Abstract
The order Mononegavirales comprises a large number of nonsegmented negative-strand RNA viruses (NNSVs). How the genome polarity is determined is a central issue in RNA virus biology. Using a prototypic species, vesicular stomatitis virus (VSV), it has been established that the negative polarity of the viral genome is defined solely by different strengths of the cis-acting replication promoters located at the 3′ ends of the genome and antigenome, resulting in the predominance of the genome over the antigenome. This VSV paradigm has long been applied for the Mononegavirales in general without concrete proof. We now found that another prototypic species, Sendai virus (SeV), undergoes a marked shift from the early antigenome-dominant to the late genome-dominant phase during the course of infection. This shift appeared to be governed primarily by the expression of the accessory C protein, because no such shift occurred in a recombinant SeV with the C gene deleted, and antigenomes were dominant throughout infection, generating antigenome-dominant and noninfectious progeny virions. Therefore, we proposed for the first time a trans-regulatory mechanism, the SeV paradigm, to dictate the genome polarity of an NNSV. A series of promoter-swapped SeV recombinants suggested the importance of the primary as well as secondary structures of the promoters in this trans-regulation.
INTRODUCTION
Single-strand RNA viruses have been roughly divided into three main classes, positive-strand, negative-strand, and ambisense viruses, according to differences in the polarity of the genome. The mechanisms determining a specific genome polarity represent one of the central questions in the biology of RNA viruses.
The order Mononegavirales, or nonsegmented negative-strand RNA viruses (NNSVs), comprise the Rhabdoviridae, Paramyxoviridae, Filoviridae, and Bornaviridae families and are characterized by the huge diversity of similar viruses (1). This similarity includes the presence of the 3′-terminal leader (Le) and 5′-terminal trailer (Tr) sequences (1). Several or more protein-encoding genes are placed in tandem between the Le and Tr, each of which is expressed principally as a monocistronic transcript using individual start and stop signals. The Le serves as the genome promoter (GP) to direct the synthesis of positive-sense antigenome RNA as well as mRNAs. The antigenome synthesized in turn serves as the template for negative-sense genome RNA synthesis. Here, the sequence complementary to the Tr is positioned at the 3′ terminus of the template and functions as the antigenome promoter (AGP).
The characteristics of NNSV have been investigated with vesicular stomatitis virus (VSV) and rabies virus (RV) in the Rhabdoviridae (2, 3). The GP and AGP of VSV differ markedly at positions 19 to 29 and 34 to 36 (4–6). Because of these differences, the AGP is much stronger at directing genome replication than the GP for antigenome synthesis; as a result, more genomes than antigenomes are produced throughout single-cycle replication in cells (2, 3). The determination of asymmetry by cis-acting elements was demonstrated by creating an RV recombinant using reverse genetics that possessed the Tr sequence at the 3′ ends of both the genome and antigenome; asymmetry was abolished in cells infected with this RV, which led to the synthesis of genomes and antigenomes in approximately equal amounts (7). The cis-acting promoter-dependent asymmetry concept, which we referred to as the VSV paradigm of NNSV replication, appears to have been applied to other NNSVs without concrete results (1, 3).
Sendai virus (SeV; also named murine parainfluenza virus type 1 or hemagglutinating virus of Japan [HVJ]) is a prototypic member of Paramyxoviridae (8, 9). It is a very valuable model paramyxovirus because studies on SeV have uncovered many paramyxovirus traits (8). SeV was shown to express two different accessory proteins from the phospho (P) protein gene, with one being a nested set of four C proteins, C′ (215 amino acids [aa]), C (204 aa), Y1 (181 aa), and Y2 (175 aa), that are translated from start codons in the +1 open reading frame (ORF) relative to the overlapping P ORF (8). These C ORFs initiated at a non-AUG codon, ACG 81, and AUGs 114, 183, and 201 and terminated at 725 (10–12). Among the four C proteins, C is the major species expressed in infected cells at a molar ratio severalfold higher than those of the other three (11, 13). Kurotani et al. succeeded in generating an SeV recombinant 4C(−) virus by reverse genetics, which expressed none of the four C proteins and demonstrated that the C proteins fell into the category of a nonessential, accessory gene product (13). However, C protein has been revealed to exert multiple important functions in viral pathogenesis as well as replication, such as evasion from host innate immunity, inhibition of virus-induced apoptosis, and promotion of efficient viral assembly and budding (14–25). C protein has been suggested also to play a role in viral RNA (vRNA) synthesis. Recently, we noted that 4C(−) virions contained more antigenomes than genomes, a situation opposite to wild-type (WT) SeV, which was shown to predominantly possess genomes (26). Together with the earlier studies described below, this led to the hypothesis that the C protein(s) could be a main player in determining SeV genome polarity. Earlier studies reported that elimination of C protein expression increased mRNA synthesis in infected cells and supplementation with C protein from cDNA suppressed this increase (27, 28) and that trans-supplied C protein inhibited viral replication (29). Detailed studies using minireplicons mimicking SeV defective interfering (DI) genomes have nicely shown that C protein inhibited vRNA synthesis specifically from the Le promoter (30). These inhibitory effects have been suggested to be exerted through physical interaction between C and L proteins, since the strength of C-L interaction correlated with the level of defects in vRNA synthesis in experiments using a series of C mutants possessing substitutions of highly conserved, charged amino acids (31, 32).
Here, we extended this line of research. Our present results showed that the amount of the antigenome was larger than that of the genome throughout the single-cycle replication of the 4C(−) virus, which indicated that the GP was intrinsically stronger than the AGP (i.e., in the absence of C proteins), an opposite situation to VSV. The greater predominance of antigenome synthesis occurred early in infection with WT SeV, which was switched to a genome-dominant phase as the synthesis of C and other viral proteins advanced late in infection. We concluded that the C protein(s) was an essential player in determining SeV negative polarity, providing for the first time the requirement of a trans-acting viral element (the SeV paradigm) for an NNSV to be an NNSV. It is intriguing that just the accessory gene products were shown to play such an important role in the NNSV life cycle.
MATERIALS AND METHODS
Cells, viruses, antibodies, and plasmids.
