Abstract
Dengue virus (DENV) interacts with host cellular factors to construct a more favorable environment for replication, and the interplay between DENV and the host cellular cytoskeleton may represent one of the potential antiviral targeting sites. However, the involvement of cellular vimentin intermediate filaments in DENV replication has been explored less. Here, we revealed the direct interaction between host cellular vimentin and DENV nonstructural protein 4A (NS4A), a known component of the viral replication complex (RC), during DENV infection using tandem affinity purification, coimmunoprecipitation, and scanning electron microscopy. Furthermore, the dynamics of vimentin-NS4A interaction were demonstrated by using confocal three-dimensional (3D) reconstruction and proximity ligation assay. Most importantly, we report for the first time the discovery of the specific region of NS4A that interacts with vimentin lies within the first 50 amino acid residues at the cytosolic N-terminal domain of NS4A (N50 region). Besides identifying vimentin-NS4A interaction, vimentin reorganization and phosphorylation by calcium calmodulin-dependent protein kinase II occurs during DENV infection, signifying that vimentin reorganization is important in maintaining and supporting the DENV RCs. Interestingly, we found that gene silencing of vimentin by small interfering RNA induced a significant alteration in the distribution of RCs in DENV-infected cells. This finding further supports the crucial role of intact vimentin scaffold in localizing and concentrating DENV RCs at the perinuclear site, thus facilitating efficient viral RNA replication. Collectively, our findings implicate the biological and functional significance of vimentin during DENV replication, as we propose that the association of DENV RCs with vimentin is mediated by DENV NS4A.
INTRODUCTION
Dengue virus (DENV) is an arthropod-borne virus classified as a member of the Flaviviridae family containing a single-stranded positive-polarity RNA genome of approximately 10.9 kb. The genomic RNA consists of a single open reading frame encoding a polyprotein, which is co- and posttranslationally processed by different host proteases and cytoplasmic viral nonstructural protein 2B (NS2B)-NS3 protease complex into three structural proteins, capsid, premembrane, and envelope, and seven NS proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 (1, 2). Flaviviral replication complex (RC) is believed to comprise the viral RNA (vRNA) template with the NS proteins and presumably some host proteins on cytoplasmic membranes (3, 4). The intracellular membranes undergo dramatic rearrangements upon induction by NS proteins to form unique membrane structures localized at the perinuclear region of infected cells. Flaviviruses are distinctive, as they induce at least two or three characteristic structures, including convoluted membranes, paracrystalline arrays, and vesicle packets (VPs)/smooth membrane structures (5, 6). The VPs induced by DENV appear as clusters of double-membrane vesicles of 80 to 150 nm in size (7). These virus-induced endoplasmic reticulum (ER)-derived membranous compartments may serve as a scaffold for the viral RCs that serve as sites of DENV RNA replication.
The limited genetic capacity of viruses led to their exploitation of host cellular factors to facilitate the completion of their life cycle, starting from entry till egress. Thus, viruses interact with cellular proteins to carry out activities not encoded in the viral genome to manipulate cellular pathways in order to create a more favorable environment for replication. However, few interactions between DENV and human proteins have been reported thus far. The major cellular contributors in virus-host interactions are the host cytoskeletal network, which serve a role for virus entry, transport to reach the replication sites, and egress (8, 9). Three cytoskeletal polymeric elements—microfilaments (5 to 6 nm in diameter), intermediate filaments (IFs) (7 to 10 nm), and microtubules (20 to 25 nm)—and a set of accessory proteins cooperate to contribute to the physical integrity and structural organization of the cytoplasm in eukaryotic cells (10). While the roles of two of the major cytoskeletal elements, microfilaments and microtubules, have been widely studied with respect to virus replication, very little is known about the third element, the IFs.
Vimentin, a 57-kDa protein, is a major component of type III IFs found in cells of mesenchymal origin and is also present in cells adapted to tissue culture and many transformed cell lines (11). Recent research has helped elucidate the significance of vimentin IFs in vesicular and organelle transport and organelle positioning and as dynamic elements (12). Vimentin reorganization in cells involves filament disassembly regulated by phosphorylation of N-terminal domains by cellular kinases (13), allowing the transport of filaments along microtubules (14). Vimentin is also redistributed in cells during virus infections. Many viruses induce the depolymerization and rearrangements of cytoskeletal filaments to alter the diffusion properties of the cytoplasm. However, evidence for the role of the dynamic vimentin IFs in the DENV replication cycle is lacking, and their association is not well characterized. As such, studies are required to understand the significance of vimentin in DENV replication.
Similarly, little is known about the function of NS4A apart from its hydrophobic nature. Its hydrophobic nature, as well as complementation analysis, potentially implicates it in proper localization of viral proteins and vRNA to sites of RNA synthesis and virion assembly (3). NS4A is a small protein comprising 150 amino acid residues with a molecular mass of approximately 16 kDa. A study by Miller and coworkers (15) has demonstrated that NS4A associates with ER membranes via four internal hydrophobic regions, where the N-terminal region resides in the cytoplasm, while the C-terminal region localizes in the ER lumen. They have also shown that NS4A lacking the 2K fragment induces intracellular membrane alterations which may harbor the DENV RC (15). Yet, definitive roles of NS4A have not yet been established, and its proper role for the DENV replication cycle is still unknown.
In order to develop new strategies to prevent and treat DENV infection, a deeper understanding of the molecular interactions of DENV proteins with host cellular factors during infection is necessary. In this study, we revealed the interaction between DENV NS4A and vimentin and further identified the region of NS4A that specifically interacts with vimentin. We also investigated the reorganization of vimentin to the perinuclear site during DENV infection, which involves phosphorylation of vimentin filaments by calcium calmodulin-dependent protein kinase II gamma (CaMKIIγ). Last, we considered the functional significance of this interaction and reorganization and the structural role vimentin plays in supporting the DENV RCs, since reorganization occurs primarily around sites of dengue viral replication and assembly.
MATERIALS AND METHODS
Cell lines and viruses.
The Huh-7 cell line was maintained in Dulbecco modified Eagle medium (DMEM) (Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS) (Nagase) and was a kind gift from Priscilla Yang, Department of Microbiology and Immunobiology, Harvard Medical School. BHK-21 cells from ATCC were grown in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% FCS. Aedes albopictus-derived C6/36 cells from ATCC were grown in L-15 medium (Leibovitz) (Sigma) supplemented with heat-inactivated 10% FCS. Huh-7 cells stably expressing different regions of truncated NS4A-EmGFP (emerald green fluorescent protein) fusion proteins were additionally supplemented with blasticidin (10 μg/ml) (Invitrogen) to maintain the expression of truncated NS4A fusion proteins. Huh-7 cells stably expressing dengue virus 2 (DENV-2) pcTAP-NS4A (TAP stands for tandem affinity purification) or pcTAP vector were supplemented with 500 μg/ml of G418 (PAA Laboratories). High-titer stock of DENV-2 New Guinea C (NGC) strain was propagated in C6/36 cells. For preparation of UV-irradiated DENV-2, DENV-2 was subjected to UV light irradiation for 2.5 h, and viral plaque assay was performed to ensure complete inactivation of all infectious DENV-2. A monolayer of Huh-7 cells was infected with DENV-2 NGC strain or UV-inactivated DENV-2 at a multiplicity of infection (MOI) of 10.
Plasmid constructs and transfection.
The construction of the plasmid pcTAPNS4A and transfection to generate a stable Huh-7 cell line expressing DENV-2 pcTAPNS4A fusion protein and pcTAP vector were performed previously by members of the laboratory (16). For the generation of different truncated NS4A-EmGFP fusion plasmid constructs, the insertion of the fragments of the gene of interest (gene encoding DENV NS4A) [amino acids 1 to 50 (N50), amino acids 1 to 74 (N74), amino acids 1 to 100 (N100), amino acids 51 to 125 (N51-127), and full-length NS4A lacking the 2K sequence consisting of amino acids 1 to 127 NS4A (−2K)] into the pcDNA6.2/C-EmGFP-GW/TOPO plasmid vector (Invitrogen) were carried out according to the manufacturer's protocol. The sequences of the primers used for cloning of different regions of NS4A are available upon request. The cDNA template used is the full-length DENV-2 NGC strain (NCBI accession no. NC_001474.2). Transfection of Huh-7 cells was carried out using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's protocol.
Western blot analysis.
Protein samples were loaded onto a 10% polyacrylamide gel. Proteins resolved by SDS-PAGE were transferred from the gel onto a nitrocellulose membrane (Bio-Rad). Thereafter, the membrane was blocked using 5% skim milk for 1 h at room temperature before incubation with desired primary antibodies of appropriate dilution (mouse monoclonal antivimentin antibody [Millipore], rat polyclonal anti-phosphorylated vimentin antibody [MBL], rabbit polyclonal anti-DENV NS4A antibody [GeneTex], rabbit polyclonal anti-CaMKIIγ antibody [Upstate], rabbit polyclonal anti-GFP antibody [Invitrogen], and mouse monoclonal antiactin antibody [Millipore]) at 4°C overnight. After three 10-min washes with Tris-buffered saline containing 0.1% Tween 20 (TBST), the membrane was probed using species-specific secondary antibodies (mouse, rabbit, or rat secondary antibodies conjugated to horseradish peroxidase [Thermo Scientific]) for 1 h at room temperature. For detection of protein bands, the membrane was treated with enhanced chemiluminescent substrate (Thermo Scientific) for 5 min and exposed to an X-ray film (Thermo Scientific), which was subsequently developed with a Kodak X-OMAT 2000 processer in the dark.
TAP.
DENV-2 NS4A-TAP (tandem affinity purification) fusion protein-containing complexes were purified using the InterPlay mammalian TAP system according to the manufacturer's protocol (Stratagene) with minor modifications. Briefly, ten 175-cm2 flasks of confluent Huh-7 cells stably expressing DENV-2 NS4A-TAP protein or control TAP protein were harvested and lysed. Then, 0.5 M EDTA and 14.4 M β-mercaptoethanol were added to the supernatant collected after the cells had been spun down for 10 min at 16,000 × g at 4°C. The supernatant was concentrated using 3-kDa Amicon Ultra centrifugal filter unit and then passed over streptavidin resin. Following an overnight incubation at 4°C, the eluted complexes were added to calmodulin resin and incubated at 4°C for 2 h. The elution of interacting proteins was performed by boiling the resin in 1× SDS-PAGE loading buffer for 5 min. The protein samples were resolved on 10% SDS-polyacrylamide gel using Hoefer SE600, fixed in gel-fixing solution, and stained using Coomassie blue staining solution. After destaining, the protein bands were excised and sent for mass spectrometric analysis using the TripleTOF 5600 LC/MS/MS (liquid chromatography coupled to tandem mass spectrometry) system (AB SCIEX) for identification of the protein partners.
Coimmunoprecipitation (Co-IP) assay.
Two T175-cm2 cell culture flasks containing confluent Huh-7 cells were infected with DENV-2 at an MOI of 10 and lysed at 24, 48, and 72 h postinfection, or transfected Huh-7 cells stably expressing different NS4A-EmGFP fusion proteins were used. Cells were lysed in cell lysis buffer (mammalian protein extraction reagent, protease and phosphatase inhibitor cocktail, and 0.5 M EDTA [Pierce]). Subsequently, supernatant was collected after clearing by centrifugation for 10 min at 16,000 × g at 4°C. Dynabeads protein G (Invitrogen) was preincubated with mouse monoclonal antivimentin antibody (Millipore) or rabbit polyclonal anti-DENV NS4A (kind gift from Ralf Bartenschlager, University of Heidelberg) or rabbit polyclonal anti-GFP antibody (Santa Cruz) for 20 min at room temperature and then incubated with cell lysate for 1 h at 4°C on a HulaMixer sample mixer. Thereafter, the Dynabeads-antibody (Ab)-antigen (Ag) complexes were washed three times using the washing buffer (Invitrogen) provided, and the target antigen was eluted in the elution buffer (Invitrogen) provided for 8 min at room temperature. The samples were analyzed by 10% SDS-PAGE, and resolved proteins were detected by Western blotting. Isotype control and mock-infected samples were included as controls for the experiments.
In situ PLA.
