Abstract
Despite the clinical relevance of latent HIV-1 infection as a block to HIV-1 eradication, the molecular biology of HIV-1 latency remains incompletely understood. We recently demonstrated the presence of a gatekeeper kinase function that controls latent HIV-1 infection. Using kinase array analysis, we here expand on this finding and demonstrate that the kinase activity profile of latently HIV-1-infected T cells is altered relative to that of uninfected T cells. A ranking of altered kinases generated from these kinome profile data predicted PIM-1 kinase as a key switch involved in HIV-1 latency control. Using genetic and pharmacologic perturbation strategies, we demonstrate that PIM-1 activity is indeed required for HIV-1 reactivation in T cell lines and primary CD4 T cells. The presented results thus confirm that kinases are key contributors to HIV-1 latency control. In addition, through mutational studies we link the inhibitory effect of PIM-1 inhibitor IV (PIMi IV) on HIV-1 reactivation to an AP-1 motif in the CD28-responsive element of the HIV-1 long terminal repeat (LTR). The results expand our conceptual understanding of the dynamic interactions of the host cell and the latent HIV-1 integration event and position kinome profiling as a research tool to reveal novel molecular mechanisms that can eventually be targeted to therapeutically trigger HIV-1 reactivation.
INTRODUCTION
Eradication of the latent HIV-1 reservoir is considered a major requirement toward the development of a cure for HIV-1 infection. Therapeutically induced reactivation of latent HIV-1 infection events will be an essential first step in this process. At present, it is widely assumed that HIV-1 latency is the result of a special restrictive histone composition or a unique restrictive chromatin environment established at the latent viral promoter. This idea has guided the majority of the therapeutic efforts to eradicate the latent HIV-1 reservoir. Histone deacetylase inhibitors (HDACi) such as valproic acid or, more recently, vorinostat/suberanilohydroxamic acid (SAHA) were used in an attempt to relieve this proposed chromatin-mediated transcriptional restriction and trigger system-wide HIV-1 reactivation (1–4). In one of these studies the authors could demonstrate vorinostat-promoted induction of viral RNA in the treated patients (4). Other reports, including a recent study by Blazkova et al. (5), using ex vivo patient material could not confirm that HDACi trigger HIV-1 reactivation (6–8). Most recently Shan et al. tested the efficacy of 17 HDAC inhibitors as HIV-1 reactivating agents in latently HIV-1-infected primary resting CD4+ T cells transduced with the antiapoptotic Bcl-2 gene (9). None of the HDAC inhibitors triggered efficient reactivation relative to CD3/CD28 monoclonal antibody (MAb) treatment during short-term treatment experiments, but some exhibited good HIV-1 reactivation efficacy in long-term treatment experiments. Notably, in these and previously published experiments, reactivated infection events reverted to a latent state when the drugs were removed from culture (10). While the value of HDAC inhibitors as HIV-1-reactivating agents in a therapeutic setting thus remains unclear, it is becoming increasingly evident that drugs that can complement or replace HDACi-based therapy approaches are needed to achieve the goal of HIV-1 eradication. A more comprehensive understanding of the dynamic interaction between the host cell and the latent virus that extends beyond the relatively static current model of latent HIV-1 infection will be needed to guide the targeted discovery and development of such HIV-1-reactivating drugs.
In support of the idea that many molecular mechanisms that control latent HIV-1 infection have yet to be identified, we recently reported that latency control starts at the level of kinase activity. We demonstrated the presence of a kinase function that acts as a master switch to control latent HIV-1 infection even in the presence of high levels of induced NF-κB activity, which was present in latently infected T cell lines and primary CD4 T cells (11). Additional evidence for a role of specific transcription factors in latency control comes from our observation that naturally occurring variations of the AP-1 motif in the CD28-responsive element (CD28RE) of the HIV-1 long terminal repeat (LTR) influence the efficacy of latency establishment (12). These data suggest that latent infection is controlled by dynamic, bi-directional interactions of the virus with the host cell at the kinase and transcription factor levels. To this end, latent HIV-1 infection can be viewed as a normal gene regulation phenomenon. Once integrated, HIV-1 acts as a cellular gene controlled by its promoter (LTR), which is structurally similar to promoters of cellular genes such as interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), and the IL-2 receptor α chain (CD25). It is worth noting that these genes, just as latent HIV-1 infection, are not expressed in CD4+ memory T cells, which are the primary cellular host of latent HIV-1 infection. Beyond the demonstration that these genes are controlled by defined kinase activities and a defined downstream transcription factor composition, there are other important reported similarities between cellular gene expression control and latent HIV-1 infection. For example, paused RNA polymerase II complex (RNAP II), which is found at the promoters of nonexpressed but inducible genes (13–16), has also been found associated with the latent LTR promoter (17–20).
Other similarities have been found at the level of nucleosome positioning. Recently, Rafati et al. reported that for latent HIV-1 infection events, the two nucleosomes that are found at the LTR are actively repositioned away from their predicted DNA binding sites as a function of the presence of brahma homolog 1 (BRG1)-associated factors (BAF) or polybromo-associated BAF (PBAF) so as to possibly restrict access of activating transcription factors to the LTR (21). Similar findings have been reported earlier for many inactive, but inducible, cellular promoters (for recent reviews, see references 22 and 23).
We here expand on our findings that kinases play a key role in the control of latent HIV-1 infection and HIV-1 reactivation. Using kinome profiling, we demonstrate that at the level of their kinase activity profiles, latently HIV-1-infected T cells phenotypically differ from uninfected cells. We demonstrate that as predicted by the protein interaction network (PIN) map generated from these data, PIM-1 kinase is involved in HIV-1 reactivation in T cell lines. This finding can be directly transferred to latent infection in primary T cells. Lastly, we provide experimental evidence that PIM-1 must act through transcription factors that bind to an AP-1 motif in the CD28RE of the latent HIV-1 LTR, linking kinase activity directly to the available transcription factor composition. In summary, our findings provide additional evidence for a key role of kinase control in HIV-1 latency and establish kinome profiling as a research tool to identify novel drug targets for HIV-1 reactivation.
MATERIALS AND METHODS
Cell culture, plasmids, and reagents.
All T cell lines were maintained in RPMI 1640 medium supplemented with 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum (FBS). The latently HIV-1-infected CA5 and EF7 T cells were generated using an NL4-3-based green fluorescent protein (GFP) reporter virus (NLENG) (11, 24). Each of these cell lines contains a single integration event within an actively expressed host gene. In CA5 T cells, the virus is integrated in the same transcriptional orientation as the host gene, while in EF7 T cells, the latent virus is integrated in the converse-sense orientation. J2574 reporter T cells have been described previously (12). Briefly, J2574 cells were generated by infecting Jurkat T cells with an HIV-1 LTR-GFP-LTR construct (p2574) and then selecting for a population that expresses no GFP in the absence of infection but expresses GFP upon Tat transduction or HIV-1 infection. The J2574 T cell population holds at least 50,000 founder cells with different integration sites. Fetal bovine serum (FBS) was obtained from HyClone (Logan, UT) and was tested on a panel of latently infected cells to ensure that the utilized FBS batch did not spontaneously trigger HIV-1 reactivation (25, 26). The phorbol ester, 13-phorbol-12-myristate acetate (PMA), was purchased from Sigma. Recombinant human TNF-α was obtained from R&D Systems (Minneapolis, MN). The Jun N-terminal protein kinase (JNK) inhibitor AS601245 and PIM-1 inhibitor IV (PIMi IV; CAS 477845-12-8) were purchased from Calbiochem (Billerica, MA). Anti-CD25 antibody was purchased from BD Biosciences-Pharmingen (San Jose, CA).
Latent HIV-1 infection of primary T cells.
Latently infected cultured central memory CD4+ T cells were prepared from primary naive cells as previously described (7, 27). Briefly, peripheral blood mononuclear cells (PBMCs) were obtained from deidentified healthy donors. Naive CD4+ T cells were isolated by MACS (magnetic cell sorting) microbead-negative sorting using a naive T cell isolation kit (Miltenyi Biotec, Auburn, CA). The purity of the sorted population was always higher than 95% with a phenotype of CD4+ CD45RA+ CD45RO− CCR7+ CD62L+ CD27+. Naive CD4+ T cells were primed with beads coated with anti-CD3 and anti-CD28 antibodies (Dynal/Invitrogen, Carlsbad, CA). Proliferating cells were expanded in medium containing 30 IU/ml recombinant IL-2 (rIL-2), and medium and IL-2 were replaced every 2 days. Defective HIV (DHIV) viruses were produced by transient transfection of HEK293T cells by calcium phosphate-mediated transfection (7, 27). To normalize infections, p24 was analyzed in virus-containing supernatants by enzyme-linked immunosorbent assay (ELISA; ZeptoMetrix, Buffalo, NY). Cells were infected by spinoculation: 1 × 106 cells were infected with 500 ng/ml p24 during 2 h at 2,900 rpm and 37°C in 1 ml.
Western blotting.