LLC-MK2, BHK-21, and 293T cells were maintained as reported previously (18). VSVs (New Jersey serotype) were propagated in BHK-21 cells. The SeV recombinants, C′/C(−) and 4C(−), lacking the expression of C′ and C, and all four C proteins (13) were provided by A. Kato (National Institute of Infectious Disease, Japan). A series of recombinants, GP24, GP30, GP33, GP42, and GP48, possessing replacements of the 3′ leader region of the viral genome, with the corresponding region of the 5′ trailer to various degrees, were provided by D. Garcin (University of Geneva, Switzerland) (33, 34). The other recombinant Cm2′ possessing the amino acid substitutions K77R and D80A (amino acid positions were assigned on the basis of those of the 204-amino-acid-length C protein, as has been typically done) within the C protein were reported previously (24). All of the recombinants as well as WT (strain Z) were recovered from cDNAs using a reverse-genetics technique in LLC-MK2 cells and were propagated in embryonated chicken eggs, as described previously (35). SeV strains Cantell, Fushimi, Hamamatsu, and Nagoya were used in addition to these recombinants. Titers were determined as described previously (36). The one-step growth kinetics of SeV recombinants were determined as described previously (24). The expression plasmid pCAG-C encoding the SeV C gene was reported previously (17).
Construction and recovery of SeV recombinants.
The pSeV(+) plasmid encoding full-length SeV cDNA (strain Z) was kindly provided by A. Kato (35). Mutations were introduced by a standard PCR technique. SeV recombinants were recovered from cDNA as described previously (35). Mutations were confirmed by direct DNA sequencing of the reverse transcription (RT)-PCR products of viral genome RNAs prepared from purified virions.
RNA preparation and strand-specific quantitative RT-PCR.
The strand-specific detection of viral genome-length RNAs by two-step quantitative real-time RT-PCR was performed as reported previously using RNA samples prepared from virus-infected LLC-MK2 or 293T cells harvested at the indicated time points (26). The amplification of a specific DNA fragment during the cycle reaction was confirmed by melting curve analysis of the PCR products.
RNA secondary structures.
Secondary structures of the 3′ 80-nucleotide (nt) regions of the genomes of the WT and a series of the GP viruses, as well as the corresponding region of the antigenomes, were estimated by computational analysis using Web-based Centroid Homfold software (www.ncrna.org/centroidhomfold).
RESULTS
SeV genome and antigenome RNA synthesis traits did not meet the VSV paradigm.
NNSVs possess the Le and Tr sequences at the 3′ and 5′ ends of their RNA genomes, respectively. The Le serves as the promoter for replication of the antigenome (genome promoter [GP]). The antigenome synthesized possesses the sequence complementary to the Tr at the 3′ end, which serves as the replication promoter for genome RNA (antigenome promoter [AGP]). Using VSV, the AGP was shown to be much stronger than the GP, leading to the larger amount of genomes over antigenomes in infected cells and progeny virions (2, 3). This VSV paradigm for determining genome polarity appears to have been applied to other NNSVs (1, 3).
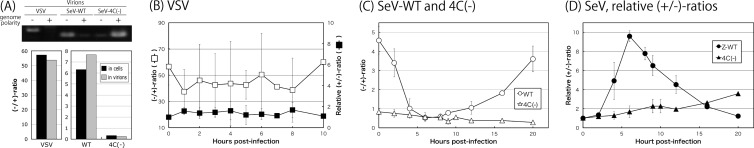
About 50 times more genomes than antigenomes were present in both VSV-infected cells and virions harvested at 10 h postinfection (p.i.), when a peak in virus titers was observed (Fig. 1A). The kinetics of the ratio of the genome to the antigenome (−/+ ratio) was found to be virtually constant at approximately 50 throughout the infection (Fig. 1B, open squares).
FIG 1.
(A) Ratio of genome (−) and antigenome (+) RNAs detected in LLC-MK2 cells (dark boxes) infected with VSV, WT SeV, and the 4C(−) virus and in their virions (gray boxes) released from these cells 10 and 20 h p.i. for VSV and SeVs, respectively. A representative gel of strand-specific RT-PCR for virion RNAs is shown. (B) Kinetics of the ratio of negative-to-positive RNAs (open squares) and the relative +/− ratio (closed squares) produced in BHK-21 cells infected with VSV. (C and D) Kinetics of the −/+ ratio (C; open symbols) and the relative +/− ratio (D; closed symbols) for WT SeV (circles) and the 4C(−) virus (triangles). Graphs represent the averages from at least three independent experiments.
At a late stage (at 20 h p.i.) of infection with WT SeV, genomes also predominated over antigenomes in cells and progeny virions, although the −/+ ratios were not as high as for VSV (∼50), at 6.3 to 7.5 (Fig. 1A). However, the kinetics of the −/+ ratio in infected cells markedly changed (Fig. 1C, open circles). Early in infection, they decreased from 4.6 to nearly 0.5 by 6 h p.i. due to vigorous antigenome synthesis relative to genome synthesis in this phase. The ratios then began to increase, ultimately reaching near-initial levels by 20 h p.i. This late phase was characterized by active synthesis of the genome. Therefore, it is clear that the genome and antigenome synthesis pattern during the course of SeV infection did not meet the VSV paradigm. There appeared to be positive-to-negative switching in SeV genomic RNA synthesis, whereas no such polarity switching was seen with VSV.
Requirements of accessory C proteins for polarity switching in SeV genome-length RNA synthesis.
We previously found that about 8 times more antigenome than genome was synthesized in cells and incorporated into the virions following 4C(−) infection, which was opposite of that found for the WT (26) (also see Fig. 1A).
The kinetics of the −/+ ratio during the course of 4C(−) infection did not significantly change without displaying such a polarity shift or switching as was found for the WT, and the ratio gradually decreased slightly (Fig. 1C, open triangles), suggesting that SeV C proteins were required for polarity switching.
In subsequent detailed studies on SeV genome-length viral RNA synthesis, we aimed to highlight the traits of antigenome synthesis, especially the predominance of antigenomes early in infection with WT SeV and throughout 4C(−) infection; therefore, we decided to primarily present results based on the +/− ratios instead of the −/+ ratios. Here, we set the initial (0 h) ratios to 1.0 for all samples, although there was a marked difference in absolute ratios between the samples. After this normalization, the +/− ratio of VSV remained unchanged at approximately 1.0, as expected (Fig. 1B, closed squares).
