Abstract
Influenza virus strains are often pleiomorphic, a characteristic that is largely attributed to specific residues in matrix protein 1 (M1). Although the mechanism by which M1 controls virion morphology has not yet been defined, it is suggested that the M1 interaction with other viral proteins plays an important role. In this study, we rescued recombinant virus WSN-AichiM1 containing the spherical A/WSN/33 (WSN) backbone and the M1 protein from A/Aichi/2/68 (Aichi). Aichi M1 differs from WSN M1 by 7 amino acids but includes those identified to be responsible for filamentous virion formation. Interestingly, Aichi virus produced spherical virions, while WSN-AichiM1 exhibited a long filamentous morphology, as detected by immunofluorescence and electron microscopy. Additional incorporation of Aichi nucleoprotein (NP) but not the hemagglutinin (HA), neuraminidase (NA), or M2 gene to WSN-AichiM1 abrogated filamentous virion formation, suggesting that specific M1-NP interactions affect virion morphology. Further characterization of viruses containing WSN/Aichi chimeric NPs identified residues 214, 217, and 253 of Aichi NP as necessary and sufficient for the formation of spherical virions. NP residues 214 and 217 localize at the minor groove between the two opposite-polarity NP helical strands of viral ribonucleocapsids, and residue 253 also localizes near the surface of the groove. These findings indicate that NP plays a critical role in influenza virus morphology, possibly through its interaction with the M1 layer during virus budding.
INTRODUCTION
Influenza virus virions are composed of the three transmembrane proteins hemagglutinin (HA), neuraminidase (NA), and M2, the matrix protein M1, and the viral nucleocapsid (vRNP), which contains nucleoprotein (NP)-encapsidated negative-strand viral RNA. All of the structural components are transported to budding sites at the apical surface of polarized cells for budding. The assembly of influenza viruses at the plasma membrane is a highly complex process involving the extensive interplay between multiple viral proteins. HA and NA concentrate in and around specialized microdomains of the plasma membrane, termed lipid rafts, while M2 is excluded from these domains (1–4). Binding of the cytoplasmic tails of HA and NA to M1 has been suggested to recruit M1 into the budding virions (5–7). Clustering of M1 on the inner bilayer and M1-M1 interactions facilitate the formation of an M1 protein patch and exclude host proteins from the assembly and budding site. Incorporation of the viral genome into budding virions is in part mediated by an interaction between M1 and various components of the vRNP complexes (8–10), as well as by an interaction between M1 and the cytoplasmic tail of the M2 ion channel protein (11). Pinching off the virus buds requires fusion of the opposing viral and cellular membranes, leading to fission and separation of the bud from the cell. M2 has been identified to play a critical role during this process of membrane fission and release (12).
Influenza A virus morphology ranges from spherical particles with a mean diameter of 100 nm to greatly elongated filamentous particles with lengths greater than several micrometers (13, 14). Although the precise mechanism that determines influenza virus morphology is currently unknown, previous studies suggest that specific M1 residues play a pivotal role. In these initial studies, lab-adapted spherical A/WSN/33 (WSN) and filamentous A/Udorn/72 (Udorn) viruses have been used to identify the genetic elements responsible for the morphological differences of influenza viruses. By genetic replacement, the Udorn M segment was identified to be a major determinant of virion morphology (15). In this same study, further analysis using chimeric M segments and site-directed mutagenesis revealed the M1 sequence to contain specific residues at positions 95 and 204 that are required for filament formation by the Udorn strain (15). Similar studies have since identified additional residues in the M1 protein that affect influenza virus morphology, including residues 30, 41, 98, 101, 102, 207, 209, and 218 (16–18).
Several other viral proteins have also been suggested to affect influenza virus morphology, including the two glycoproteins HA and NA. Evidence supporting a role for the two glycoproteins was obtained from a study originally aimed at examining the function of the highly conserved HA and NA cytoplasmic tails (7). In this study, a recombinant WSN virus containing deletions of the NA cytoplasmic tail domain produced elongated and irregularly shaped particles compared to the wild type (wt). Furthermore, while deletion of the HA cytoplasmic tail was not sufficient to induce morphological change, a recombinant virus lacking both the HA and NA cytoplasmic tails exhibited an even more profound alteration in particle shape than viruses with a deletion in NA alone (7). Similarly, a deletion or mutations at residues 74 to 79 in the WSN M2 cytoplasmic tail resulted in a filamentous morphology (19). These studies indicate that the M1 interaction with the transmembrane proteins can affect virus budding and morphology.
In addition to the transmembrane proteins, some indirect studies suggest that NP may also affect virion morphology. One of these reports showed the difference in M1-vRNP binding affinity, which was higher in spherical viruses than in filamentous viruses (20). Another report showed that a temperature-sensitive mutation at NP residue 239 results in the production of abnormally shaped virions without affecting virus RNA synthesis (21). At this stage, it is unclear how vRNPs contribute to the morphology of budding viruses. In this study, we determined the possible role of NP in virus morphology using WSN and A/Aichi/2/68 (Aichi), the latter of which includes M1 residues that confer filamentous morphology. We found that even with the M1 residues, Aichi possesses a spherical phenotype. Furthermore, recombinant WSN expressing Aichi M1 (WSN-AichiM1) induced filaments from infected cell surfaces, confirming the presence of the residues responsible for filament formation in Aichi M1. Because Aichi formed mainly spherical virions, the data suggest that an interaction between M1 and another viral protein influences Aichi morphology. Upon characterization of WSN-AichiM1 viruses expressing additional Aichi proteins, we discovered that coexpression of Aichi M1 and NP completely inhibited filament formation. We identified three NP residues, 214, 217, and 253, which play a critical role in virion morphology. Interestingly, these residues localize at the potential M1-interaction sites on the vRNP. Possible roles of the specific M1-vRNP interaction and how it determines virion morphology are discussed.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK), human kidney 293T, and human lung A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 8% fetal calf serum. A/Aichi/2/68 (H3N2) was obtained from A. Klimov (Centers for Disease Control, Atlanta, GA).
