Abstract
B cell differentiation into antibody-secreting cells (ASCs) is a tightly regulated process under the control of multiple transcription factors. One such transcription factor, Ets1, blocks the transition of B cells to ASCs via two separate activities: (i) stimulating the expression of target genes that promote B cell identity and (ii) interfering with the functional activity of the transcription factor Blimp1. Ets1 is a member of a multigene family, several members of which are expressed within the B cell lineage, including the closely related protein Ets2. In this report, we demonstrate that Ets1, but not Ets2, can block ASC formation despite the fact that Ets1 and Ets2 bind to apparently identical DNA sequence motifs and are thought to regulate overlapping sets of target genes. The DNA binding domain of Ets1 is required, but not sufficient by itself, to block ASC formation. In addition, less conserved regions within the N terminus of Ets1 play an important role in inhibiting B cell differentiation. Differences between the N termini of Ets1 and Ets2, rather than differences in the DNA binding domains, determine whether the proteins are capable of blocking ASC formation or not.
INTRODUCTION
Plasma cells or antibody-secreting cells (ASCs) are terminally differentiated B cell effectors that are specialized to secrete large amounts of immunoglobulin (Ig). The differentiation process of these cells involves expansion of the cytoplasmic compartment due to the substantial increase in the volume of the endoplasmic reticulum, which is needed for increased Ig synthesis and secretion. B cell differentiation into ASCs can be triggered by T cell-derived stimuli or by Toll-like receptor (TLR) ligands such as lipopolysaccharide (LPS), which binds TLR4, or unmethylated CpG-containing oligonucleotides, which bind TLR9.
ASC differentiation is controlled by a set of key transcription factors, some of which promote the differentiation process and others of which inhibit it. The best-known transcription factor that drives terminal differentiation of B cells into ASCs is Blimp1 (also known as PRDI-BF1). Blimp1 is a zinc finger-containing transcriptional repressor that inhibits the expression of genes characteristic of mature B cells (1, 2). Other transcription factors that promote ASC differentiation include XBP1 and IRF4 (3, 4). Transcription factors that inhibit the differentiation of B cells into ASCs include Pax5, Bcl6, Mitf, and Bach2 (5–7). We have previously demonstrated that the transcription factor Ets1 can block B cell differentiation into ASCs (8).
Ets1 is the founding member of the Ets family of transcription factors, which is comprised of 26 members in mice. Ets1 is highly expressed in B and T lymphocytes and regulates their functional responses (9, 10). Within the Ets gene family, Ets1 is most closely related to Ets2. Ets1 and Ets2 share 96% amino acid identity within their DNA binding domains (the Ets domain) and bind to indistinguishable DNA sequences in vitro (11, 12). Furthermore, both Ets1 and Ets2 share very similar domain structures outside the Ets domain, including a Pointed or SAM domain involved in protein/protein interactions, an acidic transactivation domain, and autoinhibitory domains that flank the Ets domain and suppress its ability to associate with DNA. Both Ets1 and Ets2 also share an N-terminal Erk mitogen-activated protein (MAP) kinase phosphorylation site (13). Phosphorylation of this residue in either protein stimulates transcriptional activity by promoting association with the coactivator CBP (14).
Both Ets1 and Ets2 are detected in primary B cells by gene expression profiling (15) and in B cell lines (16). Within the primary cell populations, Ets1 is found at high levels in naive and memory B cells, at low levels in germinal center B cells, and at very low levels in plasma cells (15). Consistent with this analysis, Ets1 protein levels are low in ASCs compared to naive B cells (8). Ets2 demonstrates a different pattern of expression, being found at low, but relatively constant, levels at all stages of B cell differentiation (15).
We previously demonstrated that Ets1 binds to the Blimp1 protein and inhibits its ability to bind to target DNA sequences to regulate gene expression (8). Ets1 can also activate the expression of genes that are normally repressed by Blimp1, such as the key B cell identity gene Pax5 (8). Both of these activities of Ets1 are dependent on the highly conserved Ets domain of the protein (8). Since the Ets domain is conserved among all members of the Ets gene family, it is possible that other Ets factors have a similar activity in repressing ASC formation. Indeed, we showed previously that at least two other Ets proteins (PU.1 and Ets2) are capable of interacting with Blimp1 in glutathione S-transferase (GST) pulldown assays (8). However, among the Ets proteins tested, only Ets1 could block Blimp1 binding to its target sites (8). This suggests that amino acid sequences outside the Ets domain, which are less conserved among the various Ets factors, are important for the ability of Ets1 to block Blimp1 DNA binding. In this report, we show that Ets2 cannot block ASC differentiation even when expressed at high levels within B cells, despite the fact that it is structurally very similar to Ets1 and likely regulates many of the same target genes.
MATERIALS AND METHODS
Plasmids.
Plasmids encoding various forms of Ets1, Blimp1, and Ets2 in GST and mammalian expression vectors were described previously (8). Additional plasmids encoding deletions and point mutations of Ets1 described in this report were generated by PCR and cloned into appropriate vectors. All PCR products were verified by sequencing prior to use. Plasmid pCL-Eco (17) was cotransfected with viral plasmids to increase titers. Plasmids BSAP-Luc (containing the murine Pax5 promoter) and pCMV-βgal, used for transfection, were described previously (8).
Cell lines and transfections.
The plasmacytoma cell line P3X and the B cell line A20 were cultured in RPMI 1640 with 10% fetal bovine serum (FBS). A20 B lymphoma cells were transfected by electroporation as described previously (8). Cos-1 cells and the retroviral packaging cell line Platinum-E (Plat-E) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% FBS. The medium for Plat-E cells also contained the selection drugs blasticidin (10 μg/ml) and puromycin (1 μg/ml) to maintain integrated viral structural genes. Plat-E cells were switched to regular DMEM on the day of transfection.
Mice and B cell purifications.
Ets1-deficient mice carrying an allele that deletes exons encoding the Pointed domain (exons IV and V) were described previously (9, 18, 19). This targeting event represents a neomorphic allele of Ets1, with a small amount of internally truncated protein being made. However, it is functionally identical to a null allele; hence, we refer to these mice as Ets1−/− mice in this article. Ets2-deficient mice were generated by crossing mice carrying a floxed allele of Ets2 (Ets2flox mice) with Meox2-Cre mice, which express Cre in the epiblast of the embryo (20). In the resulting mice, Cre mediates deletion of the Ets2 gene in all epiblast-derived tissues (i.e., in the entire embryo but not extraembryonic tissues). We refer to Ets2flox/flox Meox2-Cre mice as Ets2−/− mice in this report. B cells were purified from the spleens of wild-type C57BL/6 mice by depleting CD43+ non-B cells.
Coimmunoprecipitation assays.
Cos-1 cells were transfected by using Fugene-6 with 1 μg of each plasmid (pCMV-HA-Ets1 or pCMV-HA-Ets2 along with pcDNA3.1 Blimp1 Δ350–557). Forty-eight hours later, cells were lysed, and supernatants were precleared by incubation with protein G-agarose beads. Lysates were then incubated with either a mouse monoclonal anti-FLAG antibody or a control mouse IgG1 antibody followed by immunoprecipitation. Immunoprecipitates were Western blotted by using antihemagglutinin (anti-HA) antibodies.
ELISPOT.
For enzyme-linked immunosorbent spot (ELISPOT) analysis, Millipore MultiScreen 96-well plates with Immobilon-P membranes were coated with 5 μg/ml of monoclonal anti-mouse IgM or polyclonal anti-mouse Ig. Spleen and lymph node cells were isolated from wild-type, Ets1−/−, or Ets2−/− mice, plated in serial dilutions, and incubated overnight. IgM-secreting ASCs were detected by using a biotin-conjugated rat anti-mouse IgM detection antibody, and IgG-secreting ASCs were detected by using a biotin-conjugated polyclonal anti-mouse IgG antibody. ELISPOT plates were counted with an automated reader (Zellnet Consulting, Fort Lee, NJ, USA).
