Abstract
Escherichia coli adapts its lifestyle to the variations of environmental growth conditions, swapping between swimming motility or biofilm formation. The stationary-phase sigma factor RpoS is an important regulator of this switch, since it stimulates adhesion and represses flagellar biosynthesis. By measuring the dynamics of gene expression, we show that RpoS inhibits the transcription of the flagellar sigma factor, FliA, in exponential growth phase. RpoS also partially controls the expression of CsgD and CpxR, two transcription factors important for bacterial adhesion. We demonstrate that these two regulators repress the transcription of fliA, flgM, and tar and that this regulation is dependent on the growth medium. CsgD binds to the flgM and fliA promoters around their −10 promoter element, strongly suggesting direct repression. We show that CsgD and CpxR also affect the expression of other known modulators of cell motility. We propose an updated structure of the regulatory network controlling the choice between adhesion and motility.
INTRODUCTION
Microorganisms adapt to changes in their environment in many different ways. Among others, they adjust gene expression, modify their metabolism, and change their surface properties by displaying particular surface proteins. The latter phenomenon is directly linked to the choice of a particular “lifestyle,” adhesion (biofilm formation) or motility (planktonic growth). In Escherichia coli, expression of flagella at the cell surface predestines the cell for motility. More than 50 genes are involved in the synthesis of flagella. These genes are classified into three groups according to their temporal expression sequence. The master regulator of flagellar synthesis, FlhDC, is expressed first, and the corresponding genes constitute the class 1 flagellar operon. FlhDC activates the expression of class 2 genes, encoding the inner part of the flagellum as well as the flagellar sigma factor FliA (σ28 or σF) and the FlgM protein (anti-σ28). The class 3 genes are transcribed by σ28-RNA polymerase (RNAP) and encode the outer components of the flagellum as well as chemotaxis proteins (for a review, see reference 1).
Flagellar synthesis is tightly controlled by several environmental conditions, including osmolarity (2) and temperature (3). Many of these environmental influences affect the transcription of flagellar genes and control in particular the expression of the master regulator, FlhDC. The regulators of the flhDC operon include cyclic AMP (cAMP)-cAMP receptor protein (CRP), H-NS (4), OmpR (2), LrhA (5), integration host factor (IHF) (2, 6), RpoN (7), and Fur (8). Other regulators affect cell motility by controlling fliA expression. These factors include NsrR (9), signaling molecules such as cyclic diguanylic acid (c-di-GMP) (reviewed in reference 10), the alarmone polyphosphate guanosine [(p)ppGpp] (11), and quorum-sensing molecules such as autoinducer-2 (AI-2) (12–14).
Transcriptomic data show that the stationary sigma factor RpoS (σ38) represses the transcription of flagellar genes in E. coli (7, 15). An rpoS mutant has an accordingly higher motility on soft-agar LB plates but also in liquid culture (7, 16). Quantitative reverse transcription-PCR (qRT-PCR) experiments show that RpoS represses fliA transcription without affecting the transcription of flhDC in exponential phase in E. coli strain MG1655 (15). However, another recent report demonstrates that rpoS (indirectly) represses flhC transcription in E. coli strain BW25113 (16). We confirmed the negative regulation of fliA transcription by RpoS by using a green fluorescent protein (GFP) transcriptional fusion both in LB and M9 media (17). Consistently, an rpoS mutant also shows higher accumulation of the FliA (RpoF) protein during growth in LB medium (18). As a consequence, an rpoS mutant strain displays more flagella than a wild-type (WT) strain in both exponential and stationary growth phases (18). RpoS accumulates in different stress conditions and at the onset of stationary phase. This stress sigma factor directs the expression of about 500 genes (15, 19–22). RpoS also represses transcription, a counterintuitive function for a sigma factor. One hypothesis to explain the repression of motility genes by RpoS involves the competition between three sigma factors (σ38, σ70, and σ28) for the RNAP-core binding at the onset of stationary phase (23, 24). However, the relatively low affinity of RpoS for the core RNAP and the rapid degradation of RpoS are not favorable for such a competition which can be further influenced by the small protein Crl (25). Interestingly, the repression of flagellar genes occurs mostly in exponential phase in minimal medium, where RpoS accumulates to a lower concentration than in stationary phase. Thus, repression by RpoS takes place in growth conditions where the concentration of RpoS is relatively low. Another straightforward mechanistic explanation of repression by RpoS is that RpoS activates the transcription of a repressor. For instance, the global regulator FNR (fumarate and nitrate reductase), whose expression is partially induced by RpoS, represses genes of the tricarboxylic acid cycle (21). Furthermore, adhesion and biofilm formation are regulated by RpoS partially through the activation of the curli regulatory protein CsgD and CpxR (26, 27) or the transcription of diguanylate cyclases (28, 29).
In this article, we studied the expression dynamics of regulators of flagellar genes in order to investigate how RpoS represses cell motility. We confirm that RpoS represses flagellar regulators in exponential phase in different growth media. We show that fliA expression is cAMP dependent in all growth phases because CRP-cAMP activates FlhDC (4). We found that two additional RpoS-dependent transcription factors are involved in the repression of the flagellar gene cascade, CsgD and CpxR. These two regulators have antagonistic effects on adhesion and biofilm formation. Our data suggest that they regulate distinct targets of the network controlling flagellar biosynthesis and that CsgD directly represses the fliA and flgM promoters.
MATERIALS AND METHODS
Strains and plasmids.
The GFP transcriptional fusions were selected from a collection containing nearly 2,000 constructs on the low-copy-number reporter plasmid pUA66 harboring gfpmut2 (30). Each fusion was verified for accurate cloning and expression level well above the background fluorescence to allow reliable quantification. We reconstructed the rpoS::gfp reporter vector by DNA amplification of the rpoS promoter region (including parts of nlpD; for primer sequences, see Table S1 in the supplemental material) and cloning into the low-copy-number plasmid pZE (31), upstream of the gfpmut3 reporter gene (32). The fusions were transformed into E. coli K-12 strain BW25113 (rrnB3 lacI+ DElacZ4787 hsdR514 DE(araBAD)567 DE(rhaBAD)568 rph-1 [33]) and its isogenic mutants, the rpoS, csgD, cpxR, and cya mutants. The other mutant strains, lacking transcriptional regulators whose expression is positively regulated by RpoS (see Table S2), were used only for motility and Congo red (CR) binding assays. Transformations were performed by using a high-throughput method that utilizes a transformation and storage solution (TSS) solution containing polyethylene glycol (PEG), dimethyl sulfoxide (DMSO), and Mg2+ at pH 6.5 (34). The kanamycin (Kn) resistance cassette was removed from the mutant strains using FLP recombinase (35) prior to transformation of pUA66 derivatives. Mutant strains and reporter plasmids were verified by PCR. RpoS was overexpressed by cloning its coding sequence into the TA cloning pTOPO2 plasmid (Invitrogen). The functionality of this overexpression, which relies on an increased copy number of the rpoS gene, was validated by complementation of the Congo red binding phenotype of an rpoS mutant strain (see below). Table S1 in the supplemental material lists the sequences of the primers used in the present study.
