Abstract
The integrin-activated Src-focal adhesion kinase (FAK) kinase complex phosphorylates PTPα at Tyr789, initiating PTPα-mediated signaling that promotes cell migration. Recruitment of the BCAR3-Cas complex by PTPα-phospho-Tyr789 at focal adhesions is one mechanism of PTPα signaling. The adaptor protein Grb2 is also recruited by PTPα-phospho-Tyr789, although the role of the PTPα-Grb2 complex in integrin signaling is unknown. We show that silencing Grb2 expression in fibroblasts abolishes PTPα-Tyr789 phosphorylation and that this is due to two unexpected actions of Grb2. First, Grb2 promotes integrin-induced autophosphorylation of FAK-Tyr397. This is impaired in Grb2-depleted cells and prohibits FAK activation and formation of the Src-FAK complex. Grb2-depleted cells contain less paxillin, and paxillin overexpression rescues FAK-Tyr397 phosphorylation, suggesting that the FAK-activating action of Grb2 involves paxillin. A second distinct role for Grb2 in PTPα-Tyr789 phosphorylation involves Grb2-mediated coupling of Src-FAK and PTPα. This requires two phosphosites, FAK-Tyr925 and PTPα-Tyr789, for Grb2-Src homology 2 (SH2) binding. We propose that a Grb2 dimer links FAK and PTPα, and this positions active Src-FAK in proximity with other, perhaps integrin-clustered, molecules of PTPα to enable maximal PTPα-Tyr789 phosphorylation. These findings identify Grb2 as a new FAK activator and reveal its essential role in coordinating PTPα tyrosine phosphorylation to enable downstream integrin signaling and migration.
INTRODUCTION
Integrins are heterodimeric receptor proteins that link the extracellular matrix (ECM) to the cytoskeleton to regulate cell shape, migration, and survival. Binding of the integrins to ECM ligands triggers the formation of focal adhesions, multiprotein signaling complexes that link the integrin cytoplasmic tails with the actin cytoskeleton (1, 2). Reversible protein tyrosine phosphorylation, catalyzed by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs), is an important mechanism controlling focal adhesion signaling and turnover to regulate cell movement (3, 4).
Focal adhesion kinase (FAK) is a central PTK involved in integrin signaling. Its recruitment to the integrin cytoplasmic tail and phosphorylation at Tyr397 are early events upon integrin engagement by the ECM (5, 6). FAK-phospho-Tyr397 serves as a docking site for Src family tyrosine kinases (SFKs) such as Src and Fyn (7, 8). Src, the best-studied SFK in this process, phosphorylates several sites in FAK, including two within the kinase domain activation loop that promote optimal FAK activation (9–11). The fully activated Src-FAK complex phosphorylates other proteins, including p130Cas (Cas) and paxillin, to promote signaling that orchestrates focal adhesion formation and disassembly, cytoskeletal reorganization, and migration (12, 13).
PTPα (PTPRA) is a classical tyrosine-specific receptor-like PTP that is involved in integrin proximal signaling events. It transiently colocalizes with at least one integrin heterodimer, αvβ3, via association with the αv subunit following activation with fibronectin (FN) or vitronectin (14). In FN-stimulated fibroblasts, PTPα dephosphorylates and activates Src and Fyn, and this is required for FAK-Tyr397 phosphorylation, SFK-FAK association, and full activation of the SFK-FAK kinase complex. These events and the associated processes of focal adhesion formation and cytoskeletal rearrangement that are required for cell spreading and migration are impaired in PTPα-null fibroblasts (14–16). In addition to this upstream signaling role, PTPα also functions downstream of the SFK-FAK complex, as PTPα itself is phosphorylated by activated SFK-FAK at a site in its C-terminal tail region, Tyr789 (17). The expression of a catalytically active but unphosphorylatable mutant (Y789F) PTPα in PTPα-null fibroblasts rescues the defective SFK and FAK activation observed in the absence of PTPα. Nevertheless, the cells still display delayed cell spreading and migration, indicating that PTPα-Tyr789 phosphorylation is required for additional downstream signaling events that promote effective cell movement (17).
We recently identified a mechanism that links PTPα-phosphoTyr789 to integrin-stimulated cell migration, demonstrating that the protein breast cancer antiestrogen resistance 3 (BCAR3) couples phosphorylated PTPα in focal adhesions to p130Cas (Cas), a critical regulator of cell movement (18). The Src homology 2 (SH2) domain of BCAR3 directly binds to PTPα-phospho-Tyr789, promoting the recruitment of BCAR3 and BCAR3-associated Cas to focal adhesions. This situates Cas for optimal interaction with and phosphorylation by Src, enhancing Cas-mediated downstream signaling.
Two other SH2 domain-containing proteins, Src and Grb2, can bind to PTPα-phospho-Tyr789. Src-SH2 binding to PTPα-phospho-Tyr789 displaces the kinase-inhibitory intramolecular interaction between the SH2 domain of Src and the phosphoTyr527 site in the tail region of Src, exposing Src phospho-Tyr527 for dephosphorylation by PTPα and resulting in Src activation (19). This mode of Src activation may be utilized in mitosis (20); however, it is not essential for PTPα-catalyzed Src activation in integrin signaling since this is supported equally well by mutant PTPα-Y789F (17).
Grb2 is an adaptor protein with a central SH2 domain and two flanking SH3 domains. Its interaction with PTPα is mediated by the SH2 and C-terminal SH3 (C-SH3) domains, through their respective binding to PTPα-phospho-Tyr789 and a small region located near the C-terminal end of the PTPα membrane proximal (D1) catalytic domain (21–24). Interestingly, while the N-terminal SH3 (N-SH3) domain of Grb2 is a well-characterized binding partner of Sos, a Ras-guanine-nucleotide exchange factor (GEF) that activates Ras/mitogen-activated protein kinase (MAPK) signaling, PTPα-bound Grb2 does not complex with Sos (21, 24). It has been variously proposed that the binding of Grb2 to PTPα may protect PTPα-phospho-Tyr789 from being dephosphorylated, mediate PTPα downstream signaling or localization, function to block PTPα activity, or sequester Grb2 to attenuate Grb2/Sos-mediated signaling (21–24). However, the actual function of PTPα-bound Grb2 remains unclear.
We find that integrin stimulation induces the binding of Grb2 to PTPα-phospho-Tyr789. The approach of silencing Grb2 expression in fibroblasts was used to investigate its role in integrin-induced PTPα-mediated signaling. Our study reveals that the depletion of Grb2 caused a significant reduction of PTPα-Tyr789 phosphorylation that is not due to deprotection and hyperdephosphorylation of PTPα but to distinguishable requirements for Grb2 in FAK activation and ensuing PTPα-Tyr789 phosphorylation. Thus, Grb2 plays unexpected essential roles in the initiation of key integrin signaling events.
MATERIALS AND METHODS
Antibodies and immunological detection reagents.
PTPα and PTPα-phospho-Tyr789 antibodies have been described previously (17). Antibodies to FAK, Grb2, paxillin, and Shp2 were from BD Transduction Laboratories. Anti-vesicular stomatitis virus glycoprotein (anti-VSVG), -actin, and -Flag antibodies were from Sigma-Aldrich. Antibody to Src (v-Src) was from Calbiochem. Antibody to BCAR3 was from Bethyl Laboratories, Inc. Antibody specific to the SH2 domain of Grb2 was from R&D Systems. Anti-myc and phosphorylation site-specific antibodies to Src-Tyr416 were from Cell Signaling, and those to FAK-Tyr397 and -Tyr576/577 were from Invitrogen. Alexa Fluor 488-conjugated anti-rabbit IgG and Alexa Fluor 594-conjugated anti-mouse IgG were from Molecular Probes. Anti-Grb2 (polyclonal rabbit) antibody (Santa Cruz Biotechnology, Inc.) and the Clean Blot IP detection reagent (horseradish peroxidase [HRP]) from Thermo Scientific were specifically used for the detection of Grb2 in VSVG immunoprecipitates.
Cell culture and treatments.
Wild-type (PTPα+/+) and PTPα-null (PTPα−/−) mouse embryo fibroblasts (MEFs) (16), FAK-null (FAK−/−) MEFs, and FAK-null MEFs reconstituting GFP-FAK-WT, GFP-FAK-Y397F, and GFP-FAK-Y925F have been described (25, 26). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin. Prior to integrin stimulation, cells were starved in DMEM containing 0.5% FBS for 18 h. They were then trypsinized with 0.05% trypsin-EDTA (Invitrogen), and the trypsinization was stopped by the addition of 0.5 mg/ml soybean trypsin inhibitor (Invitrogen)–DMEM. The cells were pelleted and washed once again with 0.5 mg/ml soybean trypsin inhibitor, followed by another wash with serum-free medium. They were resuspended in DMEM containing 0.1% bovine serum albumin (BSA) and maintained in suspension for 1 h at 37°C. To engage integrins, suspended cells (105 cells/ml) were plated onto FN-coated plates and incubated at 37°C for 15 min (unless otherwise specified) before harvesting. In one experiment, the suspended cells were treated with 100 μM NSC 87877 (Shp1/Shp2 inhibitor; Santa Cruz) for 1 h prior to FN stimulation. The FN-coated dishes were prepared by adding 10 μg/ml FN (Chemicon International, Inc.) in phosphate-buffered saline (PBS) overnight at 4°C. For serum stimulation, adherent MEFs were starved in DMEM containing 0.5% FBS for 18 h and treated with 10% FBS for 30 s, 1 min, 2 min, and 5 min prior to harvesting.
Generation of siRNA-resistant Grb2 expression plasmids.