Cells were harvested by centrifugation, washed once with phosphate-buffered saline (PBS) buffer, and lysed in radioimmunoprecipitation assay (RIPA) buffer (Cell Signaling, Danvers, MA) according to the manufacturer's instructions. Protein concentration of the lysates was determined by the bicinchoninic acid (BCA) method according to the manufacturer's recommendations (Pierce, Rockford, IL). About 20 to 40 μg of protein per sample was separated on precast 10% Mini Protean TGX gels (Bio-Rad, Hercules, CA) and subsequently transferred to a polyvinylidene difluoride (PVDF) membrane using an iBlot gel transfer system (Invitrogen, Carlsbad, CA). Western blotting was performed according to standard protocols. IκB protein (or tubulin as a control) was detected with specific monoclonal antibodies (Cell Signaling, USA). A horseradish peroxidase-conjugated mouse anti-rabbit polyclonal antibody (Cell Signaling) was used as a secondary antibody. The blot was developed using Western Lightning Ultra Chemiluminescent Substrate from PerkinElmer, Inc. (Waltham, MA), and detected in an EpiChemi3 Darkroom (UVP BioImaging System, Upland, CA).
TransAM assays for NF-κB.
NF-κB p50 and p65 activities in nuclear extracts of cells were determined using TransAM assays (Active Motif). All experiments were performed according to the manufacturer's instructions. TransAM assays quantify the ability of activated NF-κB to bind to an NF-κB consensus sequence in solution, with a 5- to 10-fold higher sensitivity than gel shift assays.
BioPlex analysis of cytokine expression.
PBMCs were generated as previously described (11). T cells were stimulated in the presence or absence of the respective inhibitors using 3 μg/ml phytohemagglutinin L (PHA-L), and culture supernatants were harvested after 24 h. Cytokine expression was determined using MilliPlex kits for IL-2, IL-4, IL-6, IL-8, IL-17, and gamma interferon (IFN-γ) (Millipore).
Flow cytometry.
Infection levels in the cell cultures were monitored by flow cytometric (FCM) analysis of GFP expression. FCM analysis was performed on a Guava EasyCyte (Guava Technologies, Inc., Billerica, MA), a FACSCalibur, or an LSRII (Becton, Dickinson, Franklin Lakes, NJ) instrument. Cell sorting experiments were performed using a FACSAria Flow Cytometer (Becton, Dickinson). Data analysis was performed using either CellQuest (Becton, Dickinson) or Guava Express (Guava Technologies, Inc.) software.
Kinomic analysis.
Kinomic profiling of Jurkat, CA5, and EF7 cellular lysates was conducted in the University of Alabama at Birmingham (UAB) Kinome Core using the PamStation 12 platform (PamGene, ‘s-Hertogenbosch, The Netherlands). This platform consists of a high-throughput peptide microarray system analyzing either 144 individual tyrosine phosphorylatable peptides on a protein tyrosine kinase (PTK) array or 144 serine and threonine kinase-phosphorylatable peptides on a serine-tyrosine kinase (STK) array. All peptides are composed of 12 to 15 amino acids that are imprinted onto an aluminum oxide matrix, allowing exposure to kinases to measure activity in lysates that are pumped through these peptide-rich matrices. Phospho-specific fluorescein isothiocyanate (FITC)-conjugated antibodies were used to detect peptide phosphorylation. Images of FITC-dependent fluorescent signal are captured via a computer-controlled charge-coupled-device (CCD) camera with kinetic image capture over time and over multiple exposures. For PTK analysis, 10 μg of each quantified lysate was mixed to a total of 28 μl in deionized H2O (dH2O) with 4 μl of 10× protein kinase (PK)/Abl kinase buffer (New England BioLabs), 4 μl of 10× bovine serum albumin (BSA) solution, and 0.4 μl of 1 M dithiothreitol (DTT; Fluka). Immediately prior to loading onto the array, 0.3 μl of the FITC-conjugated PY20 phosphotyrosine antibody (PamGene) was added along with 4 μl of a freshly prepared 4 mM ATP solution to the lysate mixture. Lysate solution was mixed by pipette and quickly loaded at 35 μl per array after a blocking step with 2% BSA was completed. During the assay, active kinases in the lysate phosphorylate specific peptides on the array that are detected by quantitating FITC intensity for each spot on each array using a constant 50-ms camera exposure time captured every 6 s over the course of the reaction (60 min). Evolve software (PamGene) generates kinetic reaction curves for each phosphopeptide probe, with the slope referred to as initial velocity (vINI) and end-of-reaction images labeled as “end level.” An additional set of images is captured following a wash step (“postwash”) at camera exposures of 10, 20, 50, 100, and 200 ms to provide an integrated measure of peptide phosphorylation. For STK analysis, 1 μg of each quantified lysate was mixed to a total volume of 34.5 μl in dH2O with 4 μl of 10× PK and 1.6 μl of 100× BSA solution (PamGene). Immediately prior to loading onto the array, 1 μl of a freshly prepared 4 mm ATP solution was added. Lysate solution was mixed by pipette quickly and loaded at 35 μl per array after the blocking step was completed by the PamStation12. For each chip (four arrays) 1.01 μl of the stock STK primary antibody mixture (PamGene) was mixed with 13.2 μl of 10% BSA in phosphate-buffered saline (PBS) and 0.35 μl of the STK FITC-conjugated secondary antibody (PamGene) and brought up to a volume of 132 μl per chip (four arrays). This mixture was gently mixed by pipette and applied at 30 μl per array, and the PamStation STK protocol was continued. Digital images were captured only as postwash pictures at 10, 20, 50, 100 and 200 ms to allow optimization of signal level quantification. An integrated signal level (S100) was calculated in a manner similar to that for PTK analysis and was used in this study. Comparisons of kinomic profiles between samples were performed using BioNavigator software, version 5 (PamGene), to identify significantly different phosphopeptides (P < 0.05, by t test). Upstream kinases were identified by scoring potential kinases based on their prevalence in the top 10 kinase scoring lists for each phosphopeptide as mapped in the Kinexus upstream kinase database (www.phosphonet.ca). Furthermore, protein interaction networks (PINs) were generated by uploading the peptide substrate information into the MetaCore knowledge base (Thomson Reuters).
Targeting PIM-1 expression.
Short hairpin RNA (shRNA) vectors targeting PIM-1 gene expression were generated using a pSilencer 5.1-U6 Retro vector from Ambion (Austin, TX). In U6/shRNA PIMpos380 (shPIM#10) we inserted GATCTCTTCGACTTCATCATTCAAGAGATGATGAAGTCGAAGAGATCTTTTTT to target PIM-1 expression. In U6/shRNA PIMpos785 we inserted GTGTCAGCATCTCATTAGATTTCAAGAGAATCTAATGAGATGCTGACATTTTTT to target PIM-1 expression (shPIM#22). Latently HIV-1-infected CA5 T cells were retrovirally transduced with these constructs, selected with puromycin, and cloned. Overexpression of PIM-1 was achieved by retroviral transduction of latently HIV-1-infected CA5 T cells with a pMSCV-PIM-1 expression vector.
Statistics.
When indicated, experiments were performed at least in triplicates. Experimental results are presented as mean values, and the standard deviation is indicated as error bars as a descriptor of the variation from the mean. Where indicated, a Student's t test was performed to evaluate the significance of possible drug effects by comparing two experimental data sets, each following a normal distribution.
RESULTS
Kinome profiling reveals PIM-1 as a kinase involved in latent HIV-1 infection.
Previous studies from our laboratories provided evidence for a key role of kinase activity in the control of latent HIV-1 infection (11). To develop a comprehensive understanding of how kinases play a role in latent HIV-1 infection, we performed kinome profiling experiments. Using kinome array analysis, we determined the baseline kinase activity profile of parental Jurkat T cells in comparison to the kinase activity profile of two molecularly well-defined, latently HIV-1-infected Jurkat T cell clones with single integration events, i.e., CA5 and EF7 T cells (24). In these experiments, cell lysates from Jurkat, CA5, and EF7 T cells were loaded on high-throughput peptide microarray chips holding either 144 individual phosphorylatable peptides specifically recognized by tyrosine kinases or 144 phosphorylatable peptides that are specifically recognized by serine/threonine protein kinases. Peptides with the greatest increase in phosphorylation in the latently HIV-1-infected cells relative to the parental Jurkat T cells were selected for upstream kinase analysis as described in Materials and Methods. In both latently HIV-1-infected cell lines, CA5 and EF7 T cells, PIM-1 was the highest-ranked kinase relative to the parental Jurkat T cells (Table 1). In addition, we also found that PIM-1 was the highest scoring kinase in J89GFP T cells, the first latently HIV-1-infected GFP reporter cell line we established (25).
TABLE 1.
Ranking of PIM kinases based on kinomically identified kinases with increased activity in latently HIV-1-infected Jurkat T cells relative to control Jurkat T cells
| Kinase | CA5 |
EF7 |
J89GFP |
|||
|---|---|---|---|---|---|---|
| Kinase rank | Ratio score | Kinase rank | Ratio score | Kinase rank | Ratio score | |
| PIM-1 | 6 | 38.2 | 4 | 40.9 | 1 | 56.3 |
| PIM-2 | 7 | 38.2 | NRa | 5 | 37.5 | |
| PIM-3 | 8 | 38.2 | 6 | 40.9 | 2 | 56.3 |
NR, not ranked.