Growth conditions.
Gene expression was measured for cultures growing either in LB (Difco) or M9 minimal medium (12.8 g/liter Na2HPO4, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2) supplemented with 0.2% glucose and 40 μg/ml kanamycin. To test curli production, bacteria were inoculated on LB agar plates containing 0.004% (wt/vol) of Congo red (Sigma). To test bacterial motility, a fixed amount of bacteria (2 μl of a fresh cell culture in stationary phase diluted at an optical density [OD] of ∼0.05) was inoculated on soft LB plates containing 0.35 g/liter agar (Difco).
Dynamic measurements of reporter gene concentration.
Measurements of gene expression were carried out at 37°C in 96-well polystyrene microplates (white-frame clear-well Isoplate-96 [product no. 5PA028; PerkinElmer]), inoculated from a fresh overnight culture in glass tubes. After dilution of the precultures, 30-fold (M9) or 50-fold (LB) into 150 μl of the same medium, the bacteria were grown in a thermostated microplate reader (Fusion; PerkinElmer) at 37°C for 15 h. To ensure microaerobic conditions and to prevent cell sedimentation, one glass bead (1 mm in diameter) was added to each well, and the microplates were shaken for 10 s every 3 min, alternatively by circular and linear movements. Absorbance (600 nm) and fluorescence (excitation at 485 nm and emission at 520 nm) were measured about 100 times during each experimental growth in the plate reader. For our microplate reader, the optical density at 600 nm (OD600) of the culture is about 2.5 times the measured absorbance, absorbance at 600 nm. The raw data were analyzed using WellReader (36) in order to determine the reporter gene concentration and calculate the promoter activity (32). Promoter activity is the time derivative of the reporter concentration plus the sum of the degradation rate of the reporter protein and the growth rate of the culture multiplied by the reporter gene concentration. For detailed equations, please refer to the supplemental material for reference 37. Absorbance and fluorescence measurements were background corrected as described previously (32, 36). Since the plasmid copy number was found to vary less than 2-fold between exponential versus stationary phases (37), no correction was implemented. Each experiment was repeated between three to five times independently and in triplicate on each microplate. The promoter activity curves of independent experiments were slightly time shifted (less than 30 min) according to their growth curves (identical time of entry into stationary phase) and averaged. Biologically impossible, negative signal values (less than 10% of the data range) were corrected by shifting the curve to positive values. The shaded regions in the figures represent the standard errors of the means of the averaged promoter activities in the same experimental condition.
Mobility shift assay of the CsgD-DNA complexes.
Promoter probes carrying the csgD, fliA, and flgM promoter regions were generated by PCR amplification (Ex Taq DNA polymerase; TaKaRa) using one 5′-fluorescein isothiocyanate (FITC)-labeled primer and derivatives of the pRS reporter plasmid as the template. Primer sequences are given in Table S1 in the supplemental material. The FITC-labeled PCR products were purified from a polyacrylamide electrophoretic gel prior to their utilization for gel shift assays. The CsgD protein was produced from the pET21a expression vector (38), and the assays were performed under standard conditions as described in reference 39.
RESULTS
RpoS represses the transcription of flagellar regulators.
Microarray data show that a large majority of the flagellar genes are downregulated by RpoS in minimal medium (15). An rpoS mutant strain is therefore more motile than a wild-type (WT) strain, and the overexpression of RpoS reduces cell motility (15). We confirmed this observation for E. coli strain BW25113 (Fig. 1A) where RpoS can be detected in exponential phase (see Fig. S3C in the supplemental material). In order to elucidate the mechanism underlying the regulation of fliA expression by RpoS, we monitored the dynamics of the fliA::gfp transcriptional fusion during growth in a batch culture. The transcriptional fusion is barely active in the WT strain, both in minimal and rich medium (Fig. 1; see Fig. 4). This is consistent with low FliA protein levels detected in the WT strain, in exponential phase in M9 medium containing 0.2% glucose at 37°C (see Fig. S4 in the supplemental material). In the ΔrpoS strain, however, fliA is strongly expressed in exponential phase, and promoter activity declines before entry into stationary phase (Fig. 1C). The fliA promoter remains barely active in stationary phase both in the WT and the rpoS mutant strain. The induction of fliA expression in exponential phase in the rpoS strain is unchanged at lower glucose concentration and enhanced by addition of Casamino Acids (Fig. S1). Our measurements of promoter activity (Fig. 1C) show that RpoS very strongly represses the expression of fliA during exponential growth: the repression factor is on the order of 100-fold.
FIG 1.
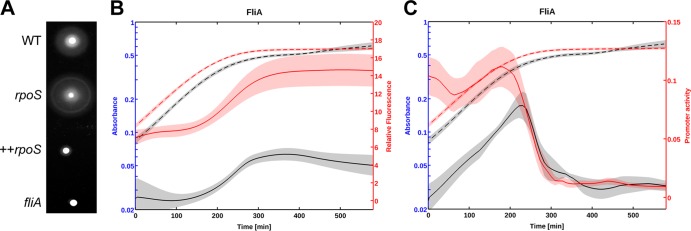
RpoS inhibits motility by repressing fliA expression in exponential phase. (A) Motility phenotypes of the wild-type (WT) E. coli strain BW25113 and the isogenic rpoS and fliA mutants. In the strain marked ++rpoS, RpoS is overexpressed by providing additional copies of the rpoS locus on a multicopy plasmid. Stationary-phase cultures were inoculated on soft LB agar (0.35%) plates and incubated overnight at 30°C. The size of the halo is an indicator of cell dispersion into soft-agar LB plates. (B and C) Relative GFP concentration (B) and computed promoter activity (C) of a fliA::gfp transcriptional fusion in a wild-type strain (black) and the rpoS mutant (red). M9 stationary-phase cultures were diluted 30-fold into 96-well plates at time zero. Absorbance at 600 nm (broken-line curves) and relative GFP fluorescence (solid-line curves) were monitored for 12 h at 37°C. The standard error of the mean is indicated by the shaded regions of the curves. A minimum of five experimental data sets were used to generate the average of each expression profile. Promoter activity is expressed in relative fluorescence units (RFU)/min (relative fluorescence per time), derived from the kinetics of the absorbance and GFP reporter data.
