Abstract
Free fatty acids are important flavor compounds in cheese. Propionibacterium freudenreichii is the main agent of their release through lipolysis in Swiss cheese. Our aim was to identify the esterase(s) involved in lipolysis by P. freudenreichii. We targeted two previously identified esterases: one secreted esterase, PF#279, and one putative cell wall-anchored esterase, PF#774. To evaluate their role in lipolysis, we constructed overexpression and knockout mutants of P. freudenreichii CIRM-BIA1T for each corresponding gene. The sequences of both genes were also compared in 21 wild-type strains. All strains were assessed for their lipolytic activity on milk fat. The lipolytic activity observed matched data previously reported in cheese, thus validating the relevance of the method used. The mutants overexpressing PF#279 or PF#774 released four times more fatty acids than the wild-type strain, demonstrating that both enzymes are lipolytic esterases. However, inactivation of the pf279 gene induced a 75% reduction in the lipolytic activity compared to that of the wild-type strain, whereas inactivation of the pf774 gene did not modify the phenotype. Two of the 21 wild-type strains tested did not display any detectable lipolytic activity. Interestingly, these two strains exhibited the same single-nucleotide deletion at the beginning of the pf279 gene sequence, leading to a premature stop codon, whereas they harbored a pf774 gene highly similar to that of the other strains. Taken together, these results clearly demonstrate that PF#279 is the main lipolytic esterase in P. freudenreichii and a key agent of Swiss cheese lipolysis.
INTRODUCTION
Flavor is a basic criterion associated with food quality. Cheese flavor results from a complex mixture of flavor compounds, which include nonvolatile and volatile compounds. Most of them are produced during cheese manufacture and ripening and result from the activity of microbial enzymes on milk components, such as fat, proteins, and carbohydrates. Therefore, to control cheese flavor, it is important to identify the main actors involved in the formation of flavor compounds, so as to modulate their activity, depending on the flavor notes targeted (1). Free fatty acids are important flavor compounds in most cheeses, where they contribute to pungent, rancid, cheesy, and fruity notes (2). They result from the partial hydrolysis of the ester linkage between a fatty acid and the glycerol moiety of milk triacylglycerides by lipolytic esterases (2). The main sources of esterases in cheese are microorganisms. However, the indigenous milk lipoprotein lipase (LPL) is involved in the early lipolysis that occurs during the first steps of cheese manufacture, and some lipases present in traditional coagulant preparations can also play an important role in the lipolysis observed in varieties such as Italian cheeses (2).
Propionibacterium freudenreichii is used as a ripening culture in the manufacture of Swiss-type cheeses and some other semihard cheeses (3, 4) and is the main agent of Swiss cheese lipolysis. For example, 96% of the free fatty acids (FFA) released during the ripening of mini-Swiss-type cheeses resulted from the activity of P. freudenreichii CIRM-BIA1T (5). The extent of lipolysis has been shown to vary significantly under the same cheesemaking conditions, depending on the strain used, with the amount of released FFA varying up to 3-fold (6–9). In particular, some strains exhibit very low lipolytic activity (8). Whether these diverse phenotypes result from differences in the number, sequence, or expression levels of esterase-encoding genes is unknown.
Twelve (putative) esterases were previously predicted from the genome sequence of P. freudenreichii CIRM-BIA1T (10), all of which are expressed regardless of the presence or absence of milk fat in the culture medium (11). The majority of lipolysis takes place early during P. freudenreichii growth (5), at a stage at which the release of intracellular enzymes from lysed cells is very unlikely. Therefore, we hypothesized that lipolysis in Swiss cheese mainly results from the activity of surface-exposed or secreted lipolytic esterases, rather than intracellular enzymes. We thus targeted two putative esterases, PF#279 and PF#774, predicted to be secreted and cell wall anchored, respectively. Our previous work showed that esterase PF#279 is effectively secreted and active on milk triglycerides (11).
The aim of the present study was to determine the respective contributions of these two enzymes to lipolysis. We assessed the activity of the putative esterase PF#774 by overexpression in P. freudenreichii CIRM-BIA1T. In the same strain, we also knocked out the genes encoding these two proteins, using recently developed genetic tools (12). In parallel, we screened a collection of previously sequenced P. freudenreichii strains for their lipolytic activity to determine whether variations in the sequences of the two targeted genes would explain the different phenotypes observed.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Twenty wild-type sequenced strains of P. freudenreichii were used (Table 1). In addition, strain LSP108 (from Laboratoires Standa, Caen, France), known for its very low lipolytic activity in Swiss cheese, was included in the present study.
TABLE 1.