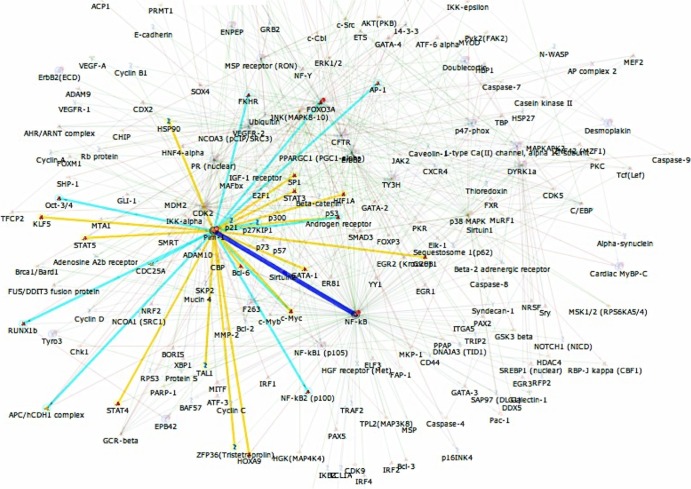
A PIM-1-centric shortest-paths PIN map derived from these experiments that describes likely relevant interactions of PIM-1 with other proteins is depicted in Fig. 1. Among other factors, PIM-1 is directly linked to NF-κB, as well as to cyclin dependent kinase 2 (CDK2). CDK2 has recently been demonstrated to be important for HIV-1 transcription by regulating the phosphorylation of HIV-1 Tat and CDK9 (28–30). The direct functional proximity of PIM-1 on the PIN to these factors can be viewed as a descriptor of the importance of PIM-1 in the context of HIV-1 latency.
FIG 1.
Shortest-paths diagram for kinase control of latent HIV-1 infection. Source Uniprot identification numbers (IDs) for phosphopeptides that were found increased in three analyzed latently HIV-1 infected T cell lines (CA5, EF7, and J89GFP) over parental Jurkat T cells along with the Uniprot ID for PIM-1 were uploaded to GeneGo MetaCore (Thomson Reuters) as seed nodes for network analysis using Dijkstra's shortest-paths algorithm to identify directed interactions among these seed nodes. PIM-1's interactions were selected (highlighted paths), with PIM-1 canonical pathway interactions highlighted in light blue, and other PIM1 interactions highlighted in yellow. NF-κB was the most interconnected node, and its interaction with PIM-1 is highlighted in dark blue.
While we demonstrate in the following that PIM-1 plays an important role in HIV-1 latency control, the results also have more general implications. Among the top 10 kinases with increased activity, not only PIM-1 but also mitogen-activated protein kinase-activated protein kinase 3 (MAPKAPK3) and PIM-3 were found altered in all three tested cell lines, suggesting that latently HIV-1-infected T cells are phenotypically altered and that these changes are essential to latency control.
PIM inhibitor IV inhibits HIV-1 reactivation in CA5 T cells.
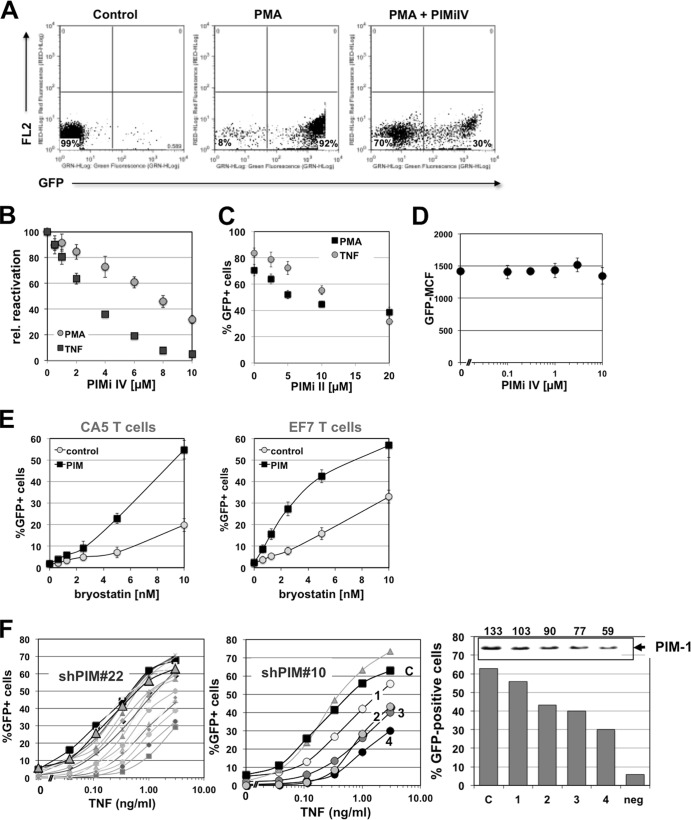
Supporting the idea that PIM-1 plays a role in HIV-1 latency control, we had identified 4-(3-(4-chlorophenyl)-2,1-benzisoxazol-5-yl)-2-pyrimidinamine, or PIM-1 inhibitor IV (PIMi IV), as an inhibitor of HIV-1 reactivation during a drug screening campaign (Fig. 2). PIMi IV prevented TNF-α-induced HIV reactivation in CA5 T cells with a 50% inhibitory concentration (IC50) of 3 μM but was somewhat less potent to inhibit PMA-induced reactivation (Fig. 2B). At a 10 μM concentration, PIMi IV was still ∼70% effective in preventing PMA-induced HIV-1 reactivation, as determined by flow cytometric analysis using the latently HIV-1-infected CA5 T cells. Optimal pretreatment time prior to stimulation was found to be 6 h. In all experiments, addition of PIMi IV at the utilized concentrations did not increase cell death relative to cell death seen under control or stimulated conditions in the absence of the inhibitor. The stimulus-dependent differences in the inhibitory capacity of PIMi IV indicated that the inhibitor could exhibit some selectivity for kinase pathways that are stimulated by TNF-α. Other commercially available PIM inhibitors were less efficient and required longer pretreatment periods prior to displaying their inhibitory activity on HIV-1 reactivation. Data for PIMi II-mediated inhibition of HIV-1 reactivation are presented in Fig. 2C. Optimal pretreatment time here was 18 h. A possible explanation for this observation is that PIMi IV is the only available PIM inhibitor that targets the active site of the enzyme, while all other available PIM inhibitors, including PIMi II, target the ATP binding site of PIM-1 (31).
FIG 2.
PIM-1 inhibitor IV prevents activation induced HIV-1 reactivation. (A) Latently HIV-1-infected CA5 reporter T cells were stimulated with the phorbol ester PMA (3 ng/ml) in the presence or absence of PIMi IV (10 μM), and reactivation was measured as the percentage of GFP-positive cells by using flow cytometric analysis. (B) PIMi IV was titrated on CA5 T cells against TNF-α (10 ng/ml) or PMA (3 ng/ml) as HIV-1-reactivating agent. The level of HIV-1 reactivation was determined as the percentage of GFP-positive cells using flow cytometric analysis and plotted over the PIMi IV concentration. CA5 T cells were preincubated for 6 h with PIMi IV prior to triggering HIV-1 reactivation. rel, relative. (C) PIMi II was titrated on CA5 T cells against TNF-α (10 ng/ml) or PMA (3 ng/ml) as HIV-1-reactivating agent. The level of HIV-1 reactivation was determined as the percentage of GFP-positive cells using flow cytometric analysis and plotted over the PIMi II concentration. CA5 T cells were preincubated for 18 h with PIMi II prior to triggering reactivation. (D) PIMi IV was titrated on chronically actively HIV-1-infected JNLG T cells. GFP mean channel fluorescence (GFP-MCF) was determined as a quantitative surrogate marker of HIV-1 expression. (E) The latently HIV-1-infected T cell lines CA5 and EF7 were retrovirally transduced to overexpress PIM-1 protein. Following retroviral transduction, bryostatin, an anticancer drug candidate that triggers PKC/NF-κB activation, was titrated on CA5-PIM or EF7-PIM cells (PIM) and the level of HIV-1 reactivation as measured by GFP expression was compared to that of the parental cells (control). (F) PIM-1 expression in CA5 T cells was knocked down using two different anti-PIM-1 shRNA constructs (shPIM#10 and shPIM#22), and PIM-1 shRNA-transduced clones were generated. For an unbiased, representative cross-section of CA5-shPIM#22 cell clones, TNF-α was then titrated on either control CA5 T cells (black symbols), a population of CA5 T cells that were transduced with a scrambled shRNA and then puromycin selected (large gray triangles), and the various generated PIM-1 shRNA transduced clones (gray symbols/lines; all left panel) and determined as the percentage of GFP-positive cells as a surrogate marker of HIV-1 reactivation. The effect of PIM-1 knockdown on concentration-dependent TNF-α-mediated HIV-1 reactivation was detailed for four CA5-shPIM#10 cell clones (middle panel), and achievable HIV-1 reactivation levels (percentage of GFP-positive cells) were correlated with PIM-1 expression as determined by Western blotting for PIM-1 (right panel). The numbers over the inset showing the Western blot data indicated the band intensities (in arbitrary units) for PIM-1 expression.