An in situ proximity ligation assay (PLA) was performed to determine protein-protein interaction event in infected cells according to the manufacturer's protocol (Olink Biosciences). Huh-7 cells or transfected stable cells were seeded onto sterile coverslips. Cells were infected with DENV-2 at an MOI of 10 and fixed at every 12-h interval from 0 h up to 72 h postinfection with cold absolute methanol (Merck) for 10 min at −20°C. The samples were incubated with primary antibodies in the antibody diluents provided overnight at 4°C. The primary antibodies were mouse monoclonal antivimentin (Millipore) and rabbit polyclonal anti-DENV NS4A or rabbit polyclonal anti-GFP (Santa Cruz). The secondary antibodies, PLA probe anti-mouse PLUS and anti-rabbit MINUS, were used at a 1:5 dilution and incubated for 1 h at 37°C. Thereafter, ligation-ligase solution was added and incubated for 30 min at 37°C. The signal development was performed by adding amplification-polymerase solution and incubating the resulting solution for 100 min at 37°C. Last, Duolink in situ mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) was used to mount the coverslips onto glass slides. Imaging was carried out using a conventional optical fluorescence microscope (Olympus IX81) with oil immersion objectives. Samples immunolabeled with rabbit polyclonal anticalnexin antibody (Cell Signaling), and mock-infected or GFP-transfected cell samples served as negative controls. The PLA signals were quantified based on the fluorescence microscopy images. Image analysis was performed using software program CellProfiler (17), and quantitative measurements by spot counting were made to quantify the number of in situ PLA signals per cell (18).
siRNA reverse transfection.
The On-TARGET plus SMARTpool small interfering RNA (siRNA) constructs targeting vimentin (NCBI accession no. NM_003380) and CaMKIIγ (NCBI accession no. NM_001222), as well as the respective scrambled siRNA constructs, were purchased from Dharmacon (Thermo Scientific). The siRNA gene sequences are vimentin (UGAAGAAACUCCACGAAGA, GCAGAAGAAUGGUACAAAU, UGAAGUGGAUGCCCUUAAA, and AACUAGAGAUGGACAGGUU) and CaMKIIγ (GGAAAGAUCCCUAUGGAAA, GCUCGGAUAUGUCGACUUC, GCACAAACAUCUC-CACUUU, and GAAGAGAUCUAUACCCUAA). Scrambled siRNA sequences were designed using InvivoGen's siRNA Wizard at http://www.sirnawizard.com/scrambled.php, and the sequences are vimentin (GCAAUUCGAACAGGCAAAA, AGACCUACUUGAGGAGAAA, GAGUACCGAUUAGCAUAGU, and AUCGAUGACGGGAUGUAAA) and CaMKIIγ (GGAGACACGUAUAGCUAAA, AUCGCUCGUACCUUGGUAG, ACCUCCGUCCAAU-AAUCAU, and AUCAUACCGCAAGAUAAUG). Specific gene-targeting siRNAs and scrambled siRNAs were dissolved in diethyl pyrocarbonate (DEPC)-treated reverse osmosis water to a stock concentration of 10 μM. The different siRNAs were diluted to desired working concentrations of 5 nM, 15 nM, 25 nM, 35 nM, 45 nM, and 50 nM with DharmaFECT cell culture reagent (Thermo Scientific) and transfection reagent (Dharmafect-4). The siRNAs targeting CaMKIIγ and vimentin were then transfected into Huh-7 cells, and the cells were subsequently subjected to DENV-2 infection 48 or 72 h after transfection, respectively. Viral supernatant was harvested 72 h postinfection for plaque assay, and cells were fixed for immunofluorescence microscopy (IFM) and harvested 48 h after transfection for Western blotting and quantitative reverse transcription-PCR (qRT-PCR).
Drug treatment assay.
Huh-7 cells were seeded on coverslips and infected with DENV-2 at an MOI of 10 for 36 h before treatment with KN93 (50 μM) (Abcam) for another 12 h (19). The treated cell monolayer was fixed for IFM in which rat polyclonal anti-phosphorylated vimentin antibody (serine-38) (MBL) or mouse monoclonal antivimentin antibody (Millipore) and rabbit polyclonal anti-DENV NS4A antibody (GeneTex) were used. Huh-7 cells that had been infected with DENV-2 and treated with 0.1% dimethyl sulfoxide (DMSO) were used as the solvent control.
Cell viability assay.
alamarBlue cell viability assay was used to assess the viability of cells following siRNA transfection and drug treatments. Huh-7 cells seeded onto a 96-well plate were treated with siRNA or drugs at desired concentrations as described above. Then, 48 or 72 h after transfection and 12 h after treatment as described above, the cells were washed before the medium was replaced with 90 μl of culture medium and 10 μl of alamarBlue reagent (Invitrogen). The plate was incubated for 2 h at 37°C in the dark before being analyzed for absorbance using an Infinite M200 microplate reader (Tecan) and an excitation wavelength of 570 nm and emission wavelength of 585 nm.
Total RNA extraction and qRT-PCR.
Upon gene silencing, Huh-7 cells were harvested for total RNA extraction using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. The samples were assayed in a 20-μl reaction mixture consisting of 10 μl of Maxima SYBR green/ROX qPCR master mix (Fermentas), 1 μl of Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega), 0.5 μl of forward and reverse primer (0.3 μM each), 1 μl of total RNA, and 7 μl of nuclease-free water. The qRT-PCR was performed using the following primers: CaMKIIγ (forward, 5′-CAGGAGTACGCAGCAAAAATCA-3′; reverse, 5′-CGAGCCTCACGTTCTAGTTTC-3′) and actin control (forward, 5′-AGCGCGGCTACAGCTTCA-3′; reverse, 5′-GGCGACGTAGCACAGCTTCT-3′). The reaction mixture was subjected to a RT step at 44°C for 30 min to generate the cDNA and initial denaturation of the RNA-DNA hybrid at 95°C for 5 min, followed by 40 cycles of PCR with denaturation occurring at 95°C for 15 s, with annealing and extension taking place at 60°C for 30 s. Thermal cycling reactions were carried out using the StepOne Plus real-time PCR system (Applied Biosystems).
Indirect immunofluorescence microscopy.
The virus-infected or mock-infected cell monolayer on coverslips was fixed with cold absolute methanol (Merck) for 10 min at −20°C or 4% paraformaldehyde (Sigma-Aldrich) and 0.1% Triton X-100 (Sigma) for 10 min at room temperature. For staining of the ER, prewarmed ER Tracker Red (BODIPY TR) glibenclamide (Invitrogen) was added before fixation and incubated at 37°C with 5% CO2 for 20 min. The cells were then incubated with desired primary antibodies of appropriate dilution (mouse monoclonal antivimentin antibody [Millipore], mouse monoclonal anti-double-stranded RNA [anti-dsRNA] antibody [English & Scientific Consulting], rat polyclonal anti-phosphorylated vimentin antibody [MBL], goat polyclonal antivimentin antibody [R & D Systems], rabbit polyclonal anti-DENV NS4A antibody [GeneTex], and rabbit polyclonal anti-DENV2 antibody [US Biologicals]) overnight at 4°C. Upon removal of primary antibodies, cells were incubated with secondary antibodies (Alexa Fluor 488-conjugated goat anti-rat IgG [Invitrogen], Alexa Fluor 633-conjugated goat anti-mouse IgG [Invitrogen], fluorescein isothiocyanate [FITC]-conjugated goat anti-rabbit IgG [Millipore], and rhodamine-conjugated donkey anti-goat IgG [Abcam]) for 2 h at room temperature. Subsequently, DAPI (Invitrogen) was used to stain cell nuclei before mounting the coverslip on a microscope slide with 1,4-diazabicyclo[2.2.2]octane (DABCO) (Invitrogen). The specimens were viewed with a 100× oil immersion lens with a numerical aperture (NA) of 1.6 of a conventional optical-fluorescence microscope (Olympus IX81) and a laser-scanning confocal microscope (Olympus Fluoview FV1000). For three-dimensional (3D) reconstruction of confocal images, cell optical sectioning by confocal microscope (z-stacks) was performed to achieve 30 optical slices (each slice 0.3 μm thick), and alignment and stacking of the individual optical sections reconstructed a 3D volume of the cell. Colocalization was quantified based on the fluorescence microscopy images. The analysis of images (n = 15) was performed using the NIH ImageJ software (Wright Cell Imaging Facility) via the colocalization analysis plug-in. Manders overlap coefficient (MOC) as a measure of colocalization represents the proportion of normalized pixels in which the two signals are colocalized (20). MOC ranges from 0 for no colocalization to 1 for perfect colocalization. We determined the percentage of colocalization between the regions of interest at respective time points based on the quantification of colocalization coefficients.
Scanning electron microscopy.
Huh-7 cells were seeded on coverslips treated with 0.1% poly-l-lysine (Sigma) and infected with DENV-2 at an MOI of 10. Forty-eight hours after infection, the cells were rinsed with phosphate-buffered saline (PBS) and extracted. Extraction of the cells was performed to obtain cytosolic free cytoskeleton structures based on procedures of Svitkina and coworkers (21) and Verkhovsky and Borisy (22) with minor modifications. For microtubule (MT)-free intermediate filament (IF)-enriched cytoskeletal preparations, cells were extracted in 100 μl of PEM (100 mM PIPES [Sigma] [pH 6.9] [adjusted with KOH], 1 mM EGTA, and 1 mM MgCl2) buffer with 1% Triton X-100 (Sigma) and 0.4 M NaCl (Merck) for 6 min at room temperature. Lysed cells were washed three times with buffer A consisting of 50 mM morpholineethanesulfonic acid (MES)-KOH (pH 6.3), 0.1 mM CaCl2, 2 mM MgC12, and 0.5 mM dithiothreitol (BDH), and incubated in 40 μl of 0.1-mg/ml gelsolin (purified from human plasma; cytoskeleton) in buffer A for 1 h at room temperature to remove any residual actin. After incubation in gelsolin, the cells were washed again with buffer A and then fixed with 4% paraformaldehyde (Sigma-Aldrich) and 0.1% glutaraldehyde (electron microscopy [EM] grade; Sigma) in PBS for 30 min at 4°C. For double immunogold labeling, the cells were washed with 0.05 M glycine (Fisher Scientific) in PBS for 20 min, blocked with 5% bovine serum albumin (BSA) (Sigma) for 30 min, and incubated with the desired primary antibodies in incubation buffer (0.1% BSA in PBS) overnight at 4°C in a humidified chamber. The primary antibodies used were mouse monoclonal antivimentin (Millipore) and rabbit polyclonal anti-DENV NS4A (GeneTex). Thereafter, the cells were probed with species-specific secondary antibodies, goat anti-mouse antibody conjugated to 5-nm gold and goat anti-rabbit conjugated to 10-nm gold (Ted Pella) for 3 h at 4°C. The secondary antibodies were washed out with incubation buffer, and the cells were fixed with 2% glutaraldehyde for 20 min. The samples were postfixed in 2% osmium tetroxide for 10 min before being dehydrated through a series of concentrations of ethanol. These preparations were then processed by critical point drying to dry sample by CO2 infiltration (Balzers critical point dryer 030). The coverslips were mounted on scanning electron microscopy (SEM) stub and transferred to a vacuum evaporator to coat the specimen with a thin layer of carbon approximately 3 nm thick. The samples were observed and photographed with a JEOL JSM-6701F field emission scanning electron microscope.
Flow cytometric analysis for annexin V binding assay.
The annexin V-FITC kit (Miltenyi Biotec) was used to detect early apoptotic events and to discriminate apoptotic, necrotic, and dead cells during DENV-2 infection. Upon harvesting by trypsinization, cells were treated with annexin V-FITC and propidium iodide (PI) (100-μg/ml) solution according to the manufacturer's recommendations. The DENV-2-infected and mock-infected samples (minimum of 10,000 events per sample) were analyzed with BD LSR Fortessa flow cytometry analyzer (Beckman Coulter), and Submit 4.3 software (Beckman Coulter) was then used to calculate the percentage of apoptotic cells and the mean percentages and standard deviations derived at each time point postinfection.
RESULTS
Vimentin is identified as a DENV NS4A-binding protein.