Retroviral production and transduction.
For production of retrovirus, the Plat-E packaging cell line was transfected with various retroviral plasmids along with helper plasmid pCL-Eco by using Fugene-6 transfection reagent, as previously described (8). Retroviral supernatants were harvested at 48 h posttransfection. For retroviral infection, purified B cells stimulated with either 5 μg/ml CpG ODN1826 (Invivogen) or 5 μg/ml LPS (Sigma-Aldrich) for 24 h were spin inoculated with retroviral supernatant in the presence of 10 μg/ml Polybrene. The cells were subsequently washed and returned to culture with fresh medium in the presence 5 μg/ml CpG ODN1826 or 5 μg/ml of LPS. Two days after infection, the green fluorescent protein-positive (GFP+) population was sorted out by using a FACs Aria cell sorter (BD Biosciences).
ELISA.
Equivalent numbers of sorted GFP-positive cells from the retrovirally transduced populations were resuspended in medium containing 5 μg/ml CpG ODN or LPS. After 48 h, supernatants were harvested and serially diluted, and an enzyme-linked immunosorbent assay (ELISA) was carried out according to standard protocols. Purified mouse IgM (clone 11E10; Southern Biotech) was used for generating a standard curve.
Western blot analysis.
Whole-cell lysates of fluorescence-activated cell sorter (FACS)-sorted GFP-positive cells were prepared, resolved on SDS-PAGE gels, and blotted with rabbit polyclonal anti-mouse Ets1 (C-20; Santa Cruz Biotechnology, Santa Cruz, CA). The same membrane was stripped and reprobed with the following antibodies: rat monoclonal anti-Blimp1 (clone 6D3; Santa Cruz Biotechnology), mouse monoclonal anti-Pax5 (clone A-11; Santa Cruz Biotechnology), mouse monoclonal anti-XBP1 (clone F-4), rabbit polyclonal anti-mouse Ets2 (a kind gift from Michael Ostrowski, Ohio State University, Columbus, OH), and mouse monoclonal anti-β-tubulin (KMX-1; Chemicon International, Temecula, CA).
Chromatin immunoprecipitation.
Mouse total splenic B cells were cross-linked by using 0.25% formaldehyde for 10 min. A chromatin immunoprecipitation (ChIP) assay was carried out by using the Magna ChIP G kit (Millipore, Billerica, MA) according to directions provided by the manufacturer. Samples were immunoprecipitated by using 2 μg of control nonspecific rabbit IgG, anti-Ets1 rabbit monoclonal antibody (Epitomics), or purified polyclonal rabbit anti-Ets2 antibody. Quantitative PCR (qPCR) was then performed on the immunoprecipitated genomic DNA with primers for regions within the Pax5 gene. Primer sequences are shown in Table 1. For each primer set, the amount of chromatin immunoprecipitated as a percentage of the input chromatin was calculated by using the following formula: percent input = 100[2∧(adjusted input CT) − (Pax5 primer CT)], where CT is the threshold cycle. Results of the ChIP assays were analyzed by using analysis of variance (ANOVA) with the Fisher least-significant-difference (LSD) test.
TABLE 1.
Primers used in this study
| Primer set | Primer sequences | Use for primers |
|---|---|---|
| P1 | TGACCCCTCCCCCTATCCTC and CTGCTCCCTCCCCAGTCTGA | ChIP of Pax5 promoter |
| P2 | AGGATCCCCTGCCTATCTGT and CAGGCCCTGAACTCAACACT | ChIP of intron 5 enhancer |
| P3 | CTTCCTCCATCCCTGGTTTC and AAATGCCTCCTCGAGGTTCT | ChIP of intron 5 enhancer |
| c-myc | GCGTGAAAGGGAAAGGACTAGCGC and GCGAGCGCTAGTCCTTTCCCTTTC | c-myc promoter Blimp1 binding site for EMSA |
| Mmp3 | AGTGGAACCAAGACAGGAAGCACTTCCTGGAGATTA and AGTGGATAATCTCCAGGAAGTGCTTCCTGTCTTGGT | Mmp3 promoter Ets1 binding site for EMSA |
| muS | CACACTGTACAATGTCTCCCT and AAAATGCAACATCTCACTCTG | Expression of secreted IgM isoform |
| muM | TCCTCCTGAGCCTCTTCTAC and CCAGACATTGCTTCAGATTG | Expression of membrane-bound IgM isoform |
| Gapdh | CATGGCCTTCCGTGTTCCTA and CCTGCTTCACCACCTTCTTGAT | Expression of housekeeping Gapdh gene |
Purification of GST fusion proteins and GST pulldown assay.
GST fusion proteins were expressed in Rosetta(DE3)/pLacI competent cells (Novagen, NJ), purified by using a standard protocol, and dialyzed prior to use. In vitro-transcribed-translated (IVT) 35S-labeled Blimp1 was generated as previously described (8). GST pulldown assays were performed by incubating IVT Blimp1 and equal amounts of GST fusion proteins, followed by incubation with glutathione-Sepharose beads, as previously described (8). The beads were washed, and bound proteins were eluted and separated on SDS-PAGE gels for visualization by autoradiography.
Electrophoretic mobility shift assays.
For electrophoretic mobility shift assays (EMSAs), double-stranded oligonucleotides containing either a Blimp1 binding site from the c-myc promoter or an Ets1 binding site from the MMP3 promoter were labeled with [α-32P]dCTP and used as probes (oligonucleotide sequences are listed in Table 1). As a source of Blimp1, whole-cell extracts from the P3X plasmacytoma cell line were isolated as described previously (8). As a source of Ets1, recombinant GST fusions of wild-type or mutant Ets1 proteins were expressed in bacteria and purified. For supershift assays, P3X extracts were preincubated with 1 μl of Blimp1 antibody (clone 6D3; Santa Cruz Biotechnology). Where indicated, GST fusion proteins were incubated with P3X extracts for 45 min on ice prior to addition of labeled probe. Images from GST pulldown assays were quantitated by using Image J.
qPCR.
Total RNA was isolated from sorted GFP+, virally infected B cells. cDNA was synthesized, and qPCR was performed by using Bio-Rad iQ SYBR green supermix. Expression levels of the membrane-bound and secreted forms of IgM were measured. As an internal control, the housekeeping gene Gapdh was used to normalize data. Differential gene expression was determined by using the ΔΔCT method. Primer sequences used are provided in Table 1.
Statistical analysis.
Data in bar graphs and scatter plots are represented as means ± standard errors of the means (SEM). Each dot in a scatter plot represents an individual mouse. Statistics in bar graphs were calculated by either paired t tests or ANOVA with the Fisher least-significant-difference test. Statistics in scatter plots were calculated by a two-tailed Mann-Whitney U test. All statistical analyses were carried out by using GraphPad Prism version 6.
RESULTS
Ets1 but not Ets2 inhibits ASC differentiation.
Ets1 is highly expressed in naive B cells, but its levels fall during activation and differentiation into ASCs. Ets1 can block the TLR-induced differentiation of mouse splenic B cells into IgM-secreting ASCs when its continued expression is driven by a viral promoter (8). This effect of Ets1 does not require expression of supraphysiological levels of Ets1, as the levels of Ets1 in freshly isolated, resting splenic B cells was similar to or higher than that found in TLR-stimulated B cells infected with a retrovirus harboring Ets1 (see Fig. S1A in the supplemental material). The block of ASC development by Ets1 is further validated by its ability to inhibit the production of secreted IgM but not membrane-bound IgM (see Fig. S1B in the supplemental material).
Ets1 possesses two separate activities that might mediate its ability to suppress ASC differentiation. First, Ets1 physically interacts with the key ASC transcription factor Blimp1 to prevent binding to Blimp1 target sites (8). Second, Ets1 can also upregulate the expression of a series of target genes that are normally repressed by Blimp1, including Pax5 (8), a transcription factor crucial for specifying and maintaining mature B cell fate (6, 21). The DNA binding domain (Ets domain) of Ets1 is required for both activities. The Ets domain is the most conserved portion of the protein, and highly related DNA binding domains are found in all other members of the Ets gene family. Thus, it is a possibility that the activity of Ets1 in blocking ASC differentiation is not unique to Ets1 but rather is a general property of the Ets gene family.