While PIMi IV prevented HIV-1 reactivation, the inhibitor had no effect on active HIV-1 expression. We titrated the compound on two chronically actively infected GFP reporter T cell lines, JNLG T cells (32) and CUCY T cells (33). Four days after the addition of the compound, we determined changes in GFP mean channel fluorescence intensity (MFI), which would be indicative of an inhibitory effect of the compound on HIV-1 expression using flow cytometry. Even at 10 μM, PIMi IV did not show any inhibitory effect on HIV-1 expression in either cell line. The data for JNLG cells are shown in Fig. 2D. We have previously demonstrated that addition of Ro24-7249, a compound previously tested as an HIV-1 transcription inhibitor, causes a decrease in GFP MFI of >80% in these cells (32, 33). Again, addition of PIMi IV at the utilized concentrations did not increase cell death relative to cell death seen under control conditions in the absence of the inhibitor. Thus, PIMi IV selectively inhibits HIV-1 reactivation without affecting active HIV-1 expression.
These results suggest that the presence of PIM-1 is essential for a stimulus to trigger HIV-1 reactivation. If this is correct, overexpression of PIM-1 will facilitate reactivation as PIM-1 is an autophosphorylating protein that is regulated primarily at the level of expression. Indeed, overexpression of PIM-1 in the latently infected CA5 and EF7 T cells did not trigger HIV-1 reactivation. As gene regulation in T cells (and other cells) generally occurs within a buffered system with multiple and often redundant levels of molecular control, it is not to be expected that every manipulation of a single factor of necessity results in an immediate phenotypic effect. However, PIM-1 overexpression facilitated reactivation by a second activating stimulus, demonstrated here for the protein kinase C (PKC) agonist bryostatin, a clinically relevant HIV-1-reactivating agent that is currently in clinical trials as an anticancer compound (Fig. 2E) (34–36). Following PIM-1 overexpression, to achieve the same level of HIV-1 reactivation, the concentration requirements for bryostatin were reduced by a factor of 5, revealing that PIM-1 regulation affected the transcriptional stability of the integrated latent HIV-1 infection event. Similar effects were observed in EF7 T cells, confirming the idea that PIM-1 presence is one key requirement to trigger HIV-1 reactivation.
PIM-1 knockdown affects HIV-1 reactivation.
Conversely, knockdown of PIM-1 should increase the concentration requirement for an activating stimulus to trigger HIV-1 reactivation. To test this idea, we transduced CA5 T cells with two different PIM-1-specific shRNA vectors. A cloning and puromycin selection step was found essential as knockdown of PIM-1 affected cell growth rates, and low-level PIM-1-expressing cells were quickly overgrown in the cell culture. Consistent with the data obtained using pharmacologic PIM-1 inhibitors, shRNA10-induced PIM-1 knockdown reduced achievable reactivation levels in the various generated CA5-PIMshRNA clones. Similar results were obtained for experiments using a second PIM-1-specific shRNA22 (shPIM#22) (Fig. 2F). Control transductions with a scrambled shRNA did not show any significant changes in the TNF-α-induced HIV-1 reactivation response. The observed inhibitory effect of PIM-1 shRNA was overall less pronounced than the inhibitory effect of PIMi IV. This could be explained by clonal effects or by the reported ability of PIM-2 and PIM-3 to at least partly compensate for PIM-1 activity that is specifically targeted by the shRNA approach. In contrast, pharmacological PIM inhibitors, at the utilized concentrations, will at least partially inhibit other PIM kinases and prevent functional compensatory escape.
PIMi IV prevents HIV-1 reactivation in latently infected primary CD4 T cells.
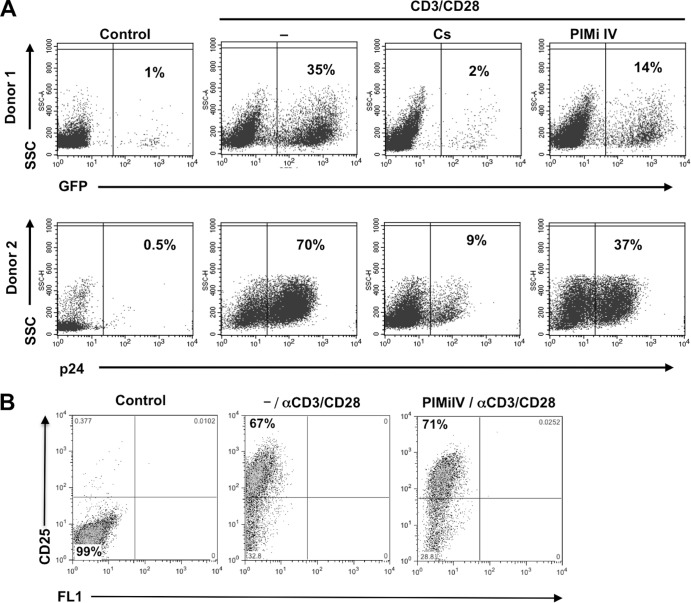
The primary goal of these studies is to provide additional evidence for the concept that a regulated kinase activity network exerts a gatekeeper function for HIV-1 latency control, and this control level even supersedes induced NF-κB activity effects. As we develop the idea of a kinase gatekeeper function for latent HIV-1 infection in T cell lines, it is informative to investigate whether even details, such as particular kinase activities, can be transferred from our T cell line models to latency control in primary CD4 T cell models. We thus tested whether PIMi IV would also inhibit reactivation of latent HIV-1 infection in a primary CD4 T cell model of HIV-1 latency. For this purpose, latently HIV-1-infected cultured central memory CD4 T cells were prepared from primary naive CD4 T cells as previously described (7, 27). Figure 3A shows the results of two independent experiments. Over active background infections at 1% and 0.5%, antibody-mediated CD3/CD28 costimulation revealed latent HIV-1 reservoirs of 35% and 70%, respectively. Cyclosporine (Cs), as a control inhibitor, abrogated CD3/CD28-mediated HIV-1 reactivation to 2% and 9%, respectively. In the presence of PIMi IV (10 μM), HIV-1 reactivation was reduced to 14% and 37%, respectively. PIMi IV at 10 μM did not affect activation-induced blast transformation (data not shown), nor did it affect upregulation of CD25 (Fig. 3B), a primary T cell activation marker (IL-2 receptor α-chain). Therefore, PIMi IV is capable of selectively reducing HIV-1 reactivation in latently infected primary CD4 T cells without affecting overall T cell activation. Following the identification of the JNK inhibitor AS601245 as an inhibitor of HIV-1 reactivation (11), PIMi IV is the second kinase inhibitor that we identified in Jurkat cell-based T cell line models that also exerts activity in primary CD4 T cell models of HIV-1 latency. This may not come as a surprise as Jurkat T cells for decades have served as one of the most reliable models for T cell signaling research.
FIG 3.
PIMi IV inhibits HIV-1 reactivation in latently HIV-1-infected primary T cells. (A) Latently HIV-1-infected cultured central memory T cells were prepared from primary naive T cells as previously described (7, 27). Active infection events were indicated by GFP fluorescence (Donor 1) or by p24 staining (Donor 2). Over low-level background infection (control), HIV-1 reactivation was triggered using a CD3/CD28 MAb combination. Cyclosporine (Cs) prevented and PIMi IV markedly inhibited CD3/CD28 MAb induced reactivation. The percentage of GFP-positive cells is indicated. (B) To test whether PIMi IV inhibits anti-CD3/CD28 MAb-mediated T cell activation, primary T cells were left untreated (control) or CD3/CD28 MAb stimulated in the absence or presence of 10 μM PIMi IV. T cell activation was determined as the induction of CD25/IL-2 receptor-α chain expression by flow cytometric analysis. The experiment is representative of results from a total of four healthy donors tested. α, anti; SSC, side scatter.
Selective effect of PIMi IV on induced cellular gene expression.
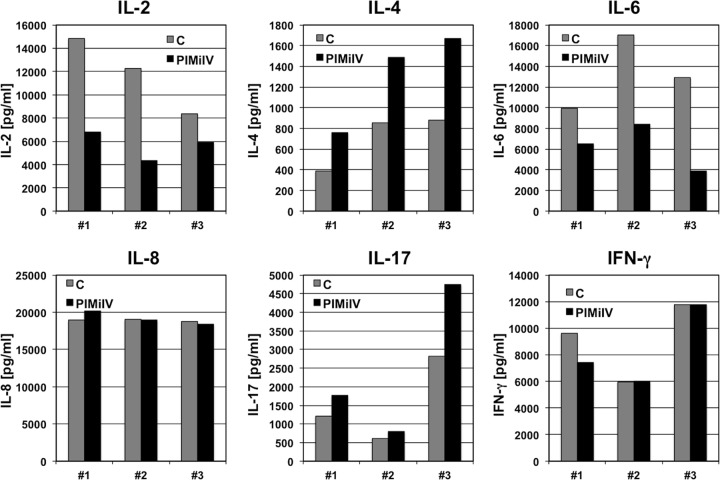
To provide additional evidence that PIMi IV specifically acts on latent HIV-1 infection, we next explored the ability of PIMi IV to regulate induced cellular gene expression. For this purpose, we stimulated peripheral blood mononuclear cells from three independent donors with PHA-L, either in the presence or the absence of PIMi IV, and determined IL-2, IL-4, IL-6, IL-8, IL-17, and IFN-γ induction. For these cytokines, we observed differences in the dynamic range of the effects between the tested donors, but all showed similar response profiles. PIMi IV inhibited IL-2 and IL-6 induction to different degrees. In contrast, the presence of PIMi IV amplified induced IL-4 and somewhat IL-17 expression. Induction of IL-8 and IFN-γ was not affected by the presence of PIMi IV (Fig. 4). Again, these data provide experimental evidence that the kinase activity targeted by PIMi IV controlled latent HIV-1 infection without impairing overall T cell function or acting as a nonspecific inhibitor of transcription. These data further imply that NF-κB activation is not affected as induction of all tested genes is NF-κB regulated. Lastly, the data suggest that PIMi IV acts as a selective transcriptional inhibitor that likely is active only in the context of a particular transcription factor binding site composition of a particular promoter.