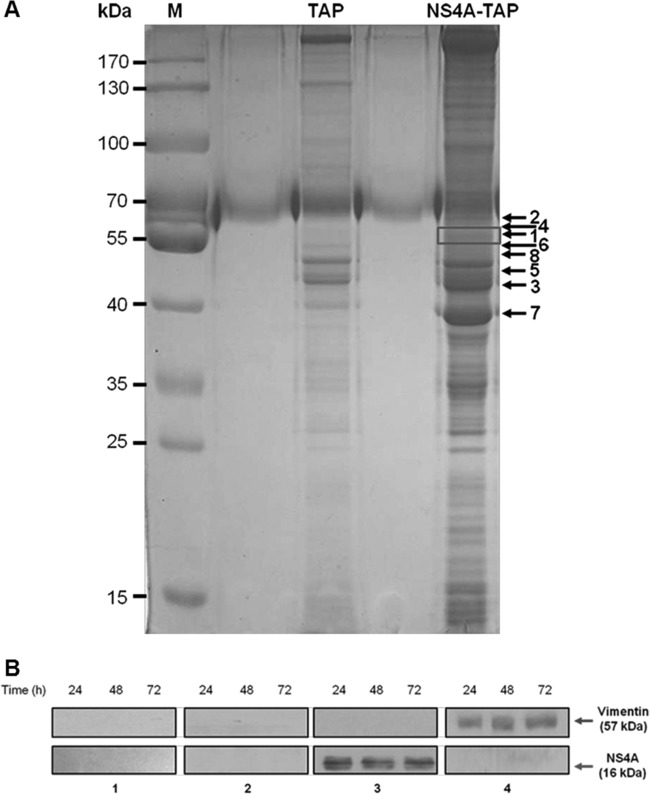
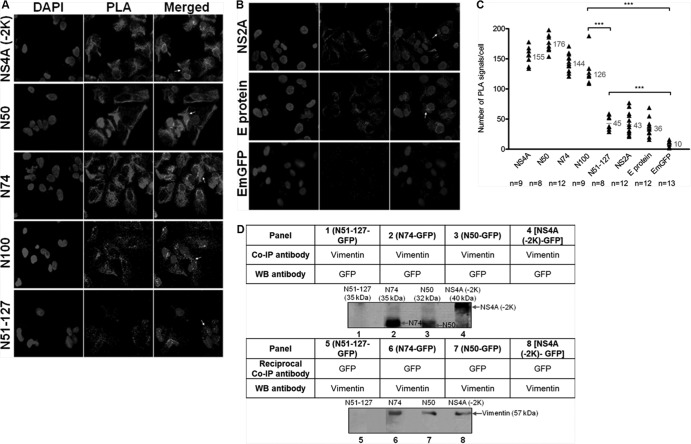
The exact role of NS4A in DENV replication has not yet been well defined, although it has been documented to be involved in the replication of flaviviruses (4, 23). Furthermore, the interactions of DENV NS4A with host cellular proteins are not well-known. To investigate which host cellular proteins interact with DENV NS4A, we performed a combination of tandem affinity purification (TAP) assay along with mass spectrometry. The purified NS4A-containing protein complexes were visualized using Coomassie blue staining (Fig. 1A). Eight prominent bands were excised, subjected to reduction and digestion in the gel, and analyzed by LC-MS/MS for protein sequencing and identification. In the mass spectrometric analysis, host cellular vimentin was identified as one of the DENV NS4A-binding proteins (Table 1).
FIG 1.
Vimentin is identified as a DENV NS4A-binding protein. (A) Huh-7 cells stably transfected with the pcTAP-NS4A or pcTAP vector (control) constructs were lysed, and the DENV-2 NS4A protein complexes were sequentially purified first by using streptavidin resin and then by using calmodulin resin. The samples were resolved on a 10% SDS-polyacrylamide gel, and bands were visualized using Coomassie blue staining. The box in the NS4A-TAP lane indicates the band identified as vimentin by mass spectrometry, and the black arrows mark the positions of the seven other proteins identified. The positions of the protein markers (M) (in kilodaltons) are indicated to the left of the gel. (B) Huh-7 cells were infected with DENV-2 at an MOI of 10 and lysed at the three stipulated time points. Infected and mock-infected total cell lysates were incubated with vimentin, NS4A, or IgG isotype antibodies and Dynabeads protein G were added to capture the protein complexes. Samples with eluted target proteins were analyzed by Western blotting using anti-NS4A or antivimentin antibodies. Mock-infected samples and IgG isotype control are shown in gels 1 and 2, respectively, and are the negative controls. Gels 3 and 4 show the infected samples in which Co-IP of NS4A with vimentin and its reciprocal assay, respectively, confirmed the interaction. The specific protein bands representing vimentin and NS4A are shown by arrows to the right of the gels.
TABLE 1.
Mass spectrometry results obtained with NS4A-TAP eluate
| Proteina | Protein identification | Accession no. | %Covb | Peptides (95%)c |
|---|---|---|---|---|
| 1 | Vimentin (Homo sapiens) | sp|P08670| | 69.1 | 43 |
| 2 | Pyruvate kinase isoenzymes M1/M2 (Homo sapiens) | sp|P14618| | 64.8 | 32 |
| 3 | Alpha-enolase (Homo sapiens) | sp|P06733| | 84.8 | 109 |
| 4 | Glucose-6-phosphate 1-dehydrogenase (Homo sapiens) | sp|P11413| | 61.4 | 28 |
| 5 | Tubulin beta chain (Homo sapiens) | sp|P07437| | 79.3 | 74 |
| 6 | Elongation factor 1-alpha (Homo sapiens) | sp|P68104| | 83.6 | 44 |
| 7 | Actin, cytoplasmic 1 (Homo sapiens) | sp|P60709| | 86.4 | 80 |
| 8 | Rab GDP dissociation inhibitor beta (Homo sapiens) | sp|P50395| | 73.3 | 26 |
Protein numbers as in Fig. 1A.
%Cov, percent sequence coverage [(number of the matched residues/total number of residues in the entire sequence) × 100%].
Peptides (95%), number of peptides identified with more than 95% confidence.
The identification of vimentin as an interacting partner of NS4A was further confirmed by performing in vitro coimmunoprecipitation (Co-IP)-Western blotting analyses. Western blotting with anti-NS4A antibody showed that vimentin coimmunoprecipitated NS4A, as an approximately 16-kDa band corresponding to NS4A being captured specifically by the antivimentin antibody was observed at all three indicated time points after DENV infection (Fig. 1B, panel 3). Furthermore, the interaction was further verified by reciprocal Co-IP using anti-NS4A antibody and Western blot analysis using antivimentin antibody. An approximately 57-kDa band corresponding to vimentin was observed at the stipulated time points, indicating that vimentin was similarly coimmunoprecipitated with NS4A (Fig. 1B, panel 4). For negative controls, no specific protein bands were detected using antivimentin or IgG isotype control antibodies against the lysates from the mock-infected cells or infected cells, respectively (Fig. 1B, panels 1 and 2, respectively). The data from Co-IP and its reciprocal assay confirm our initial results obtained with the TAP assay. Altogether, these findings indicate that vimentin directly interacts with NS4A, thus leading us to investigate further the role of vimentin-NS4A interaction.
Colocalization of vimentin with NS4A, ER, and dsRNA involved rearrangement of vimentin during DENV replication.
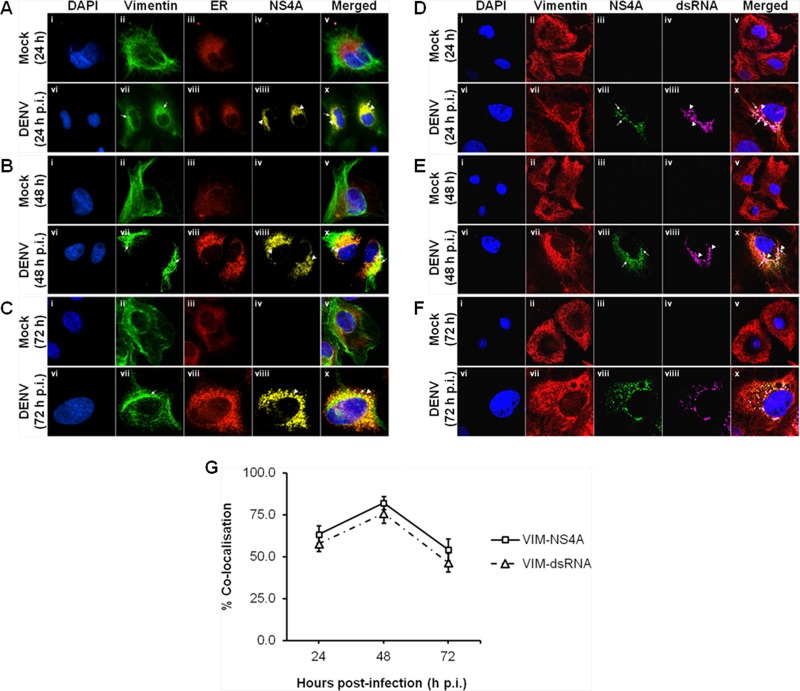
We next examine the interaction and colocalization of vimentin with NS4A using an immunofluorescence assay. Since it is believed that the virus-induced ER-derived membranous compartments serve as a scaffold for the DENV RCs and NS4A associates with ER membranes, we analyzed the colocalization of DENV RCs with vimentin as well as NS4A. The distribution patterns of vimentin and ER in mock- and DENV-infected cells were studied. In mock-infected cells, vimentin displayed an extended network arrangement from the perinuclear region outwards to the cell plasma membrane (Fig. 2A, B, and C, panels ii), and the ER presented the typical reticular structure appearance across the indicated time points (Fig. 2A, B, and C, panels iii). However, changes in the vimentin network and ER structure and intensity were observed in the DENV-infected cells (Fig. 2A, B, and C, panels vii to viii). DENV infection induced a significant change in the morphology of vimentin structure. Vimentin filaments have retracted from the cell periphery at 24 h postinfection (Fig. 2A, panel vii) and reorganized into dense structures around the perinuclear region whereby aggregation was more extensive at 48 h postinfection (Fig. 2B, panel vii). Flaviviruses induce proliferation and hypertrophy of the ER membranes during a productive infection (6). Similarly, the number of cytoplasmic foci of ER increased in infected cells, and the intensity of the ER was greater than in the mock-infected cells. In addition, it has lost the reticular structure appearance and appeared to have a punctuate staining pattern (Fig. 2A, B, and C, panels viii). A similar staining pattern was observed for DENV NS4A, and it appeared as dot-like structures which concentrated in the region occupied by the DENV RCs (Fig. 2A, B, and C, panels viiii) and displayed colocalization with ER and vimentin (Fig. 2A, B, and C, panels x). To quantify this colocalization, we determined the percentage of vimentin that colocalized with NS4A at the three stipulated time points based on MOC. The colocalization quantification showed a trend of increasing colocalization from 24 h postinfection (p.i.) to 48 h p.i. and peaked at 48 h p.i. (82%) before decreasing at 72 h p.i. (Fig. 2G). Since NS4A is assumed to serve as a scaffold for the formation of RCs (15, 24) and it localized to DENV-induced ER-derived structures, the colocalization studies suggest that substantial vimentin reorganization induced by DENV may occur jointly with the construction of DENV RCs. Furthermore, vimentin may associate with the RCs wherein it contributes to a structural role in anchoring and supporting the RCs.
FIG 2.
Colocalization of vimentin with NS4A, ER, and dsRNA involved rearrangement of vimentin during DENV replication. Huh-7 cells were infected with DENV-2 at an MOI of 10 and fixed with ice-cold methanol at 24, 48, and 72 h postinfection before processing for immunofluorescence analysis. (A, B, and C) Subcellular localization of vimentin (green), ER (red), and DENV NS4A (yellow) were visualized using LSCM, with the nuclei stained with DAPI (blue). ER Tracker Red (BODIPY TR) glibenclamide (Invitrogen) was used for the staining of ER. Colocalization was denoted by white arrowheads in panels x. The images were taken at a magnification of ×100. (D, E, and F) Subcellular localization of vimentin (red), DENV NS4A (green), and dsRNA (violet) were visualized using LSCM, with the nuclei stained with DAPI (blue). White arrows denote NS4A, and white arrowheads denote dsRNA. The images were taken at a magnification of ×200 (the images were enhanced by a ×2 magnification in addition to 100×). (G) Colocalization quantification was based on MOC using whole-cell immunofluorescence (WCIF) ImageJ software (20) and represented as percent colocalization at the respective time points postinfection.