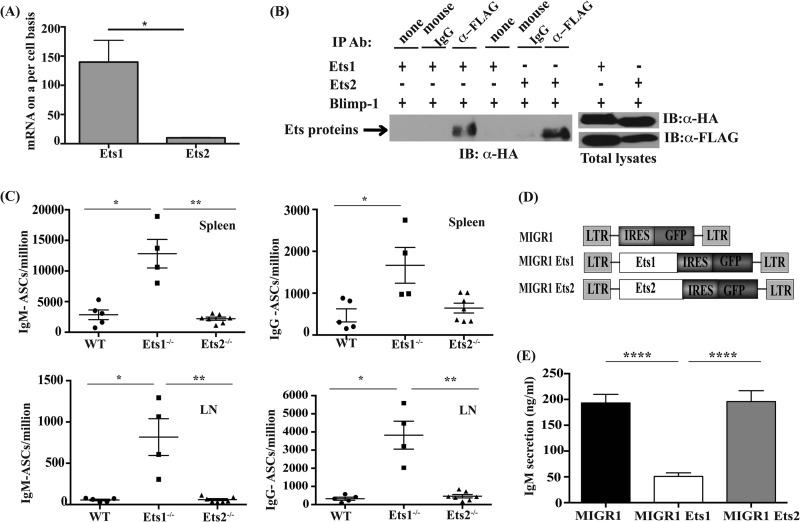
Previously, multiple Ets family members have been shown to be expressed in the Raji B cell line, including the closely related Ets1 and Ets2 genes (22). Indeed, we found that both Ets1 and Ets2 were also expressed in mouse splenic B cells, although the levels of Ets2 were about 10-fold lower than those of Ets1 (Fig. 1A). Like Ets1, Ets2 physically interacts with Blimp1 in GST pulldown assays. However, in contrast to Ets1, the interaction of Ets2 with Blimp1 fails to inhibit Blimp1 binding to a target sequence (8). To further validate the interaction of Ets2 with Blimp1, we transfected a cell line with HA-tagged Ets1 or Ets2 and a FLAG-tagged version of Blimp1. The Blimp1 expression vector that we used in these assays contains an internal deletion of sequences encoding the PEST domain of Blimp1. Deletion of the PEST sequence results in a more stable Blimp1 protein, and we previously used this version of Blimp1 in coimmunoprecipitation assays with Ets1 (8). In cells coexpressing either Ets1 or Ets2 and Blimp1, immunoprecipitation with an antibody to HA efficiently pulled down Blimp1, whereas a control rat IgG antibody failed to precipitate Blimp1 (Fig. 1B). Thus, Ets1 and Ets2 bind to Blimp1 equivalently in this coimmunoprecipitation assay, similar to what we saw previously using GST pulldown assays (8).
FIG 1.
Ets1 but not Ets2 regulates ASC formation. (A) Quantitative PCR analysis of the relative expression levels of Ets1 and Ets2 in resting B cells purified from the spleen of mice. The y axis represents total mRNA copies per cell calculated based on a standard curve with a known amount of Ets1 or Ets2 plasmid DNA. *, P < 0.05. (B) Coimmunoprecipitation (IP) of Ets1 and Ets2 with Blimp1. HA-tagged Ets1 or HA-tagged Ets2 together with FLAG-tagged Blimp1 lacking the PEST sequence (Blimp1Δ350–557) were transfected into Cos-1 cells. Forty-eight hours later, immunoprecipitation reactions were carried out by using either anti-FLAG antibody (Ab) or control mouse IgG followed by Western blotting (IB) with anti-HA antibody. Levels of transfected Ets1, Ets2, and Blimp1Δ350–557 in whole-cell lysates of the cells are shown at the side. (C) Quantitation of IgM- and IgG-secreting ASCs in the spleens and lymph nodes (LN) of wild-type (WT), Ets1−/−, or Ets2−/− mice using ELISPOT assays. Each dot represents an individual mouse. The tissue and type of ASCs analyzed are indicated at the top of each subpanel. (D) Ets1 and Ets2 retroviral constructs used for transduction experiments. LTR, long terminal repeat; IRES, internal ribosomal entry site. (E) ELISA of IgM levels in the supernatants of splenic B cells stimulated with CpG ODN and infected with empty retrovirus (MIGR1) or retroviruses encoding Ets1 or Ets2. Shown are the averages and SEM of 5 independent retroviral transduction experiments (****, P < 0.0001).
We next wished to determine the role of Ets2 in inhibiting ASC formation. Mice deficient in Ets1 have a 5- to 10-fold increase in the numbers of IgM-secreting ASCs in their lymphoid organs and elevated titers of serum IgM (23–25). If Ets2 plays a similar role in regulating ASC differentiation, we would predict elevated numbers of ASCs in the lymphoid organs of Ets2-deficient mice. To investigate this, we analyzed mice carrying a floxed allele of Ets2 crossed to Meox2-Cre mice (referred to as Ets2−/− mice in this report). Germ line Ets2-deficient mice exhibit a visible wavy-hair phenotype (20, 26), which was also found in the Meox2-Cre:Ets2flox/flox mice, confirming the deletion of Ets2 in these animals. Normal numbers of peripheral B cells of marginal zone and follicular types were present in Ets2−/− mice (not shown). ELISPOT analysis showed wild-type levels of IgM- and IgG-secreting ASCs in the spleen and lymph nodes of Ets2−/− mice, whereas levels of both IgM- and IgG-secreting ASCs were significantly elevated in Ets1−/− mice (Fig. 1C). These data indicate that loss of Ets2 in vivo does not lead to spontaneous differentiation of B cells into ASCs.
To further confirm that Ets1 plays a unique role in restraining ASC differentiation, we assessed the ability of Ets1 and Ets2 to modulate the differentiation of B cells stimulated with a synthetic TLR9 ligand (CpG ODN), a potent inducer of ASC differentiation. We infected splenic B cells stimulated with CpG ODN with retroviruses encoding Ets1 and Ets2 (Fig. 1D) and measured IgM levels in the supernatants of the cultures as a measure of ASC differentiation. Consistent with previous data, ectopic expression of Ets1 led to a significant block in IgM secretion. In stark contrast, ectopic expression of Ets2 failed to inhibit ASC differentiation and resulted in IgM secretion comparable to that of control transduced cells (Fig. 1E).
To determine whether Ets1 could block B cell differentiation in response to other TLR ligands, we stimulated splenic B cells with LPS and infected them with retroviral vectors expressing Ets1, Ets2, or a control empty vector. Similar to the results obtained with CpG ODN, Ets1 inhibited ASC formation in response to LPS (see Fig. S2 in the supplemental material). Ets2, on the other hand, showed only weak activity in inhibiting ASC differentiation in LPS-stimulated B cells. Thus, Ets1 acts downstream of multiple TLR signals to limit ASC formation.
Ets1 and Ets2 have differential effects on Pax5 expression in B cells.
