Abstract
During herpes simplex virus 1 (HSV-1) infection, empty procapsids are assembled and subsequently filled with the viral genome by means of a protein complex called the terminase, which is comprised of the HSV-1 UL15, UL28, and UL33 proteins. Biochemical studies of the terminase proteins have been hampered by the inability to purify the intact terminase complex. In this study, terminase complexes were isolated by tandem-affinity purification (TAP) using recombinant viruses expressing either a full-length NTAP-UL28 fusion protein (vFH476) or a C-terminally truncated NTAP-UL28 fusion protein (vFH499). TAP of the UL28 protein from vFH476-infected cells, followed by silver staining, Western blotting, and mass spectrometry, identified the UL15, UL28, and UL33 subunits, while TAP of vFH499-infected cells confirmed previous findings that the C terminus of UL28 is required for UL28 interaction with UL33 and UL15. Analysis of the oligomeric state of the purified complexes by sucrose density gradient ultracentrifugation revealed that the three proteins formed a complex with a molecular mass that is consistent with the formation of a UL15-UL28-UL33 heterotrimer. In order to assess the importance of conserved regions of the UL15 and UL28 proteins, recombinant NTAP-UL28 viruses with mutations of the putative UL28 metal-binding domain or within the UL15 nuclease domain were generated. TAP of UL28 complexes from cells infected with each domain mutant demonstrated that the conserved cysteine residues of the putative UL28 metal-binding domain and conserved amino acids within the UL15 nuclease domain are required for the cleavage and packaging functions of the viral terminase, but not for terminase complex assembly.
INTRODUCTION
A critical step during productive herpes simplex virus 1 (HSV-1) infection is the cleavage and packaging of replicated, concatemeric viral DNA into preformed capsids. This process has been shown to require the products of seven viral genes: UL6, UL15, UL17, UL25, UL28, UL32, and UL33 (1). Many of these genes have been assigned putative functions based on genetic and biochemical studies and on similarities between HSV-1 and double-stranded DNA (dsDNA) bacteriophage cleavage/packaging systems (2). The UL6 protein forms a dodecameric complex at one of the 12 capsid vertices and functions as a portal through which viral DNA can enter or exit the capsid (3–6). The UL17 and UL25 proteins have recently been shown to form a heterodimeric complex termed the capsid vertex-specific component (CVSC) that functions at each of the capsid vertices to stabilize DNA-filled capsids (7–9). The UL32 protein is not well characterized but may play a role in localizing capsids to sites of DNA packaging (10, 11). The remaining three proteins, UL15, UL28, and UL33, form a putative complex within infected cells that functions as the viral terminase (1).
Difficulties with the purification of the terminase proteins and the lack of an in vitro cleavage and packaging system in HSV-1 has hampered studies on the roles of the UL15, UL28, and UL33 proteins in the DNA-packaging reaction. However, cryo-electron microscopy (EM) and genetic studies have provided several lines of evidence demonstrating that these proteins function as the HSV-1 terminase. They include studies demonstrating that (i) viral DNA is not cleaved in HSV-1 mutants encoding mutations that preclude the interaction of UL15 or UL33 with UL28 (12–16), (ii) HSV-1 recombinants with mutations in the UL6 portal that interfere with the terminase-portal interaction are deficient in cleavage and packaging of replicated viral DNA (17), (iii) the transient association of UL15 and UL28 with viral capsids is also observed with dsDNA bacteriophage terminases (2, 18–21), (iv) human cytomegalovirus (HCMV) mutants displaying resistance to inhibitors of viral DNA cleavage possess mutations within homologs of UL15 and UL28 (22, 23), and (v) the UL15 and UL28 proteins contain several conserved domains that are essential for cleavage and packaging of viral DNA (Fig. 1) (13, 14, 16, 22–34).
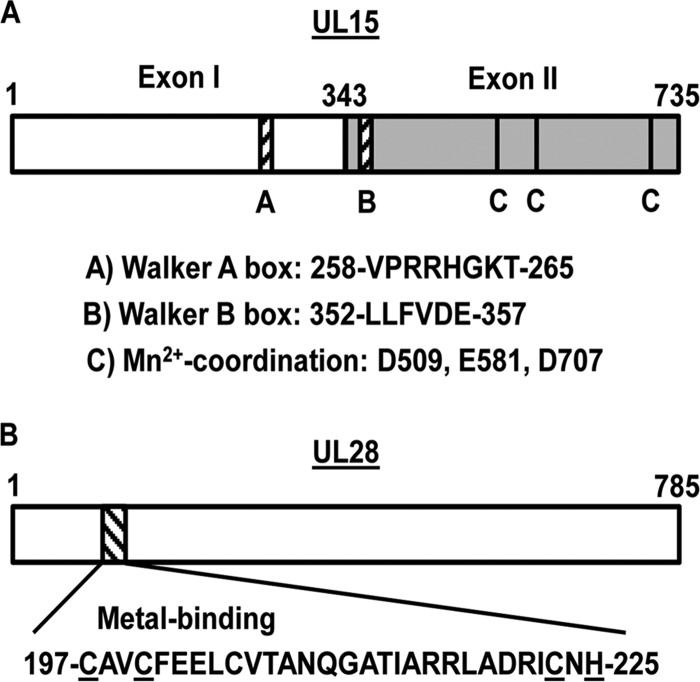
FIG 1.
Conserved UL15 and UL28 protein domains. The schematic depicts the UL15 (A) and UL28 (B) proteins, with conserved amino acid sequences indicated. All the numbers correspond to amino acid numbers. (A) The gray shading delineates UL15 exon II from exon I. (B) Conserved cysteine and histidine residues within the putative UL28 metal-binding domain are underlined.
UL15 is the most conserved gene within the family Herpesviridae and expresses a protein encoded within two exons of a spliced transcript (Fig. 1A) (35). Exon I and the N-terminal region of exon II contain domains typically found in proteins that function in ATP metabolism, suggesting that these regions provide the motor function for DNA translocation into the capsid (30, 36–38). Specifically the Walker A and B box motifs are conserved in homologs of the UL15 protein found in dsDNA bacteriophages and are essential for viral DNA cleavage and packaging (26, 30, 36, 38). Two recent studies have demonstrated that a purified, soluble C-terminal fragment of the UL15 protein and the HCMV UL89 protein (a UL15 homolog) possess nuclease activity, and the crystal structures of both fragments identified metal-coordinating amino acids that are conserved within herpesvirus UL15 homologs (32, 39). The UL28 protein has been shown to bind specific HSV-1 DNA sequences that are required for cleavage and packaging, and this function has also been observed with the HCMV UL56 protein (a UL28 homolog) (40, 41). Furthermore, UL28 encodes a putative metal-binding domain that was discovered in HCMV and shown to contain residues that are conserved among herpesvirus UL28 homologs (Fig. 1B) (22, 42). The final terminase subunit, UL33, is the least well characterized but may be required for correct assembly of the terminase complex or may act to regulate the enzymatic activity of the other subunits during complex function (13, 15, 16).
Although a great deal of information has been obtained from genetic studies of the HSV-1 cleavage/packaging genes, several of these proteins have proven difficult to isolate for biochemical and structural studies. The goal of the present study was to isolate a functional terminase complex from HSV-1-infected cells by affinity chromatography, using a virus that expressed the tandem-affinity purification (TAP) tag fused to the N terminus of the UL28 protein. TAP resulted in the isolation of soluble UL28 complexes containing the UL15 and UL33 proteins. The isolated complexes were characterized using genetic and biochemical approaches to determine the composition of the complex and the roles of conserved domains in terminase function. We view these studies as a critical step in understanding how the terminase complex functions in the context of a productive HSV-1 infection. The discovery of small organic molecules that inhibit herpesvirus replication by blocking processing and encapsidation demonstrates the validity of targeting the cleavage/packaging complex for therapeutic intervention (22, 43). These efforts have led to the discovery of an HCMV terminase inhibitor (Letermovir) that is presently in phase III studies (44).
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney (Vero) and UL15-complementing (C2) cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 5% newborn calf serum, 100 U penicillin per ml, and 100 μg streptomycin per ml (growth medium) (29). UL28-complementing (CV28) cells were maintained exactly like Vero cells but in medium supplemented with 10% newborn calf serum (15). The KOS strain of HSV-1 was used as the wild-type virus. The GCB virus, containing a 1,881-bp deletion within the UL28 open reading frame (ORF), was described previously (14).
Construction of recombinant viruses.
The vFH475 and vFH476 mutant viruses were generated by recombination of a KOS genome maintained within a bacterial artificial chromosome (BAC) (Table 1) (45). The KOS BAC clone was transferred to bacterial strain GS1783, and mutagenesis was performed using the two-step bacteriophage Red-mediated homologous-recombination system (46), as previously described (47). Each additional mutant virus was generated by the same methods but through recombination of a bFH476 or bFH503 BAC that was transformed into GS1783 bacteria (Table 1). A list of the primers and template plasmid DNA used to amplify the kanamycin resistance constructs will be provided on request. Plasmids p-EP-TAP-in and p-EP-Kan-S were obtained from P. Kinchington (University of Pittsburgh). During generation of the bFH503 BAC, sequencing of the UL28 metal-binding domain region revealed that several clones contained only the C197A mutation, resulting in the generation of the bFH543 BAC. BAC DNA was transfected into Vero cells, UL28-complementing CV28 cells, or UL15-complementing C2 cells (Table 1), and recombinant viruses were harvested from cell lysates and plaque purified. Insertions, deletions, or base pair changes were confirmed by PCR amplification of purified viral DNA using primers that flank the UL15 or UL28 open reading frames, and the PCR products were sequenced.
TABLE 1.