Plasmids and virus rescue.
pPolI vectors which include human RNA polymerase I promoter and pCAGGS expression vectors for rescue of A/WSN/33 were provided by Y. Kawaoka (University of Wisconsin, Madison, WI) (22). cDNA clones of Aichi virus were synthesized from viral RNAs extracted from infected cells by reverse transcription-PCR (RT-PCR) using a SuperScript III One-step RT-PCR Platinum Taq HIFi kit (Invitrogen). All eight viral cDNAs were subcloned into pPolI vectors. Aichi PB1, PB2, PA, and NP cDNAs were also subcloned into pCAGGS vectors. pPolI-AichiM1, which expresses Aichi M1 and WSN M2, was constructed by replacing the BsiWI-StuI fragments of the genes. This results in replacing only the M1 coding region of the WSN M gene. Mutations in the pPolI-WSN NP plasmid were made by site-directed mutagenesis and confirmed by sequence analysis. pPolI-NP WA and pPolI-NP AW, which express chimeric NP genes, were constructed by replacing SphI-AflIII fragments of pPolI-WSN NP and pPolI-Aichi NP. Influenza viruses were rescued with the 12-plasmid reverse genetics system developed by Neumann et al. (22). For rescue, pPolI vectors encoding each of the full-length viral RNA segments were transfected into MDCK/293T cocultures together with pCAGGS containing WSN PB1, PB2, PA, and NP genes using Lipofectamine 2000 (Invitrogen). Rescued viruses were plaque purified and propagated in MDCK cells cultured with DMEM containing 0.15% bovine serum albumin (BSA) and tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin at 1 μg/ml.
Immunofluorescence (IF) microscopy.
MDCK or A549 cells were infected with viruses for 1 h at 37°C and cultured in DMEM containing 0.15% BSA. After 18 h, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) containing magnesium and calcium [PBS(+)] (Corning) and treated for 10 min with 0.1% Triton X-100 if permeabilization was required. To detect viral surface proteins, unpermeabilized infected cells were incubated with anti-H1 or anti-H3 goat serum (NR-3148 and NR-3118, respectively; BEI Resources), followed by Alexa Fluor 488 anti-goat IgG (Molecular Probes). For detection of NP, permeabilized cells were incubated with mouse anti-NP monoclonal antibody (MAb) (NR-4544; BEI Resources), followed by Texas Red anti-mouse IgG (Molecular Probes). Fluorescent images were obtained using an Olympus FV1000 confocal microscope or Olympus IX50 inverted fluorescence microscope.
Electron microscopy.
For scanning electron microscopy (SEM), MDCK cells were grown on glass coverslips and infected for 18 h. After fixation with 2.5% glutaraldehyde, cell surfaces were visualized under a Zeiss Auriga Supra 40VP Field Emission microscope. To analyze virion morphology by transmission electron microscopy (TEM), supernatants of infected MDCK cells were harvested at 24 h postinfection (p.i.) and spun through a 100,000-molecular-weight-cutoff Amicon filter (Millipore) for 18 min at 2,000 × g. Concentrated virus was applied to carbon-coated 200-mesh grids for 5 min, washed once with PBS(+), and negatively stained for 1 min with filter-sterilized 2% phosphotungstic acid (PTA). Electron micrographs were taken with a Hitachi 7650 microscope.
Characterization of purified viruses and immunoprecipitation.
MDCK cells were infected at a multiplicity of infection (MOI) of 3 with WSN, WSN-AichiM1, or WSN-AichiM1 expressing Aichi NP (WSN-AichiM1/NP) virus for 1 h at 37°C. After 8 h, cells were washed once with PBS(+) and labeled for an additional 18 h with [35S]methionine-cysteine (PerkinElmer). Cell culture supernatants, cleared of cell debris, were spun through 30% glycerol cushions at 115,000 × g for 90 min at 4°C. Pellets containing virus were resuspended in SDS sample buffer and analyzed by SDS-PAGE. Cell lysates harvested in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) were used for immunoprecipitation using anti-NP MAb kindly provided by G. Whittaker (Cornell) and anti-M1 goat serum (ViroStat). Band intensities were measured using Bio-Rad Quantity One software.
Subcellular fractionation.
Subcellular fractions of infected MDCK cells were prepared at 18 h p.i. using a Nuclear/Cytosol Fractionation Kit (BioVision) or ProteoExtract Subcellular Proteome Extraction Kit (Calbiochem). Each fraction was suspended in NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) and separated by SDS-PAGE. Following transfer to a polyvinylidene difluoride (PVDF) membrane, proteins were subjected to Western blot analysis. NP and M1 were detected with anti-NP (NR-4544; BEI Resources) and anti-M1 (GA2B; Santa Cruz) MAbs. Mouse anti-lamin A/C (Upstate) and chicken anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Millipore) antibodies were used as markers for nuclear and cytoplasmic fractions, respectively. Proteins were quantified by measuring band intensities with Bio-Rad Quantity One software.