The plasmid pSVE-Grb2-myc-P49L (Addgene, Cambridge, MA) containing full-length Grb2 with a point mutation (P49L) in the N-SH3 domain, and with a C-terminal myc tag, was used as a template in a PCR employing modified primers in order to generate a small interfering RNA (siRNA)-resistant Grb2 coding sequence with a 5′ BamHI site and a 3′ HindIII site. The forward (F1, 5′-ATCATGGATCCGTCATGGAGGCGATTGCGAAATATG-3′) and reverse (R1, 5′-AGCGTAAGCTTTCACAAGTCTTCTTCAGAAATAAGCTTTTGTTCG-3′) primers contained nucleotide substitutions (underlined in the F1 primer sequences) that confer resistance to the Grb2-targeted siRNA that was used in experiments and nucleotides 5′ to the start codon (in italics in the F1 sequence above) that also did not match the Grb2 siRNA. The PCR product was cloned into pcDNA3.1(−) vector that had been digested with BamHI and HindIII, generating the expression plasmid pcDNA3.1(−)-Grb2-myc-P49L. Site-directed mutagenesis using overlap extension PCR was performed using the pSVE-Grb2-myc-P49L template to revert the N-SH3 mutation to the wild-type (WT) Grb2 sequence (changing the mutated Lys to Pro). Separate PCRs were performed to produce two pieces of the complete Grb2-myc that overlapped in sequence around the mutated region, using the primer pairs F1 and R2 (5′-TATGTAGTTCTGGGAATGAAGCCGTC-3′, with mutated bases underlined) and F2 (5′-GACGGCTTCATTCCCAAGAACTACATA-3′, with mutated bases underlined) and R1. The resulting PCR products were gel purified and combined to use as the templates in another PCR using primers F1 and R1 to generate WT-Grb2-myc. This was cloned into BamHI- and HindIII-digested pcDNA3.1(−) to produce the expression plasmid pcDNA3.1(−)-WT-Grb2-myc. The same procedure was used to generate the plasmids pcDNA3.1(−)-Grb2-myc-P206L (C-SH3 mutant) and pcDNA3.1(−)-Grb2-myc-R86K (SH2 mutant), except that pcDNA3.1(−)-WT-Grb2-myc was used as the template in first-round PCRs. The primer pairs used for first-round PCRs were F1 and R3 (5′-GACATAATTGCGCAGAAACATGCCGGT-3′, with mutated Pro → Leu underlined) or R4 (5′-GCTCTCACTCTCTTTGATCAGGAAGGC-3′, with mutated Arg → Lys underlined) and F3 (5′-ACCGGCATGTTTCTGCGCAATTATGTC-3′, with mutated Pro → Leu underlined) or F4 (5′-GCCTTCCTGATCAAAGAGAGTGAGAGC-3′, with mutated Arg → Lys underlined) and R1, followed by second-round PCR on the mixed templates using primer pairs F1 and R1. To generate pcDNA3.1(−)-Grb2-myc-P49L/P206L (N- and C-SH3 mutants), pcDNA3.1(−)-Grb2-myc-P49L was used as a template in first-round PCR with primer pairs F1 and R3 and F3 and R1, followed by second-round PCR as above.
Other expression plasmids, siRNAs, and transfection.
The pXJ41-neo-VSVG-PTPα plasmids (WT or Y789F) and the pXJ40-FLAG-BCAR3 WT and R177K expression constructs were described previously (18, 27). The pcDNA3.1-mCherry-Paxillin plasmid was a gift from C. James Lim. Transfection of MEFs was carried out using Lipofectamine LTX reagent (Invitrogen). Transfected cells were grown for 24 h in antibiotic-free medium prior to further experimentation. Control nontargeting and Grb2 targeting siRNAs were from Integrated DNA Technologies, Inc. Shp2-targeting siRNA (On-Target plus Smart pool, Mouse PTPN11) was from Dharmacon. Cells were transfected with 20 nM siRNA using Lipofectamine RNAiMax reagent (Invitrogen) and grown for 48 h before further experimentation. In experiments where cells were cotransfected with both siRNA and DNA plasmid, the cells were grown for 48 h prior to further experimentation.
Cell lysis, immunoprecipitation, and immunoblot analysis.
Cells were harvested in 1% NP-40 lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 2 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg/ml leupeptin, and 10 μg/ml aprotinin) and processed for immunoprecipitation and immunoblotting experiments as described previously (16).
Paxillin mRNA.
Total RNA was extracted from cells treated with control or Grb2 siRNA using a QIAshredder and an RNeasy Minikit (Qiagen) according to the manufacturer's instructions. For reverse transcription (RT)-PCR, 600 ng of RNA was amplified with paxillin- and GAPDH (glyceraldehyde-3-phosphate dehydrogenase)-specific primers (28) using a OneStep RT-PCR kit (Qiagen).
Wound healing assay.
Cells (5 × 106 cells/ml) were seeded into FN-coated (100 μg/ml in PBS) microfluidic channels of a BioFlux 24-well plate (Fluxion Biosciences, Inc.) by shear flow. Cells were allowed to attach and form a monolayer in the channel overnight. Then, one-half of the channel was trypsinized using 0.25% trypsin-EDTA under a flow rate of 20 dyn/cm2. Trypsinization was terminated by 10% FBS in DMEM. An image of the wounded edge of the cell monolayer was taken every 15 min over a 24-h period using an Olympus IX81 microscope equipped with a CoolSnap HQ2 charge-coupled-device (CCD) camera. A 20× objective lens was used to visualize cell movement in the wounded area. Images were analyzed using the software Metamorph and ImageJ. Alternatively, for some experiments (see Fig. 8B), cell monolayers in culture dishes were “wounded” with a sterile pipette tip to create a clear area that was photographed prior to continued culture of the cells for 14 h. Cells were then stained with 1 nM calcein AM in medium and photographed under fluorescence. The width of the initial wound and of the remaining gap at 14 h was measured in arbitrary units. The averages of three measurements each of the initial and final gaps were used as follows to calculate the distance migrated by one edge of the cells: (initial width − final width)/2.
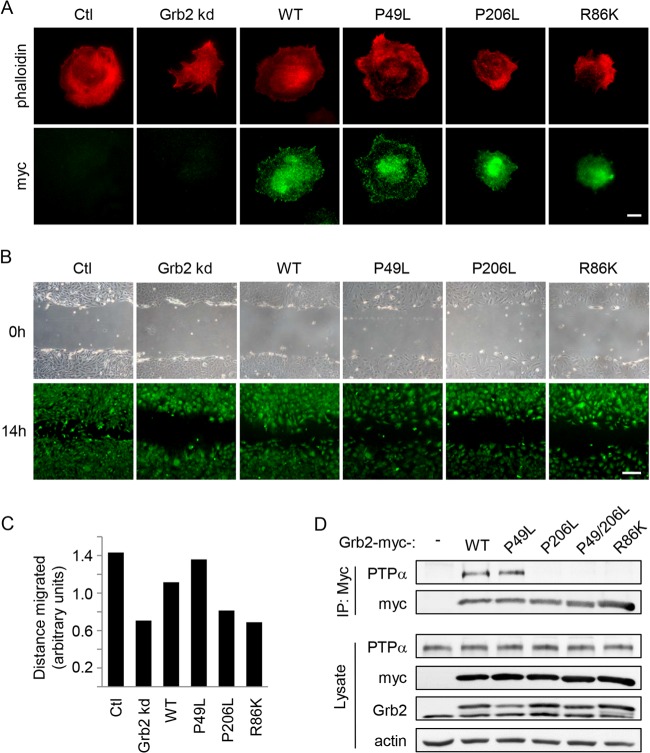
FIG 8.
Grb2 SH2 and C-terminal SH3 domains are required for cell spreading and migration and PTPα-Grb2 association. (A) MEFs treated with either control (Ctl) or Grb2 siRNA duplex or cotransfected with Grb2 siRNA and Grb2-myc-WT or Grb2-myc mutants were kept in suspension for 1 h and replated on FN-coated dishes for 15 min. Cells were fixed and immunostained with phalloidin (red) and myc (green). Bar, 20 μm. (B) Cells were plated on FN-coated coverslips for 48 h. A wound was created (0 h), and the cells were allowed to migrate into the cleared area for 14 h prior to staining with 1 nM calcein AM in medium. Bar, 250 μm. (C) The distance migrated by cells in two independent experiments as in panel B was measured as described in Materials and Methods, and the average is shown in the graph. (D) MEFs were transfected with Grb2-myc-WT or mutants. Anti-myc immunoprecipitates from adherent cell lysates were probed for PTPα and myc. Lysates were immunoblotted as shown.
In vitro kinase assay.
Cell lysates (1 mg) were precleared with 50 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology, Inc.) at 4°C for 1 h. Precleared lysates were incubated with anti-Src antibody for 1 h and then with 50 μl of protein A/G plus agarose beads for 20 min. Beads were washed 4 times for 5 min with PBS, and a kinase reaction solution (Genway Universal tyrosine kinase assay kit) containing 1:1,440 β-mercaptoethanol was added to the beads. Serial dilutions of these samples were prepared and transferred into a microplate in duplicate. The samples were incubated with 40 mM ATP-2Na (supplied with the kit) for 30 min and blocked with blocking solution (supplied with the kit) for 30 min at 37°C. After removal of blocking solution, the samples were then incubated with antiphosphotyrosine (anti-PY20) antibody conjugated with HRP for 30 min at 37°C. HRP substrate (TMBZ) was added to the samples, and after 25 min, the absorbance was measured at 450 nm.
Immunofluorescent staining.
Cells transfected with control or Grb2 siRNA and with or without Grb2 mutants using Lipofectamine RNAiMax (Invitrogen) were placed on FN-coated glass coverslips for 15 min, washed twice with cold PBS, and fixed for 30 min in 4% paraformaldehyde in PBS. Fixed cells were washed twice with PBS and permeabilized with 0.02% Triton X-100 in PBS for 15 min. They were then washed twice with PBS and blocked with 3% BSA in PBS for 1 h. After blocking, the cells were incubated with anti-Src Tyr416 (1:100) or anti-FAK Tyr397 (1:250) and antivinculin (1:250) antibodies or with anti-myc antibody (1:1,000) and Alexa Fluor 594-phalloidin (1:250) for 2 h at room temperature. This was followed by incubation with Alexa Fluor 488-conjugated anti-rabbit or anti-mouse IgG (1:250) and/or Alexa Fluor 594-conjugated anti-mouse IgG (1:250 dilution) as appropriate. Cells were imaged using an Olympus IX81 Cell^TIRF (total internal reflection fluorescent) system equipped with a CoolSnap HQ2 CCD camera. The fluorescently labeled proteins were visualized using a 60× TIRF objective lens (numerical aperture of 1.49) with the 488-nm and 561-nm lasers set to achieve a calibrated penetration depth of 85 nm for each laser. Images were processed using the software ImageJ.
RESULTS
Grb2 silencing reduces PTPα-Tyr789 phosphorylation.
Integrin stimulation induces the phosphorylation of PTPα at Tyr789 (17), a reported binding site for Grb2 (21, 22). We found that the association of Grb2 with PTPα was induced upon integrin engagement (Fig. 1A, left panels) and required PTPα-Tyr789 (Fig. 1A, right panels). To investigate the role of Grb2 in coordinating PTPα-mediated signaling events, Grb2 expression in mouse embryo fibroblasts was depleted using Grb2-targeting siRNA. Two siRNA duplexes were tested for knockdown efficiency. Duplex 1 targets the 5′ untranslated exon of Grb2 and effected <55% silencing of Grb2 expression compared to the nontargeting control siRNA over 24, 48, and 72 h posttransfection. Duplex 2, which targets the N terminus of Grb2, consistently produced >90% knockdown efficiency by 48 h posttransfection (Fig. 1B, upper panels). The expression and Tyr789 phosphorylation of PTPα were examined in the Grb2 siRNA-treated cells. While PTPα expression was unaltered, phosphorylation at Tyr789 was reduced in a manner corresponding to the extent of Grb2 depletion (Fig. 1B, lower panels). This suggests that PTPα-Tyr789 phosphorylation is modulated in a Grb2 dosage-dependent manner. The more-efficient Grb2-directed duplex 2 siRNA was used in subsequent experiments.
FIG 1.