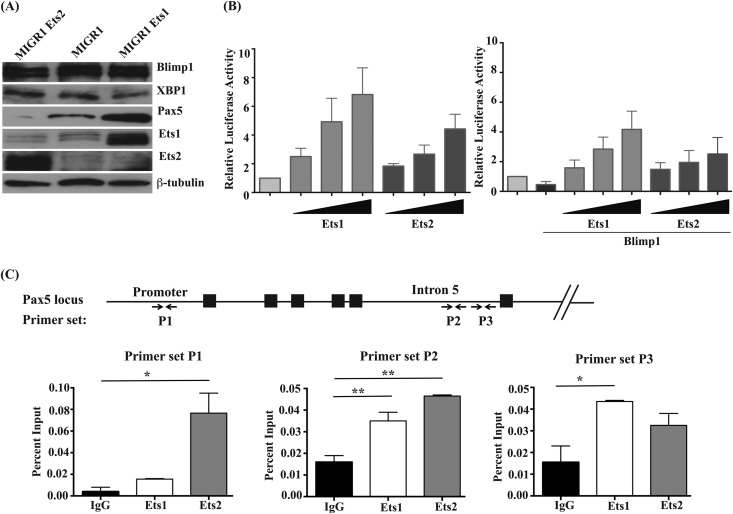
The data described above demonstrate that two closely related transcription factors have very different effects on B cell differentiation in response to TLR signals. However, Ets1 and Ets2 have nearly identical DNA binding Ets domains (96% amino acid identity) (see Fig. S3 in the supplemental material) and bind to the same target DNA sequences in EMSAs (11, 12). We sought to determine if Ets2 could regulate the expression of the Pax5 gene, which we have previously shown to be a target of Ets1 (8). As a first step, we determined whether Pax5 levels were sustained in B cells virally infected with an Ets2 construct, as we have seen previously with Ets1 viral constructs. As expected, viral expression of Ets1 did not alter the levels of Blimp1 induced by TLR9 ligand but led to a significant stimulation of Pax5 expression (Fig. 2A). In contrast, Ets2 failed to stimulate Pax5 expression in TLR9-stimulated B cells. Despite this, Ets1 and Ets2 were both capable of transactivating a reporter gene construct containing 1.8 kb of the murine Pax5 promoter in transient-transfection assays (Fig. 2B), although the degree of transactivation was somewhat weaker with Ets2 than with Ets1. Both Ets1 and Ets2 could also partially reverse Blimp1-dependent repression of the Pax5 promoter in transient-transfection assays (Fig. 2B).
FIG 2.
Ets1 but not Ets2 stimulates Pax5 expression in differentiating B cells, although both can transactivate the gene. (A) Western blot analysis of Blimp1, XBP-1, Pax5, Ets1, and Ets2 in retrovirally infected B cells. Levels of β-tubulin are shown as a loading control. (B) A20 B lymphoma cells were transfected with the BSAP-Luc reporter gene construct (containing bp −1771 to +50 of the mouse Pax5 gene promoter sequences fused to a firefly luciferase reporter). Cells were cotransfected with various concentrations of pCMV-HA-Ets1 or pCMV-HA-Ets2 and also cotransfected pCMV-βgal as an internal control. In some samples, cells were also cotransfected with pCDNA3.1 Blimp1. Shown are the averages of relative luciferase (luc) activities (after normalization to β-galactosidase) from three independent experiments. (C) Chromatin immunoprecipitation analysis using splenic B cells purified from wild-type mice. Chromatin was immunoprecipitated with polyclonal rabbit anti-Ets1 or anti-Ets2 antibodies. Quantitative PCR was performed with primers specific to the mouse Pax5 promoter sequences or the mouse Pax5 B cell-specific enhancer located in intron 5 (as shown by arrows at the bottom). The percentage of input chromatin was calculated for each primer set. Shown are the averages ± SEM of two separate chromatin immunoprecipitation experiments.
To further explore the interaction of Ets proteins with the Pax5 gene, we performed chromatin immunoprecipitation (ChIP) analyses using antibodies specific for Ets1 or Ets2 and primers located in either the promoter of Pax5 or the B cell-specific enhancer located in intron 5 of the Pax5 gene (Fig. 2C). We detected statistically significant binding of Ets2 to one site in the Pax5 promoter. In contrast, we did not observe statistically significant binding of Ets1 to the Pax5 gene promoter (although a non-statistically significant enrichment of Ets1 binding was detected with several primer sets [Fig. 2C and data not shown]). Significant Ets1 and Ets2 binding to sites in the Pax5 intronic enhancer was also found (Fig. 2C). Overall, both Ets1 and Ets2 bind to sequences in the Pax5 gene in vivo and transactivate the Pax5 promoter in transient-transfection assays. However, as noted above, only Ets1 can maintain high levels of Pax5 expression in virally infected B cells undergoing differentiation.
Conserved residues in the DNA binding domain of Ets1 are important for its activity in stimulating Pax5 expression and inhibiting ASC formation.
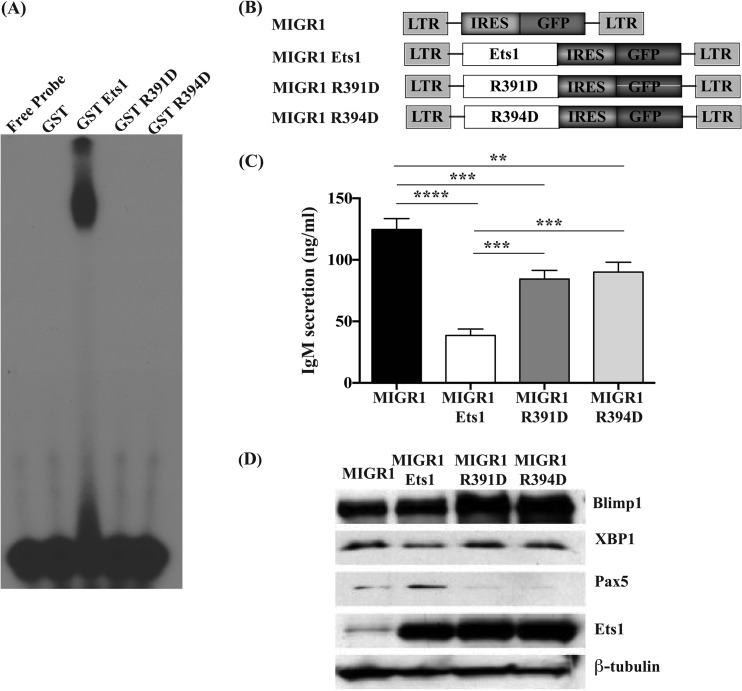
We have described two separate activities for Ets1 in regulating B cell differentiation: the ability of Ets1 to bind directly to target genes such as Pax5 and regulate their expression as well as the physical interaction of Ets1 with Blimp1 in a protein/protein complex that results in an inhibition of Blimp1's ability to bind to its target sites. It is unclear which of these activities is most crucial for Ets1 to inhibit ASC differentiation or if they both play partial roles. To explore whether or not direct binding of Ets1 to DNA is important for its activity in blocking ASC formation, we generated mutant versions of Ets1 in which we altered highly conserved arginine residues (R391 and R394) within the Ets domain that are found in helix H3 and known to interact with nucleotide bases within the core Ets target sequence GGAA/T (27–29). As expected, these mutations completely abolished Ets1 DNA binding in EMSAs (Fig. 3A).
FIG 3.
Conserved arginine residues in the Ets domain of Ets1 are important for regulation of Pax5 expression and for blocking of ASC formation. (A) EMSA using a known Ets1 binding site from the human MMP3 (stromelysin 1) promoter to confirm that the R391 and R394 mutations interfere with Ets1 binding to its target sites. (B) Constructs used in retroviral transductions. (C) Analysis of IgM secretion by CpG-stimulated B cells infected with wild-type Ets1 or the R391D or R394D mutation of Ets1. Shown are the averages and SEM of 3 independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (D) Western blot analysis of Blimp1, XBP-1, Pax5, and Ets1 in CpG-stimulated B cells infected with retroviruses encoding wild-type Ets1 or the R391D or R394D mutation of Ets1. β-Tubulin served as an internal loading control.
We cloned the mutant versions of Ets1 into a retroviral vector (Fig. 3B), infected splenic B cells, and measured IgM secretion in sorted GFP-positive cells induced to undergo differentiation with CpG ODN. Ectopic expression of wild-type Ets1 led to a strong reduction in IgM secretion, as expected (Fig. 3C). Both mutant constructs were less effective than wild-type Ets1 at inhibiting IgM secretion. Our data suggest that direct Ets1 binding to target sequences in DNA plays only a partial role in restraining ASC differentiation.