RESULTS
WSN expressing Aichi M1 forms filamentous virions.
The classical human strain Aichi shares the Udorn M1 amino acid residues at positions 41, 95, 204, and 218, which have previously been shown to induce a filamentous phenotype (15, 17). First, we utilized MDCK cells to examine Aichi virion morphology and the surface structure of infected cells to determine if these residues in Aichi M1 affect influenza virus morphology. Despite the presence of residues that cause filamentous virion formation, Aichi produced predominantly spherical virions, and few filamentous protrusions were detected in infected cells, similar to cells infected with WSN, which is a well-characterized, lab-adapted influenza virus strain known to produce uniformly spherical virions (15) (Fig. 1). To determine if Aichi M1 can alter WSN morphology, we rescued recombinant WSN viruses, which included either the whole Aichi M segment (WSN-AichiM) or a chimeric WSN/Aichi M segment (WSN-AichiM1) encoding Aichi M1 and WSN M2 proteins. We then tested whether expression of these segments was sufficient to induce a filamentous phenotype. Whereas cells infected with the WSN and Aichi viruses exhibited a punctate staining pattern characteristic of spherical-virion-producing influenza virus strains, long filamentous structures were observed on the surface of cells infected with WSN-AichiM. A similar induction of filaments could be seen on the surface of cells infected with a WSN virus expressing the Aichi M1 protein alone (WSN-AichiM1) (Fig. 1, top panels).
FIG 1.
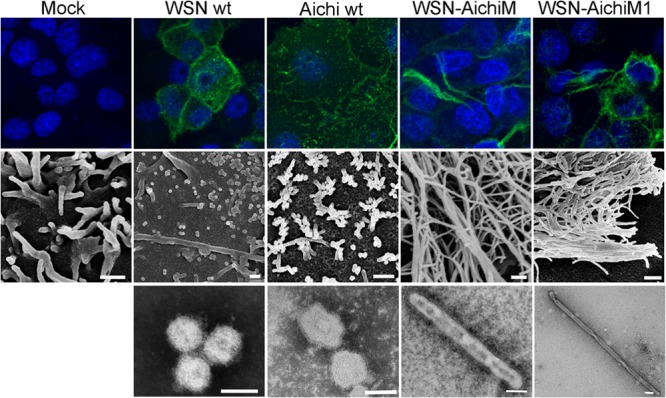
Aichi M1 can convert WSN morphology to filamentous. MDCK cells were mock infected or infected with WSN, Aichi, WSN-AichiM and WSN-AichiM1 viruses at an MOI of 1 for 18 h as indicated. Cell surfaces were stained for viral proteins using anti-H1N1 or anti-H3N2 goat serum and Alexa Fluor 488 anti-goat IgG (top panels) or visualized by SEM (middle panels). Virions released into the culture supernatants were negatively stained with 2% PTA and examined by TEM (bottom panels). White bars in SEM and TEM images measure 500 nm and 100 nm, respectively.
We further examined the morphology of budding virions by scanning electron microscopy (SEM). As expected, small spherical structures with an approximate diameter of 100 nm were present on the surfaces of cells infected with the WSN wild-type virus. These same structures were visible on cells infected with Aichi virus although short filaments were also detected. In sharp contrast, cell surfaces infected with either the WSN-AichiM or WSN-AichiM1 recombinant virus were covered with an abundance of long filaments that differed from those seen in wt WSN- or Aichi-infected cells (Fig. 1, middle panels). We next verified the morphology of virions released into cell culture supernatants using transmission electron microscopy (TEM). Consistent with our SEM data, the majority of WSN and Aichi virions released from infected cells were found to be spherical. Conversely, the majority of WSN-AichiM and WSN-AichiM1 virions were filamentous, forming virions of several micrometers in length (Fig. 1, bottom panels). These results indicate that Aichi M1 contains residues that alter the virus morphology of WSN from spherical to filamentous.
Presence of Aichi NP, but not the viral transmembrane proteins, influenced WSN-AichiM1 morphology.
Aichi forms spherical virions despite its M1 residues that cause filament formation. This finding suggests that another Aichi component or components prevent filament formation induced by the M1 protein. Previous studies have suggested that interactions between M1 and the viral transmembrane proteins altered influenza virus virion morphology (7, 19, 23). We therefore tested the role of each of the transmembrane proteins by characterizing recombinant WSN-AichiM1 viruses that also express Aichi HA, NA, or M2. We had already observed that the addition of Aichi M2 (WSN-AichiM) had no effect on filament formation (Fig. 1). The remaining recombinant viruses, WSN-AichiM1 expressing Aichi HA and NA (WSN-AichiM1/HA and WSN-AichiM1/NA, respectively), were successfully rescued, and the surface structures of infected cells were determined by IF analysis. WSN-AichiM1/HA induced long filamentous structures in infected cells, similar to those observed in WSN-AichiM1-infected cells (Fig. 2A). WSN-AichiM1/NA also induced filaments although this was less evident than in cells infected with WSN-AichiM1 or WSN-AichiM1/HA (Fig. 2A). These results suggest that the interaction of M1 with the transmembrane proteins does not significantly affect WSN-AichiM1 morphology.
FIG 2.