Recombinant viruses
| Virus no. | Virus name | BACa | Cell lineb |
|---|---|---|---|
| vFH475 | NTAP-UL28 | KOS | Vero |
| vFH476 | CMV-NTAP-UL28 | KOS | Vero |
| vFH499 | CMV-NTAP-UL28-741s | bFH476 | CV28 |
| vFH505 | CMV-NTAP-UL28(Δ197-225) | bFH476 | CV28 |
| vFH503 | CMV-NTAP-UL28(C197A,C200A) | bFH476 | CV28 |
| vFH504 | CMV-NTAP-UL28(C223A,H225A) | bFH476 | CV28 |
| vFH542 | CMV-NTAP-UL28(C197A,C200A,C223A,H225A) | bFH503 | CV28 |
| vFH543 | CMV-NTAP-UL28(C197A) | bFH476 | CV28 |
| vFH546 | CMV-NTAP-UL28(C197H) | bFH476 | CV28 |
| vFH506 | CMV-NTAP-UL28, UL15(D706A,D707A) | bFH476 | C2 |
| vFH507 | CMV-NTAP-UL28, UL15(D509A) | bFH476 | C2 |
The BAC that was modified to generate the named virus.
Cell line used for BAC transfection and plaque purification of virus.
TAP.
The TAP protocol was performed by infecting 5 × 108 Vero cells with wild-type KOS or recombinant viruses at a multiplicity of infection (MOI) of 10 PFU/cell. The infection was allowed to proceed for 18 h at 37°C, after which the cells were harvested and pelleted via centrifugation at 5,000 rpm for 10 min at 4°C. All remaining steps were performed at 4°C. The cells were washed in a total volume of 50 ml 1× phosphate-buffered saline (PBS), and the final cell pellet was resuspended in 24 ml of streptavidin binding buffer (SBB) (300 mM KCl, 40 mM Tris-HCl [pH 7.5], 2 mM EDTA, 0.1% NP-40 substitute, and 5 mM β-mercaptoethanol) containing protease inhibitors (Roche; catalog no. 1 697 498). The cell suspension was sonicated using a probe sonicator 4 times for 10 s each time at an output of 6 W with chilling on ice between the sonication steps, resulting in a total cell lysate. Benzonase (1,500 U; Novagen; catalog no. 71205-3) was added to the samples and left at 4°C for 30 min. The extract was then clarified via centrifugation at 12,000 rpm in a Sorvall SS-34 rotor for 20 min at 4°C. The supernatant was transferred to a new tube, 1.5 ml of streptavidin resin (0.75-ml packed volume; Pierce; catalog no. 53117) was added, and the samples were rotated at 4°C for 2 h. The resin was pelleted by centrifugation at 3,500 rpm for 5 min, and the supernatant was removed. The resin was washed three times by being resuspended in 5 ml of SBB, followed by centrifugation at 3,500 rpm for 5 min. Protein was eluted from the resin by adding 3 ml of streptavidin elution buffer (300 mM KCl, 40 mM Tris-HCl [pH 7.5], 2 mM EDTA, 0.1% NP-40 substitute, 5 mM β-mercaptoethanol, 2 mM d-biotin [Sigma; catalog no. 47868]) containing Roche protease inhibitors, and the sample was rotated at 4°C for 30 min. The protein-resin mixture was spun down at 7,000 rpm in a microcentrifuge for 2 min, and the supernatant was collected as the streptavidin eluate. Twelve milliliters of calmodulin-binding buffer (CBB) (150 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM magnesium acetate, 1 mM imidazole, 2 mM CaCl2, 0.1% NP-40 substitute, 10 mM β-mercaptoethanol) containing Roche protease inhibitors was added to the streptavidin eluate. An additional 11.25 μl of 1 M CaCl2 was added to the mixture, along with 1.2 ml of calmodulin resin (0.6-ml packed volume; Agilent Technologies; catalog no. 214303-52), and samples were rotated at 4°C for 2 h. The resin was pelleted by centrifugation at 3,500 rpm for 5 min, and the supernatant was removed. The resin was washed three times by resuspension in 5 ml of CBB, followed by centrifugation at 3,500 rpm for 5 min. Protein was eluted from the resin by adding 3 ml of calmodulin elution buffer (150 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM magnesium acetate, 1 mM imidazole, 0.1% NP-40 substitute, 10 mM β-mercaptoethanol, 2 mM EGTA), and the sample was rotated at 4°C for 30 min. The protein-resin mixture was spun down at 7,000 rpm in a microcentrifuge for 2 min, and the supernatant was collected as the final (calmodulin) eluate.
Sucrose gradient ultracentrifugation.
UL28 protein complexes were isolated from vFH476-infected Vero cells by a partial TAP procedure after elution from the streptavidin resin. The eluted protein sample was concentrated 10-fold by column centrifugation (Pierce; catalog no. 89884A). The concentrated proteins were separated by centrifugation on 2.5 to 20% sucrose (in 1× PBS) gradients (SW50.1 rotor at 35,000 rpm for 18 h). Fractions were collected from the bottom to the top of the gradient using a Beckman fraction recovery system (Beckman; catalog no. 34890). A total of 33 fractions (130 μl each) were collected, and protein was precipitated by adding an equal volume of 16% trichloroacetic acid. Proteins were pelleted by centrifugation at 13,000 rpm for 10 min, and the pellets were resuspended in 30 μl 2× PAGE loading buffer (Invitrogen) supplemented with 0.4 M Tris base. Odd-numbered protein fractions were resolved on a 4 to 12% SDS-polyacrylamide gel and visualized by Western blotting using anti-UL28, -UL15, or -UL33 antibodies. Molecular weight control proteins, aldolase and bovine serum albumin (BSA) (GE Healthcare Life Sciences), were processed in the same manner but were visualized by staining with Imperial protein stain (Thermo Scientific; catalog no. 24615).
Mass spectrometry.
TAP-purified proteins from vFH476-infected cells were resolved on a 4 to 12% SDS-polyacrylamide gel and stained with Imperial protein stain (Thermo Scientific; catalog no. 24615). The stained gels were sent to Lisa Jones (Washington University in St. Louis, St. Louis, MO), who performed the remaining mass spectrometry (MS) analysis by the following methods. The in-gel digest was performed as previously described (48). Briefly, gel plugs were washed with acetonitrile (Sigma, St. Louis, MO) in a 96-well plate and rocked for 10 min twice. The samples were dried in a speed vac for 10 min. A 0.2-μg/μl stock of trypsin (Sigma, St. Louis, MO) was made up in 100 μl of 1 mM triethyl ammonium bicarbonate, pH 8.2. Trypsin (5 μl) was added to each dried gel plug, and the plate was incubated at 58°C for 30 min. After digestion, 1 μl of 1% acetonitrile with 1% formic acid was added to each gel plug, and the plate was incubated at 37°C for 1 h. The solution from each 96-well plate was transferred to autosampler vials and centrifuged at 10,000 rpm for 40 min. To reduce keratin contamination, the in-gel digest was performed in a laminar flow hood. The digested samples were loaded onto a 100-μm by 2-cm Acclaim PepMap100 C18 nanotrap column (5 μm; 100 Å; Thermo Scientific, Pittsburgh, PA) with an Ultimate 3000 liquid chromatograph (Thermo Scientific, Pittsburgh, PA) at 8 μl/min. The peptides were separated on a silica capillary column that was custom packed with C18 reverse-phase material (Magic; 0.075 mm by 150 mm; 5 μm; 120 Å; Michrom Bioresources, Inc., Auburn, CA). The gradient was pumped at 260 nl/min from 0 to 80% solvent B (20% water, 80% acetonitrile, 0.1% formic acid) for 60 min and then to 80% solvent B for 7 min and reequilibrated to solvent A (water, 0.1% formic acid) for 10 min. The mass spectrometry was performed on an LTQ-FT-ICR Ultra (Thermo-Fisher, Pittsburgh, PA). The mass spectrometer was operated in data-dependent acquisition mode controlled by Xcalbur 2.0.7 software. Peptide mass spectra were acquired from an m/z range of 350 to 2,000 at high mass resolving power. The top six most abundant multiply charged ions with minimum signal intensity at 800 counts were subjected to collision-induced dissociation (CID) in the linear ion trap. Charge state rejection of +1 ions was employed. Precursor activation was performed with an isolation width of 2 Da and an activation time of 30 ms. The raw data were aligned and converted into a centroided peaklist file by Progenesis LC-MS (Nonlinear Dynamics, Durham, NC) (49). The files were searched using MASCOT 2.2.06 (Matrix Science, London, United Kingdom). The enzyme specificity was set to trypsin with 2 missed cleavages. The mass tolerances for precursor and fragment ions were 12 ppm and 0.6 Da, respectively. Oxidation of methionine was specified in Mascot as a variable modification. The data were searched against the NCBI 20120108 database. A threshold of 5% probability that protein identification was incorrect was implemented. Scaffold_3_00_08 (Proteome Software Inc., Portland, OR) was used to validate tandem mass spectrometry (MS-MS)-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability, as specified by the Peptide Prophet algorithm (50). Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (51).
Western blotting.
Protein samples were separated on a 4 to 12% SDS-polyacrylamide gel and transferred to nitrocellulose. The nitrocellulose was washed twice in Tris-buffered saline (TBS) and incubated overnight in Rockland Near Infra-Red blocking buffer (Rockland Immunochemicals; catalog no. MB-070-003). The primary antibodies used (dilutions in parentheses) include UL28 rabbit polyclonal antibody UL28–glutathione S-transferase (GST) (1:1,000) (52); VP5 rabbit polyclonal antibody NC1 (1:5,000) (53); rabbit monoclonal anti-calmodulin-binding protein epitope tag antibody, clone C16T (1:3,000) (Millipore; catalog no. 05-932); UL15 rabbit polyclonal antibody UL15-GST(1-104) (1:1,000) (54); and UL33 rabbit polyclonal antibody UL33-GST (1:500) (55). The blocked nitrocellulose was reacted with the diluted antibodies for 2 h at room temperature, washed five times in TBS with 0.5% Tween 20, and incubated with IRDye 800-conjugated goat anti-rabbit secondary antibody (Rockland Immunochemicals) diluted 1:15,000 in Rockland Near Infra-Red blocking buffer with 0.1% Tween 20. The blots were washed and scanned using an Odyssey system (Li-Cor, Lincoln, NE). Integrated intensity values were obtained using Odyssey software version 3.0 (Licor, Lincoln, NE).