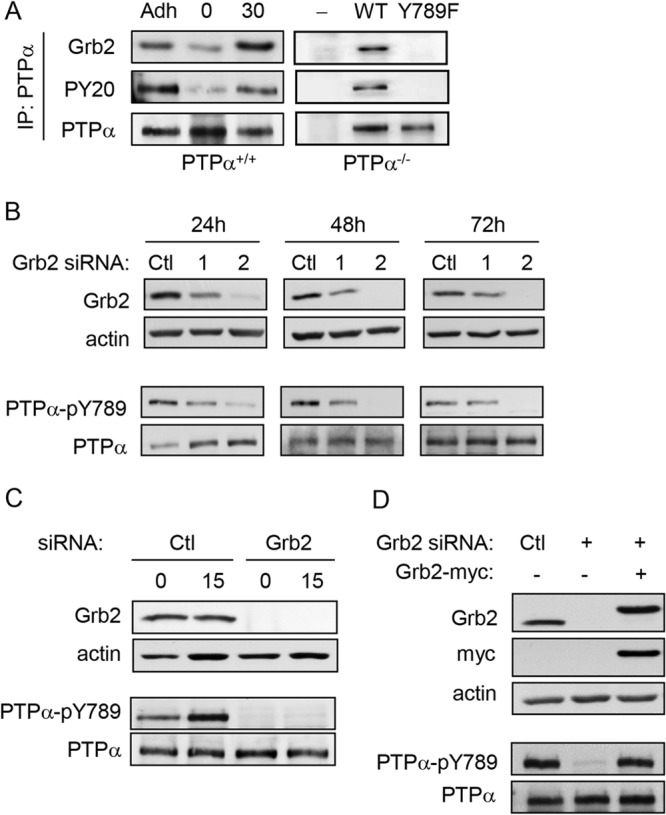
Grb2 knockdown reduces PTPα Tyr789 phosphorylation. (A) Left panel, wild-type (PTPα+/+) MEFs were starved overnight (adherent cells [Adh]) and then trypsinized and kept in suspension for 1 h (0) before plating on FN-coated plates for 30 min (30). Right panel, PTPα-null MEFs (PTPα−/−) and PTPα-null MEFs reexpressing WT or Y789F-PTPα were stimulated on FN as described above. PTPα immunoprecipitates were probed for Grb2, phosphotyrosine (PY20), and PTPα. (B) Wild-type MEFs were treated with control (Ctl) or Grb2 siRNA duplex 1 or 2 for 24, 48, and 72 h. Cell lysates were probed for Grb2, actin, PTPα-pY789, and PTPα. (C) Wild-type MEFs were treated with control (Ctl) or Grb2 siRNA for 48 h and then starved overnight. Cells were trypsinized and held in suspension for 1 h (0), and then a portion of the suspended cells were plated on FN-coated plates for 15 min (15). Lysates were probed as shown. (D) Wild-type MEFs treated with either control (Ctl) or Grb2 siRNA or cotransfected with Grb2 siRNA and myc-tagged Grb2-WT were stimulated on FN-coated dishes for 15 min. Lysates were probed as shown.
Next, we examined whether Grb2 specifically regulates integrin-induced PTPα-Tyr789 phosphorylation. Cells treated with control or Grb2 siRNA were trypsinized, kept in suspension for 1 h, and replated on fibronectin (FN)-coated dishes to stimulate integrin signaling for 15 min. As shown in Fig. 1C, PTPα-Tyr789 phosphorylation was induced in an integrin-dependent manner in control cells but was virtually undetectable in Grb2 knockdown cells upon FN stimulation. Indeed, the depletion of Grb2 also decreased the amount of residual PTPα-phospho-Tyr789 in the suspended cells. Expressing siRNA-resistant myc-tagged Grb2 in Grb2-depleted cells restored integrin-induced PTPα-Tyr789 phosphorylation (Fig. 1D), demonstrating that the effect of Grb2 depletion on PTPα was not due to off-target effects of the siRNA. The altered phosphorylation of PTPα at Tyr789 could be due to either the loss of Grb2-dependent kinase activity or Grb2-mediated inhibition of dephosphorylation of this site.
Cell migration is reduced in the absence of Grb2.
We previously showed that integrin-induced PTPα-Tyr789 phosphorylation is required for cytoskeletal reorganization and cell migration (17). To determine whether Grb2 depletion and the concomitantly reduced PTPα-phospho-Tyr789 affect cell migration, control and Grb2 siRNA-treated cells were seeded as a monolayer in a microfluidic channel and one-half of the monolayer was trypsinized under controlled shear flow to simulate wounding. Cell movement was tracked in real time over 24 h. Figure 2A and B depict the tracks of 10 randomly chosen cells along the wounded edge of the cell monolayer from each of the control and Grb2 knockdown groups, respectively. Control cells expressing Grb2 ranged more broadly into the cleared wound area than did cells depleted of Grb2, which despite exhibiting dynamic movement at the cell edges, appeared to be restricted in their range of movement. Cell tracking confirmed these observations, since the average total distance traveled by Grb2 knockdown cells was ∼30% less than by the control cells (Fig. 2C), and they exhibited an even greater reduction of 50% in the average net distance (linear distance between the original and the final positions) traveled (Fig. 2D). This suggests that the depletion of Grb2 not only reduces cell motility but also inhibits directional cell movement, hindering the cells from moving effectively into the disrupted edge and the wound. However, whether this migration defect observed in the absence of Grb2 is exclusively dependent on reduced PTPα-phospho-Tyr789 or involves other Grb2-mediated signaling events is unknown.
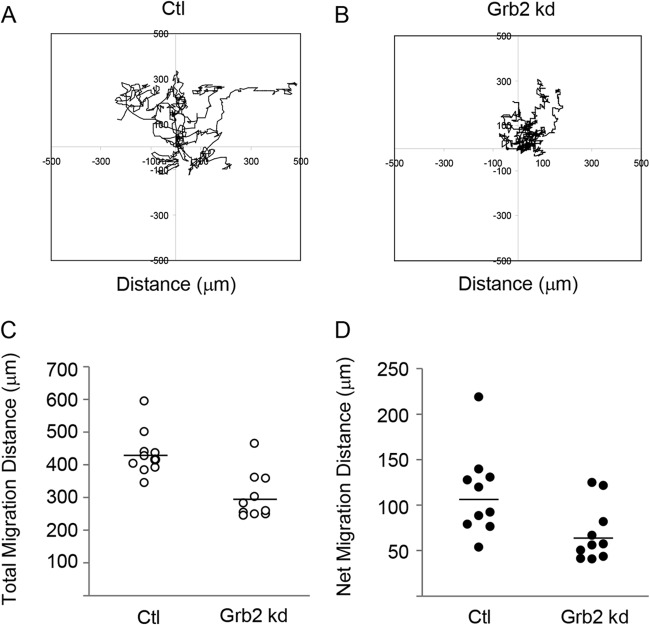
FIG 2.
Cell migration is reduced in the absence of Grb2. The migration of control (Ctl) and Grb2 siRNA-treated wild-type MEFs into the cleared area of a wounded cell monolayer was monitored for 24 h by time-lapse live-cell microscopy. Cell movement was tracked using ImageJ MtrackJ plugin. The two-dimensional (2D) random migration of 10 cells from each of the control (A) and Grb2 siRNA-treated (B) cell groups was measured and graphed. (C) The total distance of each track was measured for 10 cells in each group. The means are shown as horizontal bars and are significantly different (P = 0.001). (D) The net (linear) distance traveled was measured from the starting point of a track to the end. Results from 10 cell tracks are shown for each group. The means are shown as horizontal bars and are significantly different (P = 0.023).
Enhanced susceptibility to dephosphorylation does not account for reduced PTPα-phospho-Tyr789 in Grb2-depleted cells.
Grb2-SH2 binding to phospho-Tyr789 of PTPα has been suggested to protect this site from dephosphorylation (21). To determine whether PTPα is hyperdephosphorylated in the absence of Grb2, we attempted to detect transient PTPα phosphorylation occurring at times earlier than 15 min following integrin stimulation. As shown in Fig. 1B, no PTPα-Tyr789 phosphorylation was detectable in Grb2-depleted cells in suspension prior to integrin engagement or at 15 min following integrin stimulation, and we were likewise unable to detect any integrin-induced phospho-Tyr789-PTPα at the earlier time of 5 min after plating these cells on FN (data not shown). To determine whether Grb2 could regulate the phosphorylation of PTPα under other conditions, serum rather than FN was used to stimulate the cells. Figure 3A and B show that in unstimulated serum-starved cells (time zero), PTPα-phospho-Tyr789 was greatly reduced (∼4-fold) in the absence of Grb2. Upon stimulation with 10% serum, control cells displayed a gradual reduction in PTPα-phospho-Tyr789 within 5 min whereas Grb2-depleted cells showed an approximate 3-fold increase at 30 s poststimulation, and this gradually decreased thereafter. The lack of serum-stimulated phosphorylation of PTPα at Tyr789 may reflect already or nearly maximal phosphorylation of this site in the control adherent, albeit serum-deprived, cells. Despite the early (up to 30 s) poststimulation differences between control and Grb2 knockdown cells, the subsequent rate of PTPα dephosphorylation is similar for these cell types. These findings suggest that while PTPα can be transiently and suboptimally phosphorylated in Grb2-depleted cells, it is not being dephosphorylated more rapidly in the absence of Grb2.
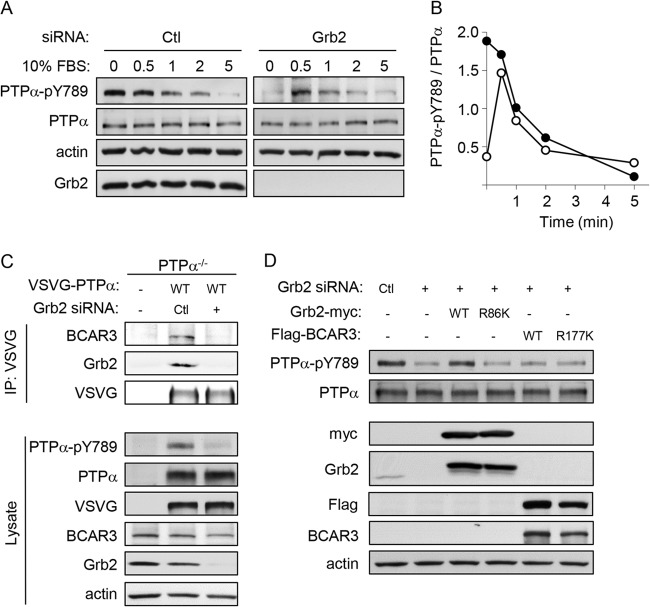
FIG 3.