Previous studies by Miller and coworkers have revealed that NS4A localizes largely in ER-derived cytoplasmic structures that contain double-stranded RNA (dsRNA) as well, suggesting that NS4A is a component of the DENV RCs (15). Since vimentin may associate with DENV RCs (Fig. 2A, B, and C), we investigated whether vimentin also colocalizes with dsRNA by performing immunofluorescence analysis, which leads to its proposed structural role during dengue virus vRNA replication. In mock-infected cells, no NS4A protein and dsRNA was detected (Fig. 2D, E, and F, panels iii and iv). On the other hand, the distribution of NS4A was observed to be analogous to what has been described earlier, in which it colocalized with vimentin across the indicated time points postinfection (Fig. 2D, E, and F, panels vii and viii). The punctuate staining pattern of dsRNA exhibited striking resemblance to the staining pattern of NS4A with only slight dissimilarities (Fig. 2D, E, and F, panels viii and viiii), thus confirming that NS4A indeed colocalized strongly with dsRNA and is a component of DENV RCs. Therefore, it was determined whether dsRNA colocalized with vimentin as well, and it was observed that reorganized vimentin not only colocalized with NS4A but also with dsRNA at 24, 48, and 72 h postinfection (Fig. 2D, E, and F, panels x). The quantification of colocalization between vimentin and dsRNA also showed a similar trend of increasing colocalization from 24 h p.i. to 48 h p.i. and peaked at 48 h p.i. (76%) before decreasing at 72 h p.i. (Fig. 2G). Taken together, vimentin colocalized with both NS4A and dsRNA at the sites of RNA replication upon DENV infection. This supports the idea of vimentin being one of the host cell factors that plays a part in the RNA amplification machinery consisting of vRNA and other viral proteins. Also, the findings from colocalization studies may suggest that DENV RCs may be tethered with vimentin in DENV-infected cells and that vimentin may consequently facilitate vRNA replication by anchoring the replication machineries in membrane compartments.
Dynamic interaction of vimentin with NS4A during DENV infection.
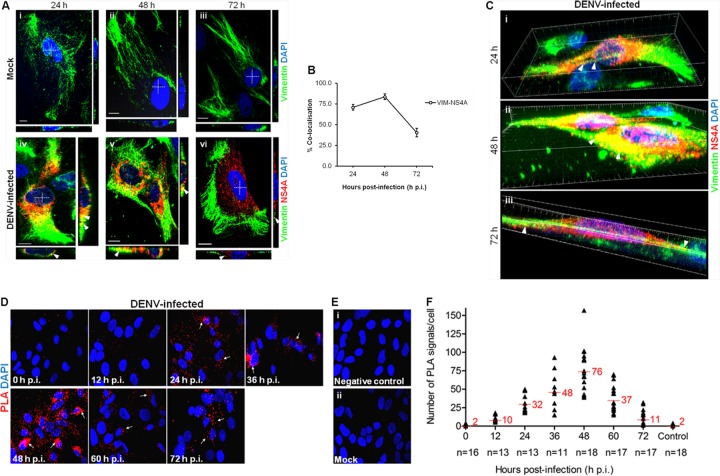
In order to further elucidate how vimentin forms a structural framework for anchoring NS4A, we examined the vimentin-NS4A interactions and vimentin reorganization by reconstructing three-dimensional (3D) confocal images of the entire vimentin network and NS4A protein distribution using laser-scanning confocal microscopy (LSCM), as it facilitates the generation of high-resolution 3D images from sections of cells. The colocalization studies led to the postulation that vimentin reorganization may lead to the formation of a physical scaffold that supports the DENV RCs, mediated by NS4A interaction. In mock-infected cells, vimentin was arranged in a filamentous network reaching the cellular periphery (Fig. 3A, panels i to iii). Upon DENV-2 infection, vimentin was observed to have lost the long filamentous shape by 24 h postinfection and showed colocalization with NS4A (Fig. 3A, panel iv). At 48 h postinfection, the vimentin was further rearranged and aggregated into dense short filaments (squiggles) that have strong colocalization with NS4A around the perinuclear site (Fig. 3A, panel v). Interestingly, remodeling of vimentin occurred at 72 h postinfection, when an enclosure surrounding the site where NS4A localized formed (Fig. 3A, panel vi). Indeed, the quantification of colocalization exhibited a parallel trend of increasing colocalization from 24 h p.i. (71%) to 48 h p.i., which peaked at 48 h p.i. (84%) before decreasing at 72 h p.i. (41%) (Fig. 3B). From the confocal 3D reconstruction images, we observed that there was indeed close association between vimentin and NS4A. Vimentin filaments had clustered together, but it was clear that NS4A aligned closely on the remaining filamentous vimentin at 24 h postinfection (Fig. 3C, panel i; see Video S1 in the supplemental material). Dynamics of vimentin allowed further aggregation of the filaments toward the cell center, constructing a physical scaffold at 48 h postinfection (Fig. 3C, panel ii). This suggests that the scaffolding framework of vimentin may hold the DENV RCs in place during RNA replication. Despite the conversion of vimentin from aggregates into cage-like structures surrounding NS4A during late infection, colocalization was still observed at 72 h postinfection (Fig. 3C, panel iii), as supported by Co-IP (Fig. 1B). This shows the dynamics of vimentin-NS4A protein interactions and thus led us to explore the pattern of interactions across various time points during the course of DENV infection.
FIG 3.
Dynamic interaction of vimentin with NS4A during DENV infection. (A) Colocalization of vimentin (green) with NS4A (red) was observed in DENV-infected cells (white arrowheads). The white cross denotes the site of xz-section and yz-section, and orthogonal sections at the indicated levels were presented. Magnification, ×100. Bar = 10 μm. (B) Colocalization quantification was based on MOC using WCIF ImageJ software (20) and represented as percent colocalization at the respective time points postinfection. VIM, vimentin. (C) Serial xy-planes were performed to obtain a 3D reconstruction of the vimentin scaffold. Snapshots of Video S1 in the supplemental material from confocal 3D reconstruction of the DENV-infected cells are shown, revealing the close interaction (white arrowheads) of vimentin and NS4A. Thirty optical slices of 0.3-μm z-section were obtained, and images were further processed using Imaris software. (D) Immunofluorescence detection of PLA signals (white arrows) in DENV-infected cells at various indicated time points. The red signals represent the vimentin-NS4A interactions, and the nuclei were stained with DAPI (blue). Vimentin- and NS4A-specific primary antibodies were used. Individual events of red PLA signal were used as reading output and quantified. (E) Cells infected and treated with anticalnexin antibody (panel i) and mock-infected cells (panel ii) served as negative controls. The images were taken at a magnification of ×100. (F) Numbers of PLA-positive foci detected in Huh-7 cells with wild-type DENV-2 infection across indicated time points. The number of in situ PLA signals represents vimentin-NS4A interactions at different time points postinfection. In situ PLA signals were quantitated on a per-cell basis using the software program CellProfiler (17) and the ImageJ program by spot counting (18). The numbers of cells counted (n) are shown, and average values are indicated in red.
From the confocal micrographs and 3D reconstructed images, we hypothesized that vimentin may assume different structural platforms across the infection time in which interaction with NS4A occurred. The postulation that dynamic vimentin-NS4A interactions may exist led us to investigate the pattern of interactions across various time points postinfection using the in situ proximity ligation assay (PLA). This recently developed proximity ligation technique allows the examination of subcellular localization of protein-protein interactions at single-molecule resolution and visualization and quantification of individual interacting pairs of protein molecules (25). The localization of interactions in situ is not lost, and the distribution of interacting proteins within individual cells can be demonstrated. It was observed that the red PLA fluorescent signals increased consistently from 12 to 36 h postinfection and that interaction between vimentin and NS4A was most prominent at 48 h postinfection when the number of PLA-positive foci representing sites of colocalization of vimentin with NS4A was greatest (Fig. 3D). In addition, the localization of vimentin-NS4A interactions was distributed around the perinuclear site, therefore further suggesting that vimentin was recruited to the perinuclear region by its reorganization mediated by NS4A interactions. As infection time progressed, the number of PLA signals declined, but there were still considerable interactions, indicating that the two proteins were still in close proximity (Fig. 3D). Interestingly, the localization of interactions at 72 h postinfection seems to be further away from the perinuclear region, which supports the observation of the cage-like structure of vimentin during late infection (Fig. 3A, panel vi). There were substantially fewer PLA-positive foci detected in cells treated with anticalnexin antibody (Fig. 3E, panel i) or in mock-infected cells, which served as controls (Fig. 3E, panel ii). From Fig. 3F, a trend was observed in the vimentin-NS4A interaction across the time after DENV infection. The association of vimentin with NS4A changed at different time points. The proximity study data and colocalization analysis showed that the strongest colocalization occurred 48 h postinfection and subsequently decreased at 72 h postinfection. Taken together, the data further imply that there is a dynamic interplay of vimentin and NS4A in DENV-infected cells and suggest a role for vimentin as a dynamic scaffold for DENV RCs.
Ultrastructural studies illustrate the close association of vimentin and NS4A.
To further examine the interaction between vimentin cytoskeleton and NS4A at an ultrastructural level of higher resolution, scanning electron microscopy (SEM) was performed. This was done using a technique which involves the extraction of cells and removal of the MTs and microfilaments (MFs) while preserving the IFs, and followed by double immunogold labeling procedure (21; see Materials and Methods). Accuracy and reliability in the visualization and identification of vimentin IFs can thus be achieved with the removal of MTs and MFs by gelsolin, followed by immunogold labeling of vimentin. The vesicles observed in Fig. 4 were likely to be the VPs that harbor the DENV RCs. DENV VPs are induced by DENV NS proteins such as NS4A, which appear as clusters of double-membrane vesicles of 80 to 150 nm in size (7). The NS4A (Fig. 4, gray arrowheads, 10-nm gold conjugate) was observed to associate with the VPs, and this observation is in agreement with previous studies that NS4A is a component of the DENV RCs. Gold particles labeling NS4A were aligned along and associated directly with the vimentin network (Fig. 4, gray arrows, 5-nm gold conjugate) or concentrated in its vicinity. These individual VPs and clusters of VPs were closely associated with the vimentin network in the absence of MTs and MFs. These results thus illustrate the strong interaction of host cellular vimentin with DENV NS4A.
FIG 4.
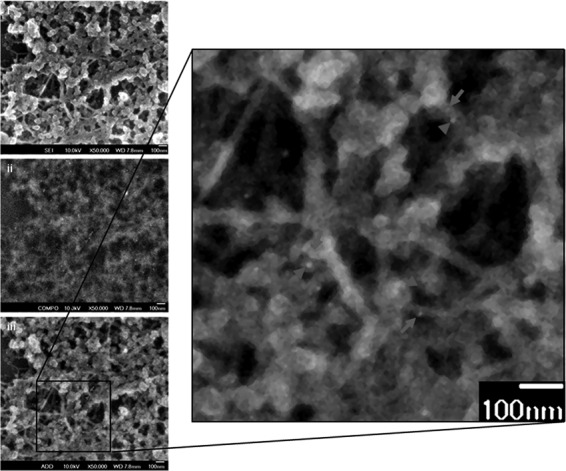
Ultrastructural studies illustrate the close association of vimentin and NS4A. Huh-7 cells were infected for 48 h and then subjected to detergent extraction, chemical fixation, and immunogold labeling for electron microscopic visualization of vimentin cytoskeleton. The enlarged image (panel iii) represents the overlay of the secondary electron image (panel i) and the backscattered electron image (panel ii). A close association of vimentin (gray arrows) with NS4A (gray arrowheads) is shown. Magnification, ×50,000. Bars = 100 nm.
N50 is the specific interacting region of DENV NS4A with vimentin.