We next performed Western blot analysis on sorted GFP-positive retrovirally infected B cells. Pax5 expression was stimulated in wild-type Ets1-transduced cells (Fig. 3D). In contrast, Pax5 was barely detectable in R391D and R394D mutant-expressing cells (Fig. 3D). In accord with previous results, Blimp1 levels remained unaltered in cells ectopically expressing wild-type Ets1. Blimp1 levels were also unaltered in cells ectopically expressing the R391D and R394D mutants of Ets1.
Mutations in R391 and R394 have dual effects on Ets1 activity.
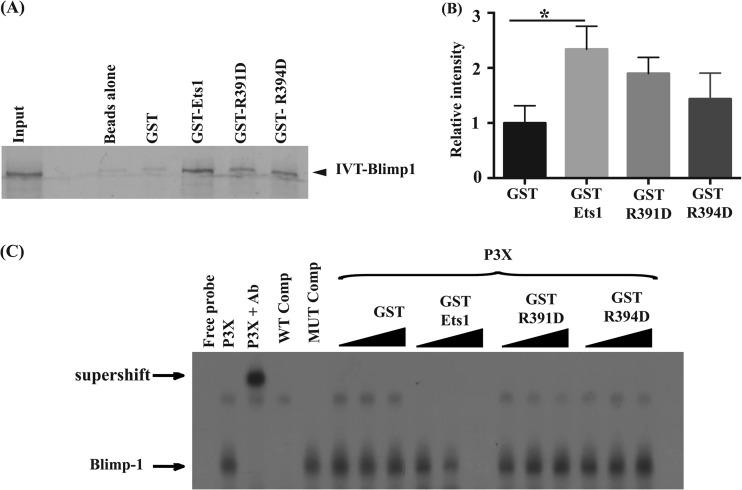
The R391 and R394 mutations completely abolish direct Ets1 binding to DNA and prevent upregulation of Pax5 expression, but intriguingly, they retained some activity in blocking ASC differentiation. This supports the idea that interaction of Ets1 with Blimp1 to block ASC formation might be important. To test this possibility, we assessed the ability of the R391D or R394D mutant version of Ets1 to interact with Blimp1 by using GST pulldown assays. As shown in Fig. 4A and B, the mutant forms of Ets1 were ∼70% as efficient as wild-type Ets1 in interacting with Blimp1 in GST pulldown assays. We next tested the ability of the mutants to inhibit Blimp1 DNA binding in EMSAs using an oligonucleotide probe harboring a Blimp1 binding site from the c-myc promoter and extracts from the plasmacytoma cell line P3X, which naturally contains large amounts of Blimp1. A specific Blimp1 DNA complex was seen with the P3X extracts, which was supershifted by the Blimp1 antibody (Fig. 4C). Wild-type Ets1 very efficiently inhibits Blimp1 binding in EMSAs. However, the mutant forms of Ets1 (R391D and R394D) failed to block Blimp1 binding, even at the highest concentrations tested in EMSAs (Fig. 4C).
FIG 4.
The conserved arginine residues of Ets1 are also required for its ability to block Blimp1 binding to target sites. (A) GST pulldown assays were performed by incubating in vitro-transcribed and -translated (IVT) 35S-labeled wild-type Blimp1 with glutathione-Sepharose alone (Beads) or with purified GST, GST-Ets1, or the GST–Ets1-R391D or GST–Ets1-R394D mutation. (B) Quantification of bands in GST pulldowns from 5 independent experiments. *, P < 0.05. (C) EMSA using the Blimp1 binding site from the c-myc promoter, using P3X lysates incubated with equal amounts of GST, GST-Ets1, GST–Ets1-R391D, or GST–Ets1-R394D.
Multiple domains of Ets1 outside the Ets domain are required to inhibit Blimp1 DNA binding activity.
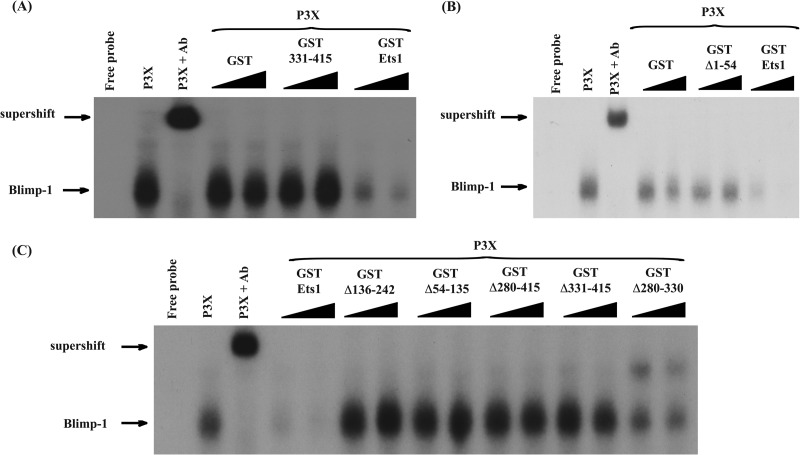
We have shown that the Ets domain is important for interactions with Blimp1, as a deletion mutant of Ets1 lacking this domain fails to bind to or inhibit Blimp1 (8). However, it is unclear if this domain alone is sufficient for the interaction. To test this, we incubated P3X extracts with increasing amounts of either a truncated form of Ets1 containing only the Ets domain (GST-Ets1 331–415), full-length GST-Ets1 as a positive control, or GST alone as a negative control (Fig. 5A). Increasing amounts of GST-Ets1 led to a reduction in Blimp1-DNA complex levels in a dose-dependent manner, whereas the complex remained unaffected in the presence of increasing amounts of GST-Ets1 331–415, indicating that the Ets domain by itself is not sufficient to block Blimp1 DNA binding (Fig. 5A). This is consistent with the fact that Ets2 (which has a nearly identical DNA binding domain) is also incapable of blocking Blimp1 binding (8).
FIG 5.
Multiple regions of Ets1 outside the Ets domain are required to inhibit Blimp1 binding to target sequences. (A) EMSA with a Blimp1 binding site from the c-myc promoter. A specific complex (Blimp1) was observed when P3X plasmacytoma extracts were used and could be supershifted with a Blimp1-specific antibody (supershift). Equivalent amounts of GST, GST-Ets1 (full-length Ets1), or GST 331–415 (encoding only the Ets domain) were used in the EMSA reactions. (B) EMSA analysis performed as described above for panel A, with equivalent amounts of GST, GST-Ets1, or GST–Ets1-Δ1–54. (C) EMSA analysis performed as described above for panel A, with equivalent amounts of GST, GST-Ets1, and various deletions of GST-Ets1 (Δ136–242 [lacking the acidic transactivation domain], Δ54–135 [lacking the Pointed domain], Δ280–415 [lacking the N-terminal autoinhibitory domain and Ets domain], Δ331–415 [lacking the Ets domain], and Δ280–330 [lacking the N-terminal autoinhibitory domain]) added to the indicated reaction mixtures. Note that addition of the Ets1 Δ280–330 form results in the appearance of a new band, which likely represents weak binding of the Ets1 protein to the c-myc probe in the absence of its autoinhibitory module.
These results imply that although the Ets domain is critical, regions outside the Ets domain that share less similarity between Ets1 and Ets2 are also involved in repressing Blimp1 DNA binding. To test this notion, we performed EMSAs using P3X extracts and recombinant proteins encoding either full-length Ets1 or various deletions of Ets1. As expected, Ets1 mutants lacking the Ets domain (GST Δ331–415 and GST Δ280–415) did not affect Blimp1 binding to DNA (Fig. 5C). Somewhat surprisingly, most of the other deletion mutants tested, including mutants lacking the N terminus of the Ets1 protein (GST Δ1–54), the Pointed domain (GST Δ54–135), and the acidic transactivation domain (GST Δ136–242), were also ineffective in inhibiting Blimp1 binding (Fig. 5B and C). A mutant lacking only the autoinhibitory module of Ets1 (GST Δ280–330) retained a partial inhibitory effect on Blimp1 binding but was not quite as effective as full-length wild-type Ets1 (Fig. 5C). Overall, our data show that the Ets domain of Ets1, while necessary for the interaction with Blimp1 in GST pulldown assays (8), is not sufficient to block Blimp1 binding to its target sites and suggest a role for multiple regions outside the Ets domain in inhibiting Blimp1 activity.