PTPα-Tyr789 dephosphorylation and phosphorylation are not altered by expression of the phospho-Tyr789 binding proteins Grb2 and BCAR3. (A) Serum stimulation induces transient PTPα-Tyr789 phosphorylation in Grb2 knockdown cells. MEFs were treated with control or Grb2 siRNA, and 48 h later they were serum starved overnight and then stimulated with 10% FBS for 0.5, 1, 2, and 5 min. Lysates were immunoblotted with PTPα-pY789, PTPα, Grb2, and actin antibodies. (B) PTPα phosphorylation per unit of PTPα (arbitrary units) in control (Ctl) siRNA-treated (closed circles) and Grb2 siRNA-treated (open circles) cells was calculated from densitometric quantification of the blots shown in panel A. The absolute amount of PTPα Tyr789 phosphorylation cannot be compared between the control and Grb2 knockdown cells since a longer exposure of the autoradiograph from the latter lysate was quantified in order to accurately detect signal. (C) BCAR3 overexpression does not rescue phospho-PTPα-Tyr789. PTPα−/− MEFs were untreated or transfected with control (Ctl) or Grb2 siRNA and VSVG-tagged WT PTPα. Anti-VSVG immunoprecipitates from adherent cell lysates were probed for BCAR3, Grb2, and VSVG (VSVG-PTPα). Lysates were probed as shown. (D) Wild-type MEFs were transfected with either control (Ctl) or Grb2 siRNA or cotransfected with Grb2 siRNA and myc-Grb2-WT, myc-Grb2-R86K, Flag-BCAR3-WT, or Flag-BCAR3-R177K. Lysates were probed as shown.
BCAR3 overexpression does not rescue phospho-PTPα-Tyr789.
Recently, we discovered that BCAR3 can also interact with PTPα-phospho-Tyr789 via its SH2 domain (18). To determine whether PTPα-BCAR3 complex formation is impaired in Grb2-depleted cells, VSVG-tagged PTPα and control or Grb2 siRNA were cotransfected into PTPα−/− cells. The presence of BCAR3 and Grb2 was detected in VSVG immunoprecipitates from cells treated with nontargeting siRNA but not in those treated with Grb2 siRNA, where phospho-Tyr789-PTPα is lacking (Fig. 3C). This confirms that PTPα-Tyr789 phosphorylation is required for BCAR3 binding to PTPα. It also supports the idea that PTPα-Tyr789 is not phosphorylated normally in Grb2-depleted cells, as otherwise the direct binding of endogenous BCAR3 to this phosphosite could potentially protect it from dephosphorylation. However, the level of endogenous BCAR3 in MEFs may be insufficient to confer detectable protection.
Accordingly, we tested whether the overexpression of wild-type BCAR3 could restore PTPα-Tyr789 phosphorylation in the absence of Grb2. Flag-tagged WT or SH2 mutant (R177K) BCAR3, which is unable to interact with PTPα (18), was overexpressed in cells treated with Grb2 siRNA. Myc-tagged WT Grb2 or Grb2 with a mutation (R86K) in the SH2 domain that mediates PTPα-phospho-Tyr789 binding was also expressed in Grb2-depleted cells. As expected, neither mutant Grb2 nor mutant BCAR3 altered the effect of Grb2 silencing on PTPα. However, WT Grb2 restored PTPα-Tyr789 phosphorylation, whereas WT BCAR3 did not (Fig. 3D). This may reflect unknown differences in their interactions with PTPα-phospho-Tyr789. Alternatively, these findings raise the possibility that defective PTPα phosphorylation in Grb2-depleted cells does not result from the absence of SH2-mediated protection from dephosphorylation but may be a distinct Grb2-specific effect.
Grb2 knockdown does not alter integrin-induced Src activation.
The similar rates of PTPα-Tyr789 dephosphorylation in cells with or without Grb2 (Fig. 3B) and the inability of BCAR3 to restore or protect PTPα-Tyr789 phosphorylation (Fig. 3D) suggest that the reduced or undetectable PTPα-Tyr789 phosphorylation in Grb2-depleted cells is not due to increased susceptibility of phospho-Tyr789 to phosphatases. Instead, this could be due to the loss of Grb2-regulated promotion of PTPα phosphorylation by upstream kinases. Hence, without Grb2, PTPα cannot be optimally phosphorylated. Both Src and FAK are required for integrin-stimulated PTPα-Tyr789 phosphorylation, as this is defective in SYF cells (lacking Src, Fyn, and Yes) and remediated by Src reexpression and is defective in FAK-null MEFs (17). During thymocyte development, Grb2 plays an upstream role in the activation of the Src family kinase Lck (29), suggesting the potential for Grb2 to regulate Src. However, silencing Grb2 expression in MEFs did not affect integrin-induced Src phosphorylation at Tyr416 (Fig. 4A and B), an event associated with Src activation. Src activity was directly determined by in vitro kinase assays of Src immunoprecipitates and was reduced by ∼25% upon Grb2 depletion (Fig. 4C). This was a minor reduction compared to that observed in Src immunoprecipitates from MEFs lacking the Src-activating phosphatase PTPα (∼90%), which were included for comparison (Fig. 4C). This suggests that reduced Src activity is unlikely to account for the drastically reduced PTPα-Tyr789 phosphorylation in Grb2-depleted cells.
FIG 4.
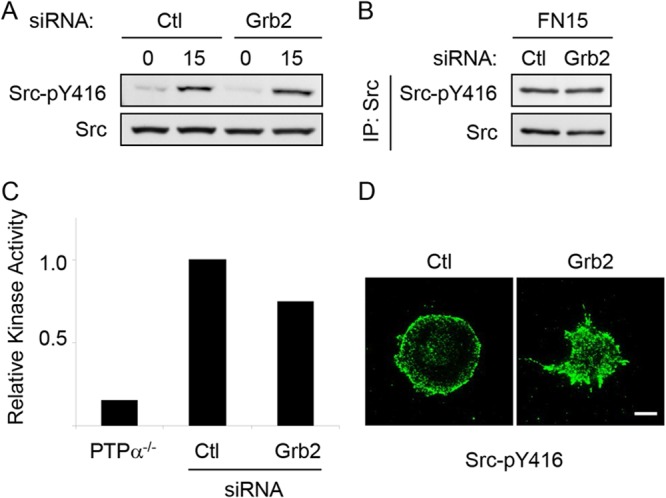
Grb2 knockdown does not prevent integrin-induced Src activation. Wild-type MEFs treated with control (Ctl) or Grb2 siRNA were kept in suspension for 1 h (0) and replated on FN-coated coverslips or dishes for 15 min (15). Cell lysates (A) and Src immunoprecipitates from lysates of FN-stimulated cells (B) were probed for Src-phospho-Tyr416 (Src-pY416) and Src. (C) Src immunoprecipitates were prepared from lysates of PTPα−/− MEFs and from control (Ctl) and Grb2 siRNA-treated wild-type MEFs after FN stimulation for 15 min and used in in vitro kinase assays. Kinase activity is shown relative to that in the control cell immunoprecipitate (PTPα−/− cells, n = 1; control and Grb2 siRNA-treated cells, n = 2). (D) Cells plated on coverslips were washed, fixed, and immunostained with Src-pY416 antibody. Bar, 20 μm.
We also examined the localization of activated Src in spreading cells after plating on FN. Immunostaining with anti-Src-phospho-Tyr416 antibody revealed that phospho-Src signal intensity and residence at the periphery of the cells appeared independent of the presence of Grb2; however, the morphologies of the spreading cells were quite different with or without Grb2 (Fig. 4D). Grb2-depleted cells display a distinctive spreading phenotype characterized by numerous pointed protrusions, in contrast to the relatively uniform and rounded morphology of the control cells. Interestingly, a similar spreading phenotype has been described for FAK-depleted rat embryo fibroblasts (REFs). These FAK-deficient cells exhibited abnormal spikelike protrusions on FN, and expression of WT FAK but not mutant Y397F FAK rescued this defect (30).
Grb2 knockdown reduces FAK activation and FAK/Src complex formation.
The resemblance between Grb2-depleted MEFs and the unphosphorylatable Y397F-FAK-expressing REFs suggests that the phosphorylation of FAK at Tyr397 may be defective in MEFs in the absence of Grb2, yielding a similar spreading phenotype and indicating a role for Grb2 in regulating PTPα-Tyr-789 phosphorylation via FAK activation. To investigate this, control and Grb2 knockdown cells were coimmunostained with FAK-phospho-Tyr397 and vinculin antibodies. As shown in Fig. 5A, FAK-phospho-Tyr397 signal is reduced and its colocalization with the early focal adhesion resident vinculin is impaired in the absence of Grb2, suggesting that Grb2 alters FAK Tyr-397 phosphorylation.
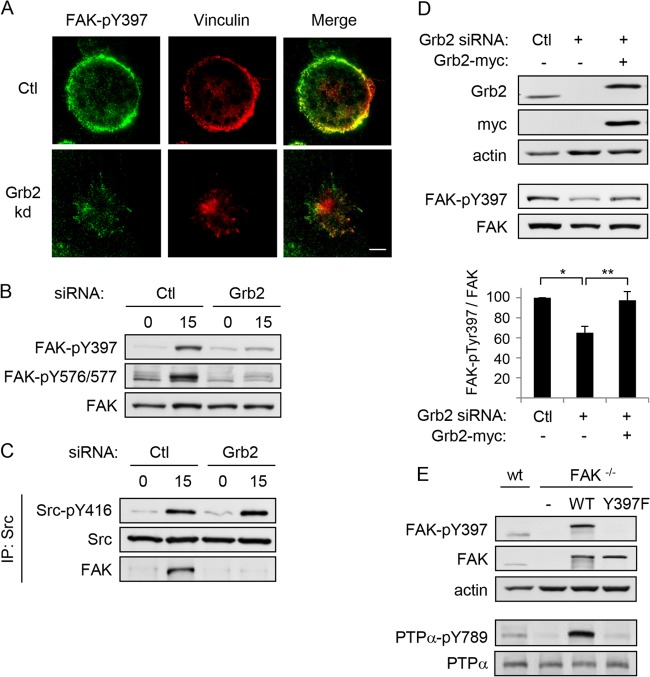
FIG 5.
Integrin-induced FAK-Tyr397 phosphorylation and FAK/Src complex formation are dependent on Grb2. Wild-type MEFs treated with control (Ctl) or Grb2 siRNA were kept in suspension (0) for 1 h and replated on FN-coated coverslips/dishes for 15 min (15). (A) Cells on coverslips were washed, fixed, and immunostained with FAK-phospho-Tyr397 (FAK-pY397, green) and vinculin (red) antibodies. Bar, 20 μm. (B) Cell lysates were collected from the plates and probed for FAK-pY397, FAK-phospho-Y576/577 (FAK-pY576/577), and FAK. (C) Anti-Src immunoprecipitates were immunoblotted with Src-pY416, Src, and FAK. (D) MEFs transfected with either control (Ctl) or Grb2 siRNA or cotransfected with Grb2 siRNA and myc-tagged Grb2 were plated on FN-coated dishes for 15 min. Lysates were immunoblotted with Grb2, myc, actin, FAK-pY397, and FAK antibodies. The graph shows the amount of FAK-pTyr397 per unit of FAK calculated from 4 experiments, and the asterisks depict significant differences (*, P = 0.00003; **, P = 0.001). (E) Wild-type MEFs (wt), FAK-null MEFs (FAK−/−), and FAK-null MEFs stably reexpressing GFP-tagged wild-type (FAK-WT) or mutant (FAK-Y397F) FAK were immunoblotted with FAK-pY397, FAK, PTPα-pY789, PTPα, and actin antibodies.