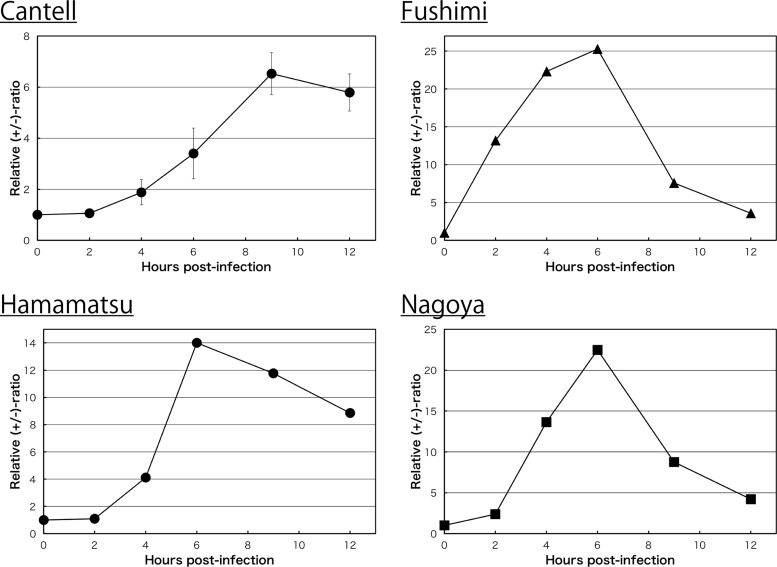
Figure 1D compared the transitional patterns of the ratios between WT SeV and 4C(−) infections. Interestingly, and as expected from Fig. 1C, the line of the kinetics for WT SeV markedly increased to approximately 10 in the initial 6 h of infection (positive phase) and then quickly reverted to the negative phase, with a continuous decrease in the ratio close to the initial level by 20 h p.i (Fig. 1D, closed circles). Similar results were observed in other SeV strains, including laboratory strains such as Cantell, Fushimi, and Nagoya, and the field isolate, Hamamatsu, although the peak values were largely different between the strains, probably a reflection of a difference in their origins (Fig. 2).
FIG 2.
Kinetics of the relative +/− ratio in LLC-MK2 cells infected with SeV strains, Cantell, Fushimi, Hamamatsu, and Nagoya.
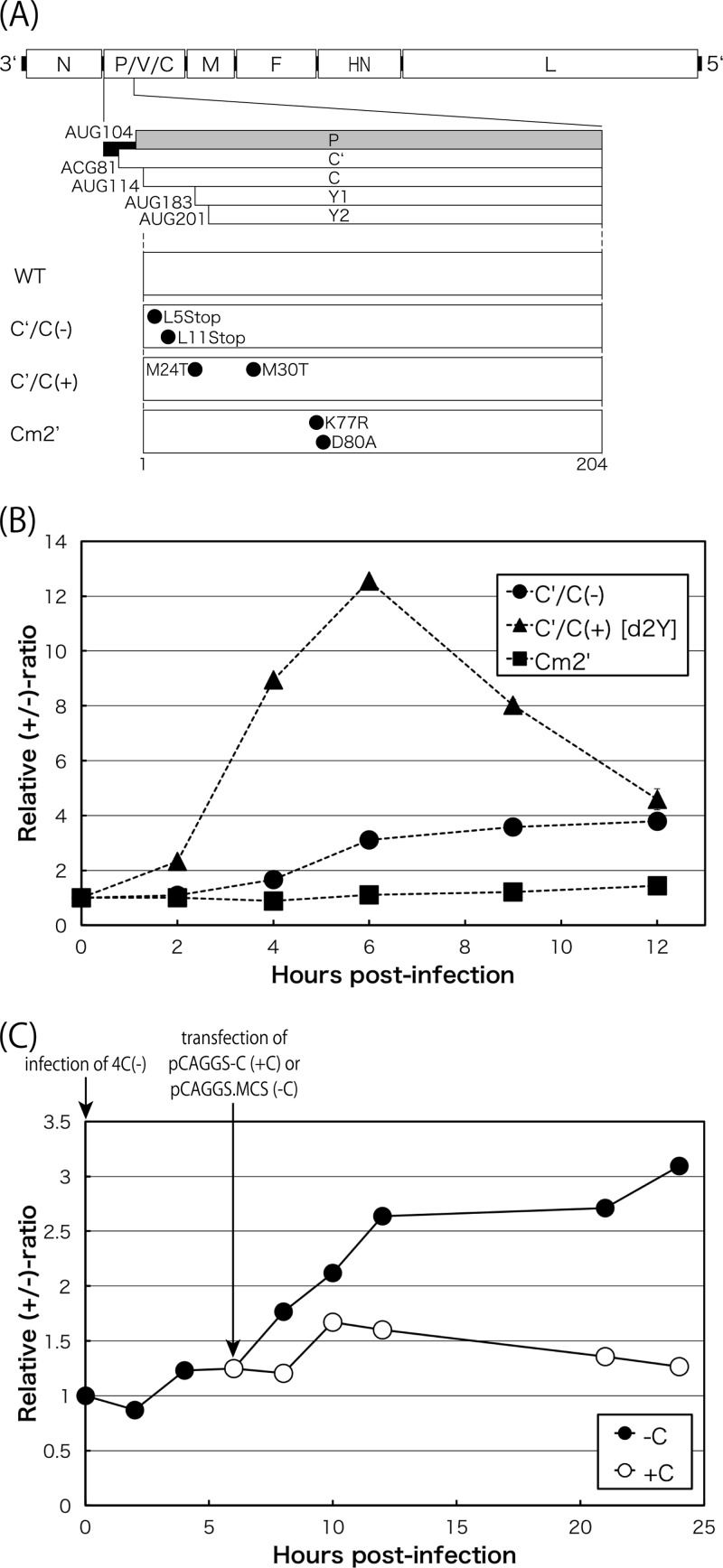
In contrast, the line of 4C(−) slowly and continuously increased to 3.5 over the experimental period without displaying such early positive and late negative phases as those observed in WT SeV (Fig. 1D, closed triangles). Differences in transitional patterns between the WT and the 4C(−) virus appeared to be caused by the presence or absence of C protein expression. We tested an additional two types of C deletion mutants, C′/C(−) [Y1/Y2(+)] (13) and C′/C(+) [Y1/Y2(−), d2Y] (24) SeVs (Fig. 3A and B). The latter clearly displayed a WT-like pattern, while the former displayed a 4C(−)-like pattern, indicating that the longer C′ and C are required for the positive-to-negative phase switching, while the shorter Y1 and Y2 could not play this role. However, the region corresponding to Y1 or Y2 in the C′ or C protein was found to be essential, because the SeV mutant Cm2′ (24) that contained two amino acid changes at positions K77R and D80A, which were mapped within the Y2, almost completely lost its switching ability and displayed a 4C(−)-like pattern (Fig. 3B). The requirement of C protein for the positive-to-negative phase control was further substantiated by the fact that its plasmid-based expression facilitated synthesis of the genome RNA of the 4C(−) virus, thereby reducing the +/− ratios (Fig. 3C).
