C in the presence of 2 volumes of 100% ethanol and 0.1 volume sodium acetate (final concentration, 0.3 M) until further processed. The amount of RNA in each pooled volume was quantified with the Qubit RNA Assay kit (Life Technologies). As an additional precaution, RNA extracts were screened for the presence of contaminating genomic DNA via a PCR with ∼1 ng of RNA serving as the template and archaeal or bacterial 16S rRNA gene primers (see Table S1 in the supplemental material) as described above. cDNA was synthesized from ∼15 ng of purified RNA with the iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA) under the following reaction cycling conditions: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. Following synthesis of cDNA, samples were purified by precipitation (with ethanol and sodium acetate as described above) and resuspended in nuclease-free water for use in pyrotag sequencing of 16S cDNA (described below).
nifH and archaeal amoA were amplified from cDNA synthesized from RNA extracted from native sediment (0 h of incubation), as well as 2- and 4-h microcosms, as previously described (5, 20). Archaeal and bacterial 16S cDNA was amplified from native sediment (0 h of incubation) only. For the 2- and 4-h microcosms, equal amounts of pooled RNA extracted from each microcosm (performed in triplicate for each time point; see details below) were combined prior to cDNA synthesis. PCR cycling conditions, primers, sequences, and annealing temperature are provided in the supplemental material (see Table S1). PCR products were quantified, cloned, and subjected to Sanger sequencing as previously described (24). Putative nifH and amoA sequences were translated with the ExPASy translate tool (http://www.expasy.ch/tools/dna.html). nifH, archaeal amoA, bacterial amoA, and archaeal/bacterial 16S cDNA plasmids were used as standards in quantitative reverse transcription-PCR (qRT-PCR) assays (see below).
qRT-PCR assays.
The abundances of 16S rRNA, nifH transcripts, and archaeal amoA transcripts in RNA extracts were determined by qRT-PCR as previously described (23), with the Power SYBR green RNA-to-CT one-step kit (Life Technologies) according to the manufacturer's protocol. qRT-PCR assays were performed on a Qiagen RotorGene-Q real-time PCR detection system. qRT-PCR assay reaction mixtures, cycling conditions, primers, and calculation of fold change (difference in the abundance of amoA or nifH transcripts between time zero and microcosm experiments as outlined below) are described in more detail in the supplemental material (see Table S1). Negative control reaction mixtures contained either no reverse transcriptase or no template RNA. Standard curves and detection limits (see Table S2) were generated with purified and quantified cDNA plasmids (see above).
Small-subunit (SSU) cDNA sequencing.
Archaeal and bacterial 16S cDNA was sequenced by the Research and Testing Laboratory (Lubbock, TX) as previously described (25), with the primers described in Table S1 in the supplemental material. Pyrotag libraries were sequenced with a 454 Genome Sequencer FLX System (Roche, Branford, CT). Postsequencing processing was performed with mother, version 1.25.1 (26) (for processing details, see the supplemental material).
Acetylene reduction assays.
The acetylene reduction assay was used as a proxy for N2 fixation (27) in order to assess the influence of amendment with NH4+ (1 mM final concentration), allylthiourea (ATU; 1 mM final concentration), or NH4+ and ATU (each at a 1 mM final concentration) on N2 fixation activity. Prior to experimentation, it was first necessary to confirm the stability of ATU at a high temperature (i.e., 86°C). ATU (pH 7.2) was added to 10 ml of autoclaved sterilized distilled water (pH 7.2) contained in 24-ml serum bottles. Triplicate incubations were prepared under an atmosphere of N2 or air. The release of NH4(T) into solution was monitored via chromatography (Metrohm, Riverview, FL) with a Metrosep C-4 150/2.0 column (Metrohm) every hour for 8 h. No NH4+ was detected (detection limit, ∼0.5 μM) in solution after 8 h (data not shown) in the presence of N2 or air.
Microcosms for the measurement of acetylene reduction were prepared as previously described (28). Briefly, ∼500 mg of sediment was transferred to presterilized 20-ml serum vials and sediments were overlaid with 10 ml of source water taken directly from the spring. Vials were sealed with butyl stoppers and purged with Ar. Microcosms were amended with NH4Cl, ATU, NH4Cl plus ATU, HgCl2 (final concentration, 500 μM; killed control), or nothing (biological controls). Assays were initiated immediately following amendment by the addition of 500 μl of high-purity, O2-free acetylene. Microcosms were incubated in situ at the thermal transect where sediments were collected. All assays were performed in triplicate, and mixtures were incubated for 4 h following the addition of acetylene. Additional replicate microcosms were prepared and incubated at PS for use in RNA-based analyses. At 2- and 4-h incubation intervals, one set of triplicate microcosms was immediately flash frozen in a dry ice-ethanol slurry for molecular analysis. Upon the completion of gas phase subsampling (described below) and dissolved inorganic N determinations (described above), all microcosms were flash frozen in a dry ice-ethanol slurry and stored at −80°C. Samples for dissolved nitrogen species in the quenched assays were determined as described above.
The presence of ethylene in the gas phase was determined by subsampling 500 μl of the headspace of each replicate assay with a gas-tight syringe and injecting it into an O2-free 3-ml serum vial containing N2. Subsamples were analyzed for the presence of ethylene immediately upon return to the laboratory at MSU as previously described (28). The presence of ethylene was determined with a model GC-8A gas chromatograph (Shimadzu Corporation, Kyoto, Japan) equipped with an 80/100 Porapak Q column (Supelco, St. Louis, MO) and a flame ionization detector with helium as the carrier gas. The temperature of the injector was held at 190°C, and the column temperature was held at 120°C. The concentration of ethylene was determined by comparison to freshly prepared standards. The reported rates of acetylene reduction (ethylene production) reflect the difference in concentration among three replicate biological controls (no treatment) and three killed controls (500 μM HgCl2) for each incubation time. Rates of acetylene reduction activity were normalized to grams of dry mass (gdm) of sediments used in each incubation.