Southern blotting.
T-175 flasks of Vero, CV28, or C2 cells (3 ×107 cells per flask) were infected with virus at an MOI of 5 PFU per cell. At 18 h postinfection (p.i.), the medium was removed and the cells were washed in 1× phosphate-buffered saline. Cells were harvested by scraping them from the plate and pelleted. The cells were lysed, and total cell DNA or DNase-resistant DNA samples were prepared as previously described (56, 57). The final DNA was digested with BamHI to assess the cleavage of viral DNA. DNA was separated by agarose gel electrophoreses, transferred to a nylon membrane, and hybridized as previously described (56). Southern blots were scanned with a Storm 840 PhosphorImager.
RESULTS
Characterization of NTAP-UL28 virus.
Previous studies have shown that the amino terminus of UL28 is not essential for virus growth and can tolerate the insertion of foreign epitopes (13). Therefore, the TAP tag was inserted at the amino terminus of UL28. The 78-amino-acid TAP tag used for these studies has been previously described (8) and consists of a streptavidin-binding peptide and a calmodulin-binding peptide. The TAP tag was inserted at the N terminus of the UL28 open reading frame through manipulation of an HSV-1 (KOS) genome maintained within a recombinant BAC (Fig. 2).
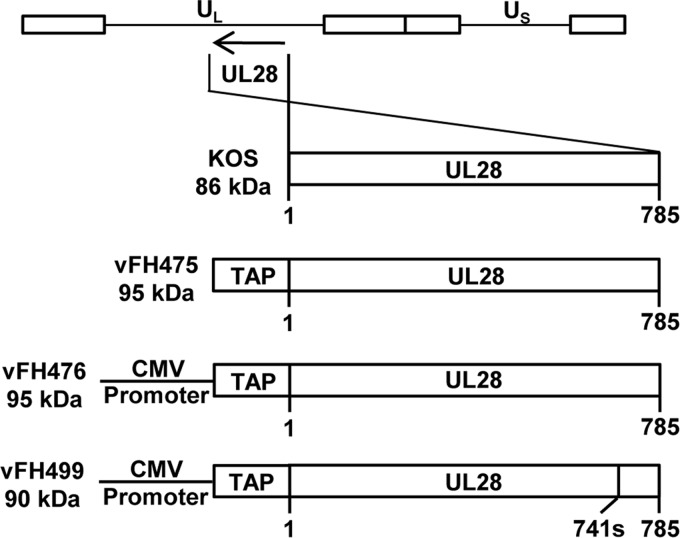
FIG 2.
Recombinant UL28 virus constructs. The HSV-1 genome is shown at the top, with the long and short unique regions represented as UL and US, respectively. The UL28 open reading frame from wild-type KOS or each recombinant virus is expanded below, with virus names and protein sizes indicated on the left of each construct. The amino acid numbers below each construct indicate protein lengths. The vFH499 virus contains a nonsense mutation of amino acid 741.
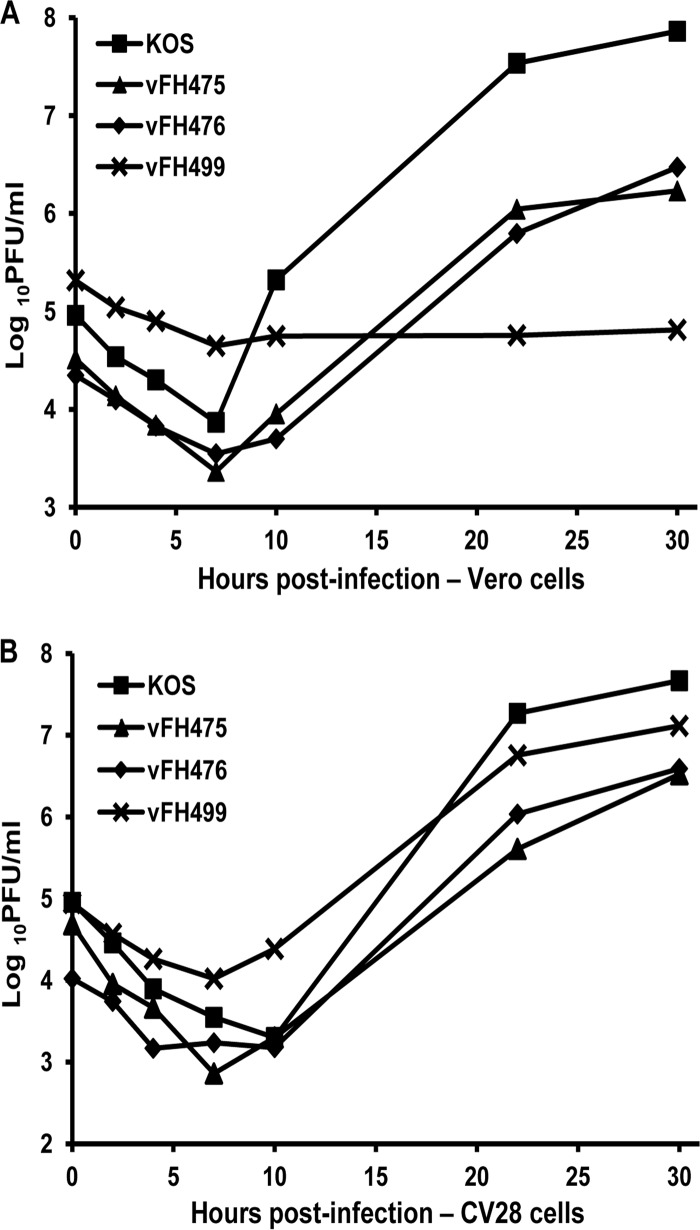
The BAC was transfected into Vero cells, and the recovered virus, vFH475, was plaque purified on Vero cells. Two additional HSV-1 UL28 recombinant viruses, vFH476 and vFH499, were generated in the same manner. vFH476 expresses an NTAP-UL28 protein under the transcriptional control of the cytomegalovirus (CMV) immediate-early promoter, and vFH499 expresses a C-terminally truncated NTAP-UL28 protein under CMV promoter control (Fig. 2). The truncation in vFH499 is the result of an insertion of a linker sequence containing an in-frame stop codon after amino acid 741 of UL28. We previously reported that this truncation precludes the interaction of both the UL15 and UL33 proteins with UL28 (13). The recovered viruses were plaque purified on Vero (vFH475 and vFH476) or UL28-complementing CV28 (vFH499) cells, virus stocks were prepared, and their titers were determined. The vFH475 and vFH476 viruses yielded similar titers on Vero and CV28 cells, while the vFH499 virus, which expresses a nonfunctional UL28 protein, formed plaques only on CV28 cells (Table 2). Intracellular replication of each recombinant virus was compared to that of wild-type KOS virus by establishing single-step growth curves in Vero and CV28 cells. Cells were infected with each virus at an MOI of 1; harvested at 0, 2, 4, 7, 10, 22, and 30 h p.i.; and assayed for infectious virus by plaque assay on CV28 cells (Fig. 3). When grown on Vero or CV28 cells, the vFH475 and vFH476 virus titers were reduced (1 to 2 log units) compared to KOS. The vFH499 virus failed to produce virus on Vero cells but grew to nearly wild-type levels on CV28 cells. Taken together, these results demonstrate that the vFH475 and vFH476 viruses grow on noncomplementing cells, but addition of the fusion protein reduces the overall virus yield, while expression from the CMV promoter does not appear to affect virus growth.
TABLE 2.
Virus growth assay
| Virus no. | Virus plating efficiency (PFU/ml) ina: |
||
|---|---|---|---|
| Vero cells | CV28 cells | C2 cells | |
| KOS | 5.9 × 109 | 9.8 × 108 | 6.7 × 109 |
| vFH475 | 3.1 × 109 | 3.8 × 108 | ND |
| vFH476 | 2.8 × 1010 | 2.1 × 109 | ND |
| vFH499 | <1,000 | 2.9 × 109 | ND |
| vFH505 | <1,000 | 3.1 × 108 | ND |
| vFH503 | <1,000 | 1.2 × 1010 | ND |
| vFH504 | <1 × 105 | 2.25 × 108 | ND |
| vFH542 | <1 × 105 | 1.55 × 108 | ND |
| vFH543 | <1,000 | 2.1 × 1010 | ND |
| vFH546 | <1,000 | 3.6 × 109 | ND |
| vFH506 | 5 × 104 | ND | 7 × 108 |
| vFH507 | <1,000 | ND | 1.75 × 109 |
ND, not determined.
FIG 3.
Recombinant NTAP-UL28 virus single-step growth curves. Vero (A) or CV28 (B) cells were infected with the indicated HSV-1 recombinant viruses or with wild-type (KOS) virus at an MOI of 1 at 4°C for 1 h and incubated at 37°C. The cultures were harvested at the indicated times postinfection and freeze-thawed, and the virus yield at each time point was determined by the plaque titer on CV28 cells.
NTAP-UL28 protein expression was examined for vFH475, vFH476, and vFH499 by Western blotting and compared to UL28 protein expression from the wild-type KOS virus. Infected Vero cell lysates were isolated at 18 h p.i. and probed with a UL28 antibody (Fig. 4A). The UL28 protein expressed by KOS resolved in SDS-PAGE with an apparent molecular mass of 86 kDa, and each recombinant virus expressed a UL28 protein with mobility corresponding to the expected molecular mass. NTAP-UL28 proteins expressed by vFH475 and vFH476 exhibited slightly lower mobility than KOS, corresponding to the additional 78 amino acids (approximately 8.6 kDa) of the TAP tag, while the NTAP-UL28 protein expressed from vFH499 was slightly smaller than that from vFH475 and vFH476 due to the truncation of 44 amino acids from the UL28 C terminus.