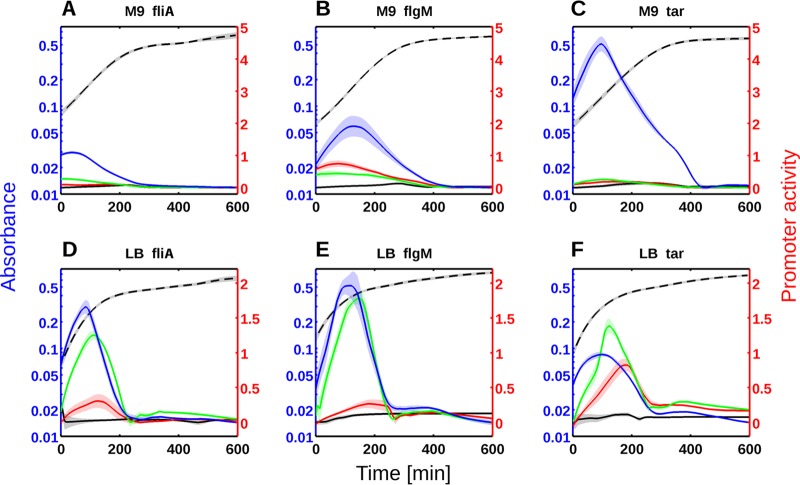
FIG 4.
Repression of flagellar class 2 and 3 genes by RpoS, CsgD, and CpxR in M9 minimal medium (A to C) or LB medium (D to F). Gene expression profiles were monitored for fliA::gfp (A and D), flgM::gfp (B and E), and tar::gfp (C and F) in the wild-type (black), rpoS (red), csgD (green), and cpxR (blue) mutant strains. Absorbance at 600 nm is shown on the blue axes, promoter activity (relative units) is shown on the red axes, and time of growth (in minutes) is shown on the black axes. The black broken-line curve represents a typical growth curve (absorbance at 600 nm) of the WT strain at 37°C. A minimum of five experimental data sets were used to generate each expression profile (average activities and standard errors of the means). The standard errors of the means are indicated by the shaded regions of the curves. Promoter activity is expressed in RFU/min (relative fluorescence per time), derived from the kinetics of the absorbance and GFP reporter data.
FliA is a sigma factor directing the transcription of flagellar class 3 genes. If RpoS affects the transcription of fliA, we should also be able to observe the (indirect) effect of RpoS on genes downstream of FliA. We tested this prediction for two σ28-dependent genes with measurable expression using GFP fusions: flgM and tar. As expected, RpoS affects the transcription of flgM and tar in a comparable way to the effect seen on fliA::gfp (Fig. 2). However, the rpoS deletion has a stronger effect on the expression of the flgM gene, probably because this gene is not only transcribed by FliA. The flgM gene is a member of both class 2 and 3 flagellar genes (40, 41) and is transcribed from multiple promoters (42). The expression dynamics of the flgM and tar genes are entirely explained by the accumulation of FliA predicted from Fig. 1B. These results demonstrate that RpoS represses flagellar genes in exponential phase, despite its low intracellular concentration (15, 23). These observations raise the question of how RpoS represses the transcription of fliA?
FIG 2.
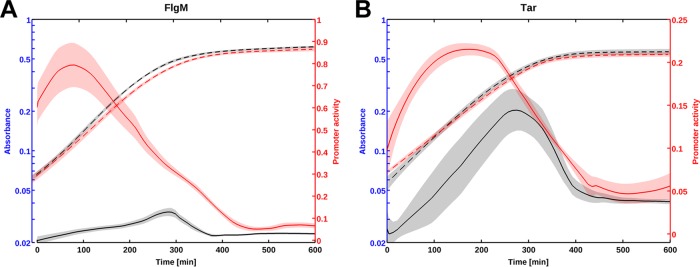
The flagellar gene cascade is repressed by RpoS at several levels. (A and B) Typical gene expression profiles for flgM::gfp (A) and tar::gfp (B). The average promoter activity computed in the wild-type strain is represented in black, and the average activity in the rpoS mutant strain is shown in red. The bacteria were grown in M9 minimal medium at 37°C (see Materials and Methods). Promoter activity is expressed in RFU/min (relative fluorescence per time), derived from the kinetics of the absorbance and GFP reporter data.
Identification of intermediate repressors activated by RpoS.
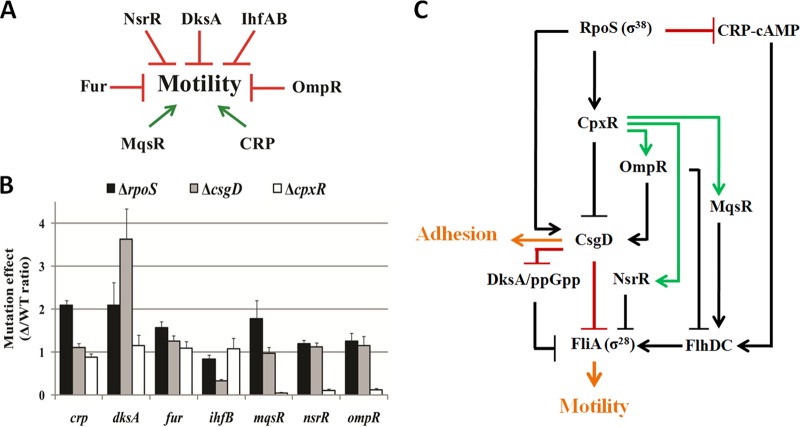
RpoS represses transcription of fliA, an unusual effect of a sigma factor. The most likely mechanistic explanation would be the activation of an “intermediate” repressor of fliA transcription by RpoS. In order to search for such an intermediate regulator, we looked for genes encoding transcriptional factors and whose transcription is affected by the rpoS mutation in microarray data performed in four growth conditions (15, 21, 22). Out of 288 transcription factor genes in E. coli (reviewed in reference 43), we found 53 genes differentially expressed in an rpoS mutant strain: 32 genes activated and 21 genes downregulated by RpoS in at least one medium (LB versus M63) or growth condition (exponential versus stationary). To identify a potential repressor(s) of flagellar genes, we selected, from the single mutant library of E. coli K-12 strain BW25113 (33), strains that carry insertion in these 32 transcription factors positively regulated by RpoS. From this selection, we eliminated mutants for master regulatory genes such as ArcA and RpoS itself, and ended up with 21 mutant strains (listed in Table S2 in the supplemental material) that were tested for swimming motility.