Origins of the wild Propionibacterium freudenreichii strains used in this studya
| Strainb | Environmental source (yr of isolation) |
|---|---|
| CIRM-BIA1 | Unknown |
| CIRM-BIA9 | Emmental cheese (1989) |
| CIRM-BIA118 | Gruyère cheese (1973) |
| CIRM-BIA119 | Gruyère cheese (1973) |
| CIRM-BIA121 | Swiss cheese (1937) |
| CIRM-BIA122 | Unknown (1992) |
| CIRM-BIA123 | Morbier cheese (1992) |
| CIRM-BIA134 | Unknown |
| CIRM-BIA135 | Ewe raw milk (1994) |
| CIRM-BIA456 | Raw milk (1992) |
| CIRM-BIA508 | Gruyère cheese (1973) |
| CIRM-BIA512 | Raw milk (1994) |
| CIRM-BIA513 | Ras cheese (1995) |
| CIRM-BIA514 | Hay (1994) |
| CIRM-BIA516 | Yak cheese (2000) |
| CIRM-BIA527 | Fribourg cheese (1992) |
| ITG P9 | Actalia (origin not given) |
| ITG P18 | Actalia (origin not given) |
| ITG P20 | Actalia (origin not given) |
| ITG P23 | Actalia (origin not given) |
| LSP108 | Laboratoires Standa (origin not given) |
Strains were identified by PCR using species-specific primers according to Tilsala-Timisjarvi and Alatossava (19).
CIRM-BIA, collection of the Centre International de Ressources Microbiennes—Bactéries d'Intérêt Alimentaire, INRA, Rennes, France; ITG, Actalia collection, Rennes, France; LSP, Laboratoires Standa, Caen, France. (Strain LSP108 is known for its weak lipolytic activity in Emmental cheese.)
The wild-type strain, CIRM-BIA1T, and its genetically modified derivatives are listed in Table 2. All strains were grown at 30°C in yeast extract-lactate (YEL) broth (13) in glass tubes without agitation. In some cases, specified in the text, YEL was supplemented with chloramphenicol (10 μg ml−1) or hygromycin B (750 μg ml−1 in YEL agar or 250 μg ml−1 in YEL broth).
TABLE 2.
Genetically modified strains and plasmids used in this study
| Strain or plasmid | Description | Origin or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning host strain | Gibco-BRL |
| P. freudenreichii | ||
| CB1Ta | P. freudenreichii subsp. shermanii type strain (wild type) | CIRM-BIA |
| CB1::pFB01:pf279 | CB1 with pf279 overexpressed downstream of Ptuf promoter | This study |
| CB1-KO-pf279 | CB1 with inactivated pf279 | This study |
| CB1-KO-pf279-C | CB1 with inactivated pf279 and complemented with pf279 | This study |
| CB1::pFB01:pf774 | CB1 with pf774 overexpressed downstream of the Ptuf promoter | This study |
| CB1-KO-pf774 | CB1 with inactivated pf774 | This study |
| Plasmids | ||
| pPK705 | E. coli-P. freudenreichii shuttle vector, 8.3 kb; Ampr Hygr | 20 |
| pFB01 | Derived from pPK705 vector, 8.5 kb; Ampr Hygr; Ptuf promoter | 14 |
| pFB01:pf279 | Derived from pFB01 vector, 9.0 kb; Ampr Hygr; pf279 under control of Ptuf promoter | This study |
| pFB01:pf774 | Derived from pFB01 vector, 9.0 kb; Ampr Hygr; pf774 under control of Ptuf promoter | This study |
| pUC:pf279:CmR | Derived from pUC18 vector, 4.7 kb, Cmr; truncated fragment of pf279 | This study |
| pUC:pf774:CmR | Derived from pUC18 vector, 4.7 kb, Cmr; truncated fragment of pf774 | This study |
CB1T = CIRM-BIA1T.
Escherichia coli DH5α was used as a cloning vector and was obtained from Gibco-BRL Life Technologies (Saint Aubin, France). It was grown at 37°C with agitation (200 rpm) in Luria-Bertani medium (LB) containing ampicillin (100 μg ml−1).
To determine the lipolytic activity of P. freudenreichii, cells were grown in a modified YEL medium containing 20 g/liter lactate and milk fat emulsified with a sodium caseinate solution (final fat concentration of 30% [wt/wt] in the broth medium), as previously described (11). Population levels of propionibacteria were monitored by measuring the optical density at 650 nm (OD650) of a culture grown in the same medium without emulsion. Cultures were harvested at the end of log-phase growth. Triplicate (CIRM-BIA1T mutants) or duplicate (wild-type strain) independent cultures were performed.
Construction of the genetically modified strains.
Reagents were purchased from New England BioLabs (Ipswich, MA), and oligonucleotides were purchased from Sigma Genosys (Haverhill, Cambridgeshire, United Kingdom). Genomic DNA was extracted using the DNeasy tissue kit (Qiagen, Courtaboeuf, France) and automated sample preparation QIAcube (Qiagen), according to the manufacturer's instructions.
(i) Overexpression of esterase in P. freudenreichii.