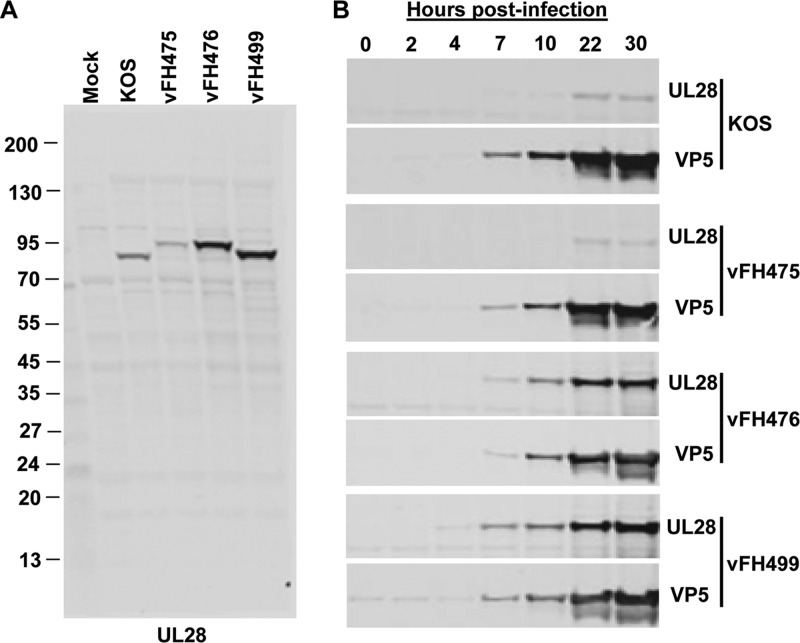
FIG 4.
Recombinant NTAP-UL28 fusion protein expression. (A) Vero cells were infected with wild-type (KOS) virus or the indicated HSV-1 recombinant viruses at an MOI of 10. At 18 h postinfection, cell lysates were harvested and resolved by SDS-PAGE, and Western blots were probed with a UL28 antibody. Protein standards (kDa) are shown on the left of the blot. (B) Vero cells were infected with wild-type (KOS) virus or the indicated HSV-1 recombinant viruses at an MOI of 1 at 4°C for 1 h and incubated at 37°C. Infected-cell lysates were harvested at the indicated times postinfection and resolved by SDS-PAGE, and Western blots were probed with UL28 and VP5 antibodies. VP5 is the major capsid protein and serves as a loading control between samples.
Total cell extracts from KOS-, vFH475-, vFH476-, and vFH499-infected Vero cells were prepared at 0, 2, 4, 7, 10, 22, and 30 h p.i., and expression of the UL28 gene product was analyzed by Western blotting (Fig. 4B). In KOS-infected Vero cells, the 86-kDa UL28 protein was first detected at 7 to 10 h p.i., and the intensity of this band showed a moderate increase at later times postinfection. The recombinant viruses expressing the 95-kDa (vFH475 and vFH476) and 90-kDa (vFH499) NTAP-UL28 fusion proteins showed a pattern of expression similar to that of the KOS UL28 protein. However, expression of the NTAP-UL28 gene from the CMV promoter (vFH476 and vFH499) resulted in increased levels of the UL28 fusion protein compared to KOS and vFH475. The blots were stripped and probed for the major capsid protein VP5, demonstrating that similar amounts of cell extracts were loaded. These results demonstrate that placing the NTAP-UL28 fusion under CMV promoter control resulted in an increase in UL28 protein levels without altering the kinetics of expression. Since the goal of these studies was to purify and characterize the HSV-1 terminase complex via affinity purification of the NTAP-UL28 protein, the recombinant viruses expressing NTAP-UL28 under CMV promoter control were utilized for the remainder of these studies.
Tandem-affinity purification of NTAP-UL28 protein complexes.
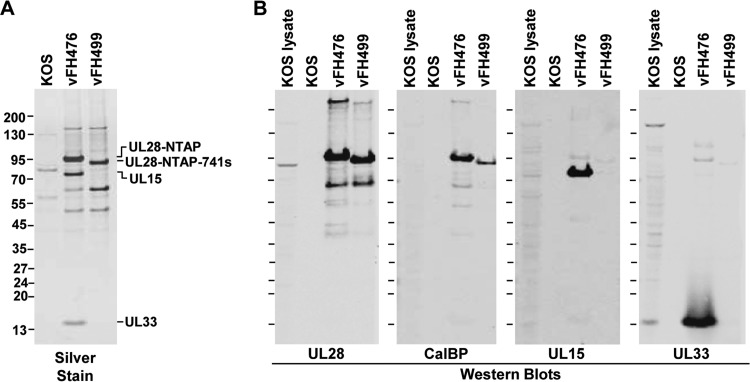
TAP is a dual purification procedure that allows the efficient isolation of protein complexes under native conditions (58). The TAP procedure was utilized to isolate the putative HSV-1 terminase complex of UL15, UL28, and UL33 from infected cells via the NTAP-UL28 fusion protein. Vero cells were infected with NTAP-UL28 viruses or KOS at an MOI of 10, and cell extracts were harvested at 18 h p.i. and subjected to the TAP procedure (Fig. 5). The TAP tag utilized in these studies encodes a calmodulin-binding domain and a streptavidin-binding domain (Stratagene-Interplay). Infected-cell extracts were incubated with streptavidin resin, and interacting complexes were eluted with biotin and then incubated with calmodulin resin in the presence of calcium and eluted with EGTA. Silver stain of the final calmodulin-eluted samples revealed a major band at approximately 95 kDa in the vFH476 lane, corresponding to the full-length NTAP-UL28 fusion protein, and a band at 90 kDa in the vFH499 lane, which corresponds to the truncated NTAP-UL28 protein (Fig. 5A). Bands observed in the KOS lane represent proteins that interact nonspecifically with the TAP resins, while additional bands in the vFH476 and vFH499 lanes represent proteins that copurify with the tagged complex. Western blot analysis identified the putative terminase subunits, UL15 and UL33, in complexes containing the full-length UL28 protein but, as expected, not in complexes containing the UL28-741s truncation mutant (Fig. 5B). Note that both the UL15 and UL33 proteins were also readily observed by silver staining in vFH476-infected cells. Blots for UL28 and the calmodulin-binding peptide confirmed that each major band observed by silver stain contained the NTAP-UL28 protein in both the vFH476 and vFH499 lanes, and additional bands at approximately 63, 50, and 43 kDa were observed that may represent cleavage or degradation products of the UL28 protein. In summary, these results indicate that TAP of the UL28 protein is an effective method for the efficient purification of the putative terminase complex of UL28, UL15, and UL33.
FIG 5.
TAP of NTAP-UL28 fusion proteins. Vero cells were infected with the indicated HSV-1 recombinant viruses or with wild-type (KOS) virus. (A) After TAP, proteins eluted from the calmodulin column were resolved by SDS-PAGE and identified by silver staining. (B) Immunoblots (antibodies are listed below each blot; CalBP, calmodulin-binding peptide antibody) demonstrating the presence of each TAP-tagged protein. Protein standards (kDa) are shown on the left of each gel or blot.
Oligomeric state of TAP-purified terminase components.
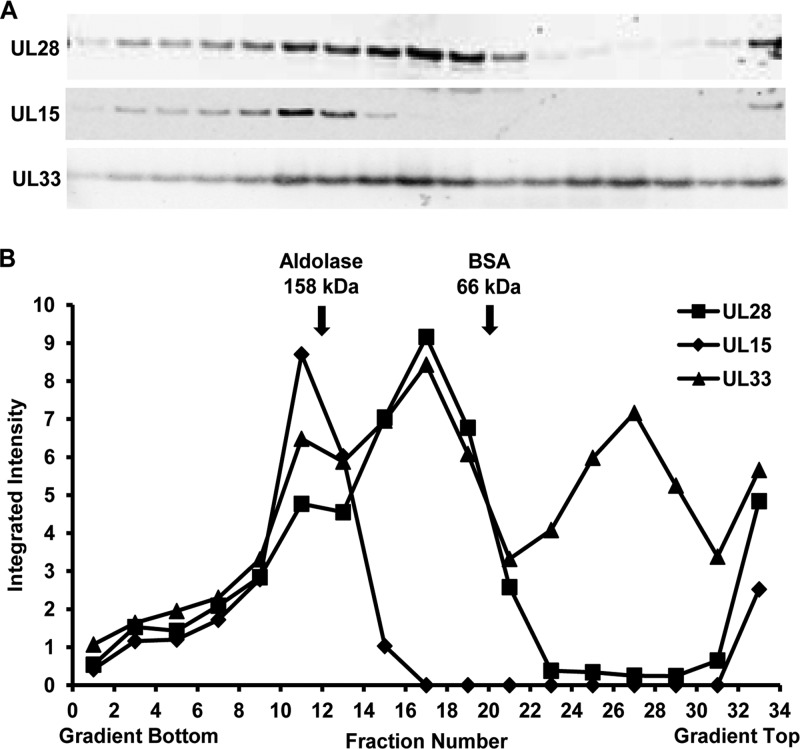
The oligomeric state of TAP-purified terminase components was examined by sucrose density gradient ultracentrifugation (Fig. 6). In order to increase the amount of input protein and minimize sample manipulation, a partial TAP procedure was performed in which UL28 complexes were isolated from vFH476-infected cells using only the initial streptavidin-binding and elution steps. The purified proteins were separated on 2.5 to 20% sucrose gradients, and fractions were collected from the bottom to the top of the gradient. A total of 33 fractions were collected and analyzed by SDS-PAGE and Western blotting for the UL15, UL28, and UL33 proteins (Fig. 6A). Molecular mass control proteins, aldolase and BSA (158 and 66 kDa, respectively), were subjected to identical centrifugation and fractionation conditions, and their relative positions in each gradient were determined by SDS-PAGE and Coomassie staining (data not shown). Immunoblots were analyzed by densitometry, and the values obtained for each fraction were plotted on the same graph, revealing three distinct peaks (Fig. 6B). In fraction 11, a protein complex composed of the UL15, UL28, and UL33 proteins in a 1:1:1 ratio would be expected to possess a molecular mass of approximately 190 kDa, while in fraction 17, a 1:1 complex of UL28 and UL33 would be approximately 109 kDa. The proposed size of each complex is consistent with the relative migration of the molecular mass standards. The observation of the UL33 protein alone in fraction 27 may suggest that the protein possesses a weaker affinity for UL28 and readily dissociates from the complex. Taken together, sucrose gradient centrifugation of UL28 complexes purified by TAP from vFH476-infected cells revealed a complex composed of the UL15, UL28, and UL33 proteins.
