Abstract
In this Review, we summarize some of the recent work in emerging computational imaging, sensing and diagnostics techniques, along with some of the complementary non-computational modalities that can potentially transform the delivery of health care globally. As computational resources are becoming more and more powerful, while also getting cheaper and more widely available, traditional imaging, sensing and diagnostic tools will continue to experience a revolution through simplification of their designs, making them compact, light-weight, cost-effective, and yet quite powerful in terms of their performance when compared to their bench-top counterparts.
Introduction
Consumer electronics industry has been experiencing a remarkable revolution, bringing high-performance computers and various tele-communication devices to users at low cost in very compact forms. This digital era has also facilitated various emerging opportunities for the development of advanced computational imaging and sensing platforms. Compared to traditional designs, these computational schemes can decrease the complexity of optical hardware, which can be compensated in the digital domain by use of novel theories and numerical algorithms. This reduction in complexity of components can also lead to light-weight and cost-effective biomedical imagers and sensors. Toward this end, there has been considerable effort to develop computational techniques in various research areas, including super-resolution microscopy[1], holographic microscopy[2**,3**,4*], fluorescence microscopy[5,6–8*], optical coherence tomography[9], endoscopy[10], spectroscopy[11–13], integral imaging[14–15], time-coded imaging[16*], giga-pixel imaging[17**,18,19], as well as magnetic resonance imaging[20*].
In parallel to these advancements in computational imaging approaches, there has been growing interest in portable and cost-effective biomedical technologies to be used as point of care (POC) diagnostic devices that can potentially improve and reform healthcare delivery in both the developed and developing countries. For this ambitious goal, the development of affordable medical testing/measurement equipment is essential since patients might not have routine access to advanced medical laboratory infrastructure in low resource settings. In developed countries, however, even if such resources are readily available, overall cost of these medical tests and diagnostic tools might become an obstacle for some patients, especially in under-represented communities. Therefore, computational imaging and sensing technologies, with their simplified and cost-effective device architectures, hold promise as field-deployable diagnostic devices for both the developed and the developing parts of the world.
To provide an overview of global health related imaging, sensing and diagnostic tools, here we review various computational imaging and sensing platforms along with some of the complementary efforts that are based on non-computational, more traditional approaches tailored for field use and/or telemedicine applications. Starting with the next section, we will provide a summary of these emerging medical diagnosis and telepathology concepts that might fundamentally impact health care delivery across the globe by combining various analog and digital resources/tools.
Computational Imaging for Global Health Applications
Optical imaging has been serving as a well-established tool in biomedical research and clinical diagnostics for several decades. Particularly over the last two decades optical microscopy has experienced a fascinating renaissance, with various fundamental advances made in spatial resolution, depth of field (DOF), field of view (FOV), imaging speed as well as effective numerical aperture (NA) of optical microscopes. However, despite these rapid advances, the basic design of conventional microscopes that are used in clinical settings has not changed much, where they still heavily rely on bulky imaging optics and costly components, limiting their use to relatively advanced laboratory settings. Therefore, there is an unmet need for field deployable and cost-effective microscopic imagers, especially for telemedicine applications.
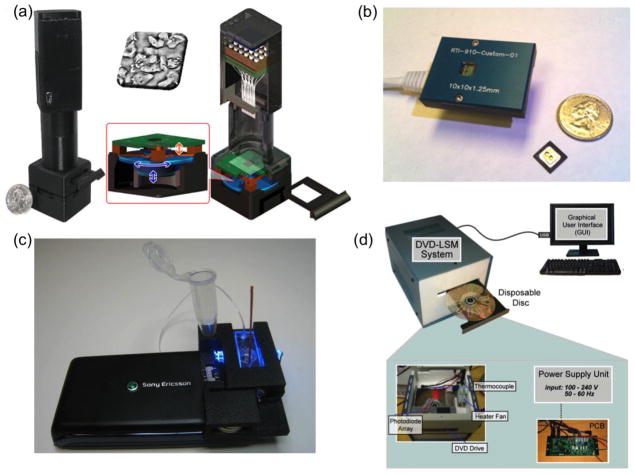
To provide solutions to this important need, there has been extensive research on the use of computational approaches to develop field-portable and cost-effective imaging tools. Along these lines, computational microscopy based on lensfree digital in-line holography[3, 21] has become an emerging technique that can provide lightweight and compact imaging devices (see Figure 1a), making them ideal for field use. These lensfree microscopes can simplify the design of optical imaging by eliminating the need for bulky and costly components such as objective lenses. Instead, computational methods are used to compensate for the lack of complexity through the help of digital reconstruction algorithms. In the holographic on-chip microscope design shown in Figure 1a, the samples are placed directly on the top of an optoelectronic sensor array (e.g., a complementary metal–oxide–semiconductor (CMOS) chip) with an object to sensor distance of e.g., 1–5 mm, and are illuminated with a partially coherent light source, created by e.g., a simple light emitting diode (LED). This illumination is then scattered