Transformed strains of P. freudenreichii CIRM-BIA1T overexpressing esterase PF#279 or PF#774 were constructed. The esterase genes pf279 (PFREUD_04340) and pf774 (PFREUD_04240) were separately cloned into the pFB01 vector downstream of the Ptuf promoter (Table 2). Ptuf was chosen because the tuf gene was shown to be strongly expressed during the growth of this strain under the same conditions (14). To obtain this construct, pf279 and pf774 were amplified by PCR from P. freudenreichii CIRM1-BIA1 genomic DNA using primers containing restriction sites (see Table S1 in the supplemental material). High-fidelity PCR was performed to ensure accurate and reliable PCR amplification. PCR mixtures contained 1 mM MgSO4, 0.3 μM each primer, 300 μM each deoxynucleoside triphosphate (dNTP), and 1.43 U of Platinum Pfx high-fidelity polymerase in 1× Pfx amplification buffer (Invitrogen, Carlsbad, CA). PCR was performed in a thermocycler (Veriti, Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 94°C for 2 min, followed by 20 cycles of 94°C for 45 s and 60°C for 45 s, with a decrease by 0.5°C at each cycle, then 72°C for 90s, followed by 10 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 90 s, followed by a final step at 72°C for 5 min. Each of the pf279 and pf774 amplicons was purified with the QIAquick PCR purification kit (Qiagen) and double digested with restriction enzymes (XbaI-HindIII), and the digested fragments were then purified. The pFB01 plasmid was digested using the same restriction enzymes, and the digestion product was subjected to 1.5% agarose gel electrophoresis. Agarose pieces containing the digested plasmid were excised and purified using PCR cleanup gel electrophoresis (Qiagen). Fragments containing pf279 and pf774 were separately introduced into the digested plasmid by ligation performed according to the manufacturer's instructions (ligase from Invitrogen, Carlsbad, CA). The resulting product was used to transform E. coli DH5α competent cells by thermal shock (15). Cells were regenerated by incubation in LB broth (90 min, 37°C, 200 rpm), and transformants were selected on LB agar-ampicillin. Recombinant plasmids were extracted with Nucleospin Multi-8-Plasmid kit (Macherey-Nagel, Hoerdt, France), according to the manufacturer's instructions and used to transform P. freudenreichii CIRM-BIA1T competent cells as previously described (16), with the following modifications. P. freudenreichii was grown in YEL medium supplemented with 0.5 M sucrose and 1% (wt/wt) glycine and incubated to an OD650 of 0.4 to 0.7. After electrotransfection (2.5 kV, 200 Ω, and 25 μF), cells were regenerated by incubation for 16 h at 30°C, and P. freudenreichii clones harboring the inserted vector were selected on YEL agar containing 250 μg/ml of hygromycin B and incubated for 4 days at 30°C under anaerobiosis (Anaerocult A; Merck, Darmstadt, Germany).
(ii) Insertional inactivation of pf279 and pf774 genes (KO mutants).
The insertional inactivation of both genes was conducted separately as previously described (12). Briefly, a suicide vector was constructed by inserting a chloramphenicol resistance gene into a pUC18 plasmid. In the resulting plasmid, internal fragments of 515 bp of the pf279 open reading frame and 516 bp of the pf774 open reading frame (accession no. AM944371 and AM944376) (10) of strain CIRM-BIA1T were separately cloned, resulting in the vectors pUC:pf279:CmR and pUC:pf774:CmR, respectively. To make these constructs, pf279 and pf774 were amplified by PCR from P. freudenreichii CIRM-BIA1T genomic DNA using primers containing restriction sites (see Table S1 in the supplemental material).
PCR mixtures contained 2 mM MgCl2, 1 μM each primer, 200 μM each dNTP, and 2.5 U of Taq polymerase (Fermentas International, Inc., Burlington, Ontario, Canada) in 1× amplification buffer (20 mM Tris HCl [pH 8.4], 50 mM KCl). PCR was performed as previously described (14). PCR products were purified, and each amplicon and the pUC18:CmR plasmid were double digested (XbaI-BamHI) and purified again as described above. Fragments containing pf279 and pf774 were separately ligated to the digested plasmid, and each ligation product was used to transform E. coli DH5α competent cells. Transformants were selected on LB agar-ampicillin, and recombinant plasmids were isolated. P. freudenreichii CIRM-BIA1T was transformed with each inactivation vector, and ad hoc transformants were selected on YEL agar containing chloramphenicol.
(iii) Complementation of KO mutants with plasmid harboring pf279.
To perform the genetic complementation of the CB1-KO-pf279 strain, pFB01:pf279 overexpression plasmids were multiplied in E. coli DH5α cells, extracted, and used to transform the CB1-KO-pf279 knockout strain, as described above. Transformants were selected on YEL agar containing hygromycin B.
Comparison of the sequences of genes encoding esterases.