FIG 4.
PIMi IV effects on activation-induced cytokine gene expression. In the absence (C, control) or presence of PIMi IV (10 μM), CD4 T cells from three healthy donors were stimulated with PHA-L (10 μg/ml). At 24 h poststimulation culture supernatants were harvested and analyzed for the presence of IL-2, IL-4, IL-6, IL-8, IL-17, and IFN-γ using multiplex analysis.
PIMi IV suppresses HIV-1 reactivation despite high levels of induced NF-κB activity.
All of the utilized HIV-1-reactivating stimulators, TNF-α, PMA, or bryostatin for T cell lines and PHA-L or anti-CD3/CD28 MAb combinations for primary CD4 T cells, converge in the NF-κB pathway. With the exception of AS601245, other reported inhibitors of HIV-1 reactivation exerted their inhibitory function on HIV-1 reactivation by preventing NF-κB activation (37). Our data on the selective effect of PIM IV on cytokine induction suggested that NF-κB activation and translocation may not be the target of PIMi IV activity. Also, if PIMi IV affected NF-κB translocation, active HIV-1 expression should be inhibited by PIMi IV, but this was not the case (Fig. 2D).
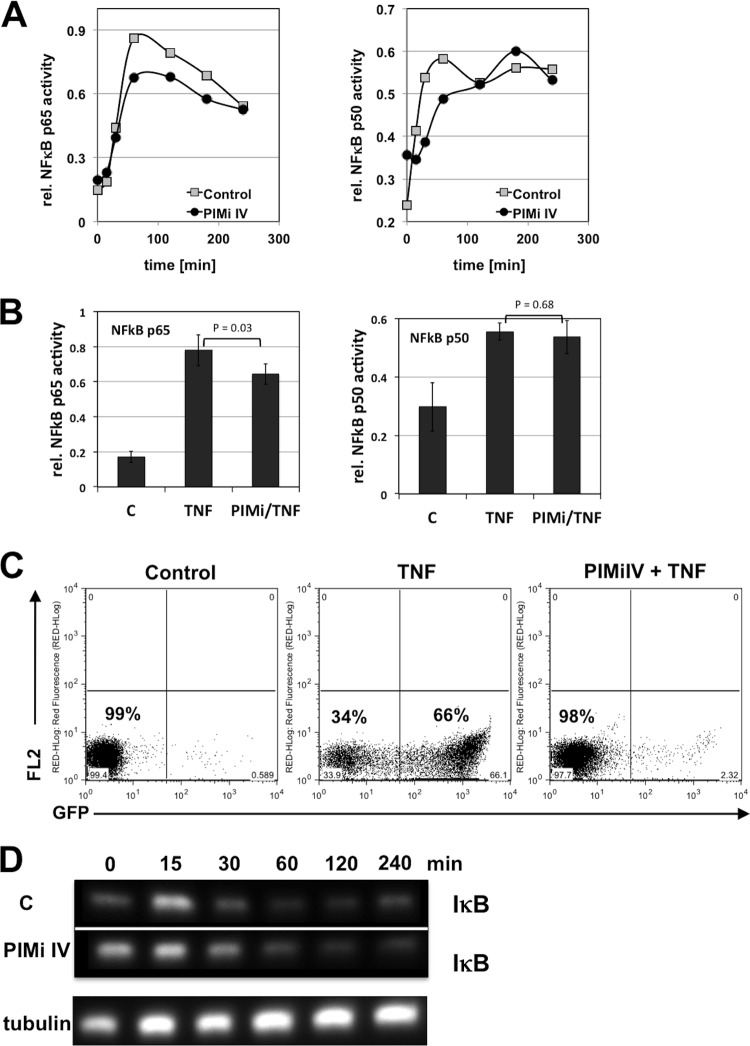
To formally demonstrate that PIMi IV prevents HIV-1 reactivation without inhibiting NF-κB activation, we stimulated the latently HIV-1-infected CA5 reporter T cells with TNF-α, either in the presence or absence of optimal inhibitory concentrations of PIMi IV (10 μM), and initially determined the kinetic NF-κB p50 and p65 activity profiles over the first 4 h of stimulation. Nuclear cell extracts from the cultures were generated at various time points for up to 4 h poststimulation, and NF-κB activity, as measured by a TransAM assay (DNA binding), was plotted over time. Possible differences in the NF-κB activation profiles were too small to account for the large inhibitory effect exerted by PIMi IV (Fig. 5A). When we compared peak NF-κB activity in the three independent experiments in the presence or absence of 10 μM PIMi IV at 60 min postactivation, we again did not detect any difference between the experimental conditions that indicated that the inhibitory effect of PIMi IV would be the result of NF-κB inhibition (Fig. 5B). In these experiments, TNF-α stimulation triggered ∼70% reactivation of latent HIV-1 infection in the control cultures, but reactivation was fully suppressed in the cultures that were treated with PIMi IV (10 μM) (Fig. 5C). In line with these data, no differences in the kinetic IκB expression profiles of TNF-α-induced control or PIMi IV treated T cells were observed during this time frame (Fig. 5D). PIMi IV thus targets a kinase activity that controls latent HIV-1 infection in the presence of high levels of NF-κB activity. The identification of a second kinase inhibitor, PIMi IV (in addition to AS601245), that prevents HIV-1 reactivation in the face of high levels of NF-κB activity confirms our recent findings that suggest a level of molecular control by a kinase network that supersedes the effect of NF-κB on latent HIV-1 infection (11).
FIG 5.
PIMi IV prevents reactivation of latent HIV-1 infection despite high levels of TNF-α-induced NF-κB activity. (A) CA5 T cells were stimulated with TNF-α (10 ng/ml) in the absence (control) or presence of PIMi IV (10 μM). Cells were harvested at the indicated time points, nuclear extracts were prepared, and NF-κB p50 and p65 activities were measured using TransAM assays. (B) Maximum initial NF-κB activation achieved in the absence or presence of PIMi IV (10 μM) at 1 h post TNF-α activation was determined in three independent experiments. The P values (Student's t test) describing the significance of possible differences between the stimulated control (C) conditions (TNF) and the PIMi IV-treated TNF-α-stimulated conditions (PIMi/TNF) are shown. (C) TNF-α-induced HIV-1 reactivation levels in CA5 T cells in the absence or presence of PIMi IV as used in the kinetic NF-κB activation experiments depicted in panel A. (D) In the absence or presence of PIMi IV, CA5 T cells were stimulated with TNF-α, and cells were harvested at the indicated time points. Western blotting was performed to determine IκB expression kinetics over 240 min. To ensure even loading of the lanes, membranes were stripped and probed for tubulin expression (shown for activated CA5 T cells treated with PIMi IV).
PIMi IV effect is dependent on the CD28RE motif of the HIV-1 LTR.
A remarkable property of PIMi IV was its differential effect on the induced expression of various cytokines. Beyond the realization that PIMi IV does not interfere with general T cell activation, these results are interesting in the context that IL-2, IL-4, IL-6, and IL-8 have all been reported to be controlled at the transcriptional level by a CD28-responsive element (CD28RE), yet functional disparity toward mitogenic stimulation for some of these promoters (IL-2, IL-6, and IL-8) and HIV-1 has been previously reported (38, 39). This raises the possibility that PIMi IV activity, which differentially acts on mitogen-induced activation of these genes, may actually be functionally linked to downstream events that interact with the CD28RE in the HIV-1 LTR (Fig. 6A). As we recently demonstrated that syngeneic virus constructs that differed in a subtype-specific manner in the region −1 to −147 relative to the transcriptional start site, which includes the CD28RE, greatly varied in their abilities to establish latent HIV-1 infection, these viral constructs provide a tool to test this hypothesis (12). Differential effects of PIMi IV on reactivation of latent infection events established with these viral constructs can link PIMi IV effects to the transcription factor binding site composition of the LTR and thus suggest that PIMi IV will affect transcription factors that interact with the respective LTR sequence.
FIG 6.