from the micro-objects to coherently interfere with the unperturbed background light, creating in-line holograms of the objects at the detector plane. Holographic reconstruction and pixel super-resolution approaches are used to partially undo the effects of diffraction and spatial undersampling occurring due to the lensfree operation and unit magnification, which can then be used to reconstruct 3D microscopic images of the specimens. At the core of this reconstruction process lies an iterative phase-recovery algorithm,[22] which is used to retrieve the 2D optical phase information that is lost during the intensity recording process at the detector plane. In addition to iterative phase-recovery, another important computational block involves pixel super-resolution (SR) [23–26,27**] that combines subpixel shifted lensfree holograms of the same object to digitally synthesize a higher resolution hologram, which is especially important to mitigate undersampling related artifacts in reconstructed holographic images. The required sub-pixel shifts for this design (Fig. 1a) is achieved through an array of LEDs that are each butt-coupled to a multi-mode fiber-optic cable with a core-diameter of ~0.1 mm. Together with its compact and light-weight imaging design, such a lensfree on-chip microscope also decouples the sample FOV from spatial resolution, providing computational microscopes that can monitor large FOVs (e.g., 24–30 mm2) without a trade-off in spatial resolution, generating for example >1 billion useful pixels in both the phase and amplitude images of the objects. The performance of these lensfree computational microscopes has been tested using e.g., blood smears[21], water-borne parasites[24], pap smears[25], viral particles[27**] and other biological specimen. Through the use of sequential LED illumination (i.e., comprising blue, green and red wavelengths), lensfree color imaging [26] has also been implemented, providing color information of the samples, while also mitigating rainbow like color artifacts that are commonly observed in holographic microscopy. Recent results using these computational holographic microscopes achieved high numerical apertures (e.g., ~ 0.9) across very large FOVs of e.g., ~20 mm2 [3] while also permitting the detection of sub-100 nm particles or viruses across the same FOV [27**]. With these rapidly improving performance metrics, these holographic on-chip microscopes could be useful, providing general-purpose wide-field microscopy and nanoscopy tools for especially POC offices and telemedicine applications.
Figure 1.
Computational imaging modalities for point of care applications. (a) Lensfree computational microscopy platform based on pixel super-resolved digital in-line holography. (b) Lensless contact imaging device. (c) Computational fluorescent imaging and cytometry on a cellphone. (d) Digital microscopy on a computer drive. Reprinted from refs. 28, 32, 38, and 34 with permission from RSC publishing (a, d), PLoS One (b), ACS publishing (c), respectively.
Another emerging lensless computational imaging approach is based on contact microscopy configuration[28] (see Figure 1b), where the samples of interest are brought almost in contact (i.e., ideally less than 1 μm distance) with the active area of the optoelectronic sensor array (e.g., a CMOS imager chip). Illuminated by an incoherent light source, these on-chip samples create shadow/diffraction patterns at the detector plane, providing 2D representations of the objects. However, these shadow patterns are also under-sampled due to the physical pixel size of the detector array, limiting the resolution of contact microscopy. There have been two main stream computational approaches employed to create higher resolution images in this contact microscopy configuration: (1) an optofluidic imaging approach[28]; and (2) a pixel SR method[29, 30]. For the first approach, an object flowing through a microfluidic channel is imaged through a tilted array of submicron metallic apertures fabricated on the image sensor, creating finely sampled shadows of the specimen as it flows through the channel. These finely sampled shadows are then used to reconstruct the 2D structure of the objects with a resolution that is better than the pixel size at the CMOS chip. Note that holographic implementations of the same opto-fluidic imaging scheme have also been reported, where the distance between the sample and the sensor-active area can be significantly increased while also achieving sub-pixel resolution and tomographic imaging on a chip[31, 32]. In the second approach, following the introduction of source shifting based pixel super-resolution in holographic on-chip imaging[29], subpixel shifted images of the same specimen are digitally combined to mitigate undersampling artifacts, creating higher resolution contact images of the samples. Being a 2D imaging modality, 3D samples (such as non-adherent cells) introduce spatial artifacts in contact microscopy, which cannot yet produce tomographic images[32,33] of microscopic samples. Similar to holographic on chip microscopy techniques discussed earlier, contact microscopy is also limited to transparent objects, and therefore both of these transmission on-chip imaging modalities would not be suitable for opaque specimen, unless a specific reflection imaging design is implemented[34].