The sequences of both pf279 and pf774 genes were extracted from the complete genomes of the 20 sequenced strains of P. freudenreichii by using the genome annotation system AGMIAL (17). In addition, the sequence of the pf279 gene was determined in the commercial strain LSP108. The sequence was also confirmed in the nonlipolytic strain CIRM-BIA514, in which a specific mutation was observed in the pf279 sequence. For this, pf279 was amplified by high-fidelity PCR from the genomic DNA of the CIRM-BIA514 and LSP108 strains using primers pf279-XbaI-F and pf279-HindIII-R (see Table S1 in the supplemental material). The PCR mixture (50 μl) contained 1.5 mM MgCl2, 200 μM dNTP, 0.5 μM each primer, ∼50 ng DNA template, 3% (vol/vol) dimethyl sulfoxide (DMSO), and 1 U high-fidelity Phusion DNA polymerase (New England BioLabs) in 1× Phusion GC buffer. A 2-step thermocycling protocol was used: initial denaturation at 98°C for 1 min, followed by 30 cycles of 98°C for 15 s and 72°C for 30 s, then 72°C for 10 min, and then a final step at 12°C for 4 min. Amplification products were purified as described above and sequenced (GATC Biotech AG, Mulhouse, France). The sequences of the pf279 and pf774 genes were analyzed using Vector NTI software (Invitrogen) and aligned at the gene and protein levels using ClustalW 2.1.
Determination of FFA.
Individual FFA were analyzed using gas chromatography (GC) according to the method of De Jong and Badings (18). Briefly, lipids were extracted from 1 g of culture medium using ether-heptane (50:50 [vol/vol]) at acidic pH in the presence of anhydrous sodium sulfate. FFA were isolated from total lipids using an aminopropyl solid-phase column and quantified using gas chromatography on a BP21 column (SGE, Ringwood, Victoria, Australia) (25 m by 0.53 mm by 0.5-μm film thickness) under the following conditions: on-column injection at 65°C; carrier gas, hydrogen, 31 kPa; temperature program, heating rate of 10°C/min from 65°C up to 240°C, maintained for 10 min; flame-ionization detector operated at 240°C.
Statistical analyses.
One-way analyses of variance were performed using R (http://www.R-project.org) to determine the effect of the strains on the concentrations of FFA and on their net production. Differences between the treatment means were compared at the 5% level of significance by using the Fisher's least significance difference (LSD) test.
Nucleotide sequence accession numbers.
The sequences of the pf279 and pf774 genes have been deposited in the EMBL data library, and the accession numbers are given in Table 3.
TABLE 3.
EMBL accession numbers of the sequences of Propionibacterium freudenreichii lipolytic esterases PF#279 and PF#774
| Accession no. | Gene | Strain |
|---|---|---|
| HG426277 | pf279 | CIRM-BIA9 |
| HG426278 | pf279 | CIRM-BIA118 |
| HG426279 | pf279 | CIRM-BIA119 |
| HG426280 | pf279 | CIRM-BIA121 |
| HG426281 | pf279 | CIRM-BIA122 |
| HG426282 | pf279 | CIRM-BIA123 |
| HG426283 | pf279 | ITG P18 |
| HG426284 | pf279 | ITG P20 |
| HG426285 | pf279 | LSP108 |
| HG426286 | pf279 | CIRM-BIA134 |
| HG426287 | pf279 | CIRM-BIA135 |
| HG426288 | pf279 | ITG P9 |
| HG426289 | pf279 | ITG P23 |
| HG426290 | pf279 | CIRM-BIA456 |
| HG426291 | pf279 | CIRM-BIA508 |
| HG426292 | pf279 | CIRM-BIA512 |
| HG426293 | pf279 | CIRM-BIA513 |
| HG426294 | pf279 | CIRM-BIA514 |
| HG426295 | pf279 | CIRM-BIA516 |
| HG426296 | pf279 | CIRM-BIA527 |
| HG426297 | pf774 | CIRM-BIA9 |
| HG426298 | pf774 | CIRM-BIA118 |
| HG426299 | pf774 | CIRM-BIA119 |
| HG426300 | pf774 | CIRM-BIA121 |
| HG426301 | pf774 | CIRM-BIA122 |
| HG426302 | pf774 | CIRM-BIA123 |
| HG426303 | pf774 | ITG P18 |
| HG426304 | pf774 | ITG P20 |
| HG426305 | pf774 | CIRM-BIA134 |
| HG426306 | pf774 | CIRM-BIA135 |
| HG426307 | pf774 | ITG P9 |
| HG426308 | pf774 | ITG P23 |
| HG426309 | pf774 | CIRM-BIA456 |
| HG426310 | pf774 | CIRM-BIA508 |
| HG426311 | pf774 | CIRM-BIA512 |
| HG426312 | pf774 | CIRM-BIA513 |
| HG426313 | pf774 | CIRM-BIA514 |
| HG426314 | pf774 | CIRM-BIA516 |
| HG426315 | pf774 | CIRM-BIA527 |
RESULTS AND DISCUSSION
This study aimed to determine the role of two putative esterases in lipolysis by P. freudenreichii, combining targeted mutation experiments and the exploration of the natural phenotypic and genomic biodiversity in 21 wild-type strains.
Phenotype of mutants of P. freudenreichii CIRM-BIA1T either overexpressing or knocked out for the two targeted genes.