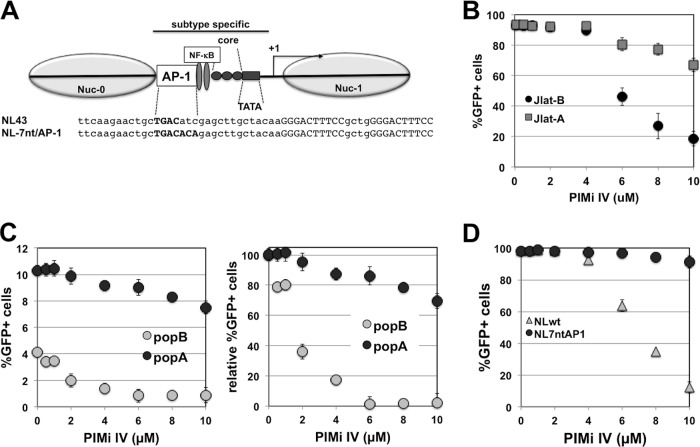
PIMi IV prevents reactivation of latent HIV-1 in an LTR sequence-dependent manner. HIV-1 LAI-B and LAI-A, two viruses that are syngeneic with the exception of the extended core/enhancer region of the LTR (from −1 to −147 nt with respect to the transcriptional start site) were used to generate latently infected T cells. (A) Schematic representation of the viral LTR indicating the extended core/enhancer region that is representative of a prototypic subtype A sequence in LAI-A and representative of a prototypic subtype B region in LAI-B. The nucleotide sequences represent the CD28RE of NL4-3 and NL-7nt/AP-1 that were used in for the experiment shown in panel D. AP-1 motifs are printed in bold capital letters, whereas NF-κB sites are indicated in capital letters only. (B) Effect of increasing amounts of PIMi IV on PMA (3 ng/ml)-induced HIV-1 reactivation of a latent LAI-B infection (Jlat-B cells) and latent LAI-A infection (Jlat-A cells). (C) Increasing concentrations of PIMi IV inhibited HIV-1 reactivation in a J2574 reporter T cell population holding ∼4% latently LAI-B infected cells (popB) but had only a minor inhibitory effect on HIV-1 reactivation in a J2574 reporter T cell population holding ∼10% latent LAI-A infection events (popA; left panel). For better comparison of the inhibitory effect of PIMi IV on the latent LAI-A and LAI-B infections in the cell populations, results were normalized to maximum achievable reactivation levels and plotted as relative level of reactivation, normalized for active background infection (0.8% for LAI-B; 1.1% for LAI-A) (right panel). (D) Using NL-7nt/AP-1, a virus that is altered in 2 nucleotides relative to NL4-3 wt to provide a subtype A prototypic AP-1 site in the CD28RE, we generated a latently infected J2574 reporter T cell clone. PIMi IV could not inhibit PMA-induced reactivation of latent NL-7nt/AP-1 infection, while it efficiently inhibited HIV-1 reactivation of latent HIV-1 NL4-3 wt infection (NL wt). All results represent the means ± standard deviations of three independent experiments.
We thus generated a panel of latently infected J2574 reporter T cells using some of these previously used HIV LAI-based viral vectors. HIV LAI-A is a viral construct in which the region −1 to −147 relative to the transcriptional start site of the parental HIV LAI (subtype B; for clarity referred to as LAI-B) was replaced by the corresponding region of a prototypic subtype A virus (40). In our experimental models, this virus established up to 5-fold higher levels of latent infection (12). The generated latently infected reporter T cell clones are henceforth referred to as Jlat-B or Jlat-A, respectively. All latent infection events in the selected T cell clones were fully reactivatable by NF-κB-activating compounds (PMA, prostratin, and TNF-α), but as in other latently infected T cells that we previously established, latent infection was refractory to treatment with histone deacetylase inhibitors (NaBu, trichostatin A, or valproic acid) (41, 42). The cell-differentiating agent hexamethylene bisacetamide (HMBA) triggered some level of HIV-1 reactivation, as did the bimodal agent SAHA/vorinostat, which acts as a cell-differentiating agent and as an HDAC inhibitor (data not shown) (41, 42).
When PIMi IV was titrated on several latently infected Jlat-B and Jlat-A clones prior to stimulation with PMA, PIMi IV inhibited reactivation of latent LAI-B infection but exerted only a marginal inhibitory effect on reactivation of latent HIV LAI-A infection (Fig. 6B). To ensure that the observed failure of PIMi IV to inhibit LAI-A reactivation was not due to some unidentified clonal effects, we next tested the inhibitory effect of PIMi IV on PMA-induced reactivation in populations of J2574 T cells either latently LAI-B infected or polyclonally infected with LAI-A. These experiments confirmed our results from the experiments in clonal T cell lines as PIMi IV inhibited HIV-1 reactivation in the latently LAI-B-infected J2574 T cell population but not in the latently LAI-A-infected T cell population (Fig. 6C).
As LAI-A and LAI-B are syngeneic with the exception of the extended core/enhancer promoter region from −1 to −147, we focused on this region to investigate whether a specific transcription factor binding motif would be responsible for this phenotype. Using a series of viruses with targeted LTR mutations that were used to establish latently infected T cells, we narrowed down the LTR region that is important for the inhibitory effect of PIMi IV to the 25 nucleotides (nt) upstream of the NF-κB element (12). This is the same region that we found to govern HIV-1 latency establishment and which holds the AP-1 motif of the CD28RE responsible for this effect (12). To test whether the AP-1 site sequence would be responsible for the selective effect of PIMi IV on reactivation, we used an NL4-3 virus in which we had mutated 2 of 3 nucleotides downstream of the 4-nt AP-1 site to generate the subtype A-specific 7-nt AP-1 site (NL-7nt/AP-1). Other than the two nucleotides, the NL4-3 wild type (wt) and the resulting NL-7nt/AP-1 were syngeneic, including the sequence of the NF-κB element (Fig. 6A). Moreover, the 2-nucleotide mutation did not attenuate the ability of NL-7nt/AP-1 to drive expression or viral replication (12; also data not shown). Using NL4-3 wt and NL-7nt/AP-1, we again generated latently infected T cells using J2574 reporter T cells. In the resulting T cell clones, no differences in responses to stimulation with PMA were observed. As shown in Fig. 6D, PIMi IV prevented reactivation of latent HIV-1 NL4-3 wt infection in J2574 cells, but PIMi IV had no tangible inhibitory effect on PMA-induced reactivation of latent NL-7nt/AP-1 infection. To the best of our knowledge, this is the first time that the activity of a kinase inhibitor that prevents HIV-1 reactivation can be functionally correlated to a specific transcription factor binding motif in the HIV-1 LTR and provides additional support for the idea that latent HIV-1 infection is a transcription factor restriction phenomenon. This selectivity of PIMi IV to a specific LTR sequence motif is similar to the observed selectivity of the inhibitor for various cytokine promoters, where PIMi IV could act as an activator, an inhibitor, or without any effect on induced gene expression (Fig. 4).
It is important to appreciate that while we refer to an AP-1 motif and have previously provided experimental evidence that AP-1 factor binding affinity is altered by these mutations (12), the respective LTR region is also targeted by other transcription factors. Among others, we have previously described a MARE half-site that overlaps with this sequence and to which c-Maf can bind (43). Thus, while these data link the PIMi IV effect to the LTR nucleotide sequence, we have yet to identify the actual transcription factor(s) that act downstream of PIM-1.
DISCUSSION
Eradication of the latent viral reservoir will be an essential component of a curative therapy for HIV-1 infection. The identification of a means to safely trigger system-wide reactivation of latent infection events is considered the crucial first step to achieve this goal. A complete and detailed understanding of the different levels of molecular control that govern latent HIV-1 infection will be essential to develop such therapeutic strategies. To this end, we have recently added to the list of molecular mechanisms controlling latent HIV-1 infection when we demonstrated that kinase control mechanisms suppress HIV-1 reactivation despite high levels of induced NF-κB activity (11). Since 2000, about 20 drugs targeting kinases have been FDA approved for a variety of diseases (for a review, see reference 44), and the number of kinase-targeting drugs in the industry pipeline is rapidly growing. A gatekeeper kinase network that controls latent HIV-1 infection should thus be an attractive druggable target to trigger HIV-1 reactivation.
Here, we expand the concept that kinase control is a crucial part of HIV-1 latency control by demonstrating that latently HIV-1-infected T cells exhibit an altered baseline kinase activity profile relative to noninfected T cells and that some of these altered kinases, as exemplified by PIM-1, can be pharmacologically or genetically targeted to alter HIV-1 latency control.
Availability of PIM-1, which by kinome profiling was identified as the top altered kinase in latently infected cells, was found to be a prerequisite to trigger latent HIV-1 infection. The role of PIM-1 in HIV-1 reactivation was confirmed using pharmacologic inhibitors, shRNA-induced knockdown, and PIM-1 overexpression. The finding was confirmed in primary CD4 T cells, where PIMi IV inhibited CD3/CD28-induced reactivation of latent HIV-1 infection.
PIM-1 is an autophosphorylating serine/threonine kinase that is primarily regulated at the protein expression level. Its expression has been reported to be regulated by cytokines such as IL-2, IL-3, IL-5, IL-6, IL-7, IL-12, IL-15, TNF-α, epidermal growth factor (EGF), and IFN-γ (reviewed in reference 45). While PIM-1 is often overexpressed in immortalized cell lines, PIM-1 is not expressed in resting primary T cells, but its expression is rapidly induced after receptor cross-linking with anti-CD3 MAbs (46). Once induced, PIM-1 has been described to phosphorylate NF-κB RelA/p65 at Ser276, thereby preventing NF-κB's ubiquitin-mediated proteolysis (47). PIM-1 has also been described to physically interact with NFATc1 and to phosphorylate NFATc1 in vitro on several serine residues (48). PIM-1 was found to enhance NFATc1-dependent transactivation and IL-2 production in Jurkat T cells, while kinase-deficient PIM-1 mutants acted as dominant negative inhibitors. NFAT, in turn, has been described early on to interact with the HIV-1 LTR (49–51) and has been shown to augment LTR transcription via binding to the dual proximal NF-κB sites (43, 52–54). NFAT further has been reported to be required for viral reactivation from latency in primary T cells (7). How PIM-1 exactly acts to control HIV-1 reactivation at the transcription factor level remains to be elucidated.