FIG 6.
Sedimentation velocity analysis of TAP-purified UL28 complexes. Complexes from vFH476-infected Vero cells were purified through the initial streptavidin column, layered onto a 2.5 to 20% sucrose gradient, and centrifuged at 35,000 rpm at 4°C for 18 h. The gradient was fractionated, and individual fractions were resolved by SDS-PAGE, followed by immunoblotting (antibodies are listed beside each blot) (A) and densitometry (B). (B) Integrated intensity values were obtained for each band using Odyssey software version 3.0 (Licor, Lincoln, NE). Control proteins, aldolase and BSA (GE Healthcare Life Sciences), were subjected to identical centrifugation and fractionation conditions, and their relative positions in each gradient, as indicated, were determined by SDS-PAGE and Coomassie staining (data not shown).
Mass spectrometry analysis of UL28 complexes.
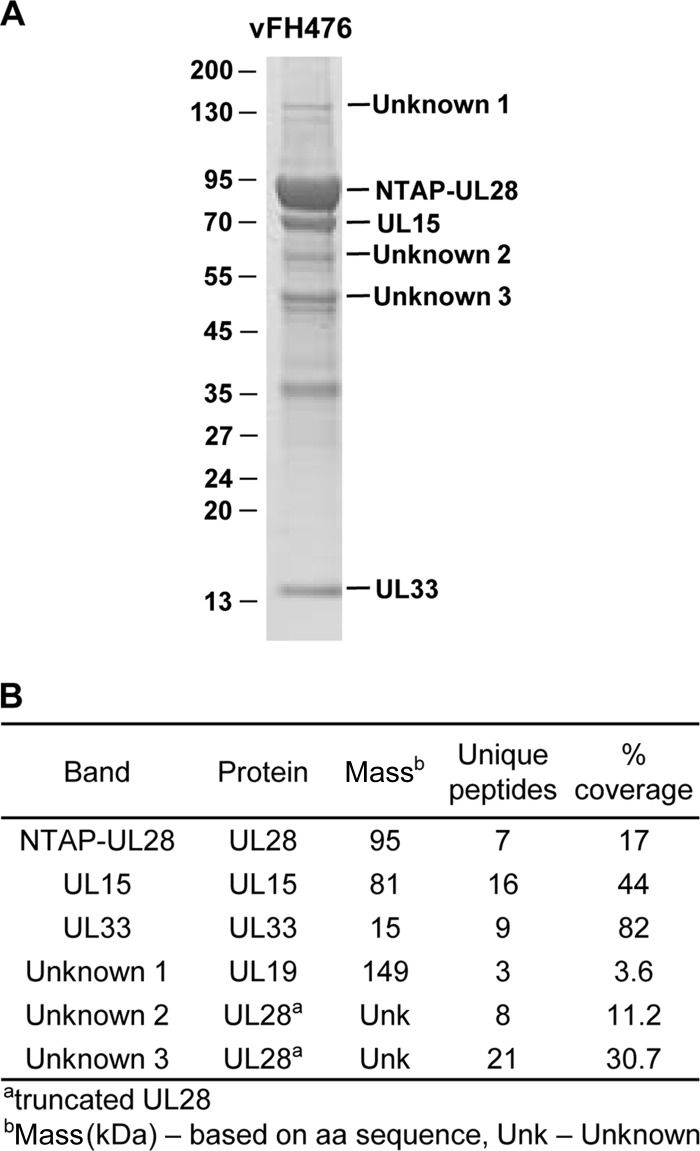
Numerous protein bands were observed that copurify with UL28 complexes by TAP (Fig. 5). One goal of this study was to characterize the proteins that were isolated as components of the terminase complex. In order to identify those proteins, UL28 complexes were purified from vFH476-infected Vero cells. The purified protein complexes were resolved by SDS-PAGE and stained, and bands corresponding to the NTAP-UL28, -UL15, and -UL33 proteins were isolated and confirmed by liquid chromatography (LC) MS-MS (Fig. 7). Also of particular interest were those bands corresponding to sizes of approximately 150, 63, and 50 kDa (unknowns 1 to 3, respectively) which were also observed as predominant bands by silver staining (Fig. 5). LC MS-MS analysis determined unknown 1 to be the VP5 major capsid protein, while unknowns 2 and 3 consisted of peptides from UL28. The VP5 band was confirmed by Western blotting (data not shown). The UL28 peptides were present in TAP preparations from both the vFH476- and vFH499-infected cells, and these peptides are most likely the result of cleavage in the C terminus of UL28, since both of the truncated proteins were found to react with UL28 and calmodulin-binding peptide antibodies (Fig. 5).
FIG 7.
Mass spectrometry analysis of isolated NTAP-UL28 fusion proteins. (A) Vero cells were infected with vFH476, and at 18 h p.i., UL28 complexes were purified by TAP. The isolated complexes were resolved by SDS-PAGE and stained using Imperial stain. Protein standards (kDa) are shown on the left of the gel. (B) The indicated protein bands in panel A were excised from the gel and analyzed by LC MS-MS with molecular masses and amino acid (aa) counts from databases.
Effects of mutations in the UL15 nuclease domain and putative UL28 metal-binding region on terminase formation and activity.
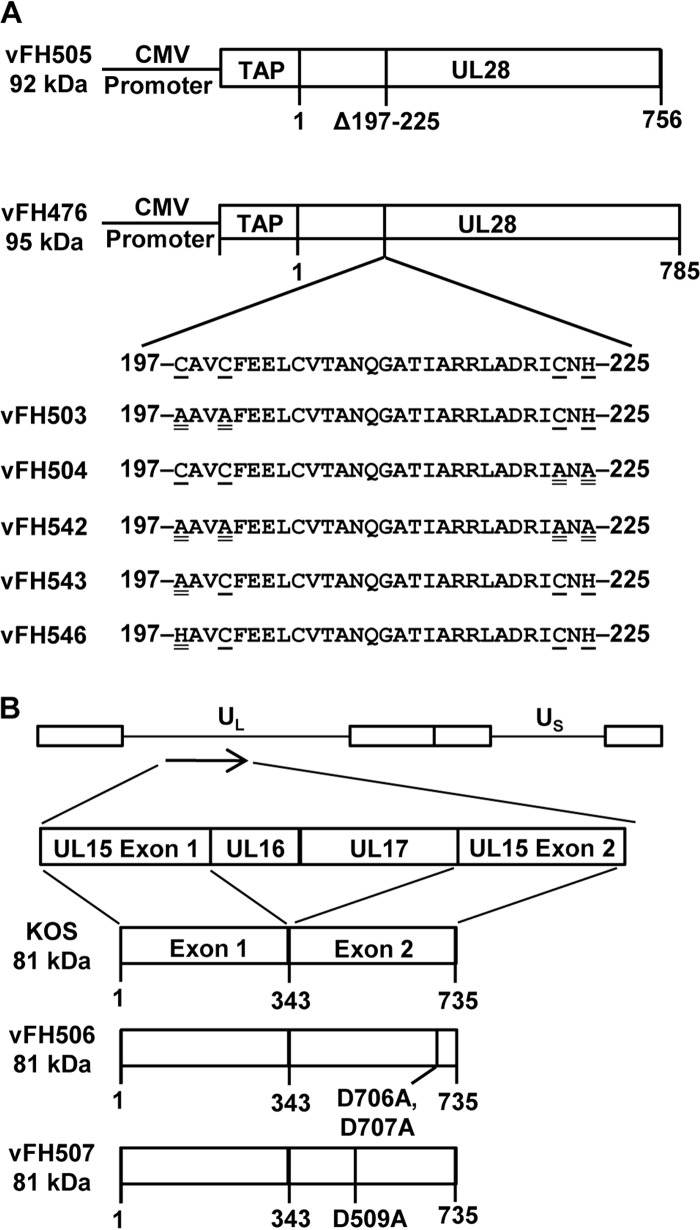
The vFH476 BAC was utilized to generate additional HSV-1 recombinants that express the full-length or C-terminally truncated NTAP-UL28 fusion protein but that also encode mutations within the nuclease domain of UL15 or the proposed metal-binding domain of UL28 (Fig. 8 and Table 1). Sequence analysis of a number of UL28 protein homologs indicated that a highly conserved region from amino acids 197 to 225 resembled a putative zinc finger motif: C197-X2-C200-X22-C223-X-H225 (22, 42). In order to determine the importance of this region, we generated recombinant viruses that contained a deletion or amino acid substitutions within amino acids 197 to 225 of UL28 (Fig. 8A). The UL28 mutations were introduced into the viral genome through genetic manipulation of the vFH476 BAC. The deletion removed the coding sequence for amino acids 197 to 225 of the UL28 ORF to generate the vFH505 mutant virus. Amino acids C197, C200, C223, and H225 are conserved and most likely correspond to the metal-coordinating amino acids within the zinc finger motif predicted for this region of UL28 (22, 42). Mutations either individually (C197A or C197H), in pairs (C197A and C200A or C223A and H225A), or as a quadruple mutant (C197A, C200A, C223A, and H225A) were introduced into this region to generate the vFH543, vFH546, vFH503, vFH504, and vFH542 mutant viruses, respectively (Fig. 8A). Two additional viruses, vFH506 and vFH507, were generated with mutations resulting in amino acid changes within the UL15 conserved nuclease domain of the vFH476 BAC (Fig. 8B). Three acidic residues, D509, E581, and D707, form the conserved triad in the UL15 nuclease active site that coordinate binding of Mg2+, which is essential for the DNA cleavage reaction carried out by the UL15 protein (32, 39). The vFH506 virus expresses a UL15 protein that contains D706A and D707A amino acids changes, and vFH507 contains a D509A amino acid substitution. All of the recombinant viruses containing UL28 or UL15 domain mutations grew only on their respective complementing cell lines, indicating that each virus expressed a nonfunctional UL15 or UL28 protein (Table 2).