All wild-type and mutant strains displayed similar growth and propionic fermentation rates. At harvest time (from 64 to 74 h of incubation), the OD650 value was 3.3 ± 0.4 (i.e., about 3.109 CFU/g).
The concentrations of the 13 main FFA observed for the wild-type strain and the five genetically modified strains in the presence of a milk fat emulsion are detailed in Table 4, and the profile of the released FFA is illustrated in Fig. 1. The concentration of total FFA in the noninoculated control medium was 946.3 ± 118.6 μg/g, and it did not significantly change during incubation (data not shown). In cultures of the wild-type strain, the concentration of FFA was about 2-fold higher than that in the control medium (Table 4).
TABLE 4.
Concentrations of FFA at the end of growth of Propionibacterium freudenreichii CIRM-BIA1T and five genetically modified strains derived from this strain in a medium containing a milk fat emulsiona
| FFA | FFA concn (μg/g) inb: |
||||||
|---|---|---|---|---|---|---|---|
| Controlc | CB1T | CB1::pFB01:pf279 | CB1-KO-pf279 | CB1-KO-pf279-C | CB1::pFB01:pf774 | CB1-KO-pf774 | |
| C4:0 | 120.7 D | 222.0 B | 250.7 A | 176.3 C | 234.3 AB | 252.3 A | 229.3 B |
| C6:0 | 11.7 C | 37.0 B | 59.0 A | 16.0 C | 56.0 A | 56.7 A | 40.7 B |
| C8:0 | 8.7 E | 26.7 C | 49.7 A | 14.7 D | 42.0 B | 46.3 AB | 29.7 C |
| C10:0 | 20.3 D | 47.7 C | 104.7 A | 30.7 D | 90.3 B | 97.3 AB | 54.0 C |
| C12:0 | 24.7 D | 57.3 C | 150.3 A | 36.3 D | 127.0 B | 133.7 B | 66.7 C |
| C14:0 | 81.7 D | 201.3 C | 643.0 A | 122.3 D | 520.0 B | 526.7 B | 224.7 C |
| C15:0 | 10.0 E | 18.0 CD | 52.3 A | 12.7 DE | 44.3 B | 44.0 B | 19.0 C |
| C16:0 | 250.3 E | 478.7 CD | 1,591.3 A | 289.3 DE | 1,353.0 B | 1,362.7 B | 562.3 C |
| C16:1 | 23.7 C | 47.7 B | 107.7 A | 32.7 C | 101.3 A | 105.3 A | 54.3 B |
| C18:0 | 108.0 D | 143.7 CD | 446.7 A | 106.7 D | 379.7 B | 387.3 B | 175.7 C |
| C18:1 | 247.7 C | 469.7 B | 1,151.0 A | 276.7 C | 1,065.7 A | 1,147.3 A | 559.3 B |
| C18:2 | 15.7 C | 29.7 B | 72.3 A | 17.7 C | 68.0 A | 70.0 A | 32.0 B |
| C18:3 | 5.7 C | 7.7 BC | 17.0 A | 6.7 BC | 17.0 A | 16.3 A | 8.3 B |
| Total | 946.3 D | 1,810.0 C | 4,744.0 A | 1,164.0 D | 4,144.0 AB | 4,290.7 B | 2,076.0 C |
See Table 2 for strain nomenclature.
Means within a row followed by a common letter do not significantly differ according to the LSD test (P > 0.05).
Noninoculated control medium.
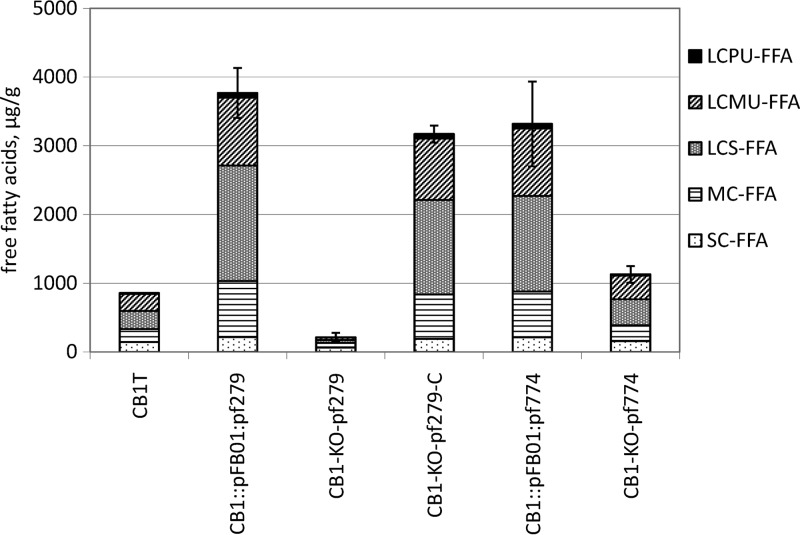
FIG 1.