Beyond the specific effect of PIMi IV on HIV-1 reactivation, our findings have implications for our understanding of latent HIV-1 infection. First, following our recent report that the JNK inhibitor AS601245 prevents reactivation of latent HIV-1 infection despite the efficient induction of NF-κB activity, we identified PIMi IV as a second kinase inhibitor that is capable of preventing HIV-1 reactivation by superseding the effect of NF-κB activity on latent HIV-1 infection. The data thus expand the concept that a kinase network is a major component of HIV-1 latency control.
The second conclusion concerns the question at what molecular level PIM-1 kinase exerts its control activity on latent HIV-1 infection. Kinases could affect many molecular mechanisms suggested to control latent HIV-1 infection. Kinase inhibitors may interfere with processes involved in histone/chromatin modifications reported to be essential for HIV-1 latency or alter the availability/activity of downstream transcription factors that are essential for HIV-1 reactivation. The functional correlation between the inhibitory activity of PIMi IV and the sequence of the AP-1 motif in the CD28RE of the LTR suggests that the gatekeeper kinase network likely exerts its downstream control of latent HIV-1 infection through the latter mechanism.
To explain that the 2-nt change in the LTR of NL-7nt/AP-1, which deprives PIMi IV of its inhibitory effect on HIV-1 reactivation, would interfere with mechanisms that affect histone modifications, nucleosome formation, nucleosome repositioning, or chromatin structure at the latent LTR, one would have to assume that regulatory mechanisms involving histone or chromatin modifications act fundamentally differently on latent NL4-3 wt infection than on NL7nt/AP-1 infection based on a 2-nt mutation that was derived from a prototypic HIV-1 subtype A LTR sequence. By extension, this would mean that the principal mechanisms governing HIV-1 latency would change as a function of the LTR nucleotide sequence. Given the uniform establishment of latent HIV-1 reservoirs in all patients tested to date, on one hand, and, on the other hand, the sequence diversity of HIV-1 LTRs, this seems unlikely.
The same considerations hold for a possible effect of PIMi IV on components of the paused RNAP II machinery at the latent LTR and its transition into active elongation following stimulation. RNAP II complex formation, P-TEFb release from its inactive complex with HEXIM-1, and availability of general transcription factors such as TFIIH could only be the target of PIMi IV when it is assumed that latent NL4-3 wt infection at the level of RNAP II pausing, release, or elongation is regulated in a fundamentally different manner than NL-7nt/AP-1 latency. To this end it is important that even the TATA boxes, the transactivation response (TAR) elements, and the polyadenylation signals in the NL4-3 and the NL-7nt/AP-1 LTRs are identical. Thus, while effects on histone composition, chromatin alterations, or RNAP II pausing are not excluded by our data, the most likely explanation of our findings should be that PIMi IV or, for that matter, changes in PIM expression by PIM-1 overexpression or knockdown affect the availability of transcription factors that bind to the AP-1 motif in the HIV-1 LTRs. There are different possibilities of how this could be achieved. One possibility is that PIMi IV may act by altering the available transcription factor composition so as to favor binding of alternative transcription factors to the 7nt/AP-1 site but not the wt AP-1 motif (Fig. 6A). However, it is more likely, based on our data, that PIMi IV may simply incompletely inhibit the activation or availability of one specific transcription factor. In this situation, higher binding affinity of the 7nt/AP-1 motif for the residual transcription factor activity would be sufficient to allow for reactivation of latent NL-7nt/AP-1 infection, but not for latent NL4-3 wt infection.
While we demonstrate that there are differences in the kinase activity profiles of uninfected and latently infected T cells and that these findings can be transferred to latently HIV-1-infected primary T cells, it remains unclear at this time why exactly these kinases are altered. It is conceivable that the observed phenotypic changes of the kinome profile are reflective of a cellular antiviral response program or that they are part of a viral program that alters cells to favor viral replication. Phenotypic (epigenetic) changes of host cells following infection or even just exposure to viruses has been recently reported in different systems (55). Specifically, for latent HIV-1 infection, a recent paper provides evidence that CD2 expression levels could be one in vivo biomarker of latent HIV-1 infection (56). Changes in the kinome profile would be the intracellular reflection of such protein expression changes.
In summary, the data thus confirm the presence of a gatekeeper kinase network that controls latent HIV-1 infection in T cells and provide experimental evidence that control is achieved at the level of restriction of specific transcription factor engagement of the HIV-1 LTR. As kinase control of latent HIV-1 infection supersedes NF-κB activity and as the data reveal that latently infected T cells phenotypically differ from uninfected T cells, our results suggest that by targeting the relevant kinase control mechanisms, it may be possible to dissociate HIV-1 reactivation from the activation of key cytokines that are particularly harmful for patients (e.g., TNF-α).
Beyond the molecular biology, the immediately apparent link between kinome analysis data and pharmacological or genetic perturbation data suggests that kinome profiling, which is now an established tool in cancer research, can also become a powerful tool to help identify the protein-protein interactions that control HIV-1 latency and guide the development of novel targeted intervention strategies. In this setting, as we begin to better understand the underlying interactions of HIV-1 latency control, kinase antagonist or agonists that can act to transition latent HIV-1 infection into an active expression state will become an important part of future effective viral eradication strategies.
ACKNOWLEDGMENTS
This work was funded in part by NIH grant R01AI064012 and NIH R56 R01AI077457 to O.K. Takao Shishido contributed to this research at the University of Alabama at Birmingham as a visiting scientist from Shionogi & Co., Ltd., Japan. Parts of the work were made possible by funding from the Alabama Drug Discovery Alliance and the UAB Center for Clinical and Translational Science, grant number UL1TR000165 from the National Center for Advancing Translational Sciences and National Center for Research Resources component of the National Institutes of Health to O.K. The work was further supported in part by NIH grant AI087508 to V.P. Some of the experiments were performed in the UAB Center for AIDS Research (CFAR) biosafety level 3 facilities and by the UAB CFAR Flow Cytometry Core/Joint UAB Flow Cytometry Core, which are funded in part by NIH/NIAID P30 AI027767 and by NIH 5P30 AR048311.
Kinome profiling was made possible through the UAB Kinome Core.
Footnotes
Published ahead of print 23 October 2013
REFERENCES
- 1.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366:549–555. 10.1016/S0140-6736(05)67098-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archin NM, Cheema M, Parker D, Wiegand A, Bosch RJ, Coffin JM, Eron J, Cohen M, Margolis DM. 2010. Antiretroviral intensification and valproic acid lack sustained effect on residual HIV-1 viremia or resting CD4+ cell infection. PLoS One 5:e9390. 10.1371/journal.pone.0009390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archin NM, Eron JJ, Palmer S, Hartmann-Duff A, Martinson JA, Wiegand A, Bandarenko N, Schmitz JL, Bosch RJ, Landay AL, Coffin JM, Margolis DM. 2008. Valproic acid without intensified antiviral therapy has limited impact on persistent HIV infection of resting CD4+ T cells. AIDS 22:1131–1135. 10.1097/QAD.0b013e3282fd6df4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. 10.1038/nature11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blazkova J, Chun TW, Belay BW, Murray D, Justement JS, Funk EK, Nelson A, Hallahan CW, Moir S, Wender PA, Fauci AS. 2012. Effect of histone deacetylase inhibitors on HIV production in latently infected, resting CD4+ T cells from infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 206:765–769. 10.1093/infdis/jis412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang HC, Xing S, Shan L, O'Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119:3473–3486. 10.1172/JCI39199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65. 10.1182/blood-2008-07-168393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duverger A, Jones J, May J, Bibollet-Ruche F, Wagner FA, Cron RQ, Kutsch O. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83:3078–3093. 10.1128/JVI.02058-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan L, Xing S, Yang HC, Zhang H, Margolick JB, Siliciano RF. 1 September 2013. Unique characteristics of histone deacetylase inhibitors in reactivation of latent HIV-1 in Bcl-2-transduced primary resting CD4+ T cells. J. Antimicrob. Chemother. 10.1093/jac/dkt338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501. 10.1016/j.immuni.2012.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolschendorf F, Bosque A, Shishido T, Duverger A, Jones J, Planelles V, Kutsch O. 2012. Kinase control prevents HIV-1 reactivation in spite of high levels of induced NF-κB activity. J. Virol. 86:4548–4558. 10.1128/JVI.06726-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duverger A, Wolschendorf F, Zhang M, Wagner F, Hatcher B, Jones J, Cron RQ, van der Sluis RM, Jeeninga RE, Berkhout B, Kutsch O. 2013. An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J. Virol. 87:2264–2277. 10.1128/JVI.01594-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunvand MW, Krumm A, Groudine M. 1993. In vivo footprinting of the human IL-2 gene reveals a nuclear factor bound to the transcription start site in T cells. Nucleic Acids Res. 21:4824–4829. 10.1093/nar/21.20.4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339:950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. 2013. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol. Cell 50:212–222. 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. 2010. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143:540–551. 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klatt A, Zhang Z, Kalantari P, Hankey PA, Gilmour DS, Henderson AJ. 2008. The receptor tyrosine kinase RON represses HIV-1 transcription by targeting RNA polymerase II processivity. J. Immunol. 180:1670–1677 [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Klatt A, Gilmour DS, Henderson AJ. 2007. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 282:16981–16988. 10.1074/jbc.M610688200 [DOI] [PubMed] [Google Scholar]