In order to select regulators affecting cell motility, we inoculated the corresponding mutants on soft-agar (0.35%) LB plates. Since the rpoS mutation leads to increased cell motility, we expected that some of these mutations should affect cell motility. Only four mutant strains presented a motility phenotype different from the wild-type strain. Two of them were known to affect RpoS protein levels, the gadX and gadW mutant strains (44, 45). The two remaining mutants are known regulators of bacterial adhesion and biofilm formation, the csgD and cpxR mutant strains. CsgD is the central regulator of curli fimbria synthesis and cellulose production (26, 27, 46), and CpxR is involved in the response to cell envelope stress and regulation of protein folding or cell adhesion (26, 47). Because CpxR is known to repress csgD transcription (26, 48, 49), the two regulators are important for both curli production and the regulation of cell motility.
Activation of csgD transcription by RpoS had been previously shown (27, 50, 51). As expected, the increase of cell motility is of comparable magnitude for the csgD and rpoS mutants, whereas the cpxR mutation has a much more drastic effect (Fig. 3A). The Congo red (CR) dye binds to curli fibers and can be used to assay curli production. RpoS and CsgD stimulate curli production (46), and their deletion reduces CR staining (Fig. 3B). In contrast, the colony formed by a cpxR mutant strain appears slightly redder or comparable to a wild-type colony (Fig. 3B). Thus, while the cpxR and csgD mutations have antagonistic effects on curli production, they both stimulate cell motility (Fig. 3C). CsgD is known to activate curli production at low temperature, and its accumulation and binding to DNA targets were studied at temperatures below 30°C (38, 52, 53). However, the stimulatory effect of the csgD deletion on swimming motility is independent of the growth temperature (see Fig. S2 in the supplemental material). As expected, overexpression of CsgD and CpxR in a wild-type strain results in reduced swimming motility (data not shown). These two regulators are therefore important intermediates, connecting RpoS to the complex regulatory network responsible for the decision between motility and adhesion.
FIG 3.
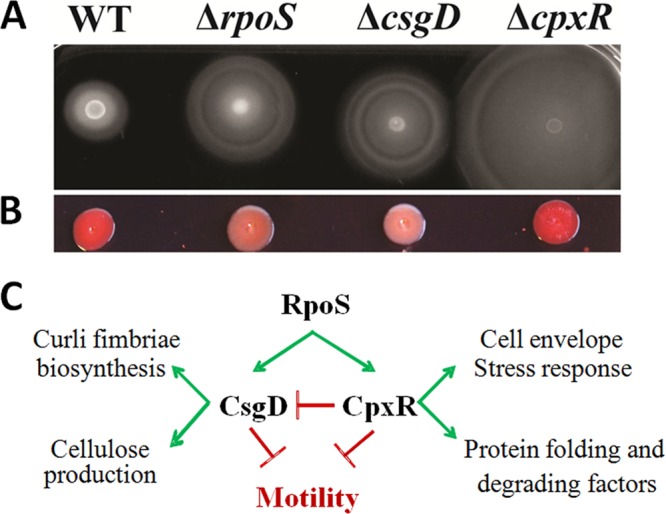
CsgD and CpxR as intermediate repressors of bacterial motility. (A and B) Motility (B) and Congo red (B) phenotypes of the wild-type E. coli BW25113 strain and rpoS, csgD, and cpxR mutant strains derived from the WT strain. Stationary-phase cultures were inoculated on the top of the LB soft-agar (0.35%) plates for motility tests and onto LB agar (1.2%) plus Congo red (0.004%) for curli fimbria staining and incubated overnight or 2 days, respectively, at 30°C. (C) Known functions and interactions between RpoS, CsgD, and CpxR.
CsgD and CpxR do not affect rpoS transcription or activity.
To confirm that CsgD and CpxR affect bacterial cell motility without affecting RpoS levels or activity, we measured rpoS transcription, RpoS protein levels, and the expression of characterized targets of RpoS in the WT and in the csgD and cpxR mutant strains. Figure S3 in the supplemental material shows that rpoS transcription is not affected by the csgD or cpxR mutations; RpoS accumulation and the expression of six known targets of RpoS induced in stationary growth phase are also not affected by these mutations. The cpxR deletion reduces the expression of only one gene, gadB, suggesting a specific regulation of this gene by CpxR. These results confirm that CsgD and CpxR repress cell motility without affecting RpoS activity.
CsgD and CpxR repress flagellar biosynthesis.
In order to elucidate the mechanism by which CsgD and CpxR repress cell motility, we measured the expression kinetics of fliA, flgM, and tar in the corresponding mutant strains. These three fusions were selected among the reporter fusions for motility genes available in our GFP collection (30), because they presented sufficient activity above the promoterless GFP vector (17). The expression of fliA is strongly increased in both mutant strains during exponential phase in minimal medium (Fig. 4A) and more drastically in rich growth medium (Fig. 4D). This regulation affects the intracellular level of FliA protein in exponential phase: as shown in Fig. S4 in the supplemental material, FliA accumulates to a greater concentration in a csgD or cpxR mutant strain compared to the WT strain.