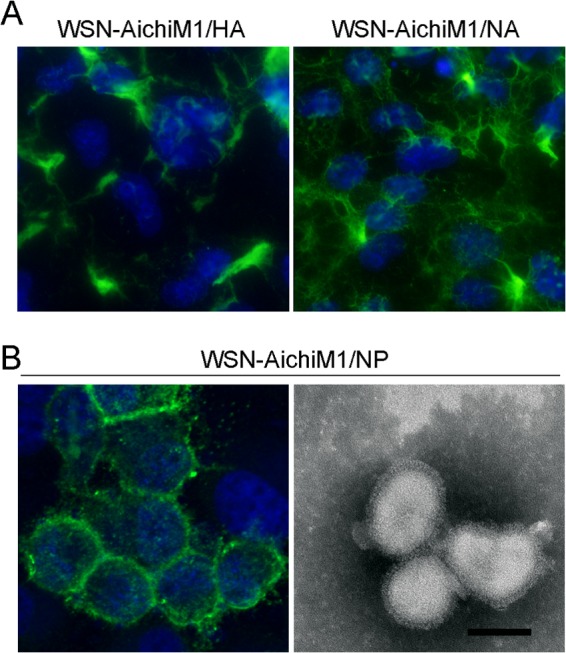
Interaction between Aichi M1 and NP influences influenza virus virion morphology. MDCK cells were infected with WSN-AichiM1/HA and WSN-AichiM1/NA (A) or WSN-AichiM1/NP (B) at an MOI of 1 for 18 h as indicated. Cell surfaces were stained for viral proteins using anti-H1N1 goat serum followed by Alexa Fluor 488 anti-goat IgG. Progeny WSN-AichiM1/NP virions released into the culture supernatant of infected cells were negatively stained with 2% PTA and examined by TEM (B, right panel). The scale bar in the TEM image measures 100 nm.
In addition to interaction of the transmembrane proteins with M1, the viral nucleocapsid interacts with the M1 protein during the assembly of influenza virus virions. Recent studies also suggest that the M1-NP interaction may be involved in determining influenza virus virion morphology (20, 21). Thus, we rescued and characterized a WSN-AichiM1 recombinant that coexpressed Aichi NP (WSN-AichiM1/NP). Examination of cell surfaces by IF revealed a complete loss of filament formation in cells following infection with the WSN-AichiM1/NP virus (Fig. 2B). Furthermore, analysis of purified virions by electron microscopy showed a spherical WSN-AichiM1/NP morphology (Fig. 2B). These results strongly suggest that specific interactions between M1 and NP can influence influenza virus virion morphology.
Characterization of M1-NP interactions in WSN-AichiM1/NP.
Recent work with cryoelectron microscopy has revealed that influenza virus virions, regardless of their shape, contain a single copy of the viral genome (24–26). Therefore, filamentous virions are expected to contain a higher M1/NP ratio due to increased surface area. To test this, MDCK cells were infected with wild-type WSN, WSN-AichiM1, or WSN-AichiM1/NP and radiolabeled with [35S]Met-Cys for 18 h. Following purification of virions by ultracentrifugation, we compared the M1 and NP contents by SDS-PAGE (Fig. 3). The relative content of NP in WSN-AichiM1 was about 50% less than that of NP in WSN, making the M1/NP ratio 2-fold greater than that of WSN. Importantly, incorporation of NP in the virions was increased by the presence of homologous Aichi NP in WSN-AichiM1/NP virions. This difference was not due to the expression of NP because all three viruses produced similar levels of NP and M1 in infected cells, as determined by immunoprecipitation (Fig. 3). These results are consistent with the virion morphology determined by IF, SEM, and TEM described above.
FIG 3.
Presence of homologous virus M1 and NP increases vRNP content in virions. MDCK cells infected with WSN, WSN-AichiM1, or WSN-AichiM1/NP virus at an MOI of 1 were labeled with [35S]methionine-cysteine for 24 h, and purified virions were analyzed by SDS-PAGE. Expression of NP and M1 in cell lysates was examined by immunoprecipitation assay. Intensities of the protein bands were quantitated, and relative ratios are shown. RIP, radioimmunoprecipitation.
Influenza virus vRNPs are produced in the nuclei of infected cells, and a specific interaction between vRNP, M1, and NEP allows nuclear export of the vRNP complexes to the cytoplasm. vRNPs are then believed to be transported to the plasma membrane through Rab11a-mediated recycling endosomes (27–29). Therefore, it is possible that an impaired interaction between M1 and NP results in reduced vRNP accumulation at the plasma membrane assembly sites, which in turn may influence influenza virus virion morphology. To examine a difference in NP and M1 localizations between the viruses, we collected cytoplasmic and nuclear fractions of infected cells at 12 and 18 h postinfection and determined the quantities of NP and M1 by Western blotting. More Aichi NP was localized in the cytoplasm than in the nucleus; however, there was no correlation between NP and M1 localization and virion morphologies (Fig. 4A). To confirm this, we also collected nuclear, cytoplasmic, and membrane fractions of infected cells, and the presence of NP was quantified by Western blotting. Again, we did not detect a significant difference in the amounts of NP recovered from membrane fractions between the viruses (Fig. 4B), suggesting that despite heterologous combination, WSN-AichiM1 vRNP was transported and associated with the plasma membrane as efficiently as wt viruses. Localization of NP at the plasma membrane was also determined by IF using MDCK cells infected with the viruses for 18 h. We used confocal microscopy to obtain three-dimensional (3D) images of NP localization at the plasma membrane. Strong punctate staining of NP was detected at the plasma membranes of cells infected with both filamentous and spherical viruses (Fig. 4C). Together, these results indicate that there was no significant difference in vRNP localization between the filamentous and spherical viruses.