The N terminus, acidic transactivation domain, and Pointed domain of Ets1 are all important for maximal activity in inhibiting ASC differentiation.
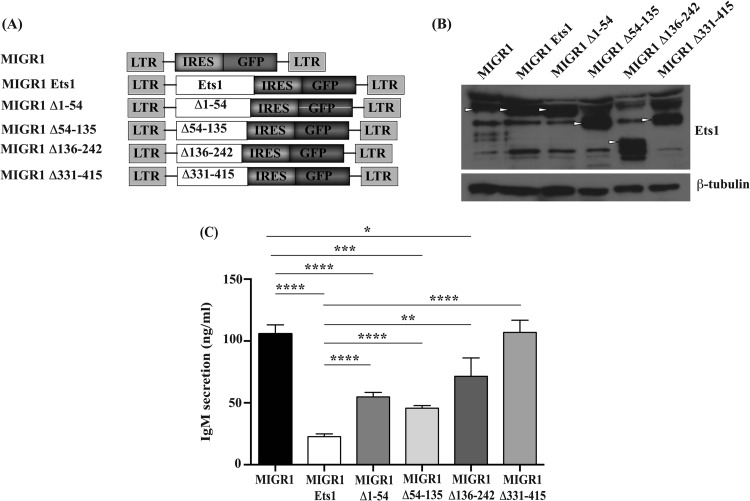
Based on the EMSA results, several regions of Ets1 are required for maximal inhibition of Blimp1 binding to target sites, including the Ets DNA binding domain, the Pointed domain, the acidic transactivation domain, and the N terminus, whereas the autoinhibitory module is largely dispensable. To determine whether the N terminus, the Pointed domain, or the acidic transactivation domain are also critical for blocking ASC formation in vivo, we subcloned versions of Ets1 lacking these domains into a retroviral vector to generate MIGR1 Δ1–54, MIGR1 Δ54–135, and MIGR1 Δ136–242, lacking the N terminus, the Pointed domain, and the transactivation domain, respectively (Fig. 6A). As a control, we also used a retroviral construct of Ets1 lacking the Ets domain (MIGR1 Δ331–415), which we have shown previously to be impaired in blocking ASC formation (8). Splenic B cells stimulated with CpG ODN were infected with retroviruses harboring full-length Ets1 or various deletions of Ets1. Western blot analysis showed that full-length Ets1 and deletions of Ets1 were expressed at similar levels (Fig. 6B).
FIG 6.
The N terminus, acidic transactivation domain, and Pointed domain of Ets1 are important for inhibiting ASC formation. (A) Diagram of various retroviral constructs used to infect B cells (Δ1–54 [lacking the flexible N terminus of Ets1], Δ54–135 [lacking the Pointed domain], Δ136–242 [lacking the acidic transactivation domain], and Δ331–415 [lacking the Ets domain]). (B) Western blot analysis to demonstrate expression levels of the various deletion mutants of Ets1. Arrowheads point to Ets1 bands in the gel. (C) IgM secretion from splenic B cells stimulated with CpG ODN and infected with a retrovirus encoding full-length Ets1 or various deletions of Ets1 or an empty retrovirus. Cells were cultured and IgM secretion was measured as described in the legend of Fig. 1E. Shown are the averages and SEM of 3 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To determine the ability of the deletions of Ets1 to suppress ASC differentiation, we sorted GFP-positive cells from each of the retrovirally transduced populations, returned them to culture for 48 h, and then measured IgM secretion by ELISA. Deletion mutants lacking the Pointed domain, the acidic transactivation domain, or the N terminus of Ets1 were less effective at inhibiting IgM secretion, suggesting that these domains are all important for maximal activity of Ets1 in blocking ASC formation (Fig. 6C). These particular mutants of Ets1 showed an intermediate ability to inhibit ASC differentiation between the effects of an empty retroviral vector (or a mutant lacking the Ets domain of Ets1) and full-length Ets1. Altogether, our data demonstrated that the Ets domain is essential for repressing ASC differentiation, whereas the N terminus, acidic transactivation domain, and Pointed domain are required for maximal activity.
Neither the MAP kinase phosphorylation site nor the SUMOylation site of Ets1 is required for inhibiting Blimp1 binding or ASC differentiation.
The results described above demonstrate that the N terminus and Pointed domains of Ets1 are important for maximal activity in blocking ASC formation. The Pointed domain of Ets1 is known to be involved in recruiting Erk MAP kinase (30), which subsequently phosphorylates a conserved threonine residue (T38) found in the N-terminal region of Ets1 (31, 32). Phosphorylation of this threonine residue results in recruitment of the coactivator protein CBP to Ets1 and stimulates its transcriptional activity (14). Thus, an Ets1 mutant lacking the Pointed domain is predicted to have reduced or absent recruitment of Erk kinases, reduced phosphorylation of T38, and a reduced ability to activate target genes such as Pax5. An Ets1 mutant lacking the N terminus of Ets1 (amino acids 1 to 54) lacks the Erk phosphorylation site and thus cannot be phosphorylated and will be less effective at activating target genes. The N terminus of Ets1 also contains a lysine residue (K15) that is subjected to modification by SUMOylation (33–35). SUMOylation of Ets1 inhibits its transactivation potential (33, 35). The recruitment of SUMOylation enzymes to Ets1 does not depend on interactions with the Pointed domain (34). Hence, the Δ54–135 mutant of Ets1 is predicted to become SUMOylated, whereas the Δ1–54 mutant of Ets1 cannot.
To determine whether phosphorylation and/or SUMOylation of Ets1 played a role in inhibiting ASC differentiation, we generated retroviruses encoding point mutations in T38 (MIGR1-T38A) and K15 (MIGR1-K15A) and performed retroviral infections in primary B cells as described above (see Fig. S4A in the supplemental material). Both the T38A and K15A mutants were expressed at levels similar to those of wild-type Ets1 (data not shown; see also Fig. S4B in the supplemental material). Expression of T38A or K15A mutants was as effective as wild-type Ets1 in blocking IgM secretion, suggesting that neither phosphorylation nor SUMOylation of Ets1 was required for this process (see Fig. S4C in the supplemental material). Therefore, both the Pointed domain and the N terminus of Ets1 are required for maximal Ets1 activity in inhibiting ASC differentiation, but known posttranslational modifications of this region appear to play no role.
DISCUSSION
We explored the differing biological roles of two closely related Ets family transcription factors (Ets1 and Ets2) in regulating B cell differentiation into ASCs. All members of the Ets gene family contain an evolutionarily conserved DNA binding domain. More than a dozen members of the Ets gene family are typically expressed in the same cell type at the same time (22), and there is an overlap in recognition sequences. Chromatin immunoprecipitation studies indicate that some Ets binding sites (EBSs) in the genome are redundantly occupied by multiple members of the Ets family (36). However, gene knockout studies have identified largely different biological roles for these Ets factors (37, 38), although in some cases, functional redundancy has also been identified (39–41). Ets1 and Ets2 share almost identical DNA binding domains, and their DNA binding specificity cannot be distinguished by in vitro assays (11, 12). Indeed, there is evidence of functional redundancy between these two factors in regulating vascular development (41). However, the reported phenotypes of the individual Ets1- and Ets2-deficient mice are largely different (23, 24, 26). In this study, we have demonstrated that Ets2 knockout mice do not exhibit the spontaneous differentiation to ASCs that is characteristic of Ets1-deficient mice.