Nucleotide sequence accession numbers.
The nifH and archaeal amoA sequences representing each operational taxonomic unit (OTU) have been deposited in the GenBank, DDBJ, and EMBL databases under accession numbers KC254651 to KC254657 and KC254659 to KC254660, respectively (see Tables S3 and S4 and Fig. S1 and S2 in the supplemental material). Sequences representing each archaeal and bacterial 16S OTU have been deposited in the GenBank, DDBJ, and EMBL databases under accession numbers KC254661 to KC254693 (see Tables S5 and S6 in the supplemental material).
RESULTS
Site characterization.
Perpetual Spouter hot spring (PS), Yellowstone National Park (YNP), WY, is a circumneutral geothermal spring characterized by a relatively low concentration of NH4(T) (Table 1). The source fluid where experiments were conducted at PS (Fig. 2) had a pH of 7.03 and a temperature of 86.4°C.
TABLE 1.
Physical and chemical data for PS (YNP thermal ID NBB 113)
| Parameter | Value |
|---|---|
| GPSa coordinates | Lat 44.7265928, long −110.7091279 |
| pH | 7.03 |
| Temp (°C) | 86.4 |
| Conductivity (mS/cm) | 6 |
| NO3− concn (μM) | 0.95 |
| NO2− concn (μM) | 0.28 |
| NH4(T) concn (μM) | 23.5 |
GPS, Global Positioning System.
FIG 2.
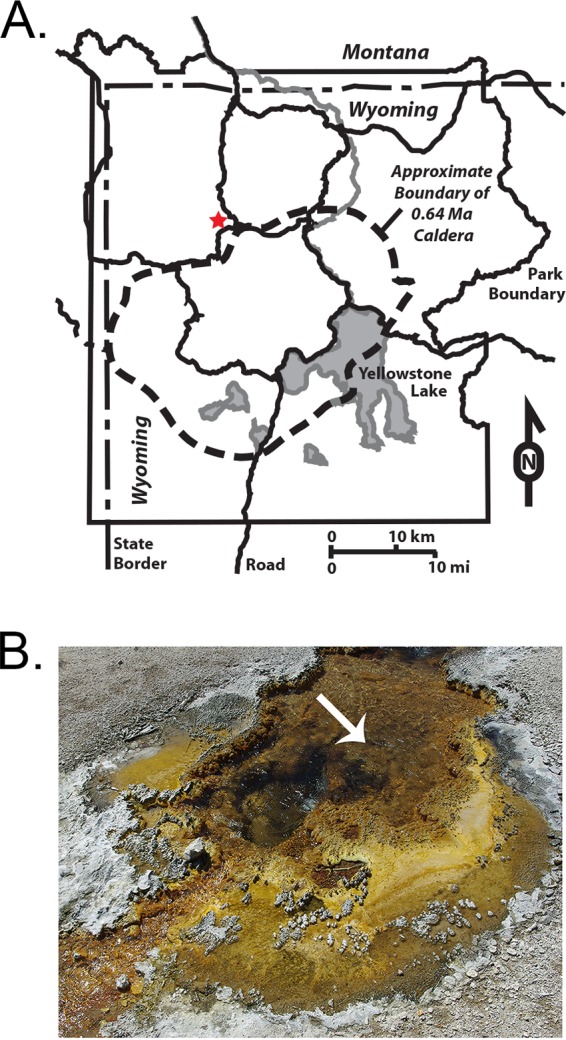
Map of the overall sampling location (A) and the location of the experimental site at PS, YNP (thermal ID NBB 113) (B). The red star indicates the approximate location of PS (A), and the white arrow indicates the sampling location at PS (B). Panel A is based on a map from the U.S. National Park Service.
Abundance and composition of SSU 16S rRNA in PS communities.
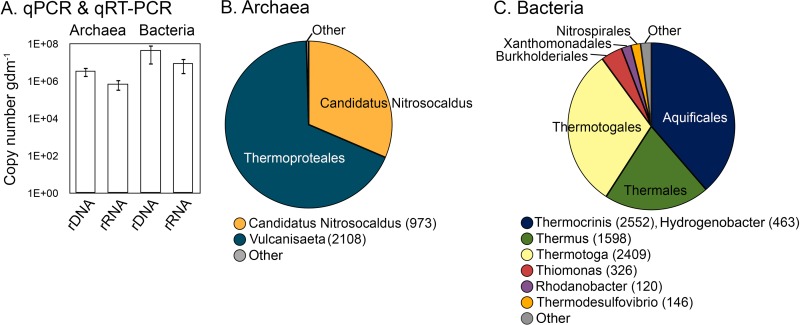
The abundance of bacterial 16S rRNA in sediment samples from the source of PS (pH 7.03, 86.4°C) was greater than the abundance of archaeal 16S rRNA (Fig. 3A). The dominant archaeal SSU 16S rRNA sequences recovered were 84% identical to those of Vulcanisaeta distributa in the order Thermoproteales (68% of all sequences) and 99% identical to those of Ca. Nitrosocaldus yellowstonii in the phylum Thaumarchaeota (31% of all sequences) (Fig. 3B). The dominant bacterial SSU 16S rRNA sequences recovered from PS sediments were 99% identical to those of T. albus in the order Aquificales (33% of all sequences), 87% identical to those of Thermotoga thermarum in the order Thermotogales (31% of all sequences), and 98% identical to those of Thermus brockianus in the order Thermales (21% of all sequences) (Fig. 3C).