To verify that integrin-induced FAK phosphorylation is defective in Grb2-depleted cells, cell lysates from suspended and FN-stimulated control and Grb2-depleted cells were probed for FAK-phospho-Tyr397 and FAK. FN-stimulated control cells show a dramatic increase in FAK-Tyr397 phosphorylation, whereas in Grb2-depleted cells this was only minimally increased upon FN-mediated integrin stimulation (Fig. 5B, upper panel). Integrin engagement stimulates PTPα-catalyzed Src activation and Src-SH2 binding to FAK-phospho-Tyr397 (14–16), enabling Src to phosphorylate sites in the kinase domain of FAK to activate FAK and generate the maximally active Src-FAK kinase complex. In accord with the defective integrin-induced phosphorylation of FAK-Tyr397, we found that Src-FAK complex formation was severely impaired in the Grb2-depleted cells (Fig. 5C), as was the phosphorylation of FAK at Tyr576/577 (Fig. 5B). FAK-Tyr397 phosphorylation was rescued by the introduction of siRNA-resistant WT Grb2 into Grb2-depleted cells (Fig. 5D) and thus was not due to off-target siRNA effects. Furthermore, experiments with FAK-null MEFs demonstrated that reexpressing WT-FAK but not mutant Y397-FAK rescued the lack of PTPα phosphorylation observed in the absence of FAK (Fig. 5E), confirming that FAK-Tyr397 phosphorylation is required for this event. Overall, our results indicate that Grb2 plays an upstream role in PTPα-Tyr789 phosphorylation through regulating FAK-Tyr397 phosphorylation to enable Src-FAK complex formation and activation.
Paxillin expression, but not Shp2 silencing, restores FAK activation in Grb2-depleted cells.
Grb2 has recently been reported to play a novel role in regulating fibroblast growth factor receptor 2 (FGFR2) phosphorylation by inhibiting the phosphatase activity of Shp2 toward the receptor (31). In Grb2 knockdown cells, Shp2 inhibition is lost and FGFR2 tyrosine phosphorylation is reduced. Since Shp2 can dephosphorylate Tyr397 of FAK (32, 33), we investigated whether reduced FAK phosphorylation at this site in Grb2 knockdown cells was likewise due to the loss of Grb2-mediated inhibition of Shp2. However, silencing Shp2 expression or inhibiting Shp2 activity concomitantly with Grb2 depletion did not restore defective phosphorylation of FAK-Tyr397 or PTPα-Tyr789 (see Fig. S1A and B in the supplemental material).
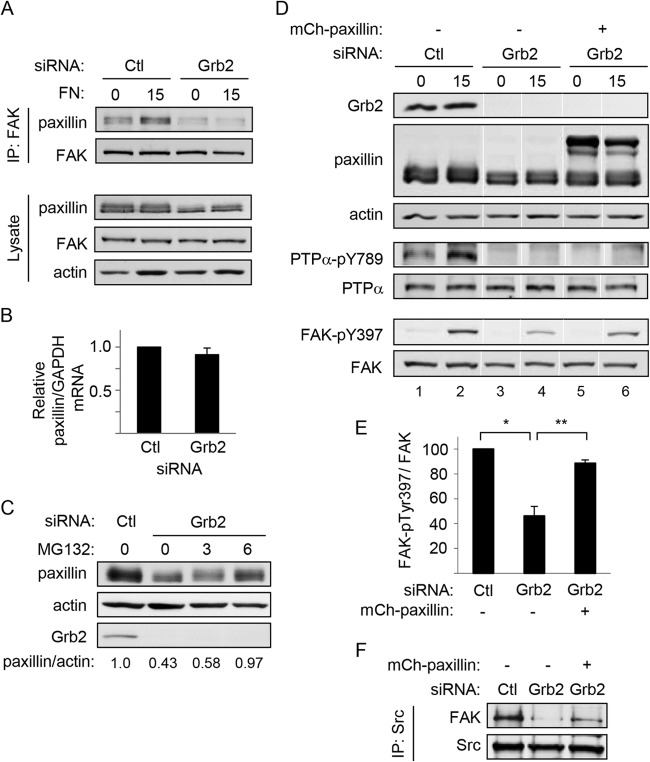
In view of the Src-FAK association defect in Grb2-depleted cells, we examined the association between FAK and another binding partner, paxillin (34, 35), that has been associated with FAK phosphorylation (36). Integrin stimulation promoted an increase in the amount of paxillin in FAK immunoprecipitates from control cells but not from Grb2 knockdown cells (respectively, 1.5-fold ± 0.23-fold versus 0.71-fold ± 0.29-fold, n = 3) (Fig. 6A, top panels). Probing cell lysates consistently revealed less paxillin to be present upon Grb2 knockdown (Fig. 6A, bottom panels). This suggested that Grb2 can mediate paxillin expression or availability, thereby affecting paxillin-FAK interaction and integrin-stimulated FAK activation. Paxillin was still reduced in SDS-solubilized lysates of Grb2-depleted cells (data not shown), suggesting that level and not merely solubility was affected. The paxillin mRNA amount was not affected by Grb2 depletion (Fig. 6B), while treatment of Grb2-depleted cells with the proteasome inhibitor MG132 for 6 h restored the paxillin level to about the same as in control cells (Fig. 6C), suggesting that paxillin was degraded more rapidly in the absence of Grb2. To investigate if the reduced paxillin was indeed affecting FAK activation, we overexpressed paxillin in Grb2 knockdown cells. Integrin-stimulated FAK-Tyr397 phosphorylation was markedly improved in these cells compared to that in Grb2-depleted cells lacking exogenous paxillin (Fig. 6D and E) and concomitantly enhanced Src-FAK association (Fig. 6F). Surprisingly, despite rescuing FAK activation, paxillin expression was unable to restore integrin-stimulated phosphorylation of PTPα at Tyr789 (Fig. 6D). This suggests that Grb2 might be important for distinct events in the process of PTPα-Tyr789 phosphorylation, with one event being paxillin-regulated FAK-Tyr397 phosphorylation to promote formation of the active Src-FAK complex and another event the regulation of subsequent PTPα phosphorylation.
FIG 6.
Paxillin expression restores FAK activation but not PTPα-Ty789 phosphorylation in Grb2-depleted cells. MEFs treated with control (Ctl) or Grb2 siRNA were kept in suspension (0) for 1 h and replated on fibronectin (FN)-coated dishes for 15 min (15). (A) Immunoprecipitates of FAK were probed with FAK and paxillin (upper panels). Cell lysates were immunoblotted for FAK, paxillin, and actin (lower panels). (B) Relative levels of paxillin mRNA per unit of GAPDH mRNA in control (Ctl) and Grb2 siRNA-treated cells were determined by RT-PCR. (C) Untreated control (Ctl) and Grb2-depleted (0) cells and Grb2-depleted cells that were incubated with 20 μM MG132 for 3 and 6 h were probed for paxillin, actin, and Grb2. The bottom numbers show the relative units of paxillin per actin in each sample. (D) MEFs were treated with control (Ctl) or Grb2 siRNA or cotransfected with Grb2 siRNA and mCherry (mCh)-paxillin for 48 h and then starved in reduced serum medium overnight. The next day, cells were trypsinized, kept in suspension (0) for 1 h, and replated on FN-coated dishes for 15 min (15). Lysates were immunoblotted with Grb2, paxillin, actin, PTPα-pY789, PTPα, FAK-pY397, and FAK antibodies. Lanes 1 to 6 in each panel are from a single gel. The white vertical lines indicate that the samples in lanes 3 and 5 were inadvertently exchanged on the original gel, and these lane images were spliced out and repositioned correctly in the final image shown here. (E) The relative amount of FAK-pY397 per unit of FAK in FN-stimulated lysates from at least 3 experiments as in panel D was quantified by densitometry and is shown in the graph. Asterisks indicate significant differences (*, P = 0.0003; **, P = 0.0008). (F) Src immunoprecipitates from FN-stimulated lysates from experiments as in panel B were probed for FAK and Src.
Grb2 domain-specific requirements for stable paxillin expression, integrin-induced FAK activation, PTPα-Tyr789 phosphorylation, and cell spreading and migration.
We investigated if specific domains of Grb2 that were responsible for these potentially distinct activities could be identified. Grb2-siRNA-resistant plasmids were used to reintroduce wild-type and mutant forms of Grb2 with function-disrupting point mutations in one or more domains (Fig. 7A) into MEFs while simultaneously depleting the cells of endogenous Grb2 using siRNA. As shown in Fig. 7B and C, reexpressed WT Grb2 and the P206L, P49L/P206L, and R86K Grb2 mutants rescued or even improved paxillin expression compared to that in the control cells. The P49L Grb2 mutant was the least effective but conferred a partial rescue of the ∼40% loss of paxillin that occurred upon Grb2 depletion, indicating that the Grb2 N-SH3 domain is important but not essential for paxillin stabilization.
FIG 7.
Differential abilities of Grb2 domain mutants to rescue paxillin expression, FAK autophosphorylation at Tyr397, and PTPα-Tyr789 phosphorylation. (A) Schematic diagram of siRNA-resistant myc-tagged Grb2-WT/mutants. Silent mutations were made in the depicted region targeted by the Grb2 siRNA. The closed circles depict the domain with the inactivating point mutation. (B to E) MEFs treated with either control (Ctl) or Grb2 siRNA duplex or cotransfected with Grb2 siRNA and Grb2-myc-WT or mutants were kept in suspension for 1 h and replated on FN-coated dishes or coverslips for 15 min. (B) Paxillin expression was analyzed by probing cell lysates, and the graph in panel C shows the relative amount of paxillin per unit of FAK as calculated from densitometric analysis of 3 experiments as in panel B. (D) FAK-Tyr397 and PTPα-Tyr789 phosphorylation was determined by probing cell lysates (D), and the graph in panel E shows the relative amount of FAK-phospho-Tyr397 per unit of FAK as calculated from densitometric analysis of 3 experiments as in panel D.
We then examined the abilities of the Grb2 mutants to rescue FAK Tyr397 and PTPα Tyr789 phosphorylation. Overall, the mutants rescued FAK-Tyr397 phosphorylation in accord with their abilities to rescue paxillin expression (Fig. 7D and E), indicating that paxillin level is a critical factor for FAK activation. The P49L Grb2 mutant was the least effective but reproducibly induced some improvement in the defective FAK-Tyr397 phosphorylation in Grb2-depleted cells. All the other Grb2 mutants, including the P49L/P206L double mutant, promoted FAK-Tyr397 phosphorylation to levels up to 40% higher than those promoted by reexpressed WT Grb2. The P49L Grb2 mutant effected an ∼50% rescue of integrin-induced PTPα-Tyr789 phosphorylation, on par with its partial rescue of paxillin expression and FAK Tyr397 phosphorylation. However, in contrast to the full restoration of paxillin expression and FAK-Tyr397 phosphorylation by the P206L, P49L/P206L, and R86K Grb2 mutants, none of these mutants were able to rescue defective PTPα-Tyr789 phosphorylation, as this was still reduced by ∼90% (Fig. 7D). Taken together, these results indicate that the Grb2 SH2 and C-SH3 domains are required, post-FAK activation, in order for PTPα to be phosphorylated.