An immediate prediction of this observation is that target genes of FliA should be transcribed more strongly in these mutants. Figure 4B and C show the kinetic measurements of flgM and tar expression in these mutants. As expected, the expression profiles of flgM and tar are entirely explained by the effects of the csgD and cpxR deletions on FliA expression. The cpxR mutation induces a massive expression peak in exponential phase and affects the expression of flgM and tar more than the csgD or rpoS deletion does. These strong effects were confirmed using quantitative RT-PCR from total RNA extracted in mid-exponential phase (see Fig. S5 in the supplemental material). In both experimental approaches, the induction factors are very strong and range from 4- to >2,000-fold. In order to confirm that CsgD and CpxR repress FliA-dependent genes, we measured expression of a selection of nine target genes of FliA using qRT-PCR. As shown in Fig. S5, both regulators exert a strong negative regulation on other class 3 motility genes (namely, aer, fliC, fliL, trg, tsr, ves, and ycgR). Interestingly, in M9 minimal medium, the expression profile of tar is much more sensitive to the cpxR deletion than to the deletion of the adhesion regulators, CsgD and RpoS (Fig. 4C). This observation is true for the other class 3 genes transcribed by the RNA polymerase associated with FliA (Fig. S5). These regulations are qualitatively independent of the growth medium, although the regulatory effects have higher amplitudes in rich medium (Fig. 4D, E, and F).
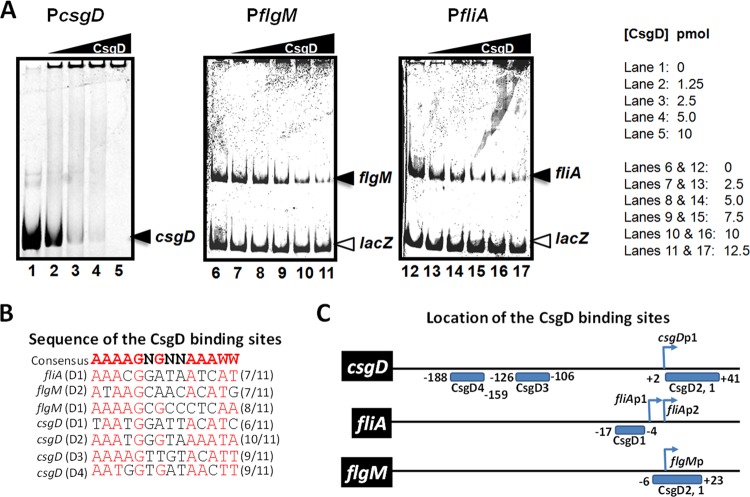
CsgD directly binds the −10 promoter element of fliA and flgM.
To test whether CsgD directly binds to the promoter regions of the genes encoding the flagellar sigma factor and its targets, we performed gel shift assays. As shown in Fig. 5A, the CsgD protein binds the promoter regions of fliA and flgM. The bound DNA is clearly shifted by CsgD, forming aggregates in the wells at the top of the gel. These interactions are consistent with known properties of CsgD and the idea of a direct repression of these regulatory genes by CsgD. To demonstrate specific binding of CsgD to these promoters, we added a competitor DNA, the lacZ promoter that does not contain a sequence similar to that of the CsgD binding site. As expected, the intensity of unbound DNA decreases proportionally to the increase of CsgD concentration in the assay for each promoter but not for the lacZ promoter DNA. This clearly demonstrates that CsgD specifically binds to the promoters of the flagellar regulators (class 2 genes). For an additional control, we tested CsgD binding to its own promoter harboring multiple CsgD binding sites (Fig. 5C). As expected, CsgD binds its promoter with a higher affinity than the one observed at the fliA and flgM promoters. Figure 5B presents the consensus sequence for CsgD binding (AAAAGNGNNAAAWW [38]) and the potential binding site overlapping the −10 promoter elements of the flagellar regulatory genes. As shown in Fig. 5C, a single putative CsgD binding site is present upstream of the −10 hexamer of fliAP2 (σ28 dependent), also overlapping the −10 box of the distal promoter fliAP1 (σ70 dependent). In addition, two putative CsgD binding sites overlap the −10 promoter element of flgM, but none were found upstream of the flhDC operon, the master regulator of class 2 genes. The two CsgD binding motifs present on the flgM promoter have more mismatches to the proposed consensus sequence for CsgD binding than the unique, high-affinity site found at the fliA promoter (Fig. 5B and C). In addition, these two sites contain cytosines instead of adenines at positions 11 and 12, as also observed at other CsgD DNA targets (38).
FIG 5.
CsgD binds the promoter region of the flagellar class 2 genes. (A) Gel shift assay of CsgD binding to its own promoter and the promoters of fliA and flgM. The protein was present at 1.25 to 12.5 pmol for 0.5 pmol of labeled DNA in a final volume of 20 μl. The concentration of CsgD is indicated by the height of the triangle above the gel, and the concentration of CsgD (in picomoles) in each lane is given to the right of the gels. (B) Consensus sequence for CsgD binding and sequences of the binding sites found around the −10 promoter element of the csgD, fliA, and flgM promoters. Individual DNA binding sites recognized by CsgD are labelled D1 to D4, which refers to specific positions within the target promoters. (C) Locations of the CsgD binding sites according to the transcription start site.
CsgD and CpxR regulate motility through several known regulators.
The control of flagellar gene expression is influenced by numerous genetic factors, summarized in Fig. 6A (for a review, see reference 54). In order to assess the roles of RpoS, CsgD, and CpxR in this network, we have tested the effects of their deletion on expression of these key modulators of cell motility. As shown in Fig. 6B, mutation of csgD and cpxR directly or indirectly affects the expression of four out of seven regulators of the cascade of flagellar genes. For example, CsgD activates ihfB, known to repress FlhD (2, 6). Therefore, CsgD can potentially exert a repressive effect on the two master regulators at two levels of the regulatory cascade: directly on fliA and indirectly on flhD. In contrast, CpxR directly or indirectly activates nsrR and ompR, both of which repress cell motility. CsgD represses the transcription of dksA, itself known to repress the transcription of flhD and fliA following starvation (11). Most modulators of E. coli cell motility are regulated by RpoS, CsgD, or CpxR both in exponential phase (data not shown) and in stationary phase (Fig. 6B). However, the magnitude of the regulatory effects is higher in stationary growth phase. Taken together, these results show that CpxR and CsgD are major actors of a very complex regulatory network (summarized in Fig. 6C) and these two regulators fine-tune the expression of cellular appendices responsible for cell motility and adhesion.
FIG 6.