FIG 3.
Abundance of archaeal and bacterial 16S rRNA genes and their transcripts extracted from sediment samples obtained at the source of PS (A). Archaeal (B) and bacterial (C) 16S rRNA composition of sediments from the source of PS. Representative OTUs from the ∼3,000 sequences in each library were binned at the order level for archaea (B) and bacteria (C). The most abundant genus in each order is indicated with the number of sequences affiliated with that genus in parentheses. Taxonomic bins that represented <5.0% of the total sequences from each assemblage were pooled and are depicted as “Other.” Results of qPCR and qRT-PCR assays are presented as the means of assays performed in triplicate on a log scale; error bars represent the standard deviations of the means.
Probing the interaction between NH4(T) oxidizers and diazotrophs.
Acetylene reduction assays (proxy for N2 fixation) and geochemical measurements were used to monitor the response of biological N2 fixation and NH4(T) oxidation activity to various amendments aimed at disrupting the interaction between these two processes. Microcosms containing PS sediments amended with 1 mM NH4+ had an average rate of acetylene reduction 63% lower than that of unamended controls (25 nmol ethylene gdm−1 h−1) (Table 2). Geochemical measurements indicated that a fraction of the NH4+ added to microcosms containing PS sediments was converted to NO2− during the course of incubation (Fig. 4). The combination of higher levels of NO2− (a proxy for NH4(T) oxidation activity) and a decreased rate of acetylene reduction activity following the addition of NH4+ suggests that the addition of exogenous NH4(T) may simultaneously stimulate the activity of NH4(T) oxidizers while also relieving fixed N limitation in diazotrophic populations.
TABLE 2.
Rates of acetylene reduction in microcosms containing sediment samples from PS
| Treatment | Avg acetylene reduction rate (C2H4 nmol gdm−1 h−1) ± SDa |
|---|---|
| Unamended | 25 ± 2.2 |
| NH4+c | 9.3 ± 0.81 |
| ATUd | BDLb |
| NH4+ + ATU | 0.60 ± 0.020 |
Assays were performed in triplicate.
BDL, below detection limit. The detection limit for C2H4 is 25 pmol gdm−1 hour−1.
NH4+ represents NH4(T) added as NH4Cl at a 1 mM final concentration.
ATU was used at a 1 mM final concentration.
FIG 4.
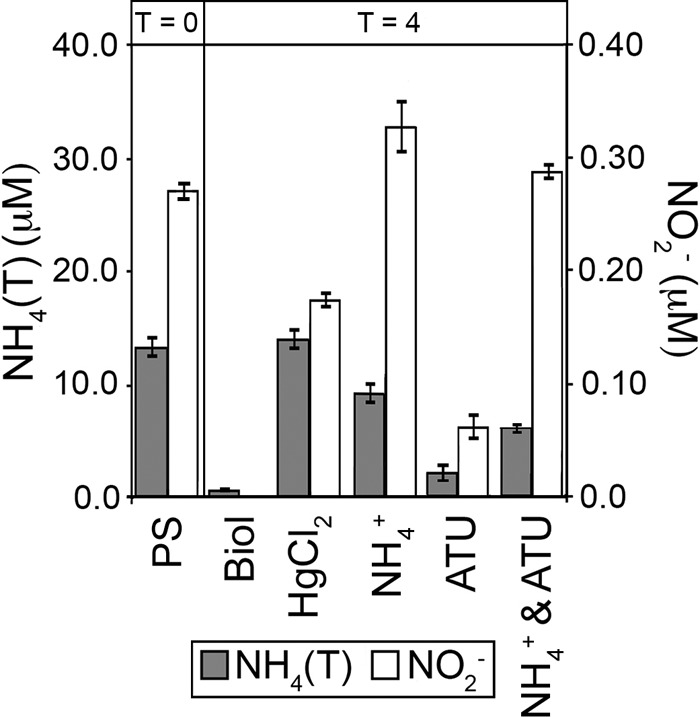
Concentrations of NH4(T) (gray bars) and NO2− (white bars) in microcosm assays. NO2− concentrations are plotted on the secondary y axis. The PS treatment reflects the starting concentrations of NH4(T) and NO2− in PS spring water in each microcosm prior to amendment (time zero). All other values were measured at 4 h postamendment. Abbreviations: Biol, unamended control; NH4+, NH4(T) added as NH4Cl (1 mM final concentration); ATU, ATU added to a final concentration of 1 mM; HgCl2, HgCl2 added to a final concentration of 500 μM.
Microcosms were amended with ATU, an inhibitor of NH4(T) oxidation, in order to examine the influence of NH4(T) oxidation on N2 fixation. ATU-amended microcosms contained measurable concentrations of NH4(T) after 4 h, in contrast to those in unamended microcosms, which were below the limit of detection (Fig. 4), indicating that NH4(T) oxidation had been at least partially inhibited by ATU. Amendment of microcosms with ATU decreased the rate of acetylene reduction activity to levels below the limit of detection, in contrast to unamended controls (Table 2). Given that ATU does not affect acetylene reduction in batch cultures of the model diazotrophic organism A. vinelandii AvOP cultivated in N-limited medium (see Table S7 in the supplemental material) and that no detectable NH4(T) is released from ATU during 24 h of incubation under conditions mimicking those of the microcosm incubations (i.e., 86°C) (data not shown), these results suggest that the decrease in acetylene reduction observed in PS in the presence of ATU is the result of decreased demand on the bioavailable NH4(T) pool due to inhibition of biological NH4(T) oxidation. Thus, the decrease in acetylene reduction activity when NH4(T) oxidation was at least partially inhibited indicates an interaction between these two guilds of organism and suggests that NH4(T)-oxidizing archaea are likely to have an affinity for NH4(T) higher than that of diazotrophs.