- 19.Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. 2006. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25:3596–3604. 10.1038/sj.emboj.7601248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi M, Karn J. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985–4995. 10.1038/sj.emboj.7601928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9:e1001206. 10.1371/journal.pbio.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang C, Pugh BF. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161–172. 10.1038/nrg2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai L, Morozov AV. 2010. Gene regulation by nucleosome positioning. Trends Genet. 26:476–483. 10.1016/j.tig.2010.08.003 [DOI] [PubMed] [Google Scholar]
- 24.Shishido T, Wolschendorf F, Duverger A, Wagner F, Kappes J, Jones J, Kutsch O. 2012. Selected drugs with reported secondary cell-differentiating capacity prime latent HIV-1 infection for reactivation. J. Virol. 86:9055–9069. 10.1128/JVI.00793-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutsch O, Benveniste EN, Shaw GM, Levy DN. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76:8776–8786. 10.1128/JVI.76.17.8776-8786.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones J, Rodgers J, Heil M, May J, White L, Maddry JA, Fletcher TM, III, Shaw GM, Hartman JL, IV, Kutsch O. 2007. High throughput drug screening for human immunodeficiency virus type 1 reactivating compounds. Assay Drug Dev. Technol. 5:181–189. 10.1089/adt.2006.040 [DOI] [PubMed] [Google Scholar]
- 27.Bosque A, Planelles V. 2011. Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods 53:54–61. 10.1016/j.ymeth.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breuer D, Kotelkin A, Ammosova T, Kumari N, Ivanov A, Ilatovskiy AV, Beullens M, Roane PR, Bollen M, Petukhov MG, Kashanchi F, Nekhai S. 2012. CDK2 regulates HIV-1 transcription by phosphorylation of CDK9 on serine 90. Retrovirology 9:94. 10.1186/1742-4690-9-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guendel I, Agbottah ET, Kehn-Hall K, Kashanchi F. 2010. Inhibition of human immunodeficiency virus type-1 by cdk inhibitors. AIDS Res. Ther. 7:7. 10.1186/1742-6405-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. 2006. Phosphorylation of HIV-1 Tat by CDK2 in HIV-1 transcription. Retrovirology 3:78. 10.1186/1742-4690-3-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce AC, Jacobs M, Stuver-Moody C. 2008. Docking study yields four novel inhibitors of the protooncogene Pim-1 kinase. J. Med. Chem. 51:1972–1975. 10.1021/jm701248t [DOI] [PubMed] [Google Scholar]
- 32.Kutsch O, Levy DN, Bates PJ, Decker J, Kosloff BR, Shaw GM, Priebe W, Benveniste EN. 2004. Bis-anthracycline antibiotics inhibit human immunodeficiency virus type 1 transcription. Antimicrob. Agents Chemother. 48:1652–1663. 10.1128/AAC.48.5.1652-1663.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kempf MC, Jones J, Heil ML, Kutsch O. 2006. A high-throughput drug screening system for HIV-1 transcription inhibitors. J. Biomol. Screen 11:807–815. 10.1177/1087057106290292 [DOI] [PubMed] [Google Scholar]
- 34.Kinter AL, Poli G, Maury W, Folks TM, Fauci AS. 1990. Direct and cytokine-mediated activation of protein kinase C induces human immunodeficiency virus expression in chronically infected promonocytic cells. J. Virol. 64:4306–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeChristopher BA, Loy BA, Marsden MD, Schrier AJ, Zack JA, Wender PA. 2012. Designed, synthetically accessible bryostatin analogues potently induce activation of latent HIV reservoirs in vitro. Nat. Chem. 4:705–710. 10.1038/nchem.1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beans EJ, Fournogerakis D, Gauntlett C, Heumann LV, Kramer R, Marsden MD, Murray D, Chun TW, Zack JA, Wender PA. 2013. Highly potent, synthetically accessible prostratin analogs induce latent HIV expression in vitro and ex vivo. Proc. Natl. Acad. Sci. U. S. A. 110:11698–11703. 10.1073/pnas.1302634110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Chen Y, Gabuzda D. 1999. ERK MAP kinase links cytokine signals to activation of latent HIV-1 infection by stimulating a cooperative interaction of AP-1 and NF-κB. J. Biol. Chem. 274:27981–27988. 10.1074/jbc.274.39.27981 [DOI] [PubMed] [Google Scholar]
- 38.Li-Weber M, Giasi M, Krammer PH. 1998. Involvement of Jun and Rel proteins in up-regulation of interleukin-4 gene activity by the T cell accessory molecule CD28. J. Biol. Chem. 273:32460–32466. 10.1074/jbc.273.49.32460 [DOI] [PubMed] [Google Scholar]
- 39.Civil A, Rensink I, Aarden LA, Verweij CL. 1999. Functional disparity of distinct CD28 response elements toward mitogenic responses. J. Biol. Chem. 274:34369–34374. 10.1074/jbc.274.48.34369 [DOI] [PubMed] [Google Scholar]
- 40.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740–3751. 10.1128/JVI.74.8.3740-3751.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richon VM, Emiliani S, Verdin E, Webb Y, Breslow R, Rifkind RA, Marks PA. 1998. A class of hybrid polar inducers of transformed cell differentiation inhibits histone deacetylases. Proc. Natl. Acad. Sci. U. S. A. 95:3003–3007. 10.1073/pnas.95.6.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richon VM, Webb Y, Merger R, Sheppard T, Jursic B, Ngo L, Civoli F, Breslow R, Rifkind RA, Marks PA. 1996. Second generation hybrid polar compounds are potent inducers of transformed cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 93:5705–5708. 10.1073/pnas.93.12.5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Clausell A, Robinson T, Yin J, Chen E, Johnson L, Weiss G, Sabbaj S, Lowe RM, Wagner FH, Goepfert PA, Kutsch O, Cron RQ. 2012. Host factor transcriptional regulation contributes to preferential expression of HIV type 1 in IL-4-producing CD4 T cells. J. Immunol. 189:2746–2757. 10.4049/jimmunol.1103129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dar AC, Shokat KM. 2011. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu. Rev. Biochem. 80:769–795. 10.1146/annurev-biochem-090308-173656 [DOI] [PubMed] [Google Scholar]
- 45.Bachmann M, Moroy T. 2005. The serine/threonine kinase Pim-1. Int. J. Biochem. Cell Biol. 37:726–730. 10.1016/j.biocel.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 46.Wingett D, Long A, Kelleher D, Magnuson NS. 1996. Pim-1 proto-oncogene expression in anti-CD3-mediated T cell activation is associated with protein kinase C activation and is independent of Raf-1. J. Immunol. 156:549–557 [PubMed] [Google Scholar]
- 47.Nihira K, Ando Y, Yamaguchi T, Kagami Y, Miki Y, Yoshida K. 2010. Pim-1 controls NF-κB signalling by stabilizing RelA/p65. Cell Death Differ. 17:689–698. 10.1038/cdd.2009.174 [DOI] [PubMed] [Google Scholar]
- 48.Rainio EM, Sandholm J, Koskinen PJ. 2002. Cutting edge: transcriptional activity of NFATc1 is enhanced by the Pim-1 kinase. J. Immunol. 168:1524–1527 [DOI] [PubMed] [Google Scholar]
- 49.Li C, Lai CF, Sigman DS, Gaynor RB. 1991. Cloning of a cellular factor, interleukin binding factor, that binds to NFAT-like motifs in the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. U. S. A. 88:7739–7743. 10.1073/pnas.88.17.7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt A, Hennighausen L, Siebenlist U. 1990. Inducible nuclear factor binding to the kappa B elements of the human immunodeficiency virus enhancer in T cells can be blocked by cyclosporin A in a signal-dependent manner. J. Virol. 64:4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. 1988. Identification of a putative regulator of early T cell activation genes. Science 241:202–205. 10.1126/science.3260404 [DOI] [PubMed] [Google Scholar]
- 52.Pessler F, Cron RQ. 2004. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 5:158–167. 10.1038/sj.gene.6364047 [DOI] [PubMed] [Google Scholar]
- 53.Cron RQ, Bartz SR, Clausell A, Bort SJ, Klebanoff SJ, Lewis DB. 2000. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 94:179–191. 10.1006/clim.1999.4831 [DOI] [PubMed] [Google Scholar]
- 54.Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. 1997. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6:235–244. 10.1016/S1074-7613(00)80326-X [DOI] [PubMed] [Google Scholar]
- 55.Ahangarani RR, Janssens W, Carlier V, Vanderelst L, Vandendriessche T, Chuah M, Jacquemin M, Saint-Remy JM. 2011. Retroviral vectors induce epigenetic chromatin modifications and IL-10 production in transduced B cells via activation of toll-like receptor 2. Mol. Ther. 19:711–722. 10.1038/mt.2010.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iglesias-Ussel M, Vandergeeten C, Marchionni L, Chomont N, Romerio F. 2013. High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J. Virol. 87:9148–9158. 10.1128/JVI.01297-13 [DOI] [PMC free article] [PubMed] [Google Scholar]