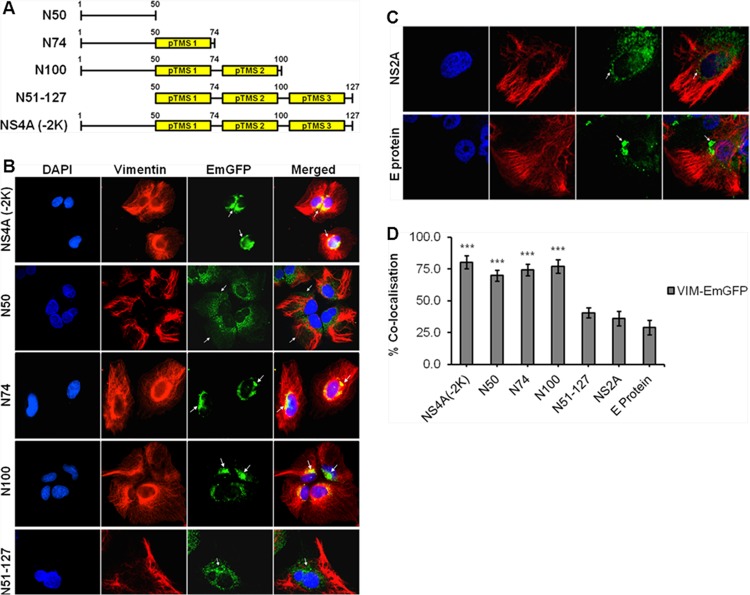
Once vimentin-NS4A interactions have been established, we proceeded to narrow down the specific region of NS4A that interacts with vimentin. Molecular cloning of different segments of DENV NS4A was performed to generate different plasmids consisting of fragments of the gene of interest in tandem with the EmGFP gene. Based on the proposed membrane topology model of DENV NS4A by Miller and coworkers (15), the following four segments of DENV NS4A were cloned: amino acids 1 to 50, 1 to 74, 1 to 100, and 51 to 127, designated N50, N74, N100, and N51-127, respectively, as well as the full-length NS4A lacking the 2K sequence consisting of amino acids 1 to 127 [NS4A (−2K)] (Fig. 5A). Subsequently, the plasmids carrying genes encoding different segments of NS4A were stably expressed in Huh-7 cells, and the subcellular localization of the EmGFP fusion proteins was verified by confocal immunofluorescence microscopy (IFM). The subcellular localization of N50-EmGFP fusion protein was observed to be distributed throughout the cell, signifying that the N50 region localized at the cytoplasmic N-terminal region lacks the transmembrane domain. In contrast, the other clones [N74, N100, N51-127, and NS4A (−2K)] having at least one predicted transmembrane segment (pTMS) displayed localization at the perinuclear region (Fig. 5B). In addition, a punctuate staining was observed, indicating that all three pTMSs are able to mediate membrane association which is consistent with the observations of Miller and coworkers (15). It was also observed that the N100, N51-127, and NS4A (−2K) clones have a stronger dot-like staining pattern, suggesting that the EmGFP fusion proteins with at least two pTMSs may induce more membrane association. Plasmids carrying genes encoding NS2A-EmGFP and E-protein–EmGFP were also transfected into Huh-7 cells, and these served as controls for subsequent biochemical assays. NS2A and E protein, which are known to associate with ER membranes, were enriched at the perinuclear region as well (Fig. 5C). From Fig. 5D, it can be observed that the average colocalization of 75% was significantly greater in the clones (NS4A (−2K), N50, N74, and N100) than in the N51-127 clone (41%) and control proteins (33%).
FIG 5.
Molecular cloning of different fragments of DENV NS4A and subcellular localization studies. (A) Schematic representations of different segments of DENV NS4A cloned to generate the respective NS4A-EmGFP fusion proteins. Yellow boxes represent the predicted transmembrane segments (pTMSs) of NS4A. (B) Huh-7 cells were stably transfected with the plasmids carrying genes encoding the NS4A-EmGFP fusion proteins before processing for immunolabeling. Confocal IFM showing subcellular localization of NS4A-EmGFP recombinant fusion proteins (green) and vimentin (red). (C) Stable cell clones expressing NS2A- and E-protein–EmGFP served as controls. The images were analyzed using LSCM and taken at a magnification of ×100. White arrows denote the subcellular localization. (D) Colocalization quantification was based on MOC using WCIF ImageJ software (20) and represented as percent colocalization of the respective NS4A-EmGFP fusion proteins and control proteins. Statistical analyses were carried out using one-way analysis of variance (ANOVA) and Bonferroni post hoc test (GraphPad Software). Values that are significantly different from those for the control proteins (P < 0.001) are indicated by three asterisks.
To delineate the region of interaction of NS4A with vimentin, we performed in situ PLA to determine the specific region of NS4A that has the strongest interaction with vimentin as determined by the number of PLA signals. The PLA signals were significantly stronger in cells expressing plasmids with genes encoding NS4A (−2K), N50, N74, and N100, compared to N51-127 (Fig. 6A and C). This indicates the importance of a sequence that is present in the former four plasmids expressed yet lacking in the latter for the interaction with vimentin, the region with amino acids 1 to 50. For controls, the PLA signals observed for cells expressing NS2A and E protein were not significantly different from those for N51-127 (Fig. 6B and C). GFP empty vector was used as a negative control to exclude the nonspecific signals, and a significant difference was observed compared to other cells expressing the protein of interest. Therefore, the region comprising the N-terminal 50 residues without the membrane-spanning segment (N50 region) may be the one interacting with vimentin, since it resides in the cytoplasm.
FIG 6.
N50 is the specific interacting region of DENV NS4A with vimentin. In situ PLA exhibits interaction of vimentin with NS4A-EmGFP fusion proteins. (A) Immunofluorescence detection of PLA signals (white arrows) in transfected cells stably expressing different regions of NS4A. Vimentin- and GFP-specific primary antibodies were used. The number of in situ PLA signals represents the vimentin-NS4A fusion protein (region of interest) interactions, and the nuclei were stained with DAPI. Individual events of PLA signal were used as reading output and quantified. (B) Plasmids expressing NS2A protein, E protein, and GFP alone were used as controls. The images were taken at a magnification of ×100. (C) Quantification of PLA-positive foci detected in stably transfected Huh-7 cells. In situ PLA signals per cell were quantitated using the software program CellProfiler (17) and ImageJ program by spot counting (18).The numbers of cells counted (n) are shown, and the average values are shown in gray. Statistical analyses were carried out using one-way ANOVA and Bonferroni post hoc test (GraphPad Software). Values that are significantly different (P < 0.001) from each other are indicated by a bar and three asterisks. (D) Co-IP of vimentin with different NS4A-GFP recombinant fusion proteins. Total cell lysates were incubated with vimentin or GFP antibodies, and Dynabeads protein G were added to capture the protein complex. Samples with eluted target proteins were analyzed by Western blotting (WB) using anti-GFP or antivimentin antibodies. Panels 1 to 4 represent Co-IP procedure, and panels 5 to 8 represent reciprocal Co-IP. The specific protein bands representing different truncated NS4A-GFP fusion proteins and the bands representing vimentin are indicated by the arrows.
To further confirm that N50 is indeed the region of DENV NS4A responsible for the interaction with vimentin, we carried out Co-IP. In cell lysate containing N51-127–GFP fusion proteins, N51-127 was not coimmunoprecipitated with vimentin using antivimentin antibody, as no protein band of approximately 35 kDa corresponding to N51-127–GFP fusion protein was observed (Fig. 6D, panel 1). In the reciprocal assay, a protein band corresponding to vimentin was also not detected (Fig. 6D, panel 5). Conversely, N74-, N50-, NS4A (−2K)-GFP fusion proteins were coimmunoprecipitated with vimentin using antivimentin antibody. Western blotting using anti-GFP antibody revealed protein bands of approximately 35 kDa, 32 kDa, and 40 kDa corresponding to N74, N50, and NS4A (−2K), respectively (Fig. 6D, columns 2 to 4). These results were verified by reciprocal Co-IP using anti-GFP antibody, and vimentin (∼57 kDa) similarly coimmunoprecipitated with N74-, N50-, NS4A (−2K)-GFP fusion proteins (Fig. 6D, panels 6 to 8). With our study, we report for the first time the discovery of the specific region of NS4A that interacts with vimentin lies within the first 50 amino acid residues at the cytosolic N-terminal domain of NS4A (N50 region).
DENV-2 infection induces vimentin reorganization by altering phosphorylation levels of vimentin.
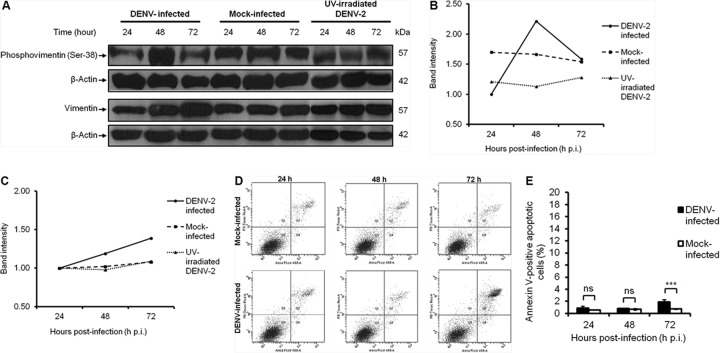
Vimentin was found to undergo reorganization and recruitment to the perinuclear site upon DENV infection (Fig. 2 and Fig. 3A). Vimentin reorganization in cells involves filament disassembly regulated by phosphorylation of the N-terminal domains by cellular kinases (13). Therefore, to further investigate the induction of vimentin reorganization, the phosphorylation levels of vimentin were examined by Western blot analysis. In view of the fact that serine-38 is one of the in vivo phosphorylation sites that are of major importance in maintaining and regulating vimentin structure (26), it was used as a reporter of vimentin phosphorylation in our study. In the mock-infected and UV-irradiated DENV-2 control setups, a constant level of phosphovimentin (serine-38) was detected across the three time points (Fig. 7A). In contrast, there was a distinct change in the phosphorylation levels of vimentin upon DENV-2 infection. We observed a transition state whereby the expression level of phosphovimentin increased markedly at 48 h postinfection from 24 h postinfection before it declined at 72 h postinfection. The significant increase in the level of phosphovimentin in DENV-infected cells at 48 h postinfection (Fig. 7A and B) was correlated with the colocalization study results (Fig. 2 and Fig. 3A) and proximity study results (Fig. 3D), as evident by the prominent reorganization and the strongest colocalization and interaction between vimentin and NS4A. These findings may indicate a relationship between vimentin phosphorylation levels and vimentin reorganization and its interaction with NS4A. An increased phosphorylation of vimentin IFs favors a depolymerized state, which allows its reassembly at the perinuclear site, and consequently forms an anchorage scaffold for the DENV RCs mediated by NS4A interaction. Interestingly, DENV-2 infection also upregulated vimentin expression, wherein the highest vimentin protein expression level occurred at 72 postinfection (Fig. 7A and C). This may further illustrate the crucial role of vimentin during the course of DENV infection.
FIG 7.
DENV-2 infection induces vimentin reorganization by altering phosphorylation levels of vimentin. (A) Western blot analysis of the phosphorylation levels of vimentin. Huh-7 cells subjected to the three treatments (DENV infection, mock infection, and UV irradiation) were lysed at 24, 48, and 72 h postinfection, and equal amounts of protein were separated by SDS-PAGE. The blots were probed with antiphosphovimentin (serine-38) or antivimentin antibodies. β-Actin was used as the loading control. (B) Quantification of protein bands detected by Western blotting. Intensity of bands representing phosphovimentin (serine-38). (C) Intensity of bands representing vimentin. Band intensities were quantitated and made reference to actin control bands (at each individual time points). (D) Bivariate annexin V/PI analysis of DENV-2-infected cells across all time points. DENV-2-infected Huh-7 cells were double labeled with annexin V (x axis) and PI (y axis). The top left quadrant (quadrant 1 [Q1]) contains the damaged (annexin V−/PI+) population, the top right quadrant (Q2) contains the necrotic (annexin V+/PI+) cells, the bottom left quadrant (Q3) contains the viable (annexin V−/PI−) cell population, and the bottom right quadrant (Q4) contains the apoptotic (annexin V+/PI−) cells. (E) Using flow cytometry, apoptosis was quantitated as the percentage of cells showing annexin V-positive staining. Three replicate experiments were carried out, and the error bars show the standard deviations (SD). Statistical analyses were performed using one-way ANOVA and Bonferroni's multiple comparison test (GraphPad Software). Values that are significantly different (P < 0.001) from each other are indicated by a bar and three asterisks. Values that are not statistically significant (ns) (P > 0.05) are indicated by bars labeled ns.