FIG 8.
Recombinant UL28 and UL15 virus constructs. (A) UL28 protein expressed from vFH505 or vFH476, with protein sizes indicated on the left of each construct. The amino acid numbers below each construct indicate deletion of the indicated amino acids (Δ) or protein length. The metal-binding domain of vFH476 is expanded to show the amino acid sequence. Recombinant viruses containing site-specific mutations within the metal-binding domain of UL28 are shown below, with virus names on the left. The underlined residues are conserved among the herpesviruses, and the double-underlined residues are mutated amino acids. (B) HSV-1 genome (top), with the long and short unique regions represented (UL and US, respectively). The UL15 protein expressed from wild-type HSV-1 KOS or each recombinant virus is expanded below, with virus names and protein sizes indicated on the left of each construct. The amino acid numbers below each construct indicate mutation of the indicated amino acid or protein length.
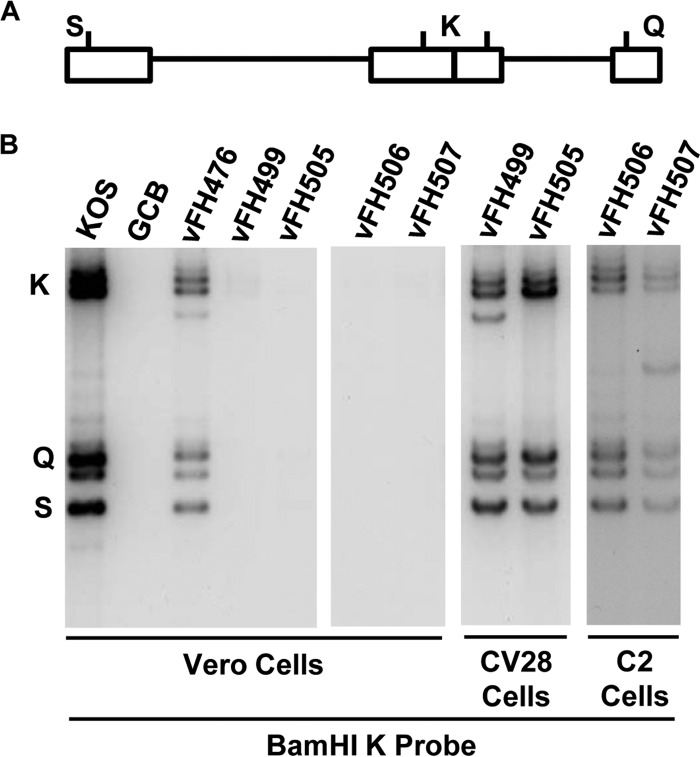
To determine whether the viruses bearing UL15 or UL28 domain mutations were impaired in DNA cleavage and packaging, viral DNA processing was investigated by Southern blotting. During viral DNA replication, concatemeric DNA is cleaved into unit length molecules and packaged into virions. The presence of chromosomal ends can be monitored by Southern blotting of total infected-cell DNA digested with BamHI and probed with the HSV-1 BamHI K fragment. Only encapsidated viral DNA with free chromosomal ends will give rise to the terminal BamHI Q and S fragments, along with the joint-spanning K fragment (Fig. 9A). If the cell extract is DNase treated prior to DNA isolation, only packaged viral DNA will be detected in the Southern blots. The viral K, Q, and S fragments were detected with DNA isolated from Vero cells infected with the wild-type KOS virus and with the NTAP-UL28 virus (vFH476). These fragments were not detected with a UL28-null virus (GCB), the C-terminally truncated UL28 virus (vFH499), or the UL28 (vFH505) and UL15 (vFH506 and vFH507) domain mutants (Fig. 9B). When the infections were performed on UL28 (CV28)- or UL15 (C2)-complementing cell lines, packaged DNA was readily detected with the UL28 or UL15 mutants (Fig. 9B), indicating that the replication defects with these viruses were due to impaired DNA cleavage and packaging.
FIG 9.
Analysis of viral DNA packaging. (A) Locations of the BamHI K joint-spanning fragment and the two end fragments, BamHI-Q and -S, in the HSV-1 genome. (B) Vero, CV28, or C2 cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h p.i., DNase I-protected total infected-cell DNA was isolated, digested with BamHI, and subjected to Southern blot analysis using the BamHI K fragment (32P labeled) as a probe.
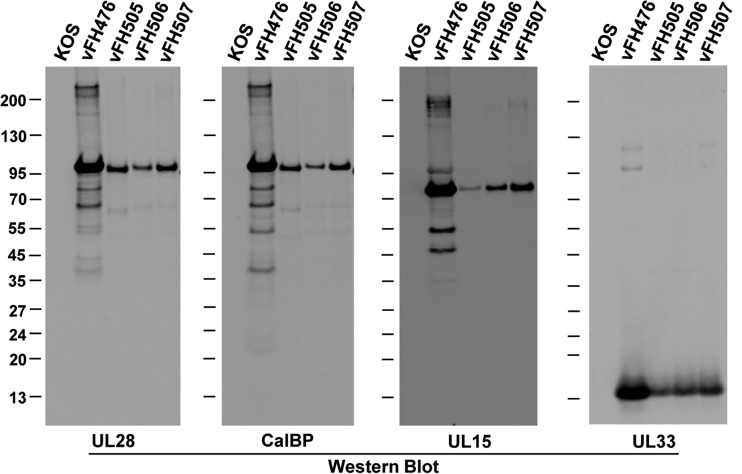
In order to determine if the UL15 or UL28 domain mutations affected terminase complex formation, UL28 protein complexes were purified from infected cells by TAP. Vero cells were infected with either the wild-type KOS virus, the parental NTAP-UL28 virus (vFH476), or the UL28 (vFH505) and UL15 (vFH506, vFH507) domain mutant viruses at an MOI of 10. At 18 h p.i., cell extracts were harvested and subjected to the TAP procedure. Western blotting of the final calmodulin-eluted samples using antibodies against UL28 revealed the full-length 95-kDa NTAP-UL28 protein for vFH476 and a smaller NTAP-UL28 protein of approximately 92 kDa for vFH505 due to the deletion of the putative metal-binding domain (Fig. 10). The UL15 domain mutants (vFH506 and vFH507) were also found to express a full-length NTAP-UL28 protein. Blots were also probed for the UL15 and UL33 proteins to determine if the domain mutations affected terminase complex formation. Both UL15 and UL33 were copurified using the full-length NTAP-UL28 virus (vFH476), as previously observed (Fig. 5), and were also associated with purified complexes containing the deletion of the putative metal-binding domain within the NTAP-UL28 fusion (vFH505). The mutated UL15 proteins encoded by vFH506 and vFH507 were shown to copurify with UL28 complexes, as well as the UL33 protein. No terminase subunits were purified from lysates of KOS-infected cells, confirming the specificity of the TAP procedure. Of note, the levels of each terminase complex subunit purified from cells infected with the UL28 and UL15 mutants (vFH505, vFH506, vFH507) appears to be decreased relative to the levels found with the full-length NTAP-UL28 virus (vFH476). These results demonstrated that the UL15 and UL33 proteins copurify with UL28 in all samples, indicating that the mutations introduced within the nuclease domain of UL15 or the metal-binding domain of UL28 do not preclude terminase complex formation.
FIG 10.
TAP and immunoblot of UL28 complexes purified from UL15 and UL28 domain mutant viruses. Vero cells were infected with the indicated HSV-1 recombinant viruses or with wild-type (KOS) virus. After TAP, proteins eluted from the calmodulin column were resolved by SDS-PAGE and identified by Western blotting (antibodies are listed below each blot; CalBP, calmodulin-binding peptide antibody). Protein standards (kDa) are shown on the left of each blot.
Finally, Western blot analysis of UL28 complexes purified by TAP revealed the purification of several truncated forms of UL28 (Fig. 5 and 7), and mass spectrometry confirmed that fragments migrating in SDS-PAGE at approximately 63 and 50 kDa were composed of UL28 peptides. In the current analysis, blotting for UL28 or the calmodulin-binding peptide demonstrated the presence of the same truncated forms of the NTAP-UL28 protein with the viruses expressing mutated UL15 proteins (vFH506 and vFH507) (Fig. 10). In contrast, the vFH505 virus contains a faint, smaller, truncated NTAP-UL28 protein of approximately 60 kDa due to the 29-amino-acid deletion of the putative metal-binding domain of UL28. The detection of these proteins in each sample using the calmodulin-binding peptide antibody suggests that the 63- and 60-kDa NTAP-UL28 proteins are the result of the loss of approximately 30 to 35 kDa (∼300 amino acids) from the C terminus of UL28.
DISCUSSION
A productive HSV-1 infection requires the UL28, UL15, and UL33 gene products for cleavage and packaging of replicated, concatemeric viral DNA into capsids. These proteins comprise a viral terminase complex similar in function to those utilized by dsDNA bacteriophages (1, 2). Terminases are powerful molecular motors that are required to drive DNA into the capsid against the strong repellent molecular charges in tightly packaged viral DNA (59). Biochemical analysis of the HSV-1 terminase has proven difficult due to the inability to purify the complex subunits, particularly UL15 and UL28. In this study, we purified the terminase complex from cells infected with HSV-1 recombinants expressing the UL28 protein fused to an N-terminal TAP tag. Additionally, mutations or deletions within the nuclease domain of UL15 or the metal-binding domain of UL28 were introduced into the genome of the NTAP-UL28 fusion virus. Biochemical and genetic analysis of isolated UL28 complexes was performed in order to determine terminase complex composition and to examine the effects of domain mutations on complex formation and function during HSV-1 infection.