FIG 3.
(A) Schematic representation of the SeV genome highlighting the region of the C gene. The amino acid changes for the mutants are indicated. (B) Kinetics of the relative +/− ratio in LLC-MK2 cells infected with the C mutant viruses, C′/C(−), C′/C(+), and Cm2′ (closed circles, triangles, and squares, respectively). (C) Kinetics of the +/− ratio for the 4C(−) virus in 293T cells received an empty (−C; closed circles) or a pCAG-C (+C; open circles) plasmid 6 h p.i. Time points for infection for the 4C(−) virus and transfection of the plasmids are indicated.
SeV C proteins downregulated leader promoter-mediated positive-strand genome RNA synthesis.
Previous studies using mini-genome replicons showed that SeV C proteins selectively inhibited viral RNA synthesis via the genomic leader promoter and, hence, the synthesis of antigenome and mRNAs (8, 27, 29, 30). The amounts of SeV proteins, including the C proteins, were shown to proportionally increase as infection advanced. Therefore, our present studies suggested that there could be a critical threshold of C proteins in cells. Below the threshold, the amount of C proteins was not high enough to exert genomic promoter inhibition, which facilitated antigenome synthesis, while sufficient C proteins become available above the threshold to exert genomic promoter inhibition, which suppressed antigenome synthesis. We then wanted to clarify the regulatory functions of SeV C proteins in determining genome polarity in the context of promoter usage.
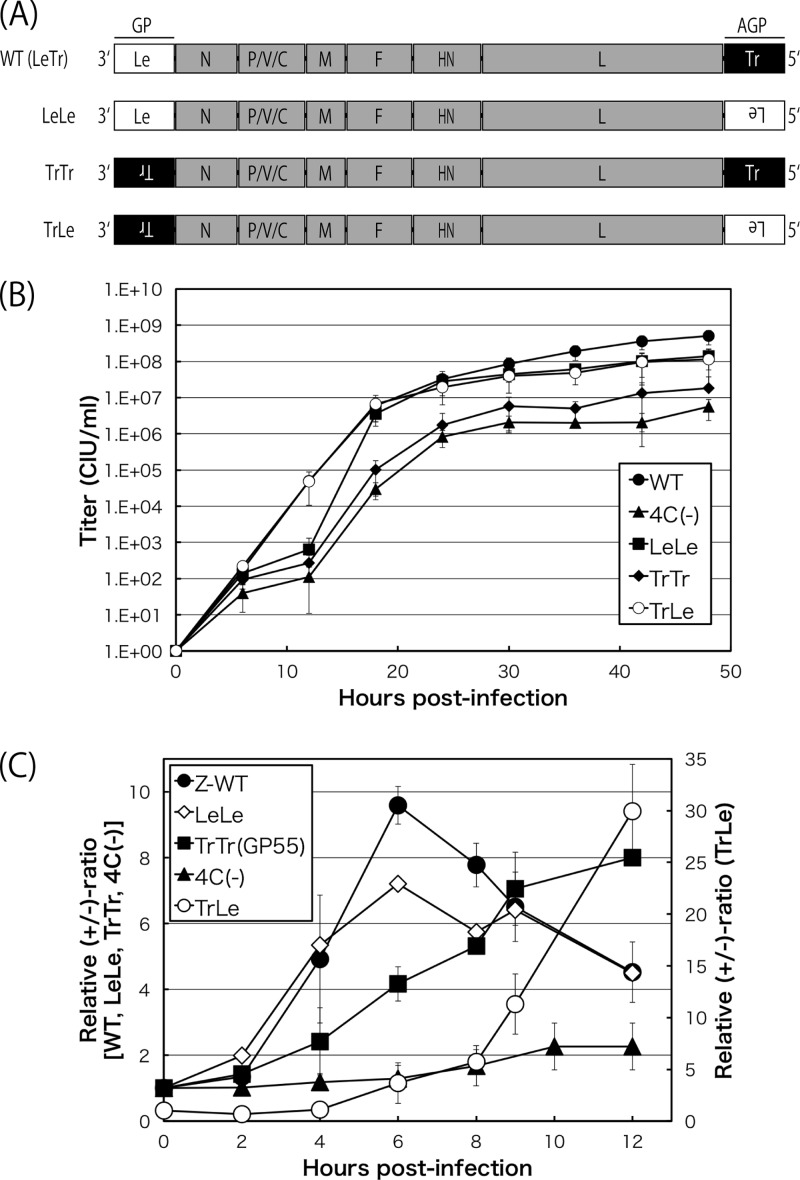
SeV recombinants possessing swaps of the 55-nt Le and Tr regions in all three possible combinations were generated in the backbone of the WT (Fig. 4). The respective complementary sequences that needed to be inserted are represented by upside-down letters in Fig. 4A. The patterns of the +/− ratio as well as growth in cells were compared with those of the WT.
FIG 4.
(A) Schematic representation of the genomic organization of the recombinant viruses, LeLe, TrTr, and TrLe, possessing swaps of the 55-nt leader and trailer regions. (B) One-step growth kinetics of the indicated SeVs, the WT, 4C(−), LeLe, TrTr, and TrLe viruses, in LLC-MK2 cells (closed circles, closed triangles, closed squares, closed diamonds, and open diamonds, respectively). Each titer represents the average from at least three independent experiments. (C) Kinetics of the relative +/− ratio in LLC-MK2 cells infected with WT, LeLe, TrTr, TrLe, and 4C(−) viruses (closed circles, open diamonds, closed squares, closed triangles, and open circles, respectively). Graphs represent the averages from at least three independent experiments.
The single-step growth curves of LeLe and TrLe viruses were comparable to that of the WT (LeTr), while the overall titers of TrTr were reduced by 1 to 2 logs (Fig. 4B). LeLe displayed WT-like positive-to-negative switching. When a sufficient amount of C proteins was not available early in infection, the leader promoter was stronger when placed in the genome than in the antigenome. However, the peak +/− ratio was significantly lower than that of the WT (Fig. 4C, open diamonds). This may have occurred because the C proteins exerted their leader-selective downregulation more or less on both the genome and antigenome. In contrast, TrTr showed no apparent polarity switching and exhibited a continuously increasing line throughout (Fig. 4C, closed squares). This suggested that the C proteins did not exert regulatory action when both the GP and AGP were the Tr type. The AGP of the TrLe virus was the Le type; therefore, it was expected to be under the suppressive control of the C proteins, while Tr-type GP could be freed from such downregulation. Therefore, the positive strand continued to markedly increase late in infection when C proteins were accumulating (Fig. 4C, open circles). A slight change in the +/− ratio was detected up to 4 h p.i., and this change was not significant at 8 h for the TrLe virus (Fig. 4C). The strength of the Tr promoter and Le promoter in this construct appeared to be nearly equivalent unless a sufficient amount of C proteins was supplied.