FIG 4.
NP localizations do not significantly differ among the recombinant viruses. MDCK cells were infected with WSN, Aichi, WSN-AichiM1, and WSN-AichiM1/NP viruses at an MOI of 1 for 18 h. (A) Localization of NP and M1 within the nuclear and cytoplasmic fractions. Mouse anti-NP MAb and goat anti-M1 serum were used for the detection of NP and M1, respectively. Lamin A/C and GAPDH antibodies were also used as markers for nuclear (Nuc) and cytoplasmic (Cyt) fractions, respectively. (B) Quantification of NP within the nuclear, cytoplasmic, and membrane fractions. NP in subcellular fractions was detected by Western blot analysis. Error bars indicate the standard deviation from three separate experiments. (C) Intracellular localization of NP was visualized by IF analysis using a confocal microscope. The z-stack images of the regions indicated by white rectangles were created and are shown below the xy-plane images.
NP residues 214, 217, and 253 are directly involved in determining influenza virus virion morphology.
The difference in virion morphology between WSN-AichiM1 and WSN-AichiM1/NP clearly indicates that the origin of NP can affect virus morphology, suggesting the presence of a specific interaction between M1 and NP that determines virion morphology. We first compared the WSN and Aichi NP protein sequences to identify the possible region of interaction. This alignment revealed a total of 37 amino acid differences, which spanned the entire length of the NP protein (Fig. 5A). To identify the region on Aichi NP responsible for the morphological change of WSN-AichiM1 virus, we constructed two chimeric NP genes: chimera NP WA encoding residues 1 to 305 from WSN fused to the rest of the sequence from Aichi, and NP AW encoding residues 1 to 305 from Aichi fused to the rest of the sequence from WSN (Fig. 5B). Using these constructs, we rescued two additional viruses with a WSN-AichiM1 background, WSN-AichiM1/NP WA and WSN-AichiM1/NP AW. MDCK cells infected with each of these viruses were observed by IF analysis. Infection with WSN-AichiM1/NP WA, which expresses the N-terminal 305 residues from WSN, did not inhibit filament formation from infected cell surfaces. In sharp contrast, long filaments were absent from cells infected with WSN-AichiM1/NP AW, indicating that the N-terminal region of Aichi NP is directly involved in determining influenza virus virion morphology (Fig. 5C).
FIG 5.
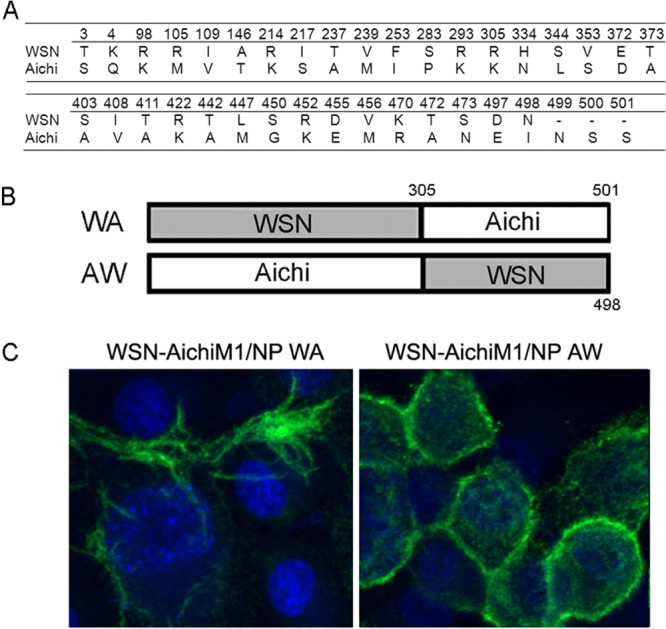
The N-terminal 305 residues of Aichi NP are directly involved in determining influenza virus virion morphology. (A) Amino acid differences between WSN and Aichi NPs. (B) Schematic diagram of chimeric NP proteins. Chimeric NP WA includes the N-terminal 305 residues from WSN and the rest of the sequence from Aichi. Chimeric NP AW contains the N-terminal 305 from Aichi and the rest of the sequence from WSN. (C) MDCK cells were infected with recombinant WSN-AichiM1 viruses expressing NP WA or NP AW at an MOI of 1 for 18 h as indicated. Cell surfaces were stained for viral proteins with anti-H1N1 goat serum followed by Alexa Fluor 488 anti-goat IgG.
Within the N-terminal 305 residues, there are 14 amino acid differences between WSN and Aichi NP sequences (Fig. 5A). Upon mapping these residues to the crystal structure of NP, we found that they localized to three very distinct regions of the protein (Fig. 6A). We therefore designed three additional NP chimeras that encode most of the WSN NP sequence but which contain the residues found in region 1 (R1), R2, or R3 of Aichi NP (Fig. 6B). Additional recombinant WSN-AichiM1 viruses that express each of these NP constructs were rescued and characterized by IF microscopy. Viruses expressing either R1 or R3 of Aichi NP were found to induce filaments similar to those seen in cells infected with WSN-AichiM1. In contrast, infection with WSN-AichiM1/NP R2 appeared to result in the production of spherical virions, as indicated by a punctate staining pattern resembling both the WSN and Aichi wild-type viruses (Fig. 6C).