Net production of free fatty acids released from a milk fat emulsion in cultures of Propionibacterium freudenreichii CIRM-BIA1T and five genetically modified strains derived from this strain. See Table 2 for strain nomenclature. FFA are expressed as μg total FFA per g culture. SC-FFA, short-chain free fatty acids (C4:0 to C8:0); MC-FFA, medium-chain free fatty acids (C10:0 to C15:0); LCS-FFA, long-chain saturated free fatty acids (sum of C16:0 and C18:0); LCMU-FFA, long-chain monounsaturated free fatty acids (sum of C16:1 and C18:1); LCPU-FFA, long-chain polyunsaturated free fatty acids (sum of C18:2 and C18:3). Values are means and standard deviations of triplicate experiments.
The two strains overexpressing either pf279 or pf774 and the complemented pf279 knockout strain exhibited similarly high lipolytic activity levels (Table 4). They released 4-fold larger FFA amounts, on average, than the wild-type strain, with marked differences for some FFA (Fig. 1). The amounts of medium- and long-chain FFA released, in particular saturated FFA, were 3.8- to 4.8-fold higher than those of the wild-type strain, whereas short-chain FFA release was less affected (1.4-fold). Consequently, the profile of released FFA by these strains differed from that of the wild-type strain, with 42.9% saturated long-chain FFA and only 6.1% short-chain FFA, versus 30.6% and 16.7%, respectively, in the wild-type strain. The results of overexpression of pf279 are in agreement with our previous results that showed that a mutant overexpressing pf279 under the control of another promoter released 5 to 8 times more FFA from milk fat than the wild-type strain (11). The results of overexpression of the pf774 gene, previously annotated as encoding a putative cell-bound esterase (11), showed that PF#774 (i) is effectively an esterase and (ii) is active on milk triglycerides. P. freudenreichii CIRM-BIA1T thus possesses a second lipolytic esterase active on milk fat, PF#774, which exhibits an apparent specificity for milk fat that did not significantly differ from that of PF#279.
The two strains knocked out for the pf279 and pf774 esterase genes (CB1-KO-pf279 and CB1-KO-pf774) exhibited very different phenotypes (Fig. 1). Inactivation of the pf279 gene induced a dramatic decrease in the lipolytic activity compared to that of the wild-type strain, with a particularly sharp decrease (−84%) in the amount of long-chain FFA released (Fig. 1). Only two FFA, butanoic and octanoic acids, were found in significantly larger amounts in cultures of CB1-KO-pf279 compared to the noninoculated control medium (Table 4). Inactivation of the pf774 gene, in contrast, did not induce any significant change in the lipolytic activity and the profiles of the released FFA compared to the wild-type strain (Table 4 and Fig. 1). These results suggest that either PF#774 was not highly expressed in the wild-type strain under our experimental conditions, or the activity of this esterase is low compared to that of PF#279.
The results of these targeted mutation experiments clearly demonstrate that PF#279 is responsible for the majority of the FFA released from an emulsion of milk fat during P. freudenreichii growth.
Exploration of phenotypic and genomic biodiversity within a collection of wild-type P. freudenreichii strains.
The lipolytic activity of 21 wild-type strains, including some strains previously characterized for their lipolytic activity in Swiss cheese, was investigated in a medium containing an emulsion of milk fat (Fig. 2).
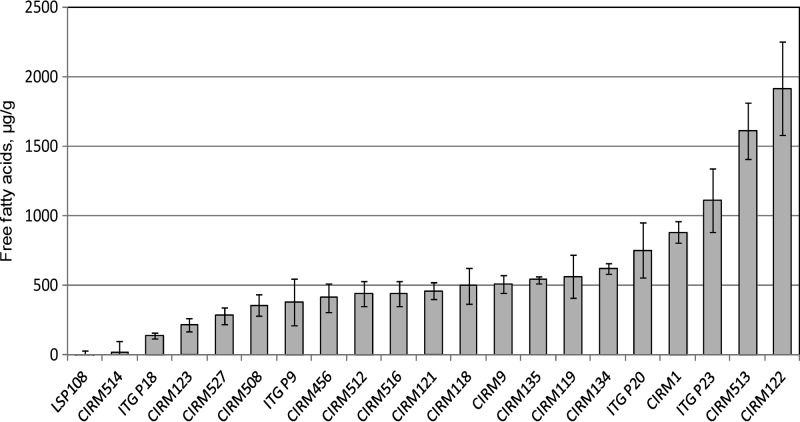
FIG 2.
Net production of free fatty acids released from a milk fat emulsion in cultures of wild Propionibacterium freudenreichii strains. Values are means and standard deviations of duplicate experiments.
At harvest time (66 to 71 h of incubation), the value of OD650 was 4.4 ± 0.3. Nineteen strains out of 21 released FFA during their growth in the presence of milk fat, with a net production of FFA ranging from 137 to 1,915 μg/g culture, whereas two strains (LSP108 and CIRM-BIA514) did not display any detectable lipolytic activity under our experimental conditions (production lower than 50 μg/g).