Some of the differences in phenotype of Ets1 and Ets2 knockout mice might be explained by differences in expression levels of Ets1 and Ets2 in various cell types. Ets1 is highly expressed within lymphoid cells and is expressed at lower levels in other tissues of adult mice (16, 42). Ets2, in contrast, is about 10 times less abundant in B cells but is more broadly expressed in a variety of tissues (16, 42). Based on the fact that Ets1 is expressed at significantly higher levels in resting B cells than is Ets2, it could be argued that Ets1 has a unique role in this population simply due to its higher expression level rather than any difference in the biochemical properties of Ets1 and Ets2. However, we have shown that retrovirus-dependent expression of Ets2 in B cells at high levels fails to block ASC formation, while Ets1 is very effective at blocking differentiation in this assay.
When B cells undergoing differentiation to ASCs are infected with a retrovirus driving expression of Ets1, the levels of the key B cell transcription factor Pax5 remain elevated instead of decreasing as expected. The maintenance of high levels of Pax5 by Ets1 is particularly significant, since it has been shown that downregulation of Pax5 is a pivotal event in the commitment to the ASC fate (43). In contrast, in cells infected with a retrovirus expressing Ets2, Pax5 levels are not sustained. Transient-transfection and ChIP assays confirm direct regulation of the Pax5 gene by Ets1. However, in these same assays, Ets2 also appeared to directly regulate Pax5 expression, despite the fact that overexpression of Ets2 in B cells failed to sustain Pax5 expression. One potential explanation for the unique role of Ets1 in maintaining Pax5 expression is that Ets1, but not Ets2, blocks DNA binding by Blimp1 (8). Blimp1 suppresses expression of the Pax5 gene (44). Since Ets2 cannot block Blimp1 DNA binding, it may be incapable of maintaining stable expression of Pax5 during B cell differentiation.
The ability of Ets1 to stably upregulate Pax5 likely contributes to its ability to block ASC differentiation. However, Ets1 can also interfere with Blimp1 activity, and Blimp1 is required for ASC differentiation. Hence, it is possible that Ets1 would be able to block ASC differentiation, even if it was unable to bind DNA and regulate target genes. To assess this possibility, we generated mutant versions of Ets1 in which two highly conserved arginine residues (R391 and R394) were mutated to aspartic acid. Mutation of these arginine residues completely blocks direct binding of Ets1 to DNA in EMSAs. The mutant versions of Ets1 have a reduced capacity to inhibit ASC formation. This initially suggested to us that direct binding of Ets1 to DNA is important for its activity in regulating ASC formation. However, subsequent analysis showed that the R391D and R394D Ets1 proteins are also less effective at interacting with Blimp1 and are unable to inhibit Blimp1 binding to its target sequences. Thus, these particular mutations do not allow us to separate the two functions of Ets1 or to test whether direct regulation of target genes by Ets1 is crucial for its activity. Separating these functions of Ets1 will require the identification of mutants that disrupt one process (i.e., Ets1 binding to its target sites in DNA or Ets1 interaction with Blimp1) without affecting the other process. Studies are under way to identify such mutants.
A deletion mutant of Ets1 that lacks the entire Ets DNA binding domain is completely ineffective in blocking B cell differentiation (8). However, the R391 and R394 mutant versions of Ets1 retain a partial activity in blocking ASC differentiation despite the fact that that they cannot bind to Ets target sites in DNA and are also slightly impaired in interactions with Blimp1 in in vitro assays. How, then, might the mutant proteins function to partially suppress ASC formation? One possibility is that mutant Ets1 proteins are recruited to promoters or enhancers of target genes in vivo through interactions with other transcription factors. Alternatively, the mutants may retain some weak activity in blocking Blimp1 binding in vivo. The mutant proteins interact more weakly with Blimp1 in GST pulldowns than does wild-type Ets1, and this appears to be insufficient to inhibit Blimp1 binding in EMSAs. However, these interactions may contribute to blocking of ASC formation in vivo. Finally, it is also possible that the Ets domain of Ets1 interacts with transcription factors (other than Blimp1) that regulate B cell differentiation in a manner that does not require either R391 or R394. Future experiments will address these various possibilities.
The above-mentioned data indicate that the conserved Ets DNA binding domain of Ets1 is crucial for the interaction with Blimp1 and modulation of its activity. However, a truncated Ets1 protein containing only the Ets domain of Ets1 failed to block Blimp1 binding activity, indicating that although the Ets domain is important, it is not sufficient for this process. By examining a series of deletion constructs, we determined that several additional domains of Ets1 (the N terminus, Pointed domain, and acidic transactivation domain) all contribute to blocking of Blimp1 binding to its target sites in EMSAs. The N-terminal 54 amino acids of Ets1 are flexible and unstructured (45) but contain a conserved threonine residue (T38) that is known to undergo phosphorylation (13, 31) and a conserved lysine (K15) that is known to undergo SUMOylation (33–35). Despite the reported functional significance of these residues, neither one is required for the ability of Ets1 to inhibit ASC formation.
In addition to the N terminus of the protein, the Pointed domain and acidic transactivation domains of Ets1 are also required for the maximal activity of Ets1 in blocking Blimp1 binding to target sequences in EMSAs and in blocking ASC formation in cultured B cells differentiating in response to TLR ligands. It is unclear why such a large portion of the protein (amino acids 1 to 242) is needed for inhibiting Blimp1 binding in EMSAs when only the Ets domain of Ets1 is required for interactions with Blimp1 in GST pulldown assays. However, we suggest that the Ets domain is the main portion of Ets1 that mediates interactions with Blimp1, while other sequences of the protein may fold in such a way to block the zinc finger region of Blimp1 from accessing its DNA substrate. Perhaps only a small segment of Ets1 outside the Ets domain is needed for blocking Blimp1 binding, but large deletions result in incorrect positioning of the required domain. Another possible model that would explain the requirement for multiple domains of Ets1 in vivo to block ASC formation is that these domains are needed for Ets1 to regulate specific target genes such as Pax5, perhaps by interacting with other transcription factors or coactivators of transcription.
We have demonstrated that two members of the Ets multigene family, Ets1 and Ets2, that share nearly identical DNA binding regions have different activities in regulating ASC formation. Recent evidence implicates a third Ets transcription factor, Spi-B, in controlling ASC formation (46). Hence, it appears that there are subsets of Ets transcription factors (including Ets1 and Spi-B) that control terminal B cell differentiation to ASCs and other subsets (including Ets2) that appear not to regulate this process. It will be important to understand how this functional dichotomy arises in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank Satrajit Sinha for helpful comments and Irene Kulik for technical assistance. We also thank Michael Ostrowski for providing antibody to Ets2 used in Western blotting and Michael Buck for advice on chromatin immunoprecipitation experiments.
These studies were supported by research grants from the Lupus Research Institute and the National Institutes of Health (AI085127).
Footnotes
Published ahead of print 25 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00612-13.