FIG 6.
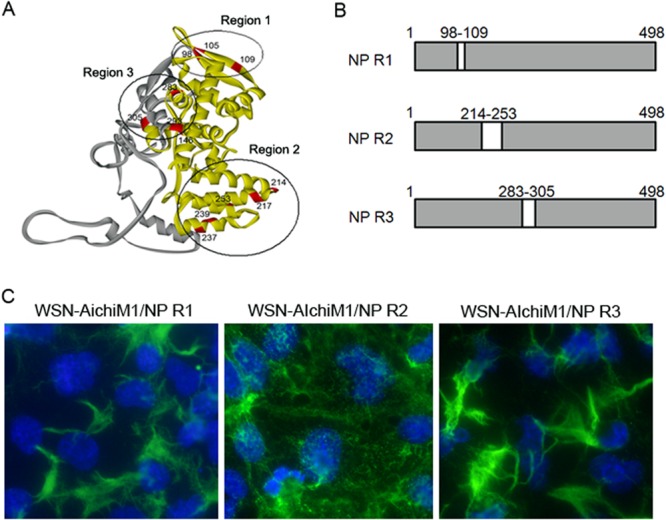
The region of Aichi NP required to inhibit filament formation by the WSN-AichiM1 virus. (A) Crystal structure of NP highlighting the locations of residues that differ between the WSN and Aichi viruses. The N-terminal 305 residues and the rest of the regions are shown in yellow and gray, respectively. Region 1 (R1), R2, and R3 are circled. (B) Schematic diagram of chimeric NP proteins. The amino acid residues of WSN NP located in R1, R2, or R3 were replaced with the corresponding Aichi NP residues to create NP R1, NP R2, and NP R3 chimeras. (C) MDCK cells were infected with recombinant WSN-AichiM1 viruses expressing the R1, R2, or R3 NP chimera at an MOI of 1, as indicated. Cell surfaces were stained for viral proteins with anti-H1N1 goat serum followed by Alexa Fluor 488 anti-goat IgG.
Region 2 of NP contains 5 amino acid differences between the WSN and Aichi sequences at positions 214, 217, 237, 239, and 253. We further identified the minimal Aichi NP residues required to inhibit filament formation of the WSN-AichiM1 virus using site-directed mutagenesis. Due to the close proximity of some of these residues, double mutations were made in the WSN NP protein at positions 214 and 217 or at 237 and 239, in which we substituted the corresponding amino acid from the Aichi NP sequence. A single substitution in the WSN NP protein was made at position 253. IF, SEM, and TEM analysis showed that while mutations at positions 237 and 239 did not alter the filamentous morphology of WSN-AichiM1, expression of residues 214 and 217 or of 253 alone from Aichi NP was sufficient for the morphological change at the surfaces of infected cells and virions (Fig. 7A). We detected the same effects in A549 cells as well (Fig. 7B). These results indicate that NP residues 214, 217, and 253 play a major role in virion formation and morphology, possibly through the interaction with M1 during virus budding at the assembly sites.
FIG 7.
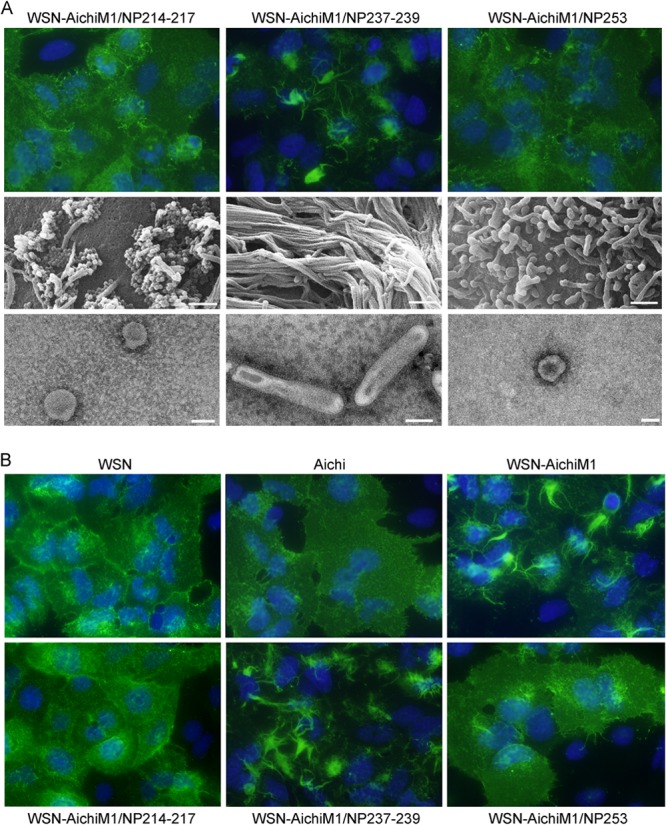
Aichi NP residues 214, 217, and 253, but not 237 and 239, are necessary and sufficient to inhibit filament formation by the WSN-AichiM1 virus. (A) MDCK cells were infected with recombinant viruses at an MOI of 1 for 18 h as indicated. Cell surfaces were stained for viral proteins with anti-H1N1 goat serum followed by Alexa Fluor 488 anti-goat IgG (top panels) or visualized by SEM (middle panels). Virions released into the culture supernatants were negatively stained with 2% PTA and examined by TEM (bottom panels). White scale bars in SEM and TEM images measure 500 nm and 100 nm, respectively. (B) A549 cells were infected with the wt or recombinant viruses, and the infected cell surface was analyzed by IF using anti-H1N1 or anti-H3N2 serum as described above.