Amendment of microcosms with both NH4+ and ATU resulted in an apparent stimulation of NH4(T) oxidation activity compared to unamended controls, as indicated by nearly complete depletion of the added NH4+ and an increase in NO2− following the 4-h incubation (Fig. 4). The concentration of NO2− in microcosms amended with both NH4+ and ATU (double amendment) was higher than that in microcosms amended with ATU only but lower than that in microcosms amended with NH4+ only. Likewise, the rate of acetylene reduction was significantly higher in microcosms amended with both ATU and NH4+ than in microcosms amended with ATU only but lower than that in microcosms amended with NH4+ only (Table 2).
To examine the effect of NH4+ and ATU amendment on diazotrophic and NH4(T)-oxidizing populations, the abundance of nifH, bacterial amoA, and archaeal amoA transcripts in RNA extracted from sediment microcosms was quantified by qRT-PCR and transcript levels were compared to those in native PS sediments (Fig. 5). Bacterial amoA transcript levels were below the limit of detection in all assays (data not shown), consistent with the absence of SSU rRNA sequences affiliated with bacterial NH4(T) oxidizers in these communities (see Table S6 in the supplemental material). Archaeal amoA transcript levels in unamended controls over the 2- and 4-h incubation periods were similar to the levels in native sediments (Fig. 5A). Likewise, nifH transcript abundances did not change significantly in unamended controls over the 2- and 4-h incubation periods (Fig. 5C). However, amendment of microcosms with NH4+ resulted in a 10-fold increase in archaeal amoA transcript levels relative to those in unamended microcosms following 2 h of incubation, consistent with geochemical data indicating active NH4(T) oxidation under these conditions (Fig. 5B). nifH transcript levels also decreased upon amendment with NH4+, resulting in 6- and 4-fold decreases relative to unamended controls in the 2- and 4-h incubations, respectively (Fig. 5D). The decrease in amoA transcripts and the increase in nifH transcripts after 4 h compared to those at the 2-h time point suggest that most of the exogenous NH4(T) had been consumed during this time, which was also indicated by the levels of NH4(T), which were lower after 4 h than at time zero (Fig. 4).
FIG 5.
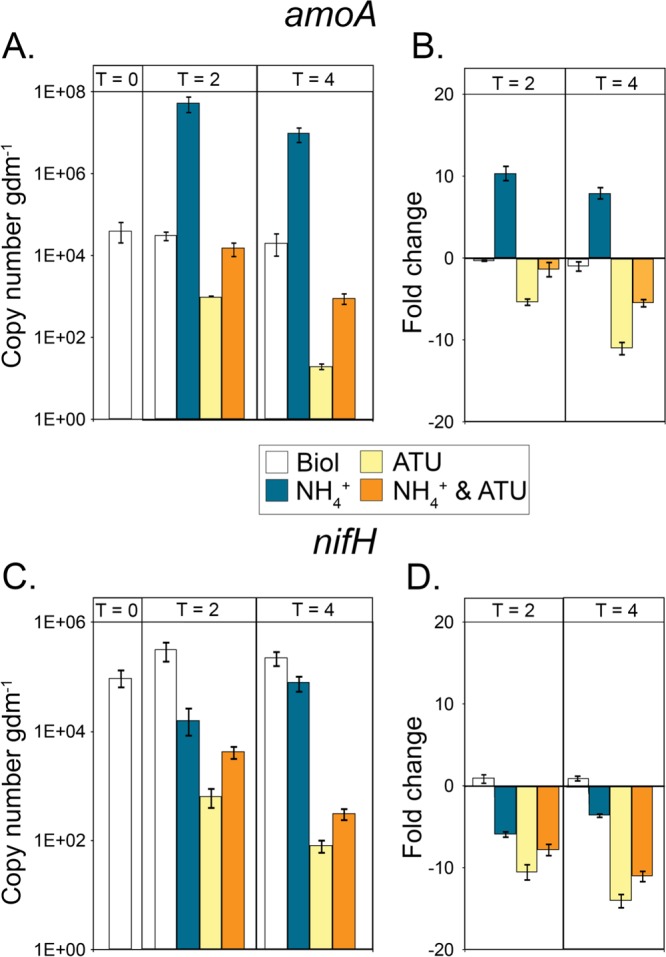
Transcript abundance and fold changes (log2) in amoA (A and B) and nifH (C and D) transcript abundance in microcosms containing sediments from the source of PS. Sediment samples were taken from replicate microcosms at time zero (sediments taken directly from the spring source) and at 2 and 4 h postamendment. Error bars represent the standard deviations of qRT-PCR assays performed in triplicate. Abbreviations: Biol, unamended control; NH4+, NH4(T) added as NH4Cl to a final concentration of 1 mM; ATU, ATU added to a final concentration of 1 mM.