Overall, the results obtained by the promoter-swapped viruses indicated that the C proteins exerted their regulatory function selectively via the Le-type promoter but not via the Tr-type promoter. The regulation of polarized viral RNA synthesis by the C proteins was achieved by suppressing leader promoter-driven viral RNA synthesis rather than enhancing that from a trailer promoter.
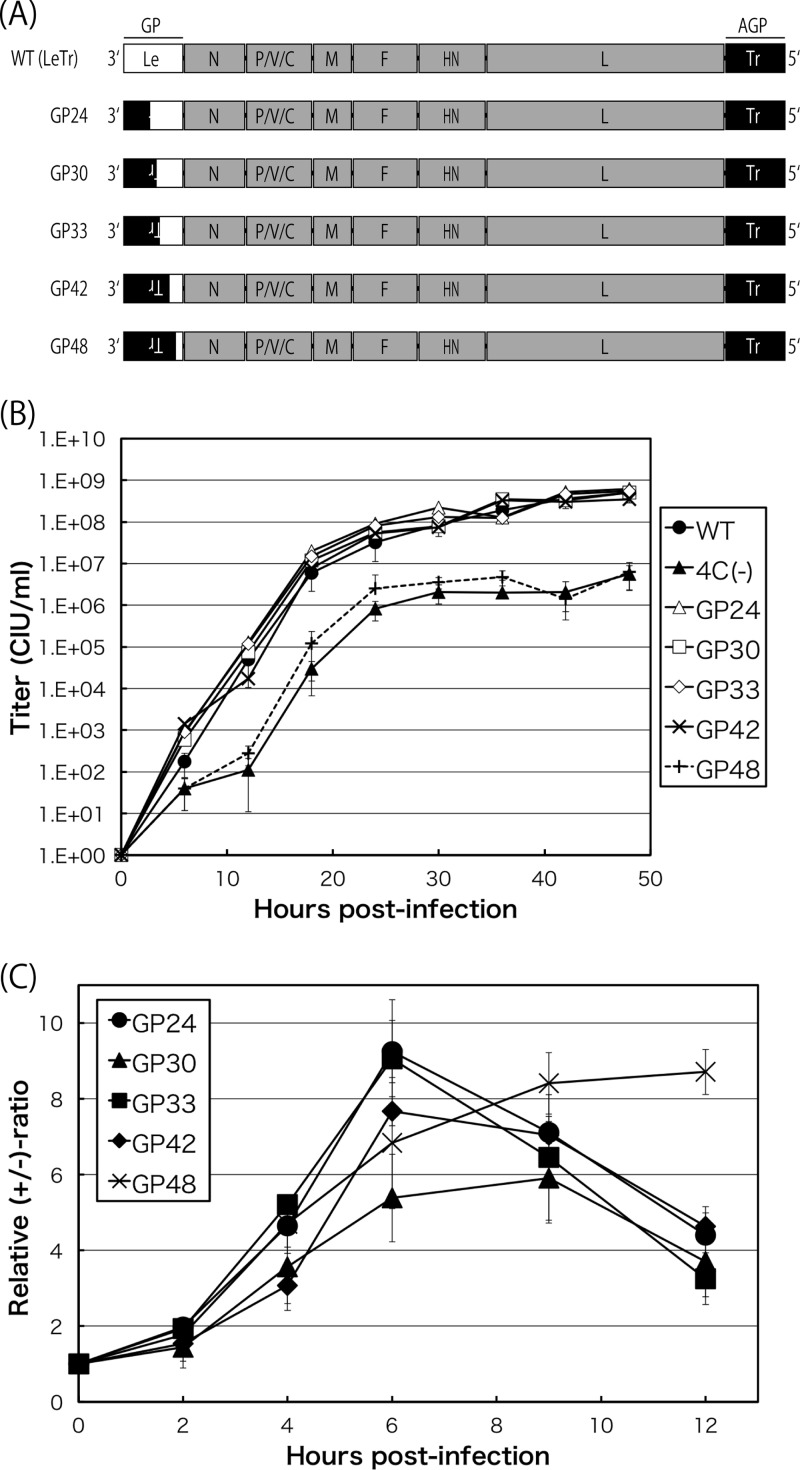
To investigate whether there was a region(s) within the 55-nt leader sequence that was crucial for polarity switching, we finally tested a series of GP viruses of various lengths (24 to 48 nt) from the 3′ terminus replaced with the corresponding trailer sequences (Fig. 5A). All of the GP viruses, except for GP48, replicated well to levels similar to those of the WT. GP48 titers were reduced by 1 to 2 logs (Fig. 5B).
FIG 5.
(A) Schematic representation of the genome organization of the GP viruses, including GP24, GP30, GP33, GP42, and GP48. (B) One-step growth kinetics of the indicated SeVs, the WT, 4C(−), GP24, GP30, GP33, GP42, and GP48 viruses, in LLC-MK2 cells (closed circles, closed triangles, open triangles, open squares, open diamonds, exes, and pluses, respectively). Each titer represents the average from at least three independent experiments. (C) Kinetics of the relative +/− ratio in LLC-MK2 cells infected with the GP viruses, GP24, GP30, GP33, GP42, and GP48 (closed circles, closed triangles, closed squares, closed diamonds, and exes, respectively). Graphs represent the averages from at least three independent experiments.
The kinetics of the +/− ratio for GP24, GP33, and GP42 were similar to those of the WT viruses (Fig. 5C, closed circles, squares, and diamonds, respectively). In contrast, GP48 showed a pattern that deviated largely from the apparently normal one shown by the above-listed three (GP24, GP33, and GP42) and resembled that of the TrTr virus (Fig. 5C, exes). The patterns of positive phase and polarity switching in GP30 were less marked than those of the greater replacement viruses, GP33 or GP42 (Fig. 5C, closed triangles). Overall, these results indicated that approximately the 5′-terminal 10 nucleotides of the 55-nt leader region were critically important for the polarized regulation of viral RNA synthesis by the C proteins.
DISCUSSION
No mechanism has been shown to selectively incorporate only the negative-strand RNA genome complex with N protein subunits (−RNP) into the virions of the Mononegavirales during the process of assembly, and this has been attributed to the indistinguishability of −RNPs and +RNPs. Genomic and antigenomic RNA-containing SeV virions were shown to be produced in proportion to their intracellular ratio, although antigenome-containing virions were not infectious (37–39).