Addition of the TAP tag to the N terminus of UL28 (vFH475) did not alter UL28 protein expression kinetics but reduced virus growth by approximately 2 log units compared to wild-type KOS virus. Western blotting of vFH475-infected cell lysates revealed relatively low-level UL28 protein expression similar to that observed with wild-type KOS virus. Therefore, a second recombinant virus, vFH476, was constructed that expresses the NTAP-UL28 fusion protein under the transcriptional control of the human CMV immediate-early gene promoter in order to increase UL28 protein expression levels. Compared to vFH475 or wild-type KOS, vFH476 expressed the NTAP-UL28 fusion protein with similar kinetics but in increased amounts more suitable for downstream biochemical and proteomic analysis. Single-step growth curves of vFH476 revealed a similar decrease in the virus titer (approximately 2 log units), as was found with the vFH475 virus, indicating that CMV promoter overexpression did not affect virus growth. It is not known why the addition of the TAP tag reduced growth of vFH475 and vFH476 relative to KOS virus growth, although it is possible that the addition of 78 amino acids to the N terminus of UL28 (the TAP tag) resulted in a steric hindrance to complex assembly or function (i.e., the terminase interaction with replicated viral DNA).
TAP of UL28 complexes from vFH476- or vFH499-infected cells followed by silver staining and Western blotting confirmed that the UL33 and UL15 proteins were copurified in complexes containing the full-length UL28 protein (vFH476) but, as expected, not from complexes containing the UL28-741s truncation mutant (vFH499). Mass spectrometry analysis of specific SDS-PAGE bands confirmed the presence of the UL15, UL28, and UL33 proteins in purified UL28 complexes from vFH476-infected Vero cells, providing further evidence that these proteins are components of the HSV-1 terminase complex.
Mass spectrometry was also utilized to identify three unknown proteins that copurified with UL28 complexes from vFH476-infected Vero cells. Two unknown proteins migrating at approximately 63 and 50 kDa were revealed to consist of UL28 peptides. These truncated UL28 bands were also detected by Western blotting of TAP-purified UL28 complexes from each of the NTAP-UL28 fusion viruses examined by immunoblotting (vFH476, vFH499, and vFH505 to vFH507), and the detection of these truncated proteins using either the UL28 or calmodulin-binding peptide antibodies suggests the loss of approximately 30 to 35 kDa (∼300 amino acids) from the C terminus of UL28. However, it is unclear whether these bands are the result of protein degradation during purification or are specific cleavage events. Identification of the VP5 major capsid protein as the third unknown protein is not surprising. The terminase associates with capsids during the DNA encapsidation process (1), and it is possible that partial or intact viral capsids are purified during TAP, which suggests that VP5 interacts only indirectly with the viral terminase. The results of the mass spectrometry analysis further confirm that only the viral UL15, UL28, and UL33 proteins appear to compose the terminase complex, maintaining the potential importance of the terminase as a target for novel antiviral compounds. However, this analysis included only proteins within specific gel slices, and a more comprehensive analysis of the entire purified sample may reveal additional terminase complex components.
Sucrose density gradient ultracentrifugation of purified complexes revealed significant amounts of the UL15, UL28, and UL33 proteins comigrating in a size range greater than 158 kDa. This size range is consistent with a 1:1:1 complex of UL15, UL28, and UL33 that possesses an approximate molecular mass of 190 kDa. These data are in agreement with a previous study demonstrating that interacting UL15 and UL28 subunits purified from HSV-1-infected cells comigrate through a sucrose gradient to a position consistent with a 1:1 heterodimer of both proteins (60). It is important to note that at the time of this study the UL33 protein was not yet implicated as an HSV-1 terminase subunit. The observation of what may be UL28-UL33 heterodimers is consistent with previous studies suggesting that the UL28-UL33 interaction enhances the interaction of UL15 with UL28 and increases the number of properly formed complexes (13, 15, 16). However, it must be noted that the presence or absence of UL15-UL28 heterodimers is difficult to determine, as these complexes would migrate to a gradient position similar to that of the UL15-UL28-UL33 heterotrimer, due to the small size of UL33 (∼14kDa). The observation of a gradient fraction containing monomeric UL33 may suggest that the protein possesses lower affinity and can readily dissociate from the complex. However, UL33 dissociation may also be the result of in vitro conditions, and it might be interesting to alter the stringency of either the TAP or gradient centrifugation method to further examine the effect on terminase complex stoichiometry.
Several amino acid domains implicated in mediating terminase complex formation and function(s) have been identified within the UL15 and UL28 subunits, and these domains are conserved across the herpesviruses (22, 32, 36, 61). In this study, mutations or deletions within the nuclease domain of UL15 or the metal-binding domain of UL28 were introduced into the genome of the NTAP-UL28 fusion virus (vFH476) in order to determine the importance of these regions in terminase complex formation and function during HSV-1 infection. Viruses encoding mutations within UL15 amino acids 509 or 706 and 707 (vFH507 and vFH506, respectively) replicated only on UL15-complementing cell lines, and both viruses were deficient in the cleavage and packaging of viral DNA, but not in assembly of the terminase complex. These results are consistent with a recent study that demonstrated that mutation of these conserved residues within the UL15 homolog in HCMV, UL89, disrupted the nuclease activity displayed by a purified C-terminal fragment of the UL89 protein (32). The crystal structure of C-terminal fragments for the UL15 and UL89 proteins revealed that these proteins use an RNase H-like, metal ion-mediated catalysis mechanism for cleavage of viral concatemeric DNA (32, 39).
In the current analysis, it seems likely that the loss of DNA cleavage and packaging activity observed in the UL15 mutant viruses (vFH506 and vFH507) is due to the disruption of metal ion coordination, and further experiments will include examining the nuclease activities of these mutants.
The UL28 protein is implicated as the DNA-binding subunit of the HSV-1 terminase and contains a domain resembling a putative zinc finger motif (C197-X2-C200-X22-C223-X-H225) (22, 42). This domain is critical for proper terminase function, as a recombinant virus encoding a deletion of the putative metal-binding domain (vFH505) replicated to high titers only on a UL28-complementing cell line and was deficient in DNA cleavage and packaging. UL28 amino acids C197, C200, C223, and H225 are conserved and most likely correspond to the metal-coordinating amino acids within the zinc finger motif predicted for this region of UL28. Point mutation of these amino acids ether singly, in pairs, or as a quadruple mutant (vFH503, vFH504, vFH542, vFH543, and vFH546) resulted in viruses that replicated to high titers only on a UL28-complementing cell line. This result is particularly surprising for vFH546, which encodes a C197H substitution, as the histidine substitution is not expected to preclude metal ion coordination. This may suggest that this domain plays a structural role, where disulfide bond formation between cysteine residues is critical to terminase assembly and/or function. The terminase complexes of several dsDNA bacteriophages consist of interacting small and large subunits, and the functional terminase holoenzyme is a multimeric complex of these proteins (62); therefore, UL28 may form intersubunit disulfide bonds to assemble and/or stabilize higher-order terminase structures. Intersubunit disulfide bonds have also been shown to be required for the stability and/or formation of UL6 portal rings (63). It is also possible that intersubunit disulfide bond formation may be occurring between UL28 and another capsid-associated protein, such as the UL6 portal, which would account for the loss of DNA cleavage and packaging observed with the UL28 metal-binding domain deletion mutant (vFH505). However, if this is the case, it does not appear that disulfide bond formation is essential for the interaction of UL15 and/or UL33 with UL28, as both subunits were shown to associate with UL28 complexes purified from the domain deletion virus (vFH505).
Several important questions remain concerning the biochemical activity and localization of the purified terminase complexes. The TAP done in the present study was performed starting with total infected-cell lysates, since this resulted in higher yields of the isolated complex. We performed the TAP starting with isolated nuclei and found the three-protein complex was present in both the cytoplasm and nuclei of cells infected with the full-length NTAP-UL28 fusion protein (data not shown). In contrast, the C-terminally truncated NTAP-UL28 fusion protein (vFH499) was found exclusively in the cytoplasm. These data are consistent with previous studies showing that the UL15-UL28-UL33 complex forms in the cytoplasm and that the nuclear localization signal in UL15 is used to import the complex into the nucleus (27).
Preliminary studies to test for biochemical activities associated with the isolated terminase complexes have demonstrated that the full-length UL28 complexes (vFH476) showed robust nuclease activity in the presence of Mn2+ or Mg2+, but not with Ca2+ ions, while no activity was found with the KOS control sample. Surprisingly, however, complexes isolated using the UL28 C-terminal truncation mutant (vFH499) also showed nuclease activity. Our present studies are focused on determining if UL28 alone possesses nuclease activity or if the activity is due to a cell nuclease that copurifies with the complex.
In conclusion, these results have demonstrated that TAP of an NTAP-UL28 fusion protein can be utilized to isolate the endogenous terminase complex from HSV-1-infected cells. This method should allow further biochemical analysis of purified complexes and demonstration of the cleavage and packaging functions attributed to the viral terminase. These results have also demonstrated the ability to purify terminase complexes from cells infected with recombinant viruses that encode deletions or mutations of specific conserved amino acids within the terminase subunits, allowing further elucidation of those domains that are essential for terminase complex assembly and function in the context of viral infection. The ability to purify endogenous terminase complexes is novel for the field and represents a critical step toward establishing an in vitro HSV-1 cleavage and packaging system.
ACKNOWLEDGMENTS
Mass spectrometry was performed at the NIH/NCRR Mass Spectrometry Resource at Washington University in St. Louis, supported by Public Health Service grant GM103422 from the National Institutes of Health.
We thank Benjamin Treat and Christopher Weiss for assisting in the isolation of several of the mutant viruses.