DISCUSSION
Influenza virus virions are known to be pleiomorphic, ranging between 100-nm spheres and filamentous particles that reach several micrometers in length (30). A set of eight segmented vRNPs is incorporated into each progeny virion regardless of spherical or filamentous morphology (24, 26, 31). Among the viral proteins, M1 is known to be the major determinant of virion morphology. Specific mutations in M1 residues, such as 41A, 95R, and 204E, alter the morphology of spherical viruses to filamentous (17, 18). However, we found that Aichi formed spherical virions in spite of its M1 residues, known to confer a filamentous morphology. By analyzing the recombinant viruses, we showed evidence that the presence of specific NP residues prevents filamentous virion formation. We identified the specific NP residues 214, 217, and 253 that are able to convert filamentous virions to spherical ones, even in the presence of filamentous M1 residues. Our data clearly indicate that NP residues are involved in the final virus assembly process that determines virion morphology.
A study of the structural organization of filamentous virions using electron cryo-tomography showed a clear image of M1 forming a structural layer beneath the viral envelope, which has a highly uniform diameter of about 100 nm (26). X-ray crystallography analysis revealed that M1 dimers stack on top of one another to create a ribbon-like structure through two separate points of interaction (32). Under acidic conditions, one of the M1-M1 interfaces gained flexibility, causing the angle of the M1 dimers to change, and thus induced curvature of the M1 ribbon that is predicted to coat the interior of influenza virus virions. These studies suggest that M1 is flexible in structure and that mutations at specific M1 residues or under various conditions, such as pH change, can alter M1-M1 interactions between the layers. Considering the major role of M1 in the structural frame, the ability of M1 to form straight or bent elongated ribbons and helices is likely to be responsible for the morphological change of influenza virus virions.
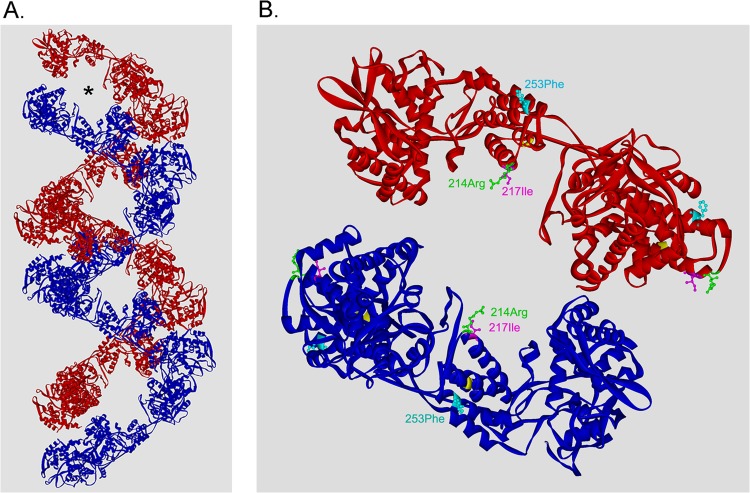
Our data indicate that specific NP residues can convert filamentous virions to spherical ones. We anticipate that the NP residues, based on their locations in vRNPs, alter virus morphology through interaction with the M1 layer. Recent studies of vRNP structure by cryogenic electron microscopy revealed that vRNPs form a double-helical conformation in which two NP strands of opposite polarity are associated with each other along the helix (33, 34). The structure of the vRNP isolated from virions showed that, in addition to a major groove that is well separated, helical vRNP possesses a minor groove that is maintained by NP associated with opposing RNA strands (Fig. 8A, asterisk) (33). NP residues 214 to 253 (Fig. 6A, R2) locate in the head domain of NP and a part of the area surrounding the minor groove. Residues 214 and 217 are on the helix α9 and locate exactly at this minor groove, which is exposed to the surface of the vRNP (Fig. 8B). NP residue 253 also locates close to this area of the surface. In contrast, residues 237 and 239, which did not affect the morphology of filamentous WSN-AichiM1 (Fig. 7), are in the middle of helix α10 and are not exposed to the surface of the vRNP (shown in yellow in Fig. 8B). Our data are consistent with the idea that the minor groove of the helical vRNP is the site of interaction with the M1 layer during virus budding. Upon incorporation of the vRNP into budding virions, these specific M1-NP interactions may induce a conformational change in the M1 protein, thus creating an angle between the M1 helical layers beneath the viral envelope, which determines the morphology of influenza viruses.
FIG 8.
Residues 214, 217, and 253 localize at the surface close to the minor groove of the vRNP. (A) A double-helical vRNP structure based on the cryo-EM reconstruction (Protein Data Bank accession number 4BBL) (33). The asterisk indicates the minor groove between the opposing NP strands. (B) Locations of the NP residues that affected virion morphology. Four NP molecules that form the minor groove are shown. Residues 237 and 239, which did not affect virus morphology, are shown in yellow.
The M1-NP interaction is unlikely to be the sole determinant of virus morphology. Deletion of the cytoplasmic tail of NA or M2 in other studies resulted in filamentous virion formation (7, 19). Similar to the case of vRNP, NA and M2 interactions with M1 may induce conformational changes of the M1 helical layers and create membrane curving at the bottom of budding virions. Interestingly, tomograms of filamentous virions showed NA clusters at the end of the virions opposite the end where the polymerase is attached (26). This localization may indicate that the M1 interaction with the NA cytoplasmic tail allows membrane curving to close the membrane of filamentous virions. Our present data and previous studies indicate that multiple protein interactions with M1 determine virus assembly and morphology. Further structural analysis of the M1-M1 interaction and the effects of other viral components are necessary to fully understand the process of virus assembly and budding.