Applying the VSV paradigm, mononegaviral genome polarity was considered to be determined by differences in promoter strength between the GP and AGP; the AGP directing the synthesis of the genome could be stronger than the GP directing the synthesis of the antigenome (1, 3).
Internal deletion DI genomes for SeV contained the authentic Le and Tr sequences present in full-length −RNA, while only the trailer and its complementary sequences were in the 3′ and 5′ ends of the copyback DI genomes. Mixed infections of eggs with SeV stocks containing internal deletion and copyback DI genomes followed by serial egg passages, together with helper SeV with the full-length RNA genome, outcompeted internal deletion-type DI genomes, resulting in the predominance of copyback DI genomes and extinction of internal deletion DI genomes (40, 41). This outcompetition was not due to size differences between the internal deletion DI and copyback DI genomes nor to whether transcription was inert (copyback DI) or active (internal deletion DI). The interpretation was that the AGP, the exact copy of the trailer, could be stronger than the GP of the leader sequence, applying the VSV paradigm. However, the interpretation could not be that simple, because the C proteins were trans-supplied from the intact helper virus required for DI genome replication in this experiment and also because the C proteins were shown to selectively inhibit RNA synthesis from the authentic leader promoter (29, 30, 32) and, hence, could facilitate copyback DI genome replication.
We have shown in this and previous reports (13, 26) that antigenome synthesis as well as mRNA synthesis predominated over genome synthesis in the case of C-deficient SeV infection and at an early phase of WT SeV infection when viral protein expression levels could be low. This suggests that the genome leader promoter was intrinsically stronger than the trailer-derived antigenome promoter, which is contrary to the VSV paradigm.
Thus, there must be a transition from the antigenome-dominant to genome-dominant phase midway in single-step replication for SeV to be an NNSV. The occurrence of this phase transition was clearly shown by the marked increase, followed by the marked decrease in positive/negative-strand ratios during single-cycle replication of WT SeV. This was not observed for VSV, whose genome polarity was determined solely by the difference in promoter strength.
The crucial requirement of the SeV accessory C proteins for the positive-to-negative shift was strongly supported by the fact that no such shift or increase then decrease in the ratio ever took place in cells infected with the 4C(−) virus that expressed none of the four C proteins (Fig. 1). The antigenome was predominant over the genome throughout in 4C(−)-infected cells. These results again strongly suggested that the intrinsic promoter activity (exhibited in the absence of the C proteins) of the genomic leader was stronger than that of the antigenomic, trailer-derived promoter and that the accumulation of the C proteins late in the life cycle may have inhibited antigenome synthesis via direct or indirect interactions with the genomic leader promoter.
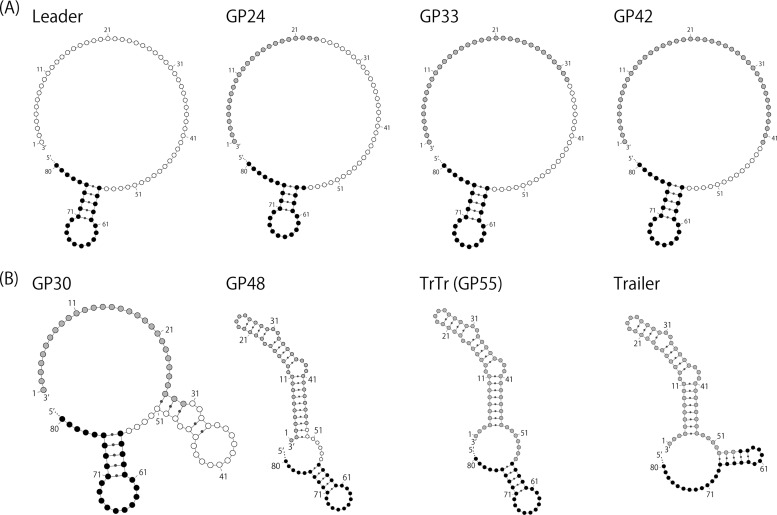
The specific inhibition of leader-driven viral RNA synthesis by C proteins was suggested through the physical interaction between the C and L proteins (32), suggesting a regulatory role of C proteins in affinity of viral RNA-dependent RNA polymerase (vRdRp) for the leader promoter region, although there is no reasonable explanation as to how vRdRp distinguishes the leader and trailer regions and how this interaction causes promoter specificity. In order to gain a deeper understanding of the promoter regions regulating genome-length viral RNA synthesis by the C proteins, we attempted to compare the secondary structures of the GP regions of GP viruses predicted by computational analysis (Fig. 6). The intact 55-nt leader region did not produce any apparent secondary structure (Fig. 6A, left). The GP24, GP33, and GP42 viruses with normal phenotypes also did not (Fig. 6A). In contrast, TrTr and GP48, which lacked polarity switching, were predicted to form large pseudoknots consisting of 45 and 49 nucleotides within their GP regions, respectively (Fig. 6B, center). The same was true for the intact trailer region (Fig. 6B, right). A smaller pseudoknot consisting of 24 nucleotides was predicted for GP30, which showed delayed polarity switching (Fig. 6B, left). Therefore, a loss or attenuation in the regulation potential of the C proteins may be dependent on the presence of the putative pseudoknots and their sizes, which suggests that the secondary structure of the promoter region may affect the activities of the promoter, interactions with the C proteins, and/or C protein-mediated regulation of SeV genome/antigenome replication.
FIG 6.
Putative secondary structures of the 3′ 80-nt regions of genome RNA for the WT (Leader), GP24, FP30, GP33, GP42, GP48, and TrTr (GP55) viruses and the intact trailer region. Nucleotides positioned at 56 to 80 are shown as closed circles, and those of the intact 55-nt leader sequence are shown as open circles. Nucleotides derived from the trailer sequence are shown as gray circles.
Since there was a reasonable association between the presence and size of the putative pseudoknot in these regions and their properties, these results seem to suggest the critical involvement of the secondary structures of the leader and trailer regions in their properties. However, the ends of the viral genomes are unlikely to form any secondary structures based on base pair matching, since genome-length mononegaviral RNAs have been considered to form RNase-resistant nucleocapsid structures due to the full and tight encapsidation of RNAs by the N proteins (1). The promoter regions at the ends of the viral genomes may be somewhat free from encapsidation to form functional secondary structures, and the difference in structures between the leader and trailer regions may cause a difference in their promoter activities. This seems to be plausible, because a similar estimation of the leader and trailer regions of VSV showed putative secondary structures opposite to those of SeV; a large pseudoknot was present in the VSV leader, and a small one was present in the trailer (data not shown).