Amendment of microcosms with ATU resulted in 5- and 11-fold decreases in the abundance of archaeal amoA transcripts relative to that in unamended controls following 2- and 4-h incubations, respectively (Fig. 5B). Likewise, amendment of microcosms with ATU resulted in 10- and 14-fold decreases in nifH transcript abundance relative to that in unamended controls following 2- and 4-h incubations, respectively (Fig. 5D). Consistent with the hypothesis that NH4+ amendment induces the expression of the NH4(T) oxidation complex to a greater extent than ATU amendment suppresses expression compared to that in unamended controls, double amendment of microcosms with both ATU and NH4+ resulted in a decrease in archaeal amoA transcripts but to a lesser extent than in microcosms treated only with ATU in both the 2- and 4-h incubations. Only a small decrease in archaeal amoA transcript levels (1-fold) was observed in microcosms amended with both ATU and NH4+ relative to those in unamended controls following 2 h of incubation. Following 4 h of incubation, a greater decrease in archaeal amoA transcript levels (5-fold) was observed in microcosms amended with both ATU and NH4+ than in unamended controls, consistent with nearly complete depletion of the available NH4(T) pools in microcosms (Fig. 4). Compared to the nifH transcript levels in unamended controls, those in microcosms amended with ATU decreased more than those that received both NH4+ and ATU. nifH transcript levels in microcosms amended with both NH4+ and ATU were 7- and 11-fold lower than those in unamended controls but 3-fold higher than those in microcosms amended with ATU only.
Composition of nifH and archaeal amoA transcripts.
The composition of nifH and archaeal amoA transcripts amplified from native sediment was compared to that obtained from unamended and NH4+-amended microcosms following 2- and 4-h incubations (Fig. 6). The composition of archaeal amoA transcripts in native PS sediments comprised two phylotypes that both clustered with Ca. Nitrosocaldus yellowstonii amoA (97 and 99% amino acid sequence identity) (Fig. 6A; see Table S4 and Fig. S2 in the supplemental material). Little variation was observed in the relative abundance of these phylotypes over the incubation period or in the presence or absence of amendments. The dominant (75% of 16 clones) nifH transcripts amplified from native sediments were closely affiliated with nifH from T. albus (97 to 100% amino acid sequence identity), with the remaining 25% of the transcripts exhibiting close affiliation with nifH from Thermodesulfovibrio yellowstonii (96 to 98% amino acid sequence identity) in the order Nitrospirales (see Table S3 and Fig. S1 in the supplemental material). Only minor variations in the composition of amoA transcripts were observed in unamended controls following 2- and 4-h incubations (Fig. 6A).
FIG 6.
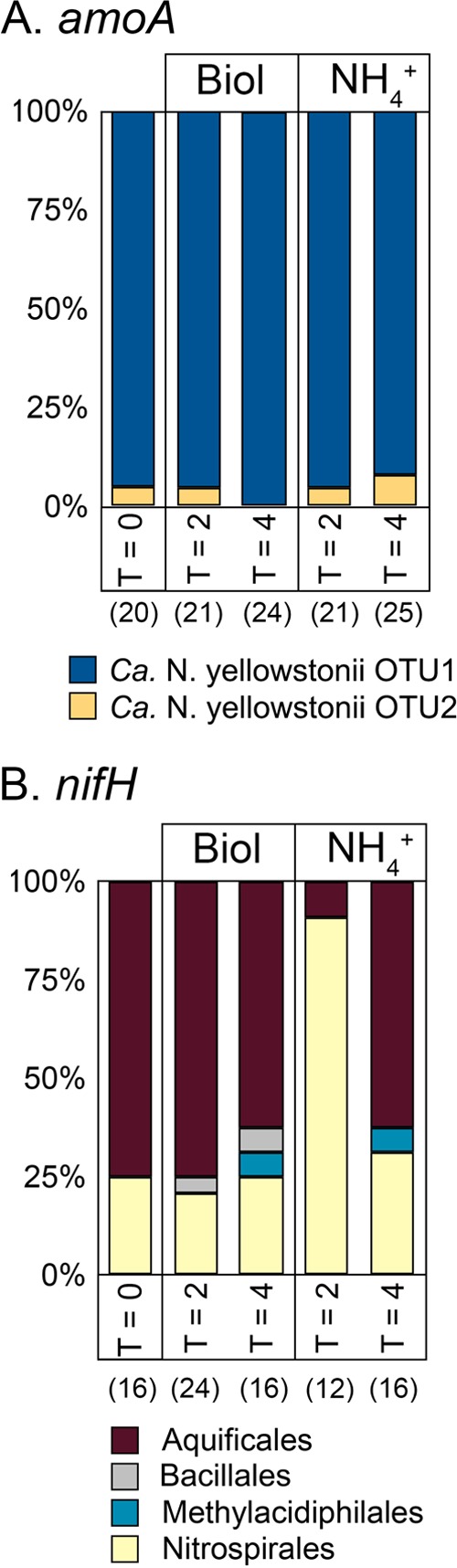
Normalized numbers of amoA (A) and nifH (B) transcript OTUs in microcosms containing sediment samples from the source of PS. Sediment samples were taken from replicate microcosms at time zero (sediments taken directly from the spring source) and at 2 and 4 h postamendment. Abbreviations; Biol, unamended control; NH4+, NH4(T) added as NH4Cl to a final concentration of 1 mM. The value in parentheses for each treatment or sampling point is the total number of clones sequenced from each clone library.
Amendment of microcosms with NH4+ resulted in a shift in the composition of nifH transcript pools with the dominant sequences (92% of 12 clones) closely affiliated with nifH from T. yellowstonii and only minor contributions from T. albus (8%) following 2 h of incubation (Fig. 6B). At 4 h of incubation, the composition of nifH transcripts again shifted, with sequences closely affiliated with T. albus (63% of 16 clones) being the most abundant and with minor contributions from T. yellowstonii (31%) and Methylacidiphilum fumariolicum (6%) (Fig. 6B). These observations suggest that T. albus is the dominant diazotroph in situ and that NH4(T) concentrations may differentially regulate the expression of nifH in T. albus and T. yellowstonii.