ACKNOWLEDGMENTS
This work was supported by the New York Influenza Center of Excellence, a member of the NIAID CEIRS network, under NIH contract HHSN266200700008C and National Institutes of Health grant R21AI078130.
We thank K. Bentley (University of Rochester Electron Microscope Research Core) for technical assistance.
Footnotes
Published ahead of print 11 December 2013
REFERENCES
- 1.Kundu A, Avalos RT, Sanderson CM, Nayak DP. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leser GP, Lamb RA. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215–227. 10.1016/j.virol.2005.09.049 [DOI] [PubMed] [Google Scholar]
- 3.Scheiffele P, Roth MG, Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501–5508. 10.1093/emboj/16.18.5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J, Pekosz A, Lamb RA. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634–4644. 10.1128/JVI.74.10.4634-4644.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali A, Avalos RT, Ponimaskin E, Nayak DP. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709–8719. 10.1128/JVI.74.18.8709-8719.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barman S, Adhikary L, Chakrabarti AK, Bernas C, Kawaoka Y, Nayak DP. 2004. Role of transmembrane domain and cytoplasmic tail amino acid sequences of influenza a virus neuraminidase in raft association and virus budding. J. Virol. 78:5258–5269. 10.1128/JVI.78.10.5258-5269.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin H, Leser GP, Zhang J, Lamb RA. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236–1247. 10.1093/emboj/16.6.1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elster C, Larsen K, Gagnon J, Ruigrok RW, Baudin F. 1997. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 78:1589–1596 [DOI] [PubMed] [Google Scholar]
- 10.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 88:2280–2290. 10.1099/vir.0.82809-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BJ, Leser GP, Jackson D, Lamb RA. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059–10070. 10.1128/JVI.01184-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossman JS, Jing X, Leser GP, Lamb RA. 2010. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142:902–913. 10.1016/j.cell.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. 10.1016/j.virol.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nayak DP, Hui EK, Barman S. 2004. Assembly and budding of influenza virus. Virus Res. 106:147–165. 10.1016/j.virusres.2004.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bourmakina SV, Garcia-Sastre A. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517–527. 10.1099/vir.0.18803-0 [DOI] [PubMed] [Google Scholar]
- 16.Bialas KM, Desmet EA, Takimoto T. 2012. Specific residues in the 2009 H1N1 swine-origin influenza matrix protein influence virion morphology and efficiency of viral spread in vitro. PLoS One 7:e50595. 10.1371/journal.pone.0050595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elleman CJ, Barclay WS. 2004. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology 321:144–153. 10.1016/j.virol.2003.12.009 [DOI] [PubMed] [Google Scholar]
- 18.Burleigh LM, Calder LJ, Skehel JJ, Steinhauer DA. 2005. Influenza a viruses with mutations in the m1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79:1262–1270. 10.1128/JVI.79.2.1262-1270.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80:5233–5240. 10.1128/JVI.00049-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Muller J, Ye Z. 2002. Association of influenza virus matrix protein with ribonucleoproteins may control viral growth and morphology. Virology 304:89–96. 10.1006/viro.2002.1669 [DOI] [PubMed] [Google Scholar]
- 21.Noton SL, Simpson-Holley M, Medcalf E, Wise HM, Hutchinson EC, McCauley JW, Digard P. 2009. Studies of an influenza A virus temperature-sensitive mutant identify a late role for NP in the formation of infectious virions. J. Virol. 83:562–571. 10.1128/JVI.01424-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350. 10.1073/pnas.96.16.9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. 2010. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84:5078–5088. 10.1128/JVI.00119-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. 2006. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature 439:490–492. 10.1038/nature04378 [DOI] [PubMed] [Google Scholar]
- 25.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103:19123–19127. 10.1073/pnas.0607614103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 107:10685–10690. 10.1073/pnas.1002123107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momose F, Sekimoto T, Ohkura T, Jo S, Kawaguchi A, Nagata K, Morikawa Y. 2011. Apical transport of influenza A virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One 6:e21123. 10.1371/journal.pone.0021123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisfeld AJ, Kawakami E, Watanabe T, Neumann G, Kawaoka Y. 2011. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J. Virol. 85:6117–6126. 10.1128/JVI.00378-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amorim MJ, Bruce EA, Read EK, Foeglein A, Mahen R, Stuart AD, Digard P. 2011. A Rab11- and microtubule-dependent mechanism for cytoplasmic transport of influenza A virus viral RNA. J. Virol. 85:4143–4156. 10.1128/JVI.02606-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palese P, Shaw M. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 31.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y. 2012. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat. Commun. 3:639. 10.1038/ncomms1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris A, Forouhar F, Qiu S, Sha B, Luo M. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289:34–44. 10.1006/viro.2001.1119 [DOI] [PubMed] [Google Scholar]
- 33.Arranz R, Coloma R, Chichon FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortin J, Martin-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. 10.1126/science.1228172 [DOI] [PubMed] [Google Scholar]
- 34.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338:1631–1634. 10.1126/science.1227270 [DOI] [PMC free article] [PubMed] [Google Scholar]