Abnormal cell morphology and impaired migration were observed in Grb2-deficient cells, as described above. Expression of the Grb2 domain mutants revealed that only the P49L Grb2 mutant showed an ability comparable to or better than that of reexpressed WT Grb2 to restore the rounded spreading morphology (Fig. 8A) and to enhance cell motility of Grb2-depleted cells (Fig. 8B and C). This suggests that the partial rescue of paxillin expression by P49L Grb2 and the consequently improved FAK-Tyr397 autophosphorylation and PTPα-Tyr789 phosphorylation are sufficient to promote these cellular processes. The inability of the P206L and R86K Grb2 mutants to restore cell spreading and migration corresponds to their failure to rescue PTPα-Tyr789 phosphorylation and indicates that phospho-PTPα is important for promoting these processes. This is consistent with our previous finding that cells expressing mutant PTPα-Y789F exhibit impaired spreading and migration (17).
Grb2 is required for FAK association with PTPα.
Since the association between Grb2 and PTPα is reported to be mediated by Grb2 C-SH3 and SH2 domains (23, 24), we speculated that the association of Grb2 with PTPα through these domains is critical for PTPα-Tyr789 phosphorylation. To verify that Grb2 association with PTPα is dependent on the Grb2 C-SH3 and SH2 domains in the MEF cell system, myc-tagged WT or mutant Grb2 was transfected into MEFs and the introduced Grb2 was immunoprecipitated using anti-myc antibody. Probing myc-Grb2 immunoprecipitates for PTPα confirmed that the interaction of PTPα and Grb2 is indeed dependent on the C-SH3 and SH2 domains of Grb2 (Fig. 8D). This suggests that Grb2-dependent PTPα-Tyr789 phosphorylation correlates with the physical interaction of Grb2 and PTPα.
Expressing most Grb2 mutants in Grb2-depleted cells can rescue FAK-Tyr397 phosphorylation, while PTPα-Tyr789 phosphorylation remains impaired, raising the possibility that, in the presence of active kinase, PTPα is phosphorylated and that Grb2 is now required to bind to phospho-Tyr789 and shield it from dephosphorylation. We therefore reexamined the ability of overexpressed BCAR3 to be substituted for Grb2 in such a protective role. In cells depleted of endogenous Grb2 and reexpressing mutant Grb2-R86K alone or together with WT-BCAR3, FAK-phospho-Tyr397 was rescued but BCAR3 failed to rescue PTPα-Tyr789 phosphorylation (see Fig. S2 in the supplemental material). This indicates that this defect is not due to hyperdephosphorylation but represents a continued failure, despite the presence of activated FAK, to achieve PTPα-Tyr789 phosphorylation.
We investigated the possibility that PTPα-associated Grb2 is required to bring activated FAK into proximity with PTPα in order to phosphorylate Tyr789. VSVG-tagged WT-PTPα was cotransfected with nontargeting control or Grb2 siRNA into PTPα-null cells, and PTPα was immunoprecipitated using anti-VSVG conjugated beads. Probing the immunoprecipitates revealed that FAK and Grb2 were complexed with PTPα in cells treated with control siRNA but not in Grb2 siRNA-treated cells (Fig. 9A, left panels). Thus, the interaction of FAK with PTPα is dependent on Grb2. Indeed, reexpressing WT-Grb2 in the presence of Grb2 siRNA restored the interactions between PTPα, FAK, and Grb2 (Fig. 9A). Furthermore, the association of FAK with PTPα requires the PTPα-binding SH2 and C-SH3 domains of Grb2 (Fig. 9A, right panels). This supports a model whereby PTPα-bound Grb2 functions as an anchor for FAK to complex with PTPα and suggests that this is permissive for PTPα phosphorylation by FAK.
FIG 9.
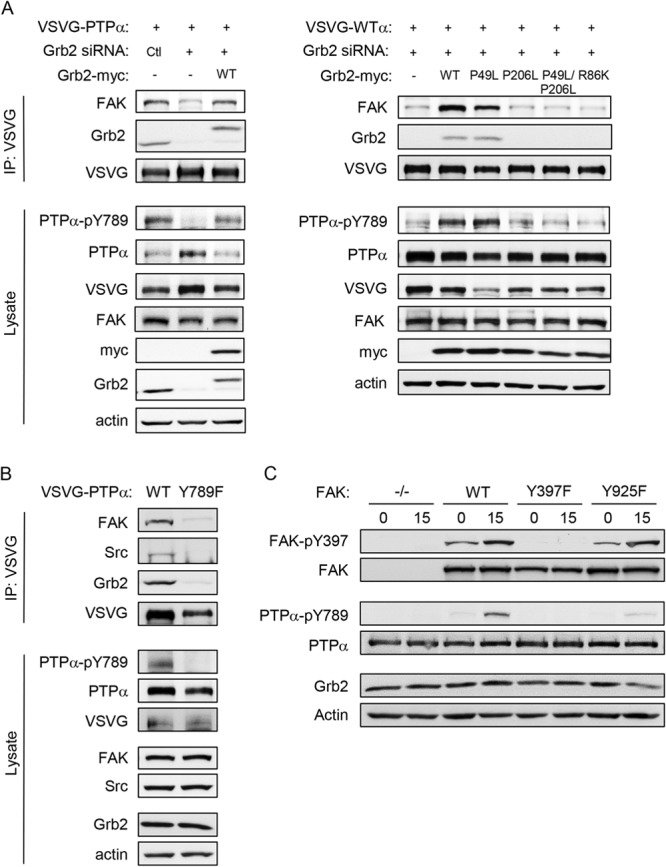
Grb2 is required for FAK association with PTPα. (A) PTPα−/− MEFs were untreated or transfected as shown with control (Ctl) or Grb2 siRNA, myc-tagged Grb2-WT or the indicated Grb2 mutants, and VSVG-WT PTPα. VSVG immunoprecipitates from adherent cell lysates and cell lysates were probed as shown. (B) PTPα−/− MEFs were transfected with VSVG-WT-PTPα and VSVG-Y789F-PTPα. VSVG immunoprecipitates from adherent cell lysates and cell lysates were probed as shown. (C) FAK-null MEFs (FAK−/−) and FAK-null MEFs stably reexpressing GFP-tagged wild-type (FAK-WT) or the indicated mutant forms of FAK (Y397F or Y925F) were kept in suspension for 1 h (lanes 0) and replated on fibronectin (FN)-coated dishes for 15 min (lanes 15). Cell lysates were probed for the indicated (phospho)proteins.
The interaction of Grb2 with PTPα is reported to depend on Grb2-SH2 binding to PTPα-phospho-Tyr789. We verified that PTPα-Tyr789 phosphorylation is indeed critical for PTPα complex formation with Grb2 and FAK, since mutant PTPα-Y789F was unable to associate with these proteins (Fig. 9B). The SH2 domain of Grb2 also binds FAK-phospho-Tyr925, a site that is phosphorylated by Src upon its association with FAK (37). We found that this FAK phosphosite was required for PTPα phosphorylation, since integrin-stimulated phosphorylation of PTPα at Tyr789 was almost abolished in FAK-null MEFs stably expressing GFP-FAK-Y925F (Fig. 9C). These findings suggest that Grb2 association with FAK and with PTPα physically links the Src-FAK kinase complex to its substrate to enable PTPα-Tyr789 phosphorylation.
DISCUSSION
In this study, we established that a PTPα-phospho-Tyr789-Grb2 complex forms upon integrin stimulation, and we employed siRNA-mediated silencing of Grb2 to investigate its role in phospho-PTPα-mediated integrin signaling. However, this revealed that PTPα is not phosphorylated at Tyr789 in cells deficient in Grb2, prompting us to address the role of Grb2 in promoting integrin-induced PTPα-Tyr789 phosphorylation rather than its action as an effector of PTPα signaling.
Both Src and FAK are required for integrin-stimulated phosphorylation of PTPα at Tyr789, indicating that this is likely catalyzed by the Src-FAK kinase complex. We found that in Grb2-depleted cells, integrin-induced Src activation is minimally affected, whereas the phosphorylation of FAK-Tyr397 is significantly abrogated. Defective Tyr397 phosphorylation would preclude Src-SH2 domain binding to Tyr397 of FAK, which enables Src phosphorylation of FAK activation loop tyrosines 576 and 577 and FAK activation, and we indeed observe impaired Src-FAK association and phosphorylation of FAK Tyr576/577. Also in accord with a defect in FAK-Tyr397 phosphorylation, the Grb2-deficient cells display an abnormal spreading phenotype on FN that is reminiscent of that of FAK-deficient fibroblasts reexpressing mutant Y397F-FAK (30), whereby the latter cells are likewise incapable of supporting integrin-induced PTPα-Tyr789 phosphorylation. Overall, silencing Grb2 expression impairs formation and activation of the Src-FAK complex that is key for numerous downstream events in integrin signaling, including PTPα-Tyr789 phosphorylation.
At present, it is unclear precisely how Grb2 regulates FAK-Tyr397 phosphorylation, particularly since the molecular mechanisms that regulate integrin-induced FAK-Tyr397 phosphorylation are complex and incompletely understood. The initial trans-autophosphorylation of Tyr397 requires alterations in FAK subcellular localization to cluster or concentrate FAK in particular cellular environments/locales such as newly forming integrin-containing adhesions, as well as the disruption of FERM (N-terminal band 4.1/ezrin/radixin/moesin) domain-mediated intramolecular interactions that inhibit FAK catalytic activity (38–41). The C-terminal FAT (focal adhesion targeting) domain of FAK is also important in FAK activation. It binds protein partners such as talin and paxillin that have long been considered to regulate FAK localization (34, 35, 42, 43). Recent findings suggest that talin is not essential for FAK recruitment to newly forming integrin-mediated adhesions; instead, FAK appears to recruit talin to these sites (44). Our finding that Grb2 silencing reduces the amount of paxillin in cell lysates and that increasing the paxillin level by expressing exogenous paxillin in the Grb2-deficient cells rescues defective FAK-Tyr397 phosphorylation and Src-FAK association suggests that paxillin-mediated FAK activation is a likely role of Grb2.