CsgD and CpxR regulate different components in the network controlling FliA expression. (A) Seven major regulators of E. coli are known to modulate flagellar biosynthesis, mostly by controlling FlhDC (see introduction). (B) Measurements of GFP transcriptional fusions in the wild-type strain and its rpoS, csgD, and cpxR mutants in M9 medium at 37°C. Gene expression values were calculated, in stationary phase (absorbance at 600 nm of 0.6), as the ratio of the relative fluorescence of the mutant versus the WT strains in at least 3 independent experiments. Very similar gene expression ratios were found in exponential phase (absorbance of 0.2). (C) Proposed model controlling cell adhesion and motility. Black arrows indicate known regulation, while green and red arrows show new stimulatory or repressive interactions, respectively, between key regulators.
fliA expression is cAMP dependent.
In the proposed network (Fig. 6C), RpoS slightly represses the transcription of CRP itself known to activate the flhDC operon (4). FlhDC is the major activator binding the fliA promoter (1), and we therefore predicted that fliA transcription should be dependent on cAMP-CRP. Indeed, fliA is not transcribed in adenylate cyclase mutants (ΔcyaA; see Fig. S6A in the supplemental material) or CRP mutants (data not shown). Supplementing a minimal medium with increasing concentrations of cAMP restores transcription of fliA in a cyaA mutant strain (Fig. S6A). The complementation by cAMP is immediate and functions at all growth phases, although it is less efficient in stationary phase (Fig. S6B). These results confirm that cAMP-CRP stimulates fliA transcription, most probably through flhDC activation. The activation of FlhDC expression by CRP is reduced by repression of crp by RpoS, a pathway independent of the csgD and cpxR genes (Fig. 6).
DISCUSSION
CsgD and CpxR regulate motility and adhesion.
Swimming motility in Escherichia coli is a highly regulated process. Here, we found that RpoS, the stationary sigma factor, indirectly represses the expression of flagellar genes during growth in both minimal and rich media. This regulation takes place in the exponential phase of growth and confirms previous observations in E. coli strains MG1655 and BW25113 (7, 16). These results also agree with previous transcriptomic studies (7, 15, 22) showing that RpoS alters a subset of genes in exponential phase, especially in minimal medium where RpoS accumulates to higher concentrations. Such specific regulatory events in fast-growing cells might rely on improved promoter recognition of a subset of genes (with better affinity for RpoS) and may be enhanced by the Crl protein (17, 27). Since direct repression by a sigma factor is a rare mechanism, we screened for potential transcriptional repressors activated by RpoS and found two regulators, CsgD and CpxR, both of which repress cell motility. They particularly repress the transcription of fliA in exponential phase and therefore indirectly downregulate the expression of genes transcribed by FliA (so-called class 3 genes of chemotaxis or encoding the outer part of the flagellum; see Fig. S5 in the supplemental material). Of these genes, FlgM, the modulator of FliA activity, is subject to a similar regulation (Fig. 4) even though the two flagellar regulators were found abundant both in exponential and stationary growth phases (55). Thus, FliA and FlgM are both negatively regulated in exponential phase by major determinants of cell adhesion and biofilm formation of members of the family Enterobacteriaceae.
Role of CsgD.
CsgD controls the secretion of curli fimbriae and cellulose production, two determinants of bacterial adhesion and biofilm formation (56). CsgD binds directly to flagellar class 2 operons (fliE and fliFGHIJK) and represses their transcription (38). Pulldown experiments suggest that CsgD interacts with FliA (σ28), probably to modulate its activity or stability (57). Here, we demonstrate that CsgD represses part of the flagellar regulatory cascade through specific inhibition of fliA transcription. This repression is logically transmitted to at least nine flagellar class 3 genes (Fig. 4 or Fig. S5 in the supplemental material). In addition, CsgD makes other connections with global regulators, thus diminishing the expression of FliA by alternative routes. For example, CsgD represses the transcription of DksA which, together with ppGpp, plays an important role in reorganizing gene expression in stationary phase, in particular by inducing the RpoS regulon (58). We also found that CsgD activates the transcription of ihfB (Fig. 6B) itself known to activate csgD transcription in exponential phase (49). Such reciprocal interactions complicate an intuitive understanding of the regulatory network controlling the decision between motility and adhesion (Fig. 6C).
Involvement of nucleoproteins in the regulatory network.
The switch in lifestyle is probably also influenced by other nucleoproteins whose concentration varies during growth in batch culture (59). For example, H-NS and IHF control csgD (and ompR) transcription (49), but only the concentration of IHF varies during the transition from exponential to stationary phase. The time-varying concentration of IHF could thus modulate CsgD accumulation. Furthermore, Fis and Dps are, respectively, the most abundant nucleoid-associated proteins (NAPS) in exponential and stationary phases of growth. It is therefore possible that the high level of Fis in exponential phase affects, indirectly and independently of CsgD, the switch between flagellar and curli expression by reducing (60) the stimulatory effect of CRP on the master activator FlhDC.
Role of CpxR.
CpxR and the inner membrane histidine kinase CpxA form a two-component system activated by an increase in osmolarity and/or following attachment to surfaces. CpxR is known to activate protein folding and degrading factors such as DegP, PpiA, and DsbA (61) and to repress curli expression (26), chemotaxis genes, and the motA gene, an essential component of the flagellar motor (62). Since CpxR represses csgD transcription (26), adhesion to hydrophobic surfaces is impaired when the Cpx pathway is altered (47). Here, we found that CpxR represses fliA more strongly than RpoS or CsgD (Fig. 4A and D). However, this strong repression of fliA is indirect, passing through several routes that involve different transcription factors. For example, we demonstrate that CpxR is essential for the transcription of the mqsR, nsrR, and ompR regulatory genes (Fig. 6B). These functional dependencies connect CpxR to genes involved in the repression of both FliA and FlhDC. In the proposed model (Fig. 6C), the regulatory path CpxR-OmpR-CsgD-FliA could explain the observed repressive effects of CpxR on fliA transcription and cell motility; another route, independent of CsgD (CpxR-OmpR-FlhDC-FliA), also explains the negative regulation of fliA by CpxR. In contrast, the straight regulatory path CpxR-CsgD-FliA may not correspond to a predominant route in exponential growth phase, probably because the inhibitory effect of CpxR on csgD is impaired by the presence of OmpR that shares the same binding region as CpxR on the csgD promoter (26, 49). Thus, CpxR may inhibit the expression of flagellar genes only during exponential growth, and CpxR may inhibit curli expression only in stationary phase. In contrast, CsgD may alternatively repress the biosynthesis of flagella in exponential phase and stimulate curli expression in stationary phase. These growth phase-dependent regulations may be further modulated by fluctuating concentrations of NAPS.