DISCUSSION
The analysis of 16S rRNA sequences obtained from PS indicates the presence of populations putatively capable of NH4(T) oxidation and N2 fixation. Moreover, the NH4(T) concentration (23.5 μM) is near the 20 μM threshold known to induce N2 fixation in pure cultures of the model diazotroph A. vinelandii (29). Suppression of acetylene reduction activity (a proxy for N2 fixation) by ATU (an inhibitor of NH4(T) oxidation) indicated that there is cross talk between the NH4(T)-oxidizing and diazotrophic populations in the PS ecosystem at the level of the bioavailable NH4(T) pool. Amendment of microcosms with NH4+ resulted in the simultaneous suppression of acetylene reduction activity and stimulation of NH4(T) oxidation activity. This observation, coupled with observations from the ATU treatment which showed a decrease in both amoA and nifH transcript levels, further indicates that the niche dimensions of Ca. Nitrosocaldus yellowstonii and the putative diazotroph Thermocrinis sp. are likely to overlap at the level of the bioavailable NH4(T) pool.
Competition for nutrients among organisms with differing nutritional requirements can be important in structuring microbial communities in natural systems (30, 32). The resource ratio model of competition proposed by Tilman (31) suggests that the individual demand for nutrients and their relative rates of utilization will dictate the frequency of taxa at equilibrium in a natural community. This model is generally used to explain the dynamics of two populations that are competing for a single resource, such as NH4(T). However, it is not known if such models are suitable for explaining the frequency dependence of NH4(T) oxidizers, which primarily use this compound as a source of energy but also must use it to satisfy their own biosynthetic N demands, and diazotrophs, which can either assimilate NH4(T) or actively generate NH4(T) from N2 for biosynthetic purposes. An intriguing possibility is that populations of Ca. Nitrosocaldus yellowstonii have adapted high-affinity NH4(T) acquisition systems, noted here and elsewhere (19), that function to meet their own energetic and biosynthetic needs, resulting in the maintenance of sufficiently low concentrations of NH4(T) in spring waters to create an environment that would select for populations capable of N2 fixation (33), such as the putative diazotroph T. albus. This hypothesis is consistent with the low concentrations of NH4(T) in many circumneutral-to-alkaline hot spring fluids (Fig. 1A), which are generally below the threshold concentration of NH4(T) for the induction of N2 fixation machinery in A. vinelandii, and the apparent ubiquity of diazotrophs in these systems (20). In further support, a recent mathematical modeling study based on the Vmax and Km values of ammonia-oxidizing bacteria and archaea, as well as diazotrophic cyanobacteria, indicated that ammonium uptake by the former may necessitate cyanobacterial nitrogen fixation (33). Importantly, while NH4 is the preferred form of bioavailable N for most microbial populations, other forms of reduced nitrogen, such as nitrate and nitrite, may also serve as sources of fixed N for the organisms inhabiting PS. Therefore, the dynamics between ammonia-oxidizing archaea (AOA) and diazotrophs in PS and other circumneutral-to-alkaline springs may be more complex than presented here, especially given the apparent presence of assimilatory nitrite and nitrate reduction systems in the genome of T. albus (Kyoto Encyclopedia of Genes and Genomes pathway tal00910).
Cooccurring populations of Ca. Nitrosocaldus yellowstonii and putative diazotrophic members of the order Aquificae (e.g., Thermocrinis sp., Hydrogenobacter sp.) have a widespread distribution in geothermal springs, in particular those in YNP that have circumneutral-to-alkaline pHs (Fig. 1B). For example, Thermocrinis species have been isolated from Octopus Spring (Lower Geyser Basin) and a spring in the Heart Lake Geyser Basin (34) and enrichments of Ca. Nitrosocaldus yellowstonii have been obtained from Octopus Spring and from several springs in the Heart Lake Geyser Basin (11). Likewise, nifH genes closely related to nifH from members of the order Aquificales have previously been detected in hot springs located in both of the aforementioned geyser basins (20) and these springs exhibit a geochemistry similar to that of springs where Thermocrinis sp. and Ca. Nitrosocaldus yellowstonii cultures were previously enriched. Early enrichments of Ca. Nitrosocaldus yellowstonii were found not to be axenic but instead to harbor several populations of bacteria, including an undescribed population affiliated with the phylum Aquificae (11). More recent metagenomic sequencing efforts conducted with this consortium have revealed the presence of a 16S rRNA gene that is closely affiliated with that of Hydrogenobacter thermophilus TK-6 (∼96% nucleotide sequence identities) (J. R. de la Torre, unpublished data). The same contig that contains this 16S rRNA gene also contains a partial nif gene cluster that includes a nifH gene closely affiliated with the nifH genes of H. thermophilus TK-6 (∼96% amino acid identity) and T. albus (∼94% amino acid identity) (see Fig. S2 in the supplemental material). The cooccurrence of NH4(T)-oxidizing Ca. Nitrosocaldus yellowstonii and members of the phylum Aquificae (which contains at least a partial nif gene cluster) in enrichment cultures, as well as N-limited circumneutral-to-alkaline geothermal springs such as PS and other YNP springs (Fig. 1B), may allude to an underlying dependence of Ca. Nitrosocaldus yellowstonii on biogenic NH4(T) in these types of hydrothermal ecosystems.