Interestingly, the lipolytic activities of P. freudenreichii were similar under the in vitro conditions used and in cheese. Specifically, ITGP18 was previously shown to exhibit low lipolytic activity, ITGP9 exhibited medium activity, and ITGP23 exhibited high activity in cheese, with net increases in FFA concentrations of 0.2, 2.1, and 3 to 4 mg/g cheese, respectively, compared to the control cheeses manufactured without propionibacteria (6, 8). Under the in vitro conditions of the present study, these three strains released 0.137, 0.379, and 1.109 mg FFA/g culture medium, respectively. Moreover, the commercial strain LSP108, known for its very low lipolytic activity in Swiss cheese, did not display any detectable activity in vitro. These results demonstrate the relevance of the conditions used in the present study to assess in vitro the lipolytic activity of P. freudenreichii in cheese.
The results of sequence alignments of the pf279 and pf774 genes showed that both genes are highly conserved in P. freudenreichii (data not shown). The predicted esterase PF#774 is a 412-residue protein in all but two strains (CIRM-BIA516 and CIRM-BIA122, for which the predicted proteins have 399 amino acid residues). In most strains (16 out of 21), the predicted sequence of the esterase PF#774 is identical. The size of the pf279 gene is 1,332 bp (for 14 strains) or 1,329 bp (for 5 strains). In the latter case, pf279 presents the same deletion of 3 consecutive bp near the 3′ end of the gene (codon TCG [Ala] or GCG [Ser] at position 1308) (results not shown). This deletion leads to a protein that is shorter by one residue (Ala or Ser) at the C-terminal end than the 443-residue protein predicted in the majority of strains.
For two strains (CIRM-BIA514 and LSP108), however, the pf279 gene shared the same single-nucleotide deletion near the 5′ end of the pf279 gene, at position 107 (see Fig. S1 in the supplemental material). This mutation results in a frameshift and the introduction of a TGA stop codon at position 190. Therefore, the translation product would consist of a 63-residue peptide with a 35-residue N-terminal sequence homologous to that of the other strains, followed by a 28-residue sequence without homology. Interestingly, these strains are the two identified as nonlipolytic (Fig. 2). This result strongly supports the conclusion that the secreted esterase PF#279 is the main agent of milk fat lipolysis in P. freudenreichii.
Significance for cheese ripening.
Taken together, the results of the present study clearly demonstrate that the secreted esterase PF#279 is the main agent of lipolysis in cheese by P. freudenreichii. The identification of this key target gene offers new screening opportunities to identify nonlipolytic strains. We also observed great variation in lipolytic activities within the pool of lipolytic strains, which should now be further investigated. The variation could result from several factors or combinations of factors, including differences in the expression levels of the pf279 gene and variations in the efficiency of secretion of PF#279. Moreover, even if the sequences of esterase genes appeared to be highly conserved within the species, we cannot exclude slight changes in sequence that could affect the secondary structure of the esterase and consequently its affinity for the substrate.
The esterases responsible for lipolysis have not been identified in most cheese-related bacteria, although the FFA released from lipolysis participate in cheese flavor in most cheeses. Lactic acid bacteria possess only intracellularly located esterases (2). Their activity on cheese fat is thus made possible only after their release in the cheese matrix and is generally very weak. The present results highlight why the contribution of P. freudenreichii to lipolysis is decisive in the internally ripened (semi-) hard cheese varieties that contain propionibacteria, such as Emmental and Maasdam cheeses. The contribution of P. freudenreichii esterases is greater than that of LPL, whose activity can be observed during the earliest steps of cheese manufacture, and far greater than that of lactic acid bacteria.
Supplementary Material
ACKNOWLEDGMENTS
M. C. Abeijón Mukdsi benefited from a grant from ECOS-MINCyT (Action no. A08B02) and a postdoctoral Bernardo Houssay grant. The sequencing was supported by INRA AIP Bioressource PropioDive 2009.
Valentin Loux and Julien Buratti created databases and implemented the AGMIAL tool. We are grateful to Alyson Yee for English language corrections. We thank Pascal Pachot, Stat-Plan, for support concerning statistical analyses. We also thank Y. Murooka, Osaka University, for providing the pPK705 vector used in this study.
Footnotes
Published ahead of print 15 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03640-13.