REFERENCES
- 1.Nutt SL, Fairfax KA, Kallies A. 2007. BLIMP1 guides the fate of effector B and T cells. Nat. Rev. Immunol. 7:923–927. 10.1038/nri2204 [DOI] [PubMed] [Google Scholar]
- 2.John SA, Garrett-Sinha LA. 2009. Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp. Cell Res. 315:1077–1084. 10.1016/j.yexcr.2008.11.015 [DOI] [PubMed] [Google Scholar]
- 3.Iwakoshi NN, Lee AH, Glimcher LH. 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194:29–38. 10.1034/j.1600-065X.2003.00057.x [DOI] [PubMed] [Google Scholar]
- 4.Lu R. 2008. Interferon regulatory factor 4 and 8 in B-cell development. Trends Immunol. 29:487–492. 10.1016/j.it.2008.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin L, Gerth AJ, Peng SL. 2004. Active inhibition of plasma cell development in resting B cells by microphthalmia-associated transcription factor. J. Exp. Med. 200:115–122. 10.1084/jem.20040612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobaleda C, Schebesta A, Delogu A, Busslinger M. 2007. Pax5: the guardian of B cell identity and function. Nat. Immunol. 8:463–470. 10.1038/ni1454 [DOI] [PubMed] [Google Scholar]
- 7.Igarashi K, Ochiai K, Muto A. 2007. Architecture and dynamics of the transcription factor network that regulates B-to-plasma cell differentiation. J. Biochem. 141:783–789. 10.1093/jb/mvm106 [DOI] [PubMed] [Google Scholar]
- 8.John SA, Clements JL, Russell LM, Garrett-Sinha LA. 2008. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J. Biol. Chem. 283:951–962. 10.1074/jbc.M705262200 [DOI] [PubMed] [Google Scholar]
- 9.Russell L, Garrett-Sinha LA. 2010. Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine 51:217–226. 10.1016/j.cyto.2010.03.006 [DOI] [PubMed] [Google Scholar]
- 10.Garrett-Sinha LA. 2013. Review of Ets1 structure, function, and roles in immunity. Cell. Mol. Life Sci. 70:3375–3390. 10.1007/s00018-012-1243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woods DB, Ghysdael J, Owen MJ. 1992. Identification of nucleotide preferences in DNA sequences recognised specifically by c-Ets-1 protein. Nucleic Acids Res. 20:699–704. 10.1093/nar/20.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ascione R, Thompson DM, Thomas R, Panayiotakis A, Ramsay R, Tymms M, Seth A. 1992. Influence of nucleotides flanking the -GGAA- core sequence on ETS1 and ETS2 DNA-binding activity and the mechanism of ETS1 autoregulation. Int. J. Oncol. 1:631–637 [DOI] [PubMed] [Google Scholar]
- 13.Wasylyk C, Bradford AP, Gutierrez-Hartmann A, Wasylyk B. 1997. Conserved mechanisms of Ras regulation of evolutionary related transcription factors, Ets1 and Pointed P2. Oncogene 14:899–913. 10.1038/sj.onc.1200914 [DOI] [PubMed] [Google Scholar]
- 14.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. 2004. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 24:10954–10964. 10.1128/MCB.24.24.10954-10964.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. 2007. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179:6808–6819 http://www.jimmunol.org/content/179/10/6808.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kola I, Brookes S, Green AR, Garber R, Tymms M, Papas TS, Seth A. 1993. The Ets1 transcription factor is widely expressed during murine embryo development and is associated with mesodermal cells involved in morphogenetic processes such as organ formation. Proc. Natl. Acad. Sci. U. S. A. 90:7588–7592. 10.1073/pnas.90.16.7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naviaux RK, Costanzi E, Haas M, Verma IM. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muthusamy N, Barton K, Leiden JM. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377:639–642. 10.1038/377639a0 [DOI] [PubMed] [Google Scholar]
- 19.Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA. 2005. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int. Immunol. 17:1179–1191. 10.1093/intimm/dxh295 [DOI] [PubMed] [Google Scholar]
- 20.Wen F, Tynan JA, Cecena G, Williams R, Munera J, Mavrothalassitis G, Oshima RG. 2007. Ets2 is required for trophoblast stem cell self-renewal. Dev. Biol. 312:284–299. 10.1016/j.ydbio.2007.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carotta S, Nutt SL. 2008. Losing B cell identity. Bioessays 30:203–207. 10.1002/bies.20725 [DOI] [PubMed] [Google Scholar]
- 22.Hollenhorst PC, Jones DA, Graves BJ. 2004. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 32:5693–5702. 10.1093/nar/gkh906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity 9:555–563. 10.1016/S1074-7613(00)80638-X [DOI] [PubMed] [Google Scholar]
- 24.Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. 1995. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature 377:635–638. 10.1038/377635a0 [DOI] [PubMed] [Google Scholar]
- 25.Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-Defranoux O, Bandeira A, Bories JC. 2010. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J. Exp. Med. 207:2113–2125. 10.1084/jem.20092153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. 1998. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 12:1315–1326. 10.1101/gad.12.9.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werner MH, Clore GM, Fisher CL, Fisher RJ, Trinh L, Shiloach J, Gronenborn AM. 1997. Correction of the NMR structure of the ETS1/DNA complex. J. Biomol. NMR 10:317–328. 10.1023/A:1018399711996 [DOI] [PubMed] [Google Scholar]
- 28.Garvie CW, Hagman J, Wolberger C. 2001. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell 8:1267–1276. 10.1016/S1097-2765(01)00410-5 [DOI] [PubMed] [Google Scholar]
- 29.Obika S, Reddy SY, Bruice TC. 2003. Sequence specific DNA binding of Ets-1 transcription factor: molecular dynamics study on the Ets domain-DNA complexes. J. Mol. Biol. 331:345–359. 10.1016/S0022-2836(03)00726-5 [DOI] [PubMed] [Google Scholar]
- 30.Seidel JJ, Graves BJ. 2002. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16:127–137. 10.1101/gad.950902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang BS, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. 1996. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 16:538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson ML, Kang HS, Lee GM, Blaszczak AG, Lau DK, McIntosh LP, Graves BJ. 2010. Ras signaling requires dynamic properties of Ets1 for phosphorylation-enhanced binding to coactivator CBP. Proc. Natl. Acad. Sci. U. S. A. 107:10026–10031. 10.1073/pnas.0915137107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji Z, Degerny C, Vintonenko N, Deheuninck J, Foveau B, Leroy C, Coll J, Tulasne D, Baert JL, Fafeur V. 2007. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 26:395–406. 10.1038/sj.onc.1209789 [DOI] [PubMed] [Google Scholar]
- 34.Macauley MS, Errington WJ, Scharpf M, Mackereth CD, Blaszczak AG, Graves BJ, McIntosh LP. 2006. Beads-on-a-string, characterization of ETS-1 sumoylated within its flexible N-terminal sequence. J. Biol. Chem. 281:4164–4172. 10.1074/jbc.M510488200 [DOI] [PubMed] [Google Scholar]
- 35.Nishida T, Terashima M, Fukami K. 2006. PIASy-mediated repression of the Ets-1 is independent of its sumoylation. Biochem. Biophys. Res. Commun. 345:1536–1546. 10.1016/j.bbrc.2006.05.065 [DOI] [PubMed] [Google Scholar]
- 36.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21:1882–1894. 10.1101/gad.1561707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallant S, Gilkeson G. 2006. ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. (Warsz.) 54:149–163. 10.1007/s00005-006-0017-z [DOI] [PubMed] [Google Scholar]
- 38.Hollenhorst PC, McIntosh LP, Graves BJ. 2011. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80:437–471. 10.1146/annurev.biochem.79.081507.103945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrett-Sinha LA, Dahl R, Rao S, Barton KP, Simon MC. 2001. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood 97:2908–2912. 10.1182/blood.V97.9.2908 [DOI] [PubMed] [Google Scholar]
- 40.Garrett-Sinha LA, Su GH, Rao S, Kabak S, Hao Z, Clark MR, Simon MC. 1999. PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity 10:399–408. 10.1016/S1074-7613(00)80040-0 [DOI] [PubMed] [Google Scholar]
- 41.Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, Muthusamy N, Man AK, Oshima RG, Leone G, Ostrowski MC. 2009. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood 114:1123–1130. 10.1182/blood-2009-03-211391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maroulakou IG, Papas TS, Green JE. 1994. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene 9:1551–1565 [PubMed] [Google Scholar]
- 43.Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM, Nutt SL. 2007. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity 26:555–566. 10.1016/j.immuni.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 44.Lin KI, Angelin-Duclos C, Kuo TC, Calame K. 2002. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol. Cell. Biol. 22:4771–4780. 10.1128/MCB.22.13.4771-4780.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slupsky CM, Gentile LN, Donaldson LW, Mackereth CD, Seidel JJ, Graves BJ, McIntosh LP. 1998. Structure of the Ets-1 pointed domain and mitogen-activated protein kinase phosphorylation site. Proc. Natl. Acad. Sci. U. S. A. 95:12129–12134. 10.1073/pnas.95.21.12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidlin H, Diehl SA, Nagasawa M, Scheeren FA, Schotte R, Uittenbogaart CH, Spits H, Blom B. 2008. Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood 112:1804–1812. 10.1182/blood-2008-01-136440 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.