Overall, a marked increase, followed by a similar decrease in the +/− ratio during SeV single-step replication, has never been described for any member of the Mononegavirales. This switching from the positive-strand-dominant to negative-strand-dominant phase is crucial for SeV to be an NNSV. It is interesting that only the accessory gene products play such a critical function in the biology of the Mononegavirales. Therefore, it can be concluded that the negative polarity of NNSV is decided not only by asymmetric promoter strength, as has been applied for VSV and other rhabdoviruses, but also via the trans-acting element, which can now be applied for the paramyxovirus SeV.
Why SeV needs to have acquired this complex RNA synthesis pattern to be an NNSV during its evolutionary process is not clearly understood. SeV and some other paramyxoviruses, such as Newcastle disease virus (NDV), have a relatively short time span of single-step growth (∼20 h or less). Nevertheless, they ultimately reach a high copy number. SeV and NDV reach final titers as high as approximately 109 PFU in tissue culture cells and 1010 PFU in chick embryos. The prerequisites for this may be the vigorous amplification of mRNAs and antigenomes early in infection and subsequent synthesis of the genome. In this context, the transitional shift from the positive-dominant to negative-dominant phase may represent a clever stratagem. Interestingly, NDV replication displayed a positive-to-negative shift pattern very similar to that of SeV (data not shown).
Why only the accessory gene products are deeply involved in this positive-to-negative switching has also not been fully elucidated. The L and P proteins constitute RNA polymerase. The N protein is essential for encapsidation of the nascent RNA chain and further serves as the scaffold for the L and P proteins to act as a polymerase. They individually possess numerous functionally critical subdomains and motifs that interact with each other (8) and may have no room to encode the switching function without opening the overlapping +1 C frame relative to the P ORF on the P gene. VSV also reach a high titer within a short time span, generally shorter than those of paramyxoviruses. Differences in the promoter activity between the genome and antigenome may be large enough such that VSV RNA polymerase favors the antigenome promoter more.
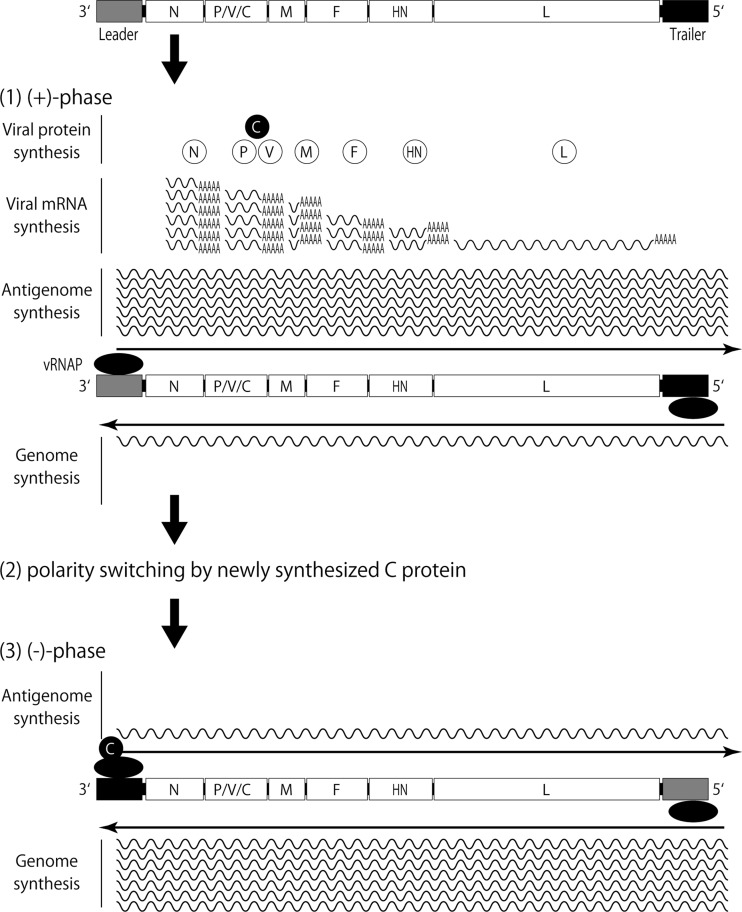
Finally, we schematically showed here a novel SeV paradigm for determining genome polarity by the participation of a trans-acting element, the C protein (Fig. 7). At an early stage of infection, where only a limited amount of C protein is available in cells, positive-sense viral RNAs (the mRNA and antigenome) are allowed to increase to support the subsequent amplification of protein synthesis and genome synthesis. Once the level of C proteins reaches and exceeds the threshold, they selectively suppress leader GP activity without affecting the trailer AGP function. Thus, the genome becomes enriched, ultimately resulting in the production of SeV with a negative-strand RNA genome. Further investigation into how much the SeV paradigm for genome polarity determination by trans-regulation can be applied to other NNSVs is warranted.
FIG 7.
A model for SeV RNA synthesis regulated by the C protein (see the text).
ACKNOWLEDGMENTS
We thank all the members of our lab for their fruitful discussions, especially Ryoko Kawabata for her excellent technical assistance. We also thank the staff of the Analysis Center of Life Science, Hiroshima University, for the use of their facilities.
This work was supported by JSPS KAKENHI grant number 23790505.