The high energetic and genetic expense of biological nitrogen fixation has selected for tight regulation of nitrogenase at the transcriptional and translational levels in response to NH4(T) (35). However, little is known regarding the regulation of NH4(T) oxidation in archaea. In the present study, treatment of microcosms with NH4+ resulted in a decrease in nifH transcripts and an increase in amoA transcripts compared to unamended controls, suggesting transcription level regulation of both processes by NH4(T) levels. With respect to nifH, the largest decrease in transcript abundance was observed between unamended and ATU-amended controls. In addition, acetylene reduction activity was not detected in ATU-amended controls, indicating that inhibition of NH4(T) oxidizers may reduce the flux of NH4(T) toward this process and partially relieve fixed-N limitation for diazotrophs. In contrast, the largest decrease in amoA transcripts was observed between microcosms amended with ATU and unamended microcosms, which may suggest that treatment of cells with ATU results in a physiological response that suppresses amoA transcription. Amendment of PS microcosms with both ATU and NH4+ yielded unexpected results, most notably, detectable acetylene reduction (in the presence of added NH4+) and NH4(T) oxidation activity (in the presence of ATU) compared to individual NH4+- or ATU-amended controls. In the latter microcosms, rates of acetylene reduction were below the limit of detection. These observations can likely be attributed to the different mechanism by which NH4(T) and ATU suppress or inhibit N2 fixation and NH4(T) oxidation, respectively, as well as the interaction/competition between these organisms for bioavailable nitrogen. ATU is thought to inhibit bacterial NH4(T) oxidation by chelating copper ions away from the active site of the ammonia monooxygenase (36), and it is possible that the same mechanism is responsible for the partial inhibition of archaeal NH4(T) oxidation observed here and in previous studies (37–41). In contrast, the data presented here indicate that NH4(T) may regulate NH4(T) oxidation at the transcriptional level. Treatment of cells with ATU would not necessarily be expected to completely suppress NH4(T) oxidation activity in the presence of ample NH4(T) (ATU plus NH4+ amendment), as new enzyme would be expected to be continuously generated if genes involved in NH4(T) oxidation were indeed transcriptionally regulated by NH4(T). In addition, elevated levels of ATU necessary for inhibition of NH4(T) oxidation have been noted in some pure cultures of AOA (37, 38), highlighting the uncertainty regarding the biochemical and/or regulatory role of ATU in inhibiting the activity of AOA.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation (NSF) grant EAR-1123689 to E.S.B. J.R.D.L.T. and J.W.P. acknowledge support for this work from NSF PIRE OISE-096842. J.R.D.L.T. acknowledges support for this work from NSF MCB-0949807. T.L.H. acknowledges support from the NAI Postdoctoral Program.
We are grateful to Christie Hendrix and Stacey Gunther for facilitating the permitting process to perform research in YNP and to Chris Allen for analytical assistance.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02577-13.
REFERENCES
- 1.Falkowski PG. 1997. Evolution of the nitrogen cycle and its influence on the biological sequestration of CO2 in the ocean. Nature 387:272–275. 10.1038/387272a0 [DOI] [Google Scholar]
- 2.Zher JP, Jenkins BD, Short SM, Steward GF. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Microb. Ecol. 7:539–554. 10.1046/j.1462-2920.2003.00451.x [DOI] [PubMed] [Google Scholar]
- 3.Ward BB, Arp DJ, Klotz MJ. 2011. Nitrification. ASM Press, Washington, DC [Google Scholar]
- 4.Kowalchuk GA, Stephen JR. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485–529. 10.1146/annurev.micro.55.1.485 [DOI] [PubMed] [Google Scholar]
- 5.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688. 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middleburg JJ, Schouten S, Damsté JSS. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322. 10.1073/pnas.0600756103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schleper C. 2010. Ammonia oxidation: different niches for bacteria and archaea? ISME J. 4:1092–1094. 10.1038/ismej.2010.111 [DOI] [PubMed] [Google Scholar]
- 8.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- 9.Bock E, Wagner M. 2006. Oxidation of inorganic nitrogen compounds as an energy source, p 457–495 In Falkow S, Rosenburg E, Schleifer KH, Stackebrandt E. (ed), Prokaryotes. Springer, New York, NY [Google Scholar]
- 10.Boyd ES, Lange RK, Mitchell AC, Havig JR, Hamilton TL, Lafrenière MJ, Shock EL, Peters JW, Skidmore M. 2011. Diversity, abundance, and potential activity of nitrifying and nitrate-reducing microbial assemblages in a subglacial ecosystem. Appl. Environ. Microbiol. 77:4778–4787. 10.1128/AEM.00376-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. 2008. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ. Microbiol. 10:810–818. 10.1111/j.1462-2920.2007.01506.x [DOI] [PubMed] [Google Scholar]
- 12.Reigstad LJ, Richter A, Daims H, Urich T, Schwark L, Schleper C. 2008. Nitrification in terrestrial hot springs of Iceland and Kamchatka. FEMS Microbiol. Lett. 64:167–174. 10.1111/j.1574-6941.2008.00466.x [DOI] [PubMed] [Google Scholar]
- 13.Dodsworth JA, Hungate BA, Hedlund BP. 2011. Ammonia oxidation, denitrification and dissimilatory nitrate reduction to ammonium in two US Great Basin hot springs with abundant ammonia-oxidizing archaea. Environ. Microbiol. 13:2371–2386. 10.1111/j.1462-2920.2011.02508.x [DOI] [PubMed] [Google Scholar]
- 14.Hatzenpichler R. 2012. Diversity, physiology and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 78:7501–7510. 10.1128/AEM.01960-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. 2008. Mesophilic crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252. 10.1038/nrmicro1852 [DOI] [PubMed] [Google Scholar]