Modifications in the cytoskeletal organization can occur during early apoptosis, and rearrangement of vimentin networks is induced, which is followed by proteolysis and disassembly (27). To exclude the possibility that vimentin reorganization occurred due to early apoptosis, we performed annexin V binding assay and flow cytometric analysis to quantitate the extent of apoptosis in DENV-2-infected cells at all three time points. As depicted in Fig. 7D, the viable cell population (bottom left quadrant [quadrant 3 {Q3}]) formed the largest fraction of the cells, and DENV-2-infected cells were comparable to mock-infected cells across all three time points. At 72 h p.i., there was an increase in the fraction of necrotic (annexin V-positive [annexin V+]/PI-positive [PI+]) cells (top right quadrant [Q2]), which was expected. Although the apoptotic (annexin V+/PI-negative [PI−]) cell fraction at 72 h postinfection was higher (with statistical significance) than its parallel mock-infected setup, this assay revealed that the extent of apoptosis at all three time points postinfection was very low, even at 72 h postinfection (lower right quadrant [Q4]). This subpopulation comprised 0.9%, 0.8%, 1.9% of the DENV-infected cells at 24, 48, and 72 h p.i., respectively (Fig. 7E). These fractions represented the early apoptotic cells which constituted only a small percentage of the total cell population. Therefore, it was concluded that indeed the observed vimentin reorganization was not induced due to early apoptotic cell death events but DENV infection of viable cells.
Calcium/calmodulin-dependent protein kinase II gamma gene knockdown significantly reduces DENV-2 titer.
Reorganization of vimentin IFs is regulated by phosphorylation in a site-specific manner (28) through cellular protein kinases such as Cdc42 and p70 S6 kinase (29), as well as stimulus-specific kinases, for example, CaMKII (30). In addition, serine-38 of vimentin is also one of the major in vitro and in vivo phosphorylation sites by CaMKII (28), and CaMKIIγ directly binds to vimentin and colocalizes with them (31). We went on further to examine the effects of gene knockdown of these cellular protein kinases, Cdc42, p70 S6 kinase, and CaMKIIγ, on DENV-2 replication. Among the three kinases, the reduction in DENV-2 titer was observed to be greatest with CaMKIIγ gene knockdown, compared to that of Cdc42 and p70 S6 kinases (data not shown). CaMKII is expressed as four isoforms: α, β, δ, and γ. The α and β isoforms are predominantly expressed in neural tissue (32), whereas the γ isoform is expressed in liver tissue (33); therefore, the γ isoform was selected for our study.
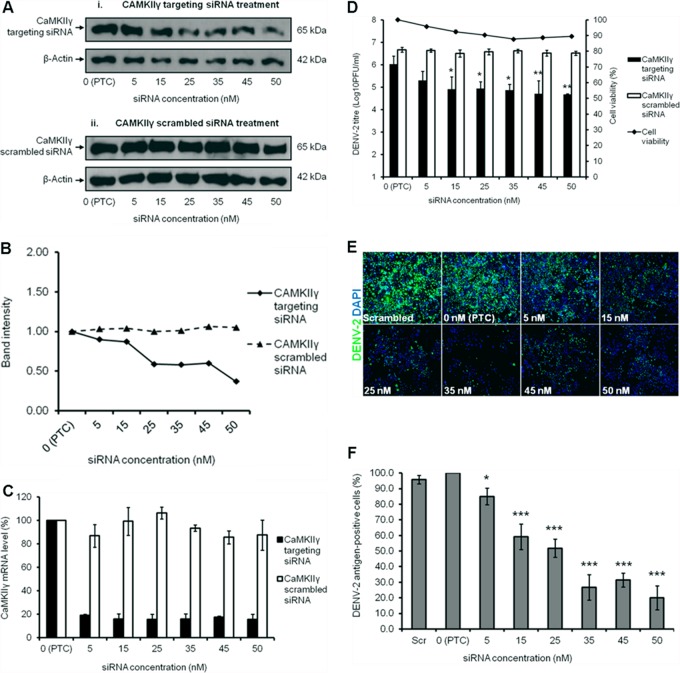
Before a viral plaque assay was performed to determine the virus titer, Western blotting and qRT-PCR were carried out to verify gene knockdown efficiency. There was a dosage-dependent reduction in the protein expression level, thus showing that the knockdown was effective despite low levels of CaMKIIγ expression still being detected (Fig. 8A, panel i, and Fig. 8B). Additionally, the decrease in DENV-2 titer despite the modest reduction in CaMKIIγ expression observed at lower siRNA concentrations may be due to the continuous siRNA knockdown effect and sustained depletion of CaMKIIγ encoded by the targeted mRNA during the 72-h infection period. This was further verified by the scrambled siRNA treatment, as no knockdown was observed throughout the spectra of siRNA concentrations used (0 nM to 50 nM) (Fig. 8A, panel ii, and Fig. 8B). The qRT-PCR data demonstrated a reduction in the mRNA level of CaMKIIγ upon knockdown, whereas with scrambled siRNA treatment, the mRNA level remained fairly constant (Fig. 8C). Knockdown of the CaMKIIγ gene indeed caused a dosage-dependent reduction in the infectious viral titer of DENV-2, with a reduction of 1.5 log units at 50 nM, relative to the positive transfection control (PTC) sample (Fig. 8D). Furthermore, minimal cellular cytotoxicity was observed across all the concentrations of siRNA used (Fig. 8D). In cells transfected with scrambled siRNA, the viral titer remained constant with minimal inhibition. Indirect IFM was used to observe the intracellular viral infectivity upon CaMKIIγ gene knockdown (Fig. 8E), and the immunofluorescence signals were quantitated. The decrease in the intracellular infectivity of DENV-2 was more significant, with a reduction of 79.9% using 50 nM siRNA compared to the PTC (Fig. 8F). These findings propose that CaMKIIγ may also be involved in DENV-2 replication through phosphorylation of vimentin at serine-38.
FIG 8.
Calcium/calmodulin-dependent protein kinase II gamma gene knockdown significantly reduces DENV-2 titer. Verification of CaMKIIγ gene knockdown with siRNA treatment. (A) Western blot analysis for detecting protein expression levels of CaMKIIγ, with β-actin as the loading control. Parallel transfection of scrambled siRNA served as a knockdown control. (B) Band intensity of CaMKIIγ gene knockdown verification. The intensities of protein bands representing CaMKIIγ protein expression level were quantitated and made reference to actin control bands (for each of the individual concentrations) and PTC. The intensities of protein bands representing protein expression level were quantitated using the ImageJ Gel Analysis program. (C) qRT-PCR data were analyzed using ABI StepOnePlus software. Cross threshold values of CaMKIIγ and β-actin were obtained and used for the calculation of the mRNA levels of CaMKIIγ, represented in percentages which made reference to the PTC. The effects of CaMKIIγ gene knockdown on replication of DENV-2 are shown. Values are means ± standard deviations (SD) (error bars). (D) Cell viability assay was performed, and the titers of virus in the supernatants of siRNA-transfected cells were analyzed by viral plaque assay. Three replicate experiments were carried out, and the SD are indicated by error bars. Statistical analyses were performed using one-way ANOVA and Dunnett's test (GraphPad Software). Values that are significantly different are indicated as follows: *, P < 0.05; **, P < 0.005. (E) siRNA-treated DENV-infected cells were fixed at the same time points, and intracellular viruses were detected by indirect IFM. Immunofluorescence detection of DENV-2 proteins (green) with the nuclei stained with DAPI (blue) is shown. The images were taken at a magnification of ×10. (F) Quantification of DENV-2 antigen-positive cells from the IFM images, represented in percentages which made reference to the PTC. Three replicate experiments were carried out, and the error bars show SD. Statistical analyses were performed using one-way ANOVA and Dunnett's test (GraphPad Software). Values that are significantly different are indicated as follows: *, P < 0.05; **, P < 0.001. Scr, scrambled siRNA.
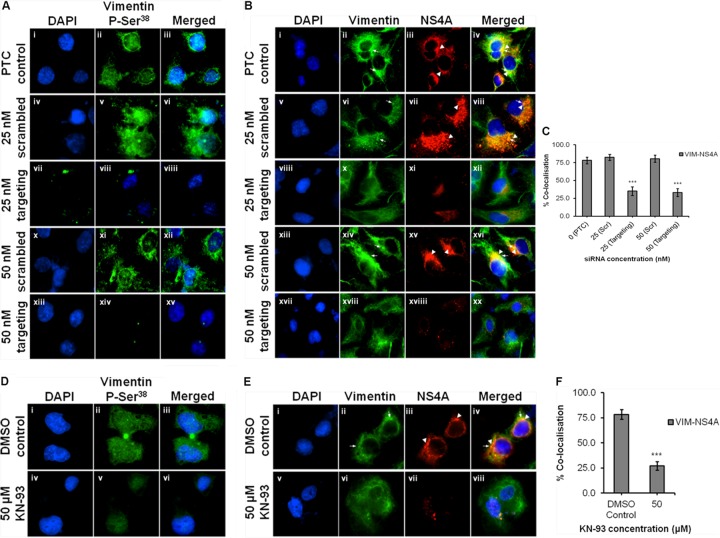
Additionally, to assess the link between CaMKIIγ, vimentin phosphorylation, and colocalization of vimentin with NS4A in DENV replication, gene silencing and inhibition of CaMKIIγ using siRNA and KN93 inhibitor were used, respectively. Upon knockdown of the CaMKIIγ gene, there was a substantial decrease in the immunofluorescence signal for phosphovimentin (serine-38) (Fig. 9A, panels vii to viiii and xiii to xv), indicating that CaMKIIγ silencing impacts vimentin phosphorylation in DENV infection. Interestingly, the colocalization of vimentin with NS4A was lost upon knockdown of CaMKIIγ (Fig. 9B, panels xii and xx), and this phenomenon was quantified wherein the colocalization coefficients showed a significant reduction from 80% to 34% (Fig. 8C). Furthermore, there was decreased vimentin reorganization (Fig. 9B, panels x and xviii) and significantly reduced immunofluorescence signal for NS4A (Fig. 9B, panels xi and xviiii). This could further support the reduction in the infectious viral titer of DENV-2 (Fig. 8D). Likewise, inhibition of vimentin phosphorylation using KN93 showed similar phenotypes wherein immunofluorescence signal for phosphovimentin (serine-38) decrease substantially (Fig. 9D, panels iv to vi) and colocalization of vimentin with NS4A was significantly decreased from 78% to 25% (Fig. 9E, panels v to viii, and Fig. 9F).
FIG 9.
Gene silencing and inhibition of CaMKIIγ reduce vimentin phosphorylation and diminish vimentin-NS4A colocalization. Huh-7 cells were transfected with CaMKIIγ targeting or scrambled siRNA and subjected to infection with DENV-2 as described in Materials and Methods. For drug inhibition of CaMKIIγ, Huh-7 cells were infected with DENV-2 at an MOI of 10 for 36 h before the treatment with KN93 (50 μM) for another 12 h (40). siRNA-treated or KN93-treated DENV-infected cells were fixed, and vimentin phosphorylation and vimentin-NS4A colocalization were investigated by indirect IFM. P-Ser38, phosphorylated serine at position 38. (A and D) Immunofluorescence detection of phosphovimentin (serine-38) (green) with the nuclei stained with DAPI (blue). Huh-7 cells infected with DENV-2 and treated with 0.1% DMSO served as a solvent control. (B and E) Subcellular localization of vimentin (green) and DENV NS4A (red) were visualized using LSCM, with the nuclei stained with DAPI (blue). Colocalization of vimentin (white arrows) with NS4A (white arrowheads) was indicated. The images were taken at a magnification of ×100. (C and F) Colocalization quantification was based on MOC using WCIF ImageJ software (20) and represented as percent colocalization which made reference to the PTC and DMSO control, respectively. Statistical analyses were carried out using one-way ANOVA and Bonferroni post hoc test (GraphPad Software). Values that are significantly different (P < 0.001) are indicated by three asterisks.