DNA binding sites.
A key functional interaction in the model proposed in Fig. 6C is the repression of fliA by CsgD. Our gel mobility shift assay shows a direct physical interaction between the CsgD protein and the promoter of this gene (Fig. 5A). This in vitro experiment also suggests a direct repression of the flgM promoter by CsgD. At least two key interactions of CsgD in the regulatory network therefore correspond to direct physical interactions. Our experiments also add three new binding sites to the list of 18 known binding sites of CsgD (38). Examination of these sequences reveals conserved A or C nucleotides at positions 11 and 12 (Fig. 5B), leading to an updated version of the consensus sequence for CsgD binding, AAAAGNGNNAMMWW. (The underlined nucleotides correspond to two modifications of the consensus sequence for CsgD binding which was previously proposed as AAAAGNGNNAAAWW in reference 38 and as shown in Fig. 5B.)
Negative regulation of cell motility by RpoS is likely mediated by direct binding of CsgD to the promoter of flagellar class 2 genes (fliE, fliF, fliA, and flgM). On the other hand, we did not find a putative CsgD box on the flhD promoter, which is consistent with the idea that RpoS represses flagellar genes only by acting on fliA transcription as demonstrated in strain MG1655 (7). However, another recent study of strain BW25113 shows that rpoS can also repress flhC in exponential phase (16). For CpxR, we identify several intermediate regulators (Fig. 6B), but sequence analysis did not reveal any putative binding site for CpxR (GTAAANNNNGTAAA) (63) within the promoters of flhD, fliA, or flgM.
Decision motility versus adhesion.
The interactions we have identified in this work further increase the complexity of the network underlying the decision between motility and adhesion. Multiple redundant connections within this network complicate a quantitative interpretation of the proposed interactions. The stress response sigma factor RpoS is at the top of this hierarchical network (Fig. 6C). This master regulator orchestrates the decision between “alternative lifestyles” by promoting the transcription of csgD and cpxR, two key regulators highly connected to the different modulators of FliA expression. Our work presents the first evidence that this differentiation process shares common transcription factors in addition to the c-di-GMP signaling molecule (28, 29, 64).
Supplementary Material
ACKNOWLEDGMENTS
We thank Herb Schellhorn for sharing transcriptomic data on the RpoS regulon, Delphine Ropers and Hidde de Jong for critical reading of the manuscript, and Corinne Pinel for technical assistance.
This work was supported by the Region Rhone-Alpes, France (Projet Cible 2011, Infectiologie), by a grant from the ANR, by the CNRS, and the Université Joseph Fourier.
Footnotes
Published ahead of print 22 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00938-13.
REFERENCES
- 1.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708. 10.1128/MMBR.64.4.694-708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin S, Park C. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler J, Templeton B. 1967. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46:175–184. 10.1099/00221287-46-2-175 [DOI] [PubMed] [Google Scholar]
- 4.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521–532. 10.1046/j.1365-2958.2002.03032.x [DOI] [PubMed] [Google Scholar]
- 6.Yona-Nadler C, Umanski T, Aizawa S, Friedberg D, Rosenshine I. 2003. Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohaemorrhagic Escherichia coli. Microbiology 149:877–884. 10.1099/mic.0.25970-0 [DOI] [PubMed] [Google Scholar]
- 7.Dong T, Yu R, Schellhorn H. 2011. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79:375–386. 10.1111/j.1365-2958.2010.07449.x [DOI] [PubMed] [Google Scholar]
- 8.Stojiljkovic I, Baumler AJ, Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531–545 [DOI] [PubMed] [Google Scholar]
- 9.Partridge JD, Bodenmiller DM, Humphrys MS, Spiro S. 2009. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol. Microbiol. 73:680–694. 10.1111/j.1365-2958.2009.06799.x [DOI] [PubMed] [Google Scholar]
- 10.Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273. 10.1038/nrmicro2109 [DOI] [PubMed] [Google Scholar]
- 11.Lemke JJ, Durfee T, Gourse RL. 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 74:1368–1379. 10.1111/j.1365-2958.2009.06939.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke MB, Sperandio V. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57:1734–1749. 10.1111/j.1365-2958.2005.04792.x [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316. 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperandio V, Torres AG, Kaper JB. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809–821. 10.1046/j.1365-2958.2002.02803.x [DOI] [PubMed] [Google Scholar]
- 15.Dong T, Schellhorn HE. 2009. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol. Genet. Genomics 281:19–33. 10.1007/s00438-008-0389-3 [DOI] [PubMed] [Google Scholar]
- 16.Ojima Y, Hakamada K, Nishinoue Y, Nguyen MH, Miyake J, Taya M. 2012. Motility behavior of rpoS-deficient Escherichia coli analyzed by individual cell tracking. J. Biosci. Bioeng. 114:652–656. 10.1016/j.jbiosc.2012.06.014 [DOI] [PubMed] [Google Scholar]
- 17.Dudin O, Lacour S, Geiselmann J. 2013. Expression dynamics of RpoS/Crl-dependent genes in Escherichia coli. Res. Microbiol. 164:838–847. 10.1016/j.resmic.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 18.Makinoshima H, Aizawa S, Hayashi H, Miki T, Nishimura A, Ishihama A. 2003. Growth phase-coupled alterations in cell structure and function of Escherichia coli. J. Bacteriol. 185:1338–1345. 10.1128/JB.185.4.1338-1345.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603. 10.1128/JB.187.5.1591-1603.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacour S, Landini P. 2004. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186–7195. 10.1128/JB.186.21.7186-7195.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580–591. 10.1007/s00438-004-1089-2 [DOI] [PubMed] [Google Scholar]
- 22.Dong T, Kirchhof MG, Schellhorn HE. 2008. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics 279:267–277. 10.1007/s00438-007-0311-4 [DOI] [PubMed] [Google Scholar]