Paxillin is important to localize FAK to focal adhesions (45, 46) but may play a more complex role in Tyr397 phosphorylation and activation. In addition to a requirement for the focal-adhesion-localizing LIM2 and LIM3 domains (47) of paxillin to promote FAK-Tyr397 phosphorylation, the non-focal-adhesion-localizing LIM1 domain is also required, as is a single LD motif that does not need to be the FAK binding LD2 or LD4 motif (48, 49). Grb2 may mediate paxillin expression and/or coordinate protein associations that promote FAK-Tyr397 phosphorylation. Treating Grb2-depleted cells with the proteasome inhibitor MG132 restored paxillin expression, suggesting that Grb2 can mediate paxillin stability. Interestingly, another adaptor protein, Nckβ, functions analogously to control neuritogenesis by maintaining paxillin stability and expression level in neurons (50). Among the Grb2 domain mutants that we tested, only the N-SH3 domain mutant was unable to fully rescue the reduced paxillin expression and FAK Tyr397 phosphorylation in Grb2-depleted cells. This indicates a more critical function of the N-SH3 domain in the Grb2 promotion of FAK activation, although the interacting ligand that may mediate this is not known.
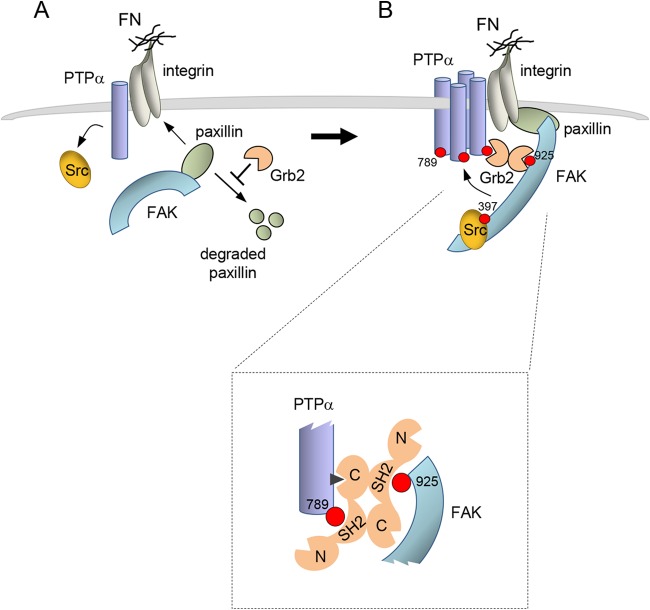
Despite the ability of heterologously expressed paxillin to restore integrin-induced FAK-Tyr397 phosphorylation and Src-FAK complexation in Grb2-depleted cells, it did not rescue PTPα-Tyr789 phosphorylation. This indicated that Grb2 was required in yet another capacity to facilitate Tyr789 phosphorylation or, in cells with rescued Src-FAK activity, prevent PTPα dephosphorylation. The inability of overexpressed BCAR3, a PTPα-phosphoTyr789-binding protein (18), to restore detectable PTPα-Tyr789 phosphorylation in Grb2-depleted cells with paxillin-remediated Src-FAK activation indicates that the low phosphorylation of PTPα is not due to enhanced susceptibility to phosphatase(s). We further distinguished the roles of Grb2 in facilitating FAK and PTPα phosphorylation based on the domains of Grb2 required to sustain each effect. While functional Grb2 SH2 and C-SH3 domains are dispensable for FAK-Tyr397 phosphorylation, they are required for PTPα-Grb2 association and PTPα-Tyr789 phosphorylation. Furthermore, these Grb2 domains are required for PTPα to associate with FAK. Phospho-Tyr925 of FAK, a Grb2-SH2 binding site (37), is also required for integrin-induced PTPα-Tyr789 phosphorylation, leading us to propose the following model (Fig. 10). Basal or low-level integrin-stimulated initial phosphorylation of PTPα at Tyr789 recruits Grb2 and promotes a second site of interaction with Grb2-C-SH3. FAK-phosphoTyr925 also recruits Grb2 via the Grb2-SH2 domain. Since both of these interactions require a Grb2-SH2 domain, we suggest that PTPα-bound Grb2 and FAK-bound Grb2 form a Grb2 dimer, thus linking Src-FAK and PTPα to bring active Src-FAK into proximity with other nearby, perhaps integrin-clustered, PTPα molecules to facilitate their phosphorylation.
FIG 10.
Two roles of Grb2 in early integrin signaling. (A) Integrin engagement stimulates PTPα-catalyzed dephosphorylation and activation of Src. In role 1, Grb2 (depicted as a dimer, but this is unclear), promotes FAK autophosphorylation at Tyr397, possibly by mediating FAK relocalization in a paxillin-dependent manner. Right, FAK autophosphorylation at Tyr397 recruits Src, with consequent Src-catalyzed phosphorylation of FAK at activation loop tyrosines (not shown) and at Tyr925. (B) In role 2, a Grb2 dimer coordinates Src-FAK and PTPα complexation via Grb2-SH2 binding to FAK-phospho-Tyr925 and to PTPα-phospho-Tyr789 (details shown in boxed inset). The latter is suggested to represent a small population of PTPα that undergoes initial phosphorylation through an unknown mechanism or has basal level phosphorylation at Tyr789. The Grb2-mediated FAK-PTPα linkage positions active Src-FAK in proximity to nearby clustered molecules of PTPα to enable their phosphorylation at Tyr789. Additional cycles of Grb2-mediated Src-FAK binding to newly phosphorylated PTPα may be utilized to achieve maximal PTPα phosphorylation. Overall, a localized concentration of PTPα-phospho-Tyr789 that acts to signal downstream is generated, for example (not shown), by recruiting the BCAR3-Cas complex (18).
Grb2 crystallizes as a dimer and in solution exists in a monomer-dimer equilibrium (51, 52). The dimer is maintained by interactions between the SH2 domain of one molecule and the C-SH3 domain of the other and vice versa, without involvement of the N-SH3 domains. In the dimeric form, the ligand binding pockets of all domains are fully exposed and accessible to bind cellular ligands (51). In support of a functional Grb2 dimer in cells, a recent study shows that both C-SH3 domains of a Grb2 dimer coordinate the binding of two molecules of FGFR2 to inhibit basal RTK signaling (53). In our model, the PTPα-bound Grb2 uses the SH2 domain and the C-SH3 domain to bind to PTPα, as indicated by our finding that each of these domains is required for Grb2-PTPα complex formation and consistent with previous work that identified PTPα residues 469 to 486 as a binding site for the Grb2 C-SH3 domain (23). Arg469 within this sequence is essential for the interaction (23), and we note that this is the N-terminal Arg of an RXXR motif that conforms to an R/KXXR/K sequence recognized by the Grb2 C-SH3 domain (54). Within the FAK-bound molecule of Grb2, the occupancies of the Grb2 N- and C-SH3 domains are not known. Occupancy of one Grb2 SH3 domain sterically hinders ligand binding to the other, such that the C-SH3 and the N-SH3 domains are not simultaneously engaged (55). Also, all known Grb2 dimer-protein complexes use either both C-SH3 domains or both N-SH3 domains of the dimer to mediate protein binding, as found not only with the Grb2-FGFR2 receptor complex (53) but also with Grb2-Sos (both N-SH3) or Grb2-Gab1 (both C-SH3 domains) complexes that form in a 2:1 stoichiometry (55). Since the PTPα-bound Grb2 molecule has its C-SH3 domain engaged, we predict that the C-SH3 domain of the FAK-bound Grb2 molecule would thus likely be engaged as well. Our model presents a new scenario, akin to the 2:2 stoichiometry of the Grb2-FGFR2 complex that juxtaposes and inhibits the FGFR2 but distinct in that each molecule of Grb2 coordinates a different ligand to form a 1:2:1 stoichiometric complex of FAK-Grb2-PTPα to promote downstream signaling.
In summary, we have identified two new roles of Grb2 in integrin signaling; one as a regulator of paxillin stability and upstream promoter of FAK-Tyr397 phosphorylation that is required for Src-FAK complex activation and another as an essential coordinator of PTPα and active Src-FAK interaction to enable the phosphorylation of a larger localized population of PTPα at Tyr789. Further studies are required to delineate the precise mechanisms of these Grb2-mediated events that culminate in PTPα phosphorylation-dependent cell spreading and migration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jason Northey, McGill University, for helpful advice about Grb2-targeting siRNA.
This work was supported by grants (MOP-49410) from the Canadian Institutes of Health Research (to C.J.P.) and the NIH (CA102310) (to D.D.S.). C.J.P. is the recipient of an Investigator Award from the Child and Family Research Institute.
Footnotes
Published ahead of print 18 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00825-13.