Vimentin is essential for anchorage of DENV replication complexes mediated by NS4A interaction.
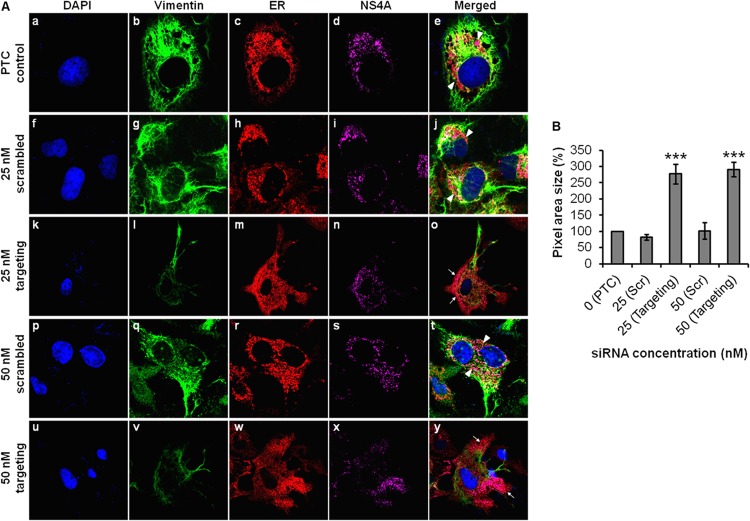
It is essential to understand the biological and functional significance of vimentin and its interaction with DENV NS4A during DENV replication. With this aim, we executed gene knockdown of vimentin using targeting siRNA and studied the effect of knockdown on the distribution of DENV RCs. In the PTC setup, vimentin was intact and reorganized to form the structural scaffold that supports the RCs. The DENV RCs, which are known to be derived from ER, were expected to be localized at the perinuclear region as observed (Fig. 10A, panels a to e). In contrast, the vimentin expression levels were substantially reduced upon vimentin knockdown. Despite having residual vimentin expression level attributed to vimentin being a housekeeping gene critical for cellular maintenance (34), the RCs were evidently redistributed to the periphery of cells with the use of 25 nM targeting siRNA (Fig. 10A, panels k to o). For a control, parallel transfection of scrambled siRNA was carried out, and vimentin organization and RC distribution were similar to that of the PTC (Fig. 10A, panels f to j). To increase the effect of vimentin knockdown, we tested a higher concentration of targeting siRNA, and the RCs appeared to be extensively diffused and spread throughout the cell cytoplasm (Fig. 10A, panels u to y). In the parallel control setup, the ER exhibited the punctuate staining pattern (Fig. 10A, panels p to t), unlike the diffused pattern observed in the vimentin knockdown cells. Upon vimentin gene knockdown, the area of distribution of DENV RCs was quantified, and the difference in area was largely significant compared to the control setups (Fig. 10B). Taken together, our findings further demonstrated the active role of vimentin as a supportive anchorage for the DENV RCs during DENV replication.
FIG 10.
Vimentin is essential for anchorage of DENV replication complexes mediated by NS4A interaction. (A) Huh-7 cells were treated with vimentin targeting or scrambled siRNA and fixed after DENV-2 infection for immunofluorescence analysis. The subcellular localization of vimentin (green), ER (red), and DENV NS4A protein (violet) were visualized by LSCM, with the nuclei stained with DAPI (blue). The images were taken at a magnification of ×100. White arrowheads denote the punctuate ER staining (concentrated localization), and white arrows denote diffused ER staining (dispersed localization). (B) Quantification analysis of the distribution of immunofluorescence signals from the IFM images, shown as percentages of pixel area size which made reference to the PTC. Statistical analyses were performed using one-way ANOVA and Dunnett's test (GraphPad Software). Values that are significantly different (P < 0.001) are indicated by three asterisks. Immunofluorescence signals were quantitated on a per-cell basis using the ImageJ software program. The number of cells counted per sample is 10. Values are means ± SD (error bars).
DISCUSSION
The molecular mechanisms of DENV replication can affect the clinical outcomes (35). It is apparent that an understanding of the DENV replication cycle will require characterization of the physical and functional interactions of the proteins that form the DENV RC, including unidentified host proteins. Here, the complex interplay of DENV NS4A and host cytoskeleton vimentin has been revealed. In this study, one of the key findings was the identification of a direct interaction between human cytoskeleton vimentin and DENV NS4A (Fig. 1). Our data suggested that there was close physical interaction between vimentin and NS4A throughout the replication cycle and thus led to the investigation of the relationship between these two proteins and their significance in DENV replication. Other studies have reported the interactions of host cellular polypyrimidine tract-binding protein with NS4A and its localization in the replication machinery (16). This may suggest a similar role for vimentin during vRNA replication. Studies on the interaction of vimentin with DENV proteins are limited, seeing that only one group has proposed the association of vimentin with DENV NS1 and its crucial role in DENV replication (36). Hence, vimentin was being worked on, as we are interested in finding out the functional significance of vimentin intermediate filaments with respect to DENV replication, which has been studied less, compared to microtubules and microfilaments. Further work is required to reveal specifically what biological functions vimentin serve and with which DENV proteins it interacts directly. This led us to explore vimentin further with various downstream experiments, as we identified vimentin as one of the DENV NS4A-binding proteins.
Subcellular colocalization studies revealed that vimentin indeed colocalized with NS4A at the perinuclear site where DENV-induced ER-derived membranous compartments localized (Fig. 2A, B, and C). The dramatic rearrangements of proliferating ER membranes were induced by NS4A to form unique membrane structures—DENV RCs, suggesting its role in RNA replication (37–39). This suggests that substantial vimentin rearrangement induced by DENV occurs jointly with the construction of DENV RCs in which compartmentalization within the induced membranes ensures a more efficient replication process, and vimentin may contribute a structural role in anchoring the RCs. Flaviviral RC is believed to comprise the vRNA template with the NS proteins and presumably some host proteins on cytoplasmic membranes (3, 4). Hence, the role of vimentin in DENV RNA replication was further elucidated, as vimentin was found to colocalize with dsRNA as well (Fig. 2D, E, and F), suggesting that vimentin is one of the host cellular factors that associates with the RNA amplification machinery and may play a direct role in DENV RNA replication. Furthermore, SEM has also demonstrated the strong association of vimentin network and DENV components at the ultrastructural level where DENV RCs aligned closely along the highly interconnected vimentin network (Fig. 4). IFs are known to control adaptor protein 3 (AP3)-dependent membrane traffic and help in vesicle formation, vesicle budding, and vesicle uncoating (40). It is also intriguing to conjecture on how vimentin might help in the formation of the ER-derived RCs, mediated by the interaction with NS4A, and potentially stabilizes the RC localization at the perinuclear site.
It is interesting to notice the dynamics of vimentin-NS4A interactions (Fig. 3) attributed to vimentin remodeling during DENV infection. These findings seem to be in accord with the fact that IFs are performing certain key cellular functions rather than being just rigid structures providing mechanical resilience to the cells, thus gaining greater significance as dynamic elements (40). We describe the pattern of interaction and show that the strongest interaction occurs at 48 h postinfection, which may imply a significance of this particular time point in the DENV replication cycle. There are some possible reasons as to why a dynamic interaction exists. During late infection, a host cellular response may be elicited to provide cytoprotective function by the remodelling of vimentin, since a greater amount of viral proteins are produced. Stefanovic and coworkers have reported the induction of vimentin rearrangement into a cytoprotective cage during African swine fever virus (ASFV) infection (19). The enclosure of viral proteins may prevent viral components spreading throughout the cytoplasm (41). Another possibility could be that the modified vimentin plays a more dynamic role, such as organizing the interior of viroplasm foci and incorporating viral proteins inside immature viral particles (42).
The amino-terminal domains of the IF proteins are thought to protrude from the surfaces of the filaments (43); thus, these sequences are likely to participate in interactions between the vimentin filaments and associated proteins. Even though the data are not sufficient to conclude the exact details of the biochemical interactions between vimentin and NS4A, our data have revealed for the first time that the specific region of NS4A that interacts with vimentin lies within the first 50 amino acid residues at the cytosolic N-terminal domain of NS4A (N50 region) (Fig. 6C and D). To complement this finding, a recent study has identified an amphipathic helix in the N-terminal region of DENV NS4A that mediates oligomerization and is essential for DENV replication (44). The oligomerization of membrane proteins can form a scaffold that can locally bend membranes (7, 45); this could mean that NS4A undergoes homo-oligomerization to form the DENV RCs. Our new findings may shed light on the function of NS4A for DENV replication, in particular the potential functional significance of the cytosolic N-terminal region, where the N50 region may serve as one of the potential antiviral targeting sites. The determination of the exact mechanism of interaction between vimentin and NS4A will require future investigations, which may lead to significant insights into antiviral strategies.
Rearrangement of vimentin was believed to be required for the formation of the virus assembly site, by providing a mechanical scaffold for the recruitment of viral proteins necessary for ASFV DNA replication (19). Risco and coworkers also showed that vimentin filaments and elements of the ER Golgi intermediate compartment concentrate in the viral factories of vaccinia virus (42). Our results indicated the evident reorganization of vimentin induced upon DENV infection. Furthermore, DENV replication seems to modulate vimentin phosphorylation in a specific way by altering the phosphorylation levels, which regulate vimentin reorganization. We detected a significant increase in the level of phosphovimentin at 48 h postinfection (Fig. 7A), which correlated strongly with the evident reorganization at the same time point (Fig. 3A). Our finding is in line with the fact that there is increased phosphorylation of vimentin when the filaments undergo marked redistribution (46). There are several ways in which the induction of vimentin reorganization during DENV infection could be explained. First, NS4A could directly induce vimentin reorganization through processes mediated by its preceding direct interaction. Alternatively, NS4A could indirectly induce vimentin reorganization through the activation of CaMKIIγ, which in turn phosphorylates vimentin at serine-38, leading to vimentin reorganization. Last, vimentin rearrangements to enclose the viral assembly site prevent the invasion of host cellular factors into the assembly site and maintain the accumulation of viral proteins (47). Thus, vimentin may emerge to be essential to confer structural integrity of the dengue virus assembly site and help organize the ER-derived membranous compartments into a more efficient 3D assemblage of replication machineries comprising interacting viral and host cell components.
Our findings point to the second hypothesis in which the vimentin-NS4A interaction leads to downstream signaling events that activate CaMKIIγ and phosphorylate vimentin. It has been reported previously that CaMKIIγ directly binds to cytoskeletal proteins such as vimentin and colocalizes with them (31). The phosphorylation of vimentin at serine-38 by CaMKIIγ was coincident with vimentin reorganization which colocalized with NS4A in place with the DENV RCs. However, it remains to be resolved whether phosphorylation was a bystander effect of CaMKIIγ activation during DENV infection. Another speculation to demonstrate a potential link between DENV NS4A and CaMKII activation relates to the cellular unfolded protein response (UPR) exploited by viruses. A study identified rotavirus gene-encoded hydrophobic viroporin nonstructural protein 4 that oligomerizes to form a transmembrane aqueous pore which releases Ca2+ from the ER into the cytoplasm, thereby activating a calcium/calmodulin-dependent kinase kinase β signaling (48). Interestingly, some of the flaviviral nonstructural proteins are hypothesized to be viroporins (49) and may cause homeostasis imbalance of Ca2+ and other ions in the ER, thus eliciting a stronger UPR. Other studies have also reported that DENV infection induces and regulates the UPR signaling cascades (50), and Flavivirus West Nile virus strain KUNV hydrophobic NS4A is a potent inducer of the UPR (51). As such, it can be hypothesized that the hydrophobic DENV NS4A which induces the local bending of ER membranes may lead to the release of Ca2+ from the ER Ca2+ store via the ER stress response, also called UPR. As a result, the released Ca2+ activates CaMKII via Ca2+-bound calmodulin, leading to vimentin phosphorylation and reorganization.