- 16.Stahl DA, de la Torre JR. 2012. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66:83–101. 10.1146/annurev-micro-092611-150128 [DOI] [PubMed] [Google Scholar]
- 17.Boyd ES, Hamilton TL, Wang J, He L, Zhang CL. 2013. The role of tetraether lipid composition in the adaptation of thermophilic archaea to acidity. Front. Microbiol. 4:62. 10.3389/fmicb.2013.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holloway JM, Nordstrom DK, Böhlke JK, McCleskey RB, Ball JW. 2011. Ammonium in thermal waters of Yellowstone National Park: processes affecting speciation and isotope fractionation. Geochim. Cosmochim. Acta 75:4611–4636. 10.1016/j.gca.2011.05.036 [DOI] [Google Scholar]
- 19.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979. 10.1038/nature08465 [DOI] [PubMed] [Google Scholar]
- 20.Hamilton TL, Boyd ES, Peters JW. 2011. Environmental constraints underpin the distribution and phylogenetic diversity of nifH in the Yellowstone geothermal complex. Microb. Ecol. 61:860–870. 10.1007/s00248-011-9824-9 [DOI] [PubMed] [Google Scholar]
- 21.Loiacono ST, Meyer-Dombard DR, Havig JR, Poret-Peterson AT, Hartnett HE, Shock EL. 2012. Evidence for high-temperature in situ nifH transcription in an alkaline hot spring of Lower Geyser Basin, Yellowstone National Park. Environ. Microbiol. 14:1272–1283. 10.1111/j.1462-2920.2012.02710.x [DOI] [PubMed] [Google Scholar]
- 22.Inskeep WP, Ackerman GG, Taylor WP, Kozubal M, Korf S, Macur RE. 2005. On the energetics of chemolithotrophy in nonequilibrium systems: case studies of geothermal springs in Yellowstone National Park. Geobiology 3:297–317. 10.1111/j.1472-4669.2006.00059.x [DOI] [Google Scholar]
- 23.Hamilton TL, Ludwig M, Dixon R, Boyd ES, Santos Dos PC, Setubal JC, Bryant DA, Dean DR, Peters JW. 2011. Transcriptional profiling of nitrogen fixation in Azotobacter vinelandii. J. Bacteriol. 193:4477–4486. 10.1128/JB.05099-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd ES, Cummings DE, Geesey GG. 2007. Mineralogy influences structure and diversity of bacterial communities associated with geological substrata in a pristine aquifer. Microb. Ecol. 54:170–182. 10.1007/s00248-006-9187-9 [DOI] [PubMed] [Google Scholar]
- 25.Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. 2008. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial Tag Encoded FLX Amplicon Pyrosequencing (bTEFAP). PLoS One 3:e3326. 10.1371/journal.pone.0003326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesiniewski RA, Oakley BB, Parks DH, Rosbinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart WD, Fitzgerald GP, Burris RH. 1967. In situ studies on N2 fixation using the acetylene reduction technique. Proc. Natl. Acad. Sci. U. S. A. 58:2071–2078. 10.1073/pnas.58.5.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton TL, Lange RK, Boyd ES, Peters JW. 2011. Biological nitrogen fixation in acidic high-temperature geothermal springs in Yellowstone National Park, Wyoming. Environ. Microbiol. 13:2204–2215. 10.1111/j.1462-2920.2011.02475.x [DOI] [PubMed] [Google Scholar]
- 29.Kleiner D. 1975. Ammonium uptake by nitrogen fixing bacteria. Arch. Microbiol. 104:163–169. 10.1007/BF00447319 [DOI] [PubMed] [Google Scholar]
- 30.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. 2008. Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62:375–401. 10.1146/annurev.micro.030608.101423 [DOI] [PubMed] [Google Scholar]
- 31.Tilman D. 1977. Resource competition between plankton algae: an experimental and theoretical approach. Ecology 58:338–348. 10.2307/1935608 [DOI] [Google Scholar]
- 32.Schink B. 2002. Synergistic interactions in the microbial world. Extremophiles 81:257–261. 10.1023/A:1020579004534 [DOI] [PubMed] [Google Scholar]
- 33.Boyett MR, Tavakkoli A, Sobolev D. 2013. Mathematical modeling of competition for ammonium among Bacteria, Archaea and cyanobacteria within cyanobacterial mats: can ammonia-oxidizers force nitrogen fixation? Ocean Sci. J. 48:269–277 http://link.springer.com/article/10.1007%2Fs12601-013-0025-y#. 10.1007/s12601-013-0025-y [DOI] [Google Scholar]
- 34.Eder W, Huber R. 2002. New isolates and physiological properties of the Aquificales and description of Thermocrinis albus sp. nov. Extremophiles 6:309–318. 10.1007/s00792-001-0259-y [DOI] [PubMed] [Google Scholar]
- 35.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2:621–631. 10.1038/nrmicro954 [DOI] [PubMed] [Google Scholar]
- 36.Bédard C, Knowles R. 1989. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Mol. Biol. Rev. 53:68–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. 2008. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc. Natl. Acad. Sci. U. S. A. 105:2134–2139. 10.1073/pnas.0708857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen T, Stieglmeier M, Dai J, Urich T, Schleper C. 2013. Responses of the terrestrial ammonia-oxidizing archaeon Ca. Nitrososphaera viennensis and the ammonia-oxidizing bacterium Nitrosospira multiformis to nitrification inhibitors. FEMS Microbiol. Lett. 344:121–129. 10.1111/1574-6968.12164 [DOI] [PubMed] [Google Scholar]
- 39.Santoro AE, Casciotti KL. 2011. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. 5:1796–1808. 10.1038/ismej.2011.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santoro AE, Casciotti KL, Francis CA. 2010. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12:1989–2006. 10.1111/j.1462-2920.2010.02205.x [DOI] [PubMed] [Google Scholar]
- 41.Lehtovirta-Morley LE, Verhamme DT, Nicol GW, Prosser JI. 2013. Effect of nitrification inhibitors on the growth and activity of Nitrosotalea devanaterra in culture and soil. Soil Biol. Biochem. 62:129–133. 10.1016/j.soilbio.2013.01.020 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.