REFERENCES
- 1.Urbach G. 1997. The flavour of milk and dairy products. II. Cheese: contribution of volatile compounds. Int. J. Dairy Technol. 50:79–89 [Google Scholar]
- 2.Collins YF, McSweeney PLH, Wilkinson MG. 2003. Lipolysis and free fatty acid catabolism in cheese: a review of current knowledge. Int. Dairy J. 13:841–866. 10.1016/S0958-6946(03)00109-2 [DOI] [Google Scholar]
- 3.Thierry A, Falentin H, Deutsch SM, Jan G. 2011. Bacteria, beneficial: Propionibacterium spp., p 403–411 In Fuquay JW, Fox PF, McSweeney P. (ed), Encyclopedia of dairy science. Elsevier, London, United Kingdom [Google Scholar]
- 4.Thierry A, Maillard MB, Bonnarme P, Roussel E. 2005. The addition of Propionibacterium freudenreichii to Raclette cheese induces biochemical changes and enhances flavour development. J. Agric. Food Chem. 53:4157–4165. 10.1021/jf0481195 [DOI] [PubMed] [Google Scholar]
- 5.Dherbécourt J, Bourlieu C, Maillard MB, Aubert-Frogerais L, Richoux R, Thierry A. 2010. Time course and specificity of lipolysis in Swiss cheese. J. Agric. Food Chem. 58:11732–11739. 10.1021/jf102572z [DOI] [PubMed] [Google Scholar]
- 6.Thierry A, Maillard MB, Richoux R, Kerjean JR, Lortal S. 2005. Propionibacterium freudenreichii strains quantitatively affect production of volatile compounds in Swiss cheese. Lait 85:57–74. 10.1051/lait:2004036 [DOI] [Google Scholar]
- 7.Fröhlich-Wyder MT, Bachmann HP. 2004. Cheeses with propionic acid fermentation, p 141–156 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese. Chemistry, physics and microbiology. Elsevier, London, United Kingdom [Google Scholar]
- 8.Chamba JF, Perréard E. 2002. Contribution of propionibacteria to lipolysis of Emmental cheese. Lait 82:33–44. 10.1051/lait:2001003 [DOI] [Google Scholar]
- 9.Bachmann HP. 1998. Lipolysis in cheese: not too much, not too little. Agrarforschung 5:293–295 [Google Scholar]
- 10.Dherbécourt J, Falentin H, Canaan S, Thierry A. 2008. A genomic search approach to identify esterases in Propionibacterium freudenreichii involved in the formation of flavour in Emmental cheese. Microb. Cell Fact. 7:16. 10.1186/1475-2859-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dherbécourt J, Falentin H, Jardin J, Maillard MB, Baglinière F, Barloy-Hubler F, Thierry A. 2010. Identification of a secreted lipolytic esterase in Propionibacterium freudenreichii, a ripening process bacterium involved in Emmental cheese lipolysis. Appl. Environ. Microbiol. 76:1181–1188. 10.1128/AEM.02453-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deutsch SM, Le Bivic P, Herve C, Madec MN, LaPointe G, Jan G, Le Loir Y, Falentin H. 2010. Correlation of the capsular phenotype in Propionibacterium freudenreichii with the level of expression of gtf, a unique polysaccharide synthase-encoding gene. Appl. Environ. Microbiol. 76:2740–2746. 10.1128/AEM.02591-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malik AC, Reinbold GW, Vedamuthu ER. 1968. An evaluation of the taxonomy of Propionibacterium. Can. J. Microbiol. 14:1185–1191. 10.1139/m68-199 [DOI] [PubMed] [Google Scholar]
- 14.Deutsch SM, Parayre S, Bouchoux A, Guyomarc'h F, Dewulf J, Dols-Lafargue M, Baglinière F, Cousin FJ, Falentin H, Jan G, Foligné B. 2012. Contribution of surface β-glucan polysaccharide to physicochemical and immunomodulatory properties of Propionibacterium freudenreichii. Appl. Environ. Microbiol. 78:1765–1775. 10.1128/AEM.07027-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 16.Gautier M, Rouault A, Sommer P, Briandet R, Cassin R. 1995. Bacteriophages infecting dairy propionibacteria. Lait 75:427–434. 10.1051/lait:19954-532 [DOI] [Google Scholar]
- 17.Bryson K, Loux V, Bossy R, Nicolas P, Chaillou S, van de Guchte M, Penaud S, Maguin E, Hoebeke M, Bessieres P, Gibrat JF. 2006. AGMIAL: implementing an annotation strategy for prokaryote genomes as a distributed system. Nucleic Acids Res. 34:3533–3545. 10.1093/nar/gkl471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Jong C, Badings HT. 1990. Determination of free fatty acids in milk and cheese. Procedures for extraction clean up and capillary gas chromatographic analysis. J. High Resolut. Chromatogr. 13:94–98 [Google Scholar]
- 19.Tilsala-Timisjarvi A, Alatossava T. 2001. Characterization of the 16S-23S and 23S-5S rRNA intergenic spacer regions of dairy propionibacteria and their identification with species-specific primers by PCR. Int. J. Food Microbiol. 68:45–52. 10.1016/S0168-1605(01)00462-7 [DOI] [PubMed] [Google Scholar]
- 20.Kiatpapan P, Hashimoto Y, Nakamura H, Piao YZ, Ono H, Yamashita M, Murooka Y. 2000. Characterization of pRGO1, a plasmid from Propionibacterium acidipropionici, and its use for development of a host-vector system in propionibacteria. Appl. Environ. Microbiol. 66:4688–4695. 10.1128/AEM.66.11.4688-4695.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.