Prior to this work, several studies employed the use of an acrylamide drug after treatment for chemical disruption of the vimentin network (52); however, few or no studies have shown the effect of siRNA-mediated vimentin gene knockdown on DENV replication. Here, we employed siRNA-mediated gene knockdown for our functional studies to investigate the role of vimentin in DENV replication. Subcellular localization studies further unraveled the structural role of vimentin and its interaction with DENV replication machinery. The dispersed localization of DENV ER-derived RCs upon vimentin knockdown suggests its active role as supportive anchorage for the RCs during viral replication (Fig. 10A). Our findings indicate that an intact vimentin network is indeed essential in DENV-2 replication. Finally, this leads to the proposed role of vimentin in dengue virus replication and promoting efficient vRNA replication.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by start-up grant R182-000-165-133 from the National University of Singapore.
Footnotes
Published ahead of print 27 November 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01249-13.
REFERENCES
- 1.Lindenbach BD, Rice CM. 2001. Flaviviridae: the viruses and their replication, p 991–1041 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Roizman B, Strauss SE. (ed), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 2.Perera R, Kuhn RJ. 2008. Structural proteomics of dengue virus. Curr. Opin. Microbiol. 11:369–377. 10.1016/j.mib.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Rice CM. 2003. Molecular biology of flaviviruses. Adv. Virus Res. 59:23–61. 10.1016/S0065-3527(03)59002-9 [DOI] [PubMed] [Google Scholar]
- 4.Westaway EG, Mackenzie JM, Khromykh AA. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 59:99–140. 10.1016/S0065-3527(03)59004-2 [DOI] [PubMed] [Google Scholar]
- 5.Ng ML. 1987. Ultrastructural studies of Kunjin virus-infected Aedes albopictus cells. J. Gen. Virol. 68:577–582. 10.1099/0022-1317-68-2-577 [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie JM, Westaway EG. 2001. Assembly and maturation of the Flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787–10799. 10.1128/JVI.75.22.10787-10799.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374. 10.1038/nrmicro1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radtke K, Dohner K, Sodeik B. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell. Microbiol. 8:387–400. 10.1111/j.1462-5822.2005.00679.x [DOI] [PubMed] [Google Scholar]
- 9.Smith GA, Enquist LW. 2002. Break ins and break outs: viral interactions with the cytoskeleton of mammalian cells. Annu. Rev. Cell Dev. Biol. 18:135–161. 10.1146/annurev.cellbio.18.012502.105920 [DOI] [PubMed] [Google Scholar]
- 10.Hartwig JM. 1992. An ultrastructural approach to understanding the cytoskeleton, p 23–46 In Carraway KL, Carraway CAC. (ed), The cytoskeleton: a practical approach. IRL Press, New York, NY [Google Scholar]
- 11.Azumi N, Battifora H. 1987. The distribution of vimentin and keratin in epithelial and nonepithelial neoplasms. A comprehensive immunohistochemical study on formalin- and alcohol-fixed tumors. Am. J. Clin. Pathol. 88:286–296 [DOI] [PubMed] [Google Scholar]
- 12.Styers ML, Salazar G, Love R, Peden AA, Kowalczyk AP, Faundez V. 2004. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol. Biol. Cell 15:5369–5382. 10.1091/mbc.E04-03-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki M, Nishi Y, Nishizawa K, Matsuyama M, Sato C. 1987. Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature 328:649–652. 10.1038/328649a0 [DOI] [PubMed] [Google Scholar]
- 14.Gyoeva FK, Gelfand VI. 1991. Coalignment of vimentin intermediate filaments with microtubules depends on kinesin. Nature 353:445–448. 10.1038/353445a0 [DOI] [PubMed] [Google Scholar]
- 15.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882. 10.1074/jbc.M609919200 [DOI] [PubMed] [Google Scholar]
- 16.Anwar A, Leong KM, Ng ML, Chu JJ, Garcia-Blanco MA. 2009. The polypyrimidine tract-binding protein is required for efficient dengue virus propagation and associates with the viral replication machinery. J. Biol. Chem. 284:17021–17029. 10.1074/jbc.M109.006239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative membrane complexes are formed at the plasma membrane. J. Virol. 84:11679–11695. 10.1128/JVI.01441-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefanovic S, Windsor M, Nagata KI, Inagaki M, Wileman T. 2005. Vimentin rearrangement during African swine fever virus infection involves retrograde transport along microtubules and phosphorylation of vimentin by calcium calmodulin kinase II. J. Virol. 79:11766–11775. 10.1128/JVI.79.18.11766-11775.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manders EEM, Verbeek FJ, Aten JA. 1993. Measurement of co-localisation of objects in dual-colour confocal images. J. Microsc. 169:375–382. 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- 21.Svitkina TM, Verkhovsky AB, Borisy GG. 1995. Improved procedures for electron microscopic visualization of the cytoskeleton of cultured cells. J. Struct. Biol. 115:290–303. 10.1006/jsbi.1995.1054 [DOI] [PubMed] [Google Scholar]
- 22.Verkhovsky AB, Borisy GG. 1993. Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J. Cell Biol. 123:637–652. 10.1083/jcb.123.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach BD, Prágai BM, Montserret R, Beran RK, Pyle AM, Penin F, Rice CM. 2007. The C terminus of hepatitis C virus NS4A encodes an electrostatic switch that regulates NS5A hyperphosphorylation and viral replication. J. Virol. 81:8905–8918. 10.1128/JVI.00937-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 80:4623–4632. 10.1128/JVI.80.9.4623-4632.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 26.Eriksson JE, He T, Trejo-Skalli AV, Härmälä-Braskén AS, Hellman J, Chou YH, Goldman RD. 2004. Specific in vivo phosphorylation sites determine the assembly dynamics of vimentin intermediate filaments. J. Cell Sci. 117:919–932. 10.1242/jcs.00906 [DOI] [PubMed] [Google Scholar]
- 27.van Engeland M, Kuijpers HJH, Ramaekers FCS, Reutelingsperger CPM, Schutte B. 1997. Plasma membrane alterations and cytoskeletal changes in apoptosis. Exp. Cell Res. 235:421–430. 10.1006/excr.1997.3738 [DOI] [PubMed] [Google Scholar]
- 28.Inagaki N, Tsujimura K, Tanaka J, Sekimata M, Kamei Y, Inagaki M. 1996. Visualization of protein kinase activities in single cells by antibodies against phosphorylated vimentin and GFAP. Neurochem. Res. 21:795–800. 10.1007/BF02532302 [DOI] [PubMed] [Google Scholar]
- 29.Chan W, Kozma R, Yasui Y, Inagaki M, Leung T, Manser E, Lim L. 2002. Vimentin intermediate filament reorganization by Cdc42: involvement of PAK and p70 S6 kinase. Eur. J. Cell Biol. 81:692–701. 10.1078/0171-9335-00281 [DOI] [PubMed] [Google Scholar]
- 30.Ando S, Tokui T, Yamauchi T, Sugiura H, Tanabe K, Inagaki M. 1991. Evidence that Ser-82 is a unique phosphorylation site on vimentin for Ca2+-calmodulin-dependent protein kinase II. Biochem. Biophys. Res. Commun. 175:955–962. 10.1016/0006-291X(91)91658-Y [DOI] [PubMed] [Google Scholar]
- 31.Marganski WA, Gangopadhyay SS, Je HD, Gallant C, Morgan KG. 2005. Targeting of a novel Ca2+/calmodulin-dependent protein kinase II is essential for extracellular signal-regulated kinase-mediated signaling in differentiated smooth muscle cells. Circ. Res. 97:541–549. 10.1161/01.RES.0000182630.29093.0d [DOI] [PubMed] [Google Scholar]
- 32.Yamauchi T, Fujisawa H. 1980. Evidence for three distinct forms of calmodulin-dependent protein kinases from rat brain. FEBS Lett. 116:141–144. 10.1016/0014-5793(80)80628-4 [DOI] [PubMed] [Google Scholar]
- 33.Swami M. 2012. Calcium-mediated control. Nat. Med. 18:670. 10.1038/nm.2781 [DOI] [Google Scholar]
- 34.Mahadevappa M, Warrington JA. 26 March 2003. Housekeeping genes. eLS. John Wiley & Sons, Hoboken, NJ. 10.1038/npg.els.0000846 [DOI] [Google Scholar]
- 35.Urcuqui-Inchima S, Patino C, Torres S, Haenni A, Diaz FJ. 2010. Recent developments in understanding dengue virus replication. Adv. Virus Res. 77:1–39. 10.1016/B978-0-12-385034-8.00001-6 [DOI] [PubMed] [Google Scholar]
- 36.Kanlaya R, Pattanakitsakul SN, Sinchaikul S, Chen ST, Thongboonkerd V. 2010. Vimentin interacts with heterogeneous nuclear ribonucleoproteins and dengue nonstructural protein 1 and is important for viral replication and release. Mol. Biosyst. 6:795–806. 10.1039/b923864f [DOI] [PubMed] [Google Scholar]
- 37.Chu PW, Westaway EG. 1992. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch. Virol. 125:177–191. 10.1007/BF01309636 [DOI] [PubMed] [Google Scholar]
- 38.Mackenzie JM, Jones MK, Young PR. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232–240. 10.1006/viro.1996.0307 [DOI] [PubMed] [Google Scholar]
- 39.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. 1998. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 245:203–215. 10.1006/viro.1998.9156 [DOI] [PubMed] [Google Scholar]
- 40.Styers ML, Kowalczyk AP, Faundez V. 2005. Intermediate filaments and vesicular membrane traffic: the odd couple's first dance? Traffic 6:359–365. 10.1111/j.1600-0854.2005.00286.x [DOI] [PubMed] [Google Scholar]
- 41.Nozawa N, Yamauchi Y, Ohtsuka K, Kawaguchi Y, Nishiyama Y. 2004. Formation of aggresome-like structures in herpes simplex virus type 2-infected cells and a potential role in virus assembly. Exp. Cell Res. 299:486–497. 10.1016/j.yexcr.2004.06.010 [DOI] [PubMed] [Google Scholar]
- 42.Risco C, Rodríguez JR, López-Iglesias C, Carrascosa JL, Esteban M, Rodríguez D. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839–1855. 10.1128/JVI.76.4.1839-1855.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steinert PM, Steven AC, Roop DR. 1985. The molecular biology of intermediate filaments. Cell 42:411–419. 10.1016/0092-8674(85)90098-4 [DOI] [PubMed] [Google Scholar]
- 44.Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. 2013. An N-terminal amphipathic helix in the dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J. Virol. 87:4080–4085. 10.1128/JVI.01900-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438:590–596. 10.1038/nature04396 [DOI] [PubMed] [Google Scholar]
- 46.Evans RM, Fink LM. 1982. An alteration in the phosphorylation of vimentin-type intermediate filaments is associated with mitosis in cultured mammalian cells. Cell 29:43–52. 10.1016/0092-8674(82)90088-5 [DOI] [PubMed] [Google Scholar]
- 47.Murti KG, Goorha R, Klymkowsky MW. 1988. A functional role for intermediate filaments in the formation of frog virus 3 assembly sites. Virology 162:264–269. 10.1016/0042-6822(88)90420-5 [DOI] [PubMed] [Google Scholar]
- 48.Crawford SE, Hyser JM, Utama B, Estes MK. 2012. Autophagy hijacked through viroporin-activated calcium/calmodulin-dependent kinase kinase-β signaling is required for rotavirus replication. Proc. Natl. Acad. Sci. U. S. A. 109:E3405–E3413. 10.1073/pnas.1216539109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang YS, Liao CL, Tsao CH, Chen MC, Liu CI, Chen LK, Lin YL. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73:6257–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umareddy I, Pluquet O, Wang QY, Vasudevan SG, Chevet E, Gu F. 2007. Dengue virus serotype infection specifies the activation of the unfolded protein response. Virol. J. 4:91. 10.1186/1743-422X-4-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85:2723–2732. 10.1128/JVI.02050-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Gao N, Wang JL, Tian YP, Chen ZT, An J. 2008. Vimentin is required for dengue virus serotype 2 infection but microtubules are not necessary for this process. Arch. Virol. 153:1777–1781. 10.1007/s00705-008-0183-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.