This work was supported by Public Health Service grants AI060836 and A1089803 from the National Institutes of Health to F. L. Homa and T32 training grant support (AI049820) for J. Heming.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Conway JF, Homa FL. 2011. Nucleocapsid structure, Assembly and DNA packaging of herpes simplex virus, p 175–193 In Weller SK. (ed), Alphaherpesviruses: molecular virology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 2.Catalano CE. 2005. Viral genome packaging machines: genetics, structure, and mechanism. Kluwer Academic Publishers, New York, NY [Google Scholar]
- 3.Cardone G, Winkler DC, Trus BL, Cheng N, Heuser JE, Newcomb WW, Brown JC, Steven AC. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426–434. 10.1016/j.virol.2006.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang JT, Schmid MF, Rixon FJ, Chiu W. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 81:2065–2068. 10.1128/JVI.02053-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newcomb WW, Juhas RM, Thomsen DR, Homa FL, Burch AD, Weller SK, Brown JC. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923–10932. 10.1128/JVI.75.22.10923-10932.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trus BL, Cheng N, Newcomb WW, Homa FL, Brown JC, Steven AC. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668–12671. 10.1128/JVI.78.22.12668-12671.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. 2011. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J. Virol. 85:4875–4887. 10.1128/JVI.00242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toropova K, Huffman JB, Homa FL, Conway JF. 2011. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J. Virol. 85:7513–7522. 10.1128/JVI.00837-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trus BL, Newcomb WW, Cheng N, Cardone G, Marekov L, Homa FL, Brown JC, Steven AC. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479–489. 10.1016/j.molcel.2007.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang YE, Poon AP, Roizman B. 1996. Properties of the protein encoded by the UL32 open reading frame of herpes simplex virus 1. J. Virol. 70:3938–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamberti C, Weller SK. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beilstein F, Higgs MR, Stow ND. 2009. Mutational analysis of the herpes simplex virus type 1 DNA packaging protein UL33. J. Virol. 83:8938–8945. 10.1128/JVI.0104-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobson JG, Yang K, Baines JD, Homa FL. 2006. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J. Virol. 80:12312–12323. 10.1128/JVI.01766-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tengelsen LA, Pederson NE, Shaver PR, Wathen MW, Homa FL. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 67:3470–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang K, Baines JD. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 80:5733–5739. 10.1128/JVI.00125-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang K, Poon AP, Roizman B, Baines JD. 2008. Temperature-sensitive mutations in the putative herpes simplex virus type 1 terminase subunits pUL15 and pUL33 preclude viral DNA cleavage/packaging and interaction with pUL28 at the nonpermissive temperature. J. Virol. 82:487–494. 10.1128/JVI.01875-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang K, Wills E, Baines JD. 2009. The putative leucine zipper of the UL6-encoded portal protein of herpes simplex virus 1 is necessary for interaction with pUL15 and pUL28 and their association with capsids. J. Virol. 83:4557–4564. 10.1128/JVI.00026-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beard PM, Duffy C, Baines JD. 2004. Quantification of the DNA cleavage and packaging proteins U(L)15 and U(L)28 in A and B capsids of herpes simplex virus type 1. J. Virol. 78:1367–1374. 10.1128/JVI.78.3.1367-1374.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687–698. 10.1128/JVI.75.2.687-698.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taus NS, Baines JD. 1998. Herpes simplex virus 1 DNA cleavage/packaging: the UL28 gene encodes a minor component of B capsids. Virology 252:443–449. 10.1006/viro.1998.9475 [DOI] [PubMed] [Google Scholar]
- 21.Yu D, Weller SK. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428–7439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krosky PM, Underwood MR, Turk SR, Feng KW, Jain RK, Ptak RG, Westerman AC, Biron KK, Townsend LB, Drach JC. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721–4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Underwood MR, Harvey RJ, Stanat SC, Hemphill ML, Miller T, Drach JC, Townsend LB, Biron KK. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baines JD, Poon AP, Rovnak J, Roizman B. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118–8124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poon AP, Roizman B. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Przech AJ, Yu D, Weller SK. 2003. Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging. J. Virol. 77:9613–9621. 10.1128/JVI.77.17.9613-9621.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang K, Homa F, Baines JD. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 81:6419–6433. 10.1128/JVI.00047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang K, Wills EG, Baines JD. 2011. A mutation in UL15 of herpes simplex virus 1 that reduces packaging of cleaved genomes. J. Virol. 85:11972–11980. 10.1128/JVI.00857-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu D, Sheaffer AK, Tenney DJ, Weller SK. 1997. Characterization of ICP6::lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J. Virol. 71:2656–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu D, Weller SK. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32–44. 10.1006/viro.1998.9041 [DOI] [PubMed] [Google Scholar]
- 31.Baines JD, Cunningham C, Nalwanga D, Davison A. 1997. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J. Virol. 71:2666–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nadal M, Mas PJ, Blanco AG, Arnan C, Sola M, Hart DJ, Coll M. 2010. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc. Natl. Acad. Sci. U. S. A. 107:16078–16083. 10.1073/pnas.1007144107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Addison C, Rixon FJ, Preston VG. 1990. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J. Gen. Virol. 71:2377–2384. 10.1099/0022-1317-71-10-2377 [DOI] [PubMed] [Google Scholar]
- 34.Cavalcoli JD, Baghian A, Homa FL, Kousoulas KG. 1993. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant (KOS) tsZ47: evidence for allelic complementation in the UL28 gene. Virology 197:23–34. 10.1006/viro.1993.1563 [DOI] [PubMed] [Google Scholar]
- 35.Costa RH, Draper KG, Kelly TJ, Wagner EK. 1985. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J. Virol. 54:317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davison AJ. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9–14. 10.1016/0042-6822(92)90056-U [DOI] [PubMed] [Google Scholar]
- 37.Draper B, Rao VB. 2007. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J. Mol. Biol. 369:79–94. 10.1016/j.jmb.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 38.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SelvarajanSigamani S, Zhao H, Kamau YN, Baines JD, Tang L. 2013. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J. Virol. 87:7140–7148. 10.1128/JVI.00311-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adelman K, Salmon B, Baines JD. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. U. S. A. 98:3086–3091. 10.1073/pnas.061555698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogner E, Radsak K, Stinski MF. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Champier G, Couvreux A, Hantz S, Rametti A, Mazeron MC, Bouaziz S, Denis F, Alain S. 2008. Putative functional domains of human cytomegalovirus pUL56 involved in dimerization and benzimidazole D-ribonucleoside activity. Antivir. Ther. 13:643–654 [PubMed] [Google Scholar]
- 43.Buerger I, Reefschlaeger J, Bender W, Eckenberg P, Popp A, Weber O, Graeper S, Klenk HD, Ruebsamen-Waigmann H, Hallenberger S. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077–9086. 10.1128/JVI.75.19.9077-9086.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldner T, Hewlett G, Ettischer N, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. 2011. The novel anticytomegalovirus compound AIC246 (Letermovir) inhibits human cytomegalovirus replication through a specific antiviral mechanism that involves the viral terminase. J. Virol. 85:10884–10893. 10.1128/JVI.05265-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gierasch WW, Zimmerman DL, Ward SL, Vanheyningen TK, Romine JD, Leib DA. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197–206. 10.1016/j.jviromet.2006.03.014 [DOI] [PubMed] [Google Scholar]
- 46.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 47.Cockrell SK, Sanchez ME, Erazo A, Homa FL. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47–57. 10.1128/JVI.01889-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newcomb WW, Jones LM, Dee A, Chaudhry F, Brown JC. 2012. Role of a reducing environment in disassembly of the herpesvirus tegument. Virology 431:71–79. 10.1016/j.virol.2012.05.017 [DOI] [PubMed] [Google Scholar]
- 49.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. 2009. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 8:3752–3759. 10.1021/pr900080y [DOI] [PubMed] [Google Scholar]
- 50.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 51.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 52.Beard PM, Taus NS, Baines JD. 2002. DNA cleavage and packaging proteins encoded by genes U(L)28, U(L)15, and U(L)33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785–4791. 10.1128/JVI.76.10.4785-4791.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Costa RH, Cohen G, Eisenberg R, Long D, Wagner E. 1984. Direct demonstration that the abundant 6-kilobase herpes simplex virus type 1 mRNA mapping between 0.23 and 0.27 map units encodes the major capsid protein VP5. J. Virol. 49:287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salmon B, Nalwanga D, Fan Y, Baines JD. 1999. Proteolytic cleavage of the amino terminus of the U(L)15 gene product of herpes simplex virus type 1 is coupled with maturation of viral DNA into unit-length genomes. J. Virol. 73:8338–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds AE, Fan Y, Baines JD. 2000. Characterization of the U(L)33 gene product of herpes simplex virus 1. Virology 266:310–318. 10.1006/viro.1999.0090 [DOI] [PubMed] [Google Scholar]
- 56.Homa FL, Otal TM, Glorioso JC, Levine M. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stow ND. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755–10765. 10.1128/JVI.75.22.10755-10765.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030–1032. 10.1038/13732 [DOI] [PubMed] [Google Scholar]
- 59.Bauer DW, Huffman JB, Homa FL, Evilevitch A. 2013. Herpes virus genome, the pressure is on. J. Am. Chem. Soc. 135:11216–11221. 10.1021/ja404008r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koslowski KM, Shaver PR, Casey JT, II, Wilson T, Yamanaka G, Sheaffer AK, Tenney DJ, Pederson NE. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davison AJ, Dargan DJ, Stow ND. 2002. Fundamental and accessory systems in herpesviruses. Antivir. Res. 56:1–11. 10.1016/S0166-3542(02)00107-9 [DOI] [PubMed] [Google Scholar]
- 62.Feiss M, Rao VB. 2012. The bacteriophage DNA packaging machine. Adv. Exp. Med. Biol. 726:489–509. 10.1007/978-1-4614-0980-9_22 [DOI] [PubMed] [Google Scholar]
- 63.Albright BS, Nellissery J, Szczepaniak R, Weller SK. 2011. Disulfide bond formation in the herpes simplex virus 1 UL6 protein is required for portal ring formation and genome encapsidation. J. Virol. 85:8616–8624. 10.1128/JVI.00123-11 [DOI] [PMC free article] [PubMed] [Google Scholar]