- 23.Jishage M, Iwata A, Ueda S, Ishihama A. 1996. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 178:5447–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda H, Fujita N, Ishihama A. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497–3503. 10.1093/nar/28.18.3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.England P, Westblade LF, Karimova G, Robbe-Saule V, Norel F, Kolb A. 2008. Binding of the unorthodox transcription activator, Crl, to the components of the transcription machinery. J. Biol. Chem. 283:33455–33464. 10.1074/jbc.M807380200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213–7223. 10.1128/JB.183.24.7213-7223.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbe-Saule V, Jaumouille V, Prevost MC, Guadagnini S, Talhouarne C, Mathout H, Kolb A, Norel F. 2006. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:3983–3994. 10.1128/JB.00033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446. 10.1101/gad.475808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. 2009. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331. 10.1099/mic.0.024257-0 [DOI] [PubMed] [Google Scholar]
- 30.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3:623–628. 10.1038/nmeth895 [DOI] [PubMed] [Google Scholar]
- 31.Elowitz MB, Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature 403:335–338. 10.1038/35002125 [DOI] [PubMed] [Google Scholar]
- 32.de Jong H, Ranquet C, Ropers D, Pinel C, Geiselmann J. 2010. Experimental and computational validation of models of fluorescent and luminescent reporter genes in bacteria. BMC Syst. Biol. 4:55. 10.1186/1752-0509-4-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172–2175. 10.1073/pnas.86.7.2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161. 10.1016/S0378-1119(02)00551-6 [DOI] [PubMed] [Google Scholar]
- 36.Boyer F, Besson B, Baptist G, Izard J, Pinel C, Ropers D, Geiselmann J, de Jong H. 2010. WellReader: a MATLAB program for the analysis of fluorescence and luminescence reporter gene data. Bioinformatics 26:1262–1263. 10.1093/bioinformatics/btq016 [DOI] [PubMed] [Google Scholar]
- 37.Berthoumieux S, de Jong H, Baptist G, Pinel C, Ranquet C, Ropers D, Geiselmann J. 2013. Shared control of gene expression in bacteria by transcription factors and global physiology of the cell. Mol. Syst. Biol. 9:634. 10.1038/msb.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 193:2587–2597. 10.1128/JB.01468-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, Ishihama A. 2007. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189:4791–4799. 10.1128/JB.00319-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macnab RM. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131–158. 10.1146/annurev.ge.26.120192.001023 [DOI] [PubMed] [Google Scholar]
- 41.Stafford GP, Ogi T, Hughes C. 2005. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 151:1779–1788. 10.1099/mic.0.27879-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salgado H, Peralta-Gil M, Gama-Castro S, Santos-Zavaleta A, Muniz-Rascado L, Garcia-Sotelo JS, Weiss V, Solano-Lira H, Martinez-Flores I, Medina-Rivera A, Salgado-Osorio G, Alquicira-Hernandez S, Alquicira-Hernandez K, Lopez-Fuentes A, Porron-Sotelo L, Huerta AM, Bonavides-Martinez C, Balderas-Martinez YI, Pannier L, Olvera M, Labastida A, Jimenez-Jacinto V, Vega-Alvarado L, Del Moral-Chavez V, Hernandez-Alvarez A, Morett E, Collado-Vides J. 2013. RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 41:D203–D213. 10.1093/nar/gks1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishihama A. 2012. Prokaryotic genome regulation: a revolutionary paradigm. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 88:485–508. 10.2183/pjab.88.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phadtare S, Inouye M. 2001. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183:1205–1214. 10.1128/JB.183.4.1205-1214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, Conway T. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190–3201. 10.1128/JB.185.10.3190-3201.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264. 10.1046/j.1365-2958.1998.00791.x [DOI] [PubMed] [Google Scholar]
- 47.Otto K, Silhavy TJ. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:2287–2292. 10.1073/pnas.042521699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038–2049. 10.1128/JB.187.6.2038-2049.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogasawara H, Yamada K, Kori A, Yamamoto K, Ishihama A. 2010. Regulation of the Escherichia coli csgD promoter: interplay between five transcription factors. Microbiology 156:2470–2483. 10.1099/mic.0.039131-0 [DOI] [PubMed] [Google Scholar]
- 50.Arnqvist A, Olsen A, Normark S. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021–1032. 10.1111/j.1365-2958.1994.tb00493.x [DOI] [PubMed] [Google Scholar]
- 51.Dong T, Schellhorn HE. 2009. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10:349. 10.1186/1471-2164-10-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brombacher E, Baratto A, Dorel C, Landini P. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 188:2027–2037. 10.1128/JB.188.6.2027-2037.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brombacher E, Dorel C, Zehnder AJ, Landini P. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847–2857. 10.1099/mic.0.26306-0 [DOI] [PubMed] [Google Scholar]
- 54.McCarter LL. 2006. Regulation of flagella. Curr. Opin. Microbiol. 9:180–186. 10.1016/j.mib.2006.02.001 [DOI] [PubMed] [Google Scholar]
- 55.Ishihama A. 1999. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4:135–143. 10.1046/j.1365-2443.1999.00247.x [DOI] [PubMed] [Google Scholar]
- 56.Zogaj X, Bokranz W, Nimtz M, Romling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151–4158. 10.1128/IAI.71.7.4151-4158.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arifuzzaman M, Maeda M, Itoh A, Nishikata K, Takita C, Saito R, Ara T, Nakahigashi K, Huang HC, Hirai A, Tsuzuki K, Nakamura S, Altaf-Ul-Amin M, Oshima T, Baba T, Yamamoto N, Kawamura T, Ioka-Nakamichi T, Kitagawa M, Tomita M, Kanaya S, Wada C, Mori H. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16:686–691. 10.1101/gr.4527806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landini P, Egli T, Wolf J, Lacour S. 2013. SigmaS, a major player in the response to environmental stresses in Escherichia coli: role, regulation and mechanisms of promoter recognition. Environ. Microbiol. Rep. 10.1111/1758-2229.12112 [DOI] [PubMed] [Google Scholar]
- 59.Dillon SC, Dorman CJ. 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8:185–195. 10.1038/nrmicro2261 [DOI] [PubMed] [Google Scholar]
- 60.Gonzalez-Gil G, Kahmann R, Muskhelishvili G. 1998. Regulation of crp transcription by oscillation between distinct nucleoprotein complexes. EMBO J. 17:2877–2885. 10.1093/emboj/17.10.2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387–398. 10.1101/gad.9.4.387 [DOI] [PubMed] [Google Scholar]
- 62.De Wulf P, Kwon O, Lin ECC. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto K, Ishihama A. 2006. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70:1688–1695. 10.1271/bbb.60024 [DOI] [PubMed] [Google Scholar]
- 64.Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62:1014–1034. 10.1111/j.1365-2958.2006.05440.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.