Footnotes
Published ahead of print 30 October 2013
REFERENCES
- 1.Lamb RA. 2007. Mononegavirales, p 1357–1362 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Lyles DS, Rupprecht CE. 2007. Rhabdoviridae, p 1363–1408 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61–119. 10.1007/978-3-662-06099-5_3 [DOI] [PubMed] [Google Scholar]
- 4.Li T, Pattnaik AK. 1999. Overlapping signals for transcription and replication at the 3′ terminus of the vesicular stomatitis virus genome. J. Virol. 73:444–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li T, Pattnaik AK. 1997. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology 232:248–259. 10.1006/viro.1997.8571 [DOI] [PubMed] [Google Scholar]
- 6.Whelan SP, Wertz GW. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J. Virol. 73:297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finke S, Conzelmann KK. 1997. Ambisense gene expression from recombinant rabies virus: random packaging of positive- and negative-strand ribonucleoprotein complexes into rabies virions. J. Virol. 71:7281–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai Y, Takakura A, Irie T, Yonemitsu Y, Gotoh B. 2011. Sendai virus: evolution from mouse pathogen to a state-of-art tool in virus research and biotechnology, p 115–173 In Samal SK. (ed), The biology of paramyxoviruses. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 9.Lamb RA, Parks GD. 2006. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 10.Curran J, Kolakofsky D. 1988. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 7:245–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran J, Kolakofsky D. 1989. Scanning independent ribosomal initiation of the Sendai virus Y proteins in vitro and in vivo. EMBO J. 8:521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta KC, Patwardhan S. 1988. ACG, the initiator codon for a Sendai virus protein. J. Biol. Chem. 263:8553–8556 [PubMed] [Google Scholar]
- 13.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. 1998. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells 3:111–124. 10.1046/j.1365-2443.1998.00170.x [DOI] [PubMed] [Google Scholar]
- 14.Garcin D, Latorre P, Kolakofsky D. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205–210. 10.1016/S0014-5793(99)01241-7 [DOI] [PubMed] [Google Scholar]
- 16.Koyama AH, Irie H, Kato A, Nagai Y, Adachi A. 2003. Virus multiplication and induction of apoptosis by Sendai virus: role of the C proteins. Microbes Infect. 5:373–378. 10.1016/S1286-4579(03)00043-1 [DOI] [PubMed] [Google Scholar]
- 17.Irie T, Nagata N, Yoshida T, Sakaguchi T. 2008. Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology 371:108–120. 10.1016/j.virol.2007.09.020 [DOI] [PubMed] [Google Scholar]
- 18.Irie T, Shimazu Y, Yoshida T, Sakaguchi T. 2007. The YLDL sequence within Sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J. Virol. 81:2263–2273. 10.1128/JVI.02218-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irie T, Inoue M, Sakaguchi T. 2010. Significance of the YLDL motif in the M protein and Alix/AIP1 for Sendai virus budding in the context of virus infection. Virology 405:334–341. 10.1016/j.virol.2010.06.031 [DOI] [PubMed] [Google Scholar]
- 20.Sakaguchi T, Kato A, Sugahara F, Shimazu Y, Inoue M, Kiyotani K, Nagai Y, Yoshida T. 2005. AIP1/Alix is a binding partner of Sendai virus C protein and facilitates virus budding. J. Virol. 79:8933–8941. 10.1128/JVI.79.14.8933-8941.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. 2004. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology 325:1–10. 10.1016/j.virol.2004.04.019 [DOI] [PubMed] [Google Scholar]
- 22.Yoshida A, Sakaguchi T, Irie T. 2012. Passage of a Sendai virus recombinant in embryonated chicken eggs leads to markedly rapid accumulation of U-to-C transitions in a limited region of the viral genome. PLoS One 7:e49968. 10.1371/journal.pone.0049968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasan MK, Kato A, Muranaka M, Yamaguchi R, Sakai Y, Hatano I, Tashiro M, Nagai Y. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619–5628. 10.1128/JVI.74.12.5619-5628.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irie T, Nagata N, Igarashi T, Okamoto I, Sakaguchi T. 2010. Conserved charged amino acids within Sendai virus C protein play multiple roles in the evasion of innate immune responses. PLoS One 5:e10719. 10.1371/journal.pone.0010719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irie T, Yoshida A, Sakaguchi T. 2013. Clustered basic amino acids of the small Sendai virus C protein Y1 are critical to its Ran GTPase-mediated nuclear localization. PLoS One 8:e73740. 10.1371/journal.pone.0073740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irie T, Nagata N, Yoshida T, Sakaguchi T. 2008. Paramyxovirus Sendai virus C proteins are essential for maintenance of negative-sense RNA genome in virus particles. Virology 374:495–505. 10.1016/j.virol.2008.01.004 [DOI] [PubMed] [Google Scholar]
- 27.Curran J, Marq JB, Kolakofsky D. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647–656. 10.1016/0042-6822(92)90588-G [DOI] [PubMed] [Google Scholar]
- 28.Curran J, Boeck R, Kolakofsky D. 1991. The Sendai virus P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J. Virol. 70:5067–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. 1997. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J. Virol. 71:9588–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grogan CC, Moyer SA. 2001. Sendai virus wild-type and mutant C proteins show a direct correlation between L polymerase binding and inhibition of viral RNA synthesis. Virology 288:96–108. 10.1006/viro.2001.1068 [DOI] [PubMed] [Google Scholar]
- 32.Horikami SM, Hector RE, Smallwood S, Moyer SA. 1997. The Sendai virus C protein binds the L polymerase protein to inhibit viral RNA synthesis. Virology 235:261–270. 10.1006/viro.1997.8702 [DOI] [PubMed] [Google Scholar]
- 33.Le Mercier P, Garcin D, Garcia E, Kolakofsky D. 2003. Competition between the Sendai virus N mRNA start site and the genome 3′-end promoter for viral RNA polymerase. J. Virol. 77:9147–9155. 10.1128/JVI.77.17.9147-9155.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Mercier P, Garcin D, Hausmann S, Kolakofsky D. 2002. Ambisense Sendai viruses are inherently unstable but are useful to study viral RNA synthesis. J. Virol. 76:5492–5502. 10.1128/JVI.76.11.5492-5502.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes to cells: devoted to molecular & cellular mechanisms. 1:569–579. 10.1046/j.1365-2443.1996.d01-261.x [DOI] [PubMed] [Google Scholar]
- 36.Kiyotani K, Takao S, Sakaguchi T, Yoshida T. 1990. Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity. Virology 177:65–74. 10.1016/0042-6822(90)90460-9 [DOI] [PubMed] [Google Scholar]
- 37.Kolakofsky D, Boy de la Tour E, Bruschi A. 1974. Self-annealing of Sendai virus RNA. J. Virol. 14:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolakofsky D, Bruschi A. 1975. Antigenomes in Sendai virions and Sendai virus-infected cells. Virology 66:185–191. 10.1016/0042-6822(75)90189-0 [DOI] [PubMed] [Google Scholar]
- 39.Mottet G, Roux L. 1989. Budding efficiency of Sendai virus nucleocapsids: influence of size and ends of the RNA. Virus Res. 14:175–187. 10.1016/0168-1702(89)90037-3 [DOI] [PubMed] [Google Scholar]
- 40.Re GG, Kingsbury DW. 1986. Nucleotide sequences that affect replicative and transcriptional efficiencies of Sendai virus deletion mutants. J. Virol. 58:578–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Re GG, Morgan EM, Kingsbury DW. 1985. Nucleotide sequences responsible for generation of internally deleted Sendai virus defective interfering genomes. Virology 146:27–37. 10.1016/0042-6822(85)90050-9 [DOI] [PubMed] [Google Scholar]