REFERENCES
- 1.Geiger B, Yamada KM. 2011. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 3:pii a005033. 10.1101/cshperspect.a005033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenlocher A, Horwitz AR. 2011. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3:a005074. 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burridge K, Sastry SK, Sallee JL. 2006. Regulation of cell adhesion by protein-tyrosine phosphatases. I. Cell-matrix adhesion. J. Biol. Chem. 281:15593–15596. 10.1074/jbc.R500030200 [DOI] [PubMed] [Google Scholar]
- 4.Zaidel-Bar R, Itzkovitz S, Ma'ayan A, Iyengar R, Geiger B. 2007. Functional atlas of the integrin adhesome. Nat. Cell Biol. 9:858–867. 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons JT. 2003. Focal adhesion kinase: the first ten years. J. Cell Sci. 116:1409–1416. 10.1242/jcs.00373 [DOI] [PubMed] [Google Scholar]
- 6.Schlaepfer DD, Mitra SK, Ilic D. 2004. Control of motile and invasive cell phenotypes by focal adhesion kinase. Biochim. Biophys. Acta 1692:77–102. 10.1016/j.bbamcr.2004.04.008 [DOI] [PubMed] [Google Scholar]
- 7.Schaller MD, Hildebrand JD, Shannon JD, Fox JW, Vines RR, Parsons JT. 1994. Autophosphorylation of the focal adhesion kinase, pp125FAK, directs SH2-dependent binding of pp60src. Mol. Cell. Biol. 14:1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing Z, Chen HC, Nowlen JK, Taylor SJ, Shalloway D, Guan JL. 1994. Direct interaction of v-Src with the focal adhesion kinase mediated by the Src SH2 domain. Mol. Biol. Cell 5:413–421. 10.1091/mbc.5.4.413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calalb MB, Polte TR, Hanks SK. 1995. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell. Biol. 15:954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Owen JD, Ruest PJ, Fry DW, Hanks SK. 1999. Induced focal adhesion kinase (FAK) expression in FAK-null cells enhances cell spreading and migration requiring both auto- and activation loop phosphorylation sites and inhibits adhesion-dependent tyrosine phosphorylation of Pyk2. Mol. Cell. Biol. 19:4806–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruest PJ, Roy S, Shi E, Mernaugh RL, Hanks SK. 2000. Phosphospecific antibodies reveal focal adhesion kinase activation loop phosphorylation in nascent and mature focal adhesions and requirement for the autophosphorylation site. Cell Growth Differ. 11:41–48 http://cgd.aacrjournals.org/cgi/content/full/11/1/41 [PubMed] [Google Scholar]
- 12.Zhao X, Guan JL. 2011. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 63:610–615. 10.1016/j.addr.2010.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra SK, Hanson DA, Schlaepfer DD. 2005. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6:56–68. 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- 14.von Wichert G, Jiang G, Kostic A, De Vos K, Sap J, Sheetz MP. 2003. RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages. J. Cell Biol. 161:143–153. 10.1083/jcb.200211061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su J, Muranjan M, Sap J. 1999. Receptor protein tyrosine phosphatase alpha activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol. 9:505–511. 10.1016/S0960-9822(99)80234-6 [DOI] [PubMed] [Google Scholar]
- 16.Zeng L, Si X, Yu WP, Le HT, Ng KP, Teng RM, Ryan K, Wang DZ, Ponniah S, Pallen CJ. 2003. PTP alpha regulates integrin-stimulated FAK autophosphorylation and cytoskeletal rearrangement in cell spreading and migration. J. Cell Biol. 160:137–146. 10.1083/jcb.200206049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, Chen SC, Pallen CJ. 2006. Integrin-induced tyrosine phosphorylation of protein-tyrosine phosphatase-alpha is required for cytoskeletal reorganization and cell migration. J. Biol. Chem. 281:11972–11980. 10.1074/jbc.M600561200 [DOI] [PubMed] [Google Scholar]
- 18.Sun G, Cheng SY, Chen M, Lim CJ, Pallen CJ. 2012. Protein tyrosine phosphatase alpha phosphotyrosyl-789 binds BCAR3 to position Cas for activation at integrin-mediated focal adhesions. Mol. Cell. Biol. 32:3776–3789. 10.1128/MCB.00214-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng XM, Resnick RJ, Shalloway D. 2000. A phosphotyrosine displacement mechanism for activation of Src by PTPalpha. EMBO J. 19:964–978. 10.1093/emboj/19.5.964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng XM, Shalloway D. 2001. Two mechanisms activate PTPalpha during mitosis. EMBO J. 20:6037–6049. 10.1093/emboj/20.21.6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Hertog J, Tracy S, Hunter T. 1994. Phosphorylation of receptor protein-tyrosine phosphatase alpha on Tyr789, a binding site for the SH3-SH2-SH3 adaptor protein GRB-2 in vivo. EMBO J. 13:3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su J, Batzer A, Sap J. 1994. Receptor tyrosine phosphatase R-PTP-alpha is tyrosine-phosphorylated and associated with the adaptor protein Grb2. J. Biol. Chem. 269:18731–18734 [PubMed] [Google Scholar]
- 23.den Hertog J, Hunter T. 1996. Tight association of GRB2 with receptor protein-tyrosine phosphatase alpha is mediated by the SH2 and C-terminal SH3 domains. EMBO J. 15:3016–3027 [PMC free article] [PubMed] [Google Scholar]
- 24.Su J, Yang LT, Sap J. 1996. Association between receptor protein-tyrosine phosphatase RPTPalpha and the Grb2 adaptor. Dual Src homology (SH) 2/SH3 domain requirement and functional consequences. J. Biol. Chem. 271:28086–28096 [DOI] [PubMed] [Google Scholar]
- 25.Schlaepfer DD, Hou S, Lim ST, Tomar A, Yu H, Lim Y, Hanson DA, Uryu SA, Molina J, Mitra SK. 2007. Tumor necrosis factor-alpha stimulates focal adhesion kinase activity required for mitogen-activated kinase-associated interleukin 6 expression. J. Biol. Chem. 282:17450–17459. 10.1074/jbc.M610672200 [DOI] [PubMed] [Google Scholar]
- 26.Tomar A, Lim ST, Lim Y, Schlaepfer DD. 2009. A FAK-p120RasGAP-p190RhoGAP complex regulates polarity in migrating cells. J. Cell Sci. 122:1852–1862. 10.1242/jcs.046870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhandari V, Lim KL, Pallen CJ. 1998. Physical and functional interactions between receptor-like protein-tyrosine phosphatase alpha and p59fyn. J. Biol. Chem. 273:8691–8698. 10.1074/jbc.273.15.8691 [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Wang R, Li QF, Tang DD. 2009. Abl knockout differentially affects p130 Crk-associated substrate, vinculin, and paxillin in blood vessels of mice. Am. J. Physiol. Heart Circ. Physiol. 297:H533–H539. 10.1152/ajpheart.00237.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang IK, Zhang J, Chiang YJ, Kole HK, Cronshaw DG, Zou Y, Gu H. 2010. Grb2 functions at the top of the T-cell antigen receptor-induced tyrosine kinase cascade to control thymic selection. Proc. Natl. Acad. Sci. U. S. A. 107:10620–10625. 10.1073/pnas.0905039107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilghman RW, Slack-Davis JK, Sergina N, Martin KH, Iwanicki M, Hershey ED, Beggs HE, Reichardt LF, Parsons JT. 2005. Focal adhesion kinase is required for the spatial organization of the leading edge in migrating cells. J. Cell Sci. 118:2613–2623. 10.1242/jcs.02380 [DOI] [PubMed] [Google Scholar]
- 31.Ahmed Z, Lin CC, Suen KM, Melo FA, Levitt JA, Suhling K, Ladbury JE. 2013. Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity. J. Cell Biol. 200:493–504. 10.1083/jcb.201204106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsutsumi R, Takahashi A, Azuma T, Higashi H, Hatakeyama M. 2006. Focal adhesion kinase is a substrate and downstream effector of SHP-2 complexed with Helicobacter pylori CagA. Mol. Cell. Biol. 26:261–276. 10.1128/MCB.26.1.261-276.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartman ZR, Schaller MD, Agazie YM. 2013. The tyrosine phosphatase SHP2 regulates focal adhesion kinase to promote EGF-induced lamellipodia persistence and cell migration. Mol. Cancer Res. 11:651–664. 10.1158/1541-7786.MCR-12-0578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hildebrand JD, Schaller MD, Parsons JT. 1995. Paxillin, a tyrosine phosphorylated focal adhesion-associated protein binds to the carboxyl terminal domain of focal adhesion kinase. Mol. Biol. Cell 6:637–647. 10.1091/mbc.6.6.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tachibana K, Sato T, D'Avirro N, Morimoto C. 1995. Direct association of pp125FAK with paxillin, the focal adhesion-targeting mechanism of pp125FAK. J. Exp. Med. 182:1089–1099. 10.1084/jem.182.4.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade R, Vande Pol S. 2006. Minimal features of paxillin that are required for the tyrosine phosphorylation of focal adhesion kinase. Biochem. J. 393:565–573. 10.1042/BJ20051241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlaepfer DD, Hanks SK, Hunter T, van der Geer P. 1994. Integrin-mediated signal transduction linked to Ras pathway by GRB2 binding to focal adhesion kinase. Nature 372:786–791. 10.1038/372786a0 [DOI] [PubMed] [Google Scholar]
- 38.Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. 2010. The FERM domain: organizing the structure and function of FAK. Nat. Rev. Mol. Cell Biol. 11:802–814. 10.1038/nrm2996 [DOI] [PubMed] [Google Scholar]
- 39.Arold ST. 2011. How focal adhesion kinase achieves regulation by linking ligand binding, localization and action. Curr. Opin. Struct. Biol. 21:808–813. 10.1016/j.sbi.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper LA, Shen TL, Guan JL. 2003. Regulation of focal adhesion kinase by its amino-terminal domain through an autoinhibitory interaction. Mol. Cell. Biol. 23:8030–8041. 10.1128/MCB.23.22.8030-8041.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lietha D, Cai X, Ceccarelli DF, Li Y, Schaller MD, Eck MJ. 2007. Structural basis for the autoinhibition of focal adhesion kinase. Cell 129:1177–1187. 10.1016/j.cell.2007.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen HC, Appeddu PA, Parsons JT, Hildebrand JD, Schaller MD, Guan JL. 1995. Interaction of focal adhesion kinase with cytoskeletal protein talin. J. Biol. Chem. 270:16995–16999. 10.1074/jbc.270.28.16995 [DOI] [PubMed] [Google Scholar]
- 43.Hall JE, Fu W, Schaller MD. 2011. Focal adhesion kinase: exploring Fak structure to gain insight into function. Int. Rev. Cell Mol. Biol. 288:185–225. 10.1016/B978-0-12-386041-5.00005-4 [DOI] [PubMed] [Google Scholar]
- 44.Lawson C, Lim ST, Uryu S, Chen XL, Calderwood DA, Schlaepfer DD. 2012. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J. Cell Biol. 196:223–232. 10.1083/jcb.201108078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hagel M, George EL, Kim A, Tamimi R, Opitz SL, Turner CE, Imamoto A, Thomas SM. 2002. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 22:901–915. 10.1128/MCB.22.3.901-915.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheswohl DM, Harrell JR, Rajfur Z, Gao G, Campbell SL, Schaller MD. 2008. Multiple paxillin binding sites regulate FAK function. J. Mol. Signal. 3:1. 10.1186/1750-2187-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown MC, Perrotta JA, Turner CE. 1996. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135:1109–1123. 10.1083/jcb.135.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas JW, Cooley MA, Broome JM, Salgia R, Griffin JD, Lombardo CR, Schaller MD. 1999. The role of focal adhesion kinase binding in the regulation of tyrosine phosphorylation of paxillin. J. Biol. Chem. 274:36684–36692. 10.1074/jbc.274.51.36684 [DOI] [PubMed] [Google Scholar]
- 49.Hoellerer MK, Noble ME, Labesse G, Campbell ID, Werner JM, Arold ST. 2003. Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 11:1207–1217. 10.1016/j.str.2003.08.010 [DOI] [PubMed] [Google Scholar]
- 50.Guan S, Chen M, Woodley D, Li W. 2007. Nckbeta adapter controls neuritogenesis by maintaining the cellular paxillin level. Mol. Cell. Biol. 27:6001–6011. 10.1128/MCB.01807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maignan S, Guilloteau JP, Fromage N, Arnoux B, Becquart J, Ducruix A. 1995. Crystal structure of the mammalian Grb2 adaptor. Science 268:291–293. 10.1126/science.7716522 [DOI] [PubMed] [Google Scholar]
- 52.McDonald CB, Seldeen KL, Deegan BJ, Lewis MS, Farooq A. 2008. Grb2 adaptor undergoes conformational change upon dimerization. Arch. Biochem. Biophys. 475:25–35. 10.1016/j.abb.2008.04.008 [DOI] [PubMed] [Google Scholar]
- 53.Lin CC, Melo FA, Ghosh R, Suen KM, Stagg LJ, Kirkpatrick J, Arold ST, Ahmed Z, Ladbury JE. 2012. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell 149:1514–1524. 10.1016/j.cell.2012.04.033 [DOI] [PubMed] [Google Scholar]
- 54.Jia CY, Nie J, Wu C, Li C, Li SS. 2005. Novel Src homology 3 domain-binding motifs identified from proteomic screen of a Pro-rich region. Mol. Cell. Proteomics 4:1155–1166. 10.1074/mcp.M500108-MCP200 [DOI] [PubMed] [Google Scholar]
- 55.McDonald CB, El Hokayem J, Zafar N, Balke JE, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Farooq A. 2013. Allostery mediates ligand binding to Grb2 adaptor in a mutually exclusive manner. J. Mol. Recognit. 26:92–103. 10.1002/jmr.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.