Another emerging trend in the development of computational imaging platforms is to implement various micro-analysis and measurement techniques/designs on consumer grade electronic devices such as cellphones[35], computer drives[36*], as well as flatbed scanners[37,38,39*]. For this end, a digital microscopy and cytometry platform has been demonstrated through the use of an opto-mechanical attachment installed on a consumer grade cellphone (see Figure 1c). This cellphone based microscope[35, 40, 41*], achieving either static imaging or flow based video-rate imaging, can monitor fluorescently labeled cells or micro-objects over a wide FOV of up to 81 mm2 with a spatial resolution of ~10 μm, which is further improved to ~2 μm over a smaller FOV of ~1–2 mm2. In this design, compressive decoding of acquired raw images is the key to boost the spatial resolution of this fluorescent imager installed on the cellphone. As another example, a computational microscopy approach has also been implemented on a consumer grade DVD driver[36*] by modifying the standard DVD architecture with an additional detector (see Figure 1d). The performance of this platform is validated through the use of surface chemistry protocols to specifically detect and count CD4+/CD8+ T cells that could be valuable for monitoring of HIV+ patients. This approach integrates a microfluidic sample handling method with digital imaging on the same platform, making it highly advantageous for cellular diagnostics testing in resource limited settings.
Conventional (Non-Computational) Imaging Techniques for Global Health Applications
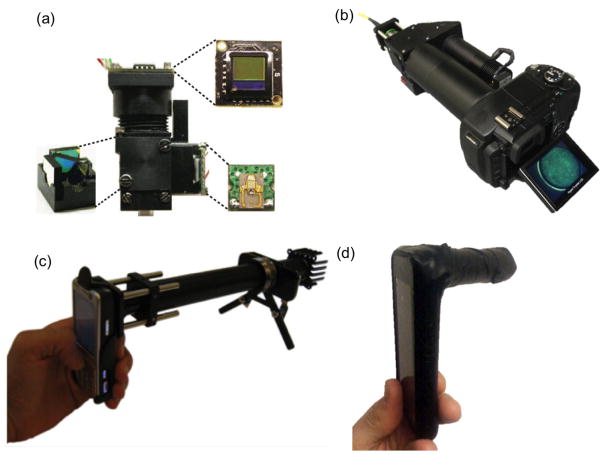
As a complementary effort to computational imaging platforms, there is an ample amount of literature addressing global health problems through the use of non-computational imaging designs. Toward this end, a widespread approach is to miniaturize conventional biomedical imaging architectures into compact units for field-portable micro-analysis. For instance, a miniaturized fluorescent microscope[42*] is designed through the integration of low-cost and compact components such as LEDs and CMOS imagers (see Figure 2a). This light-weight microscope can achieve 0.5 mm2 imaging FOV with ~2.5 μm spatial resolution. The imaging performance of such a miniature fluorescence microscope is tested by imaging mouse brain, however, this integrated imaging platform could also be utilized as a portable biomedical diagnostics tool.
Figure 2.
Non-computational, conventional imaging methods implemented on consumer electronics devices. (a) Miniaturized fluorescence microscope. (b) Fiber optic fluorescence microscope on a digital camera for in vivo imaging. (c) Fluorescent microscopy on a cellphone. (d) Bright field microscopy and spectroscopy on a cellphone. Reprinted from refs. 40, 41, 42, and 43 with permission from Nature Publishing Group (a) and PLoS One (b–d), respectively.
Following the same motivation presented in the previous section, implementation of non-computational conventional imaging modalities on consumer-grade electronics products has also gained significant attention recently. For example, a fiber-optic fluorescence microscope[43**] has been installed on an off-the-shelf digital camera (see Figure 2b) to perform in-vivo cellular imaging for inspection of tissue samples in low resource settings. This microscope, achieving ~5 μm spatial resolution over ~0.66 mm FOV in width, has successfully imaged early neoplastic changes in human epithelial tissues. Furthermore, bright field and fluorescence microscopic imaging modalities[44**,45] implemented on cellphone cameras have also been widely explored to perform microscopic investigation of e.g., blood or tissue specimens (see Figures 2c–d). These non-computational, conventional imaging and sensing platforms, some of which are highlighted in Figure 2, along with their computational counterparts could be useful for telemedicine applications in general.
Computational Sensing for Global Health Applications
In parallel to these recent advances in computational imaging techniques that are discussed earlier, sensing technologies also leverage computation and digital processing approaches, achieving performance improvements beyond traditional sensor systems. For this end, much research has been dedicated to develop new computational sensing technologies, including digital sensors running on cellphones[46*, 47] or computer drives[48–51], as well as sensing approaches based on digital biochemical assays[52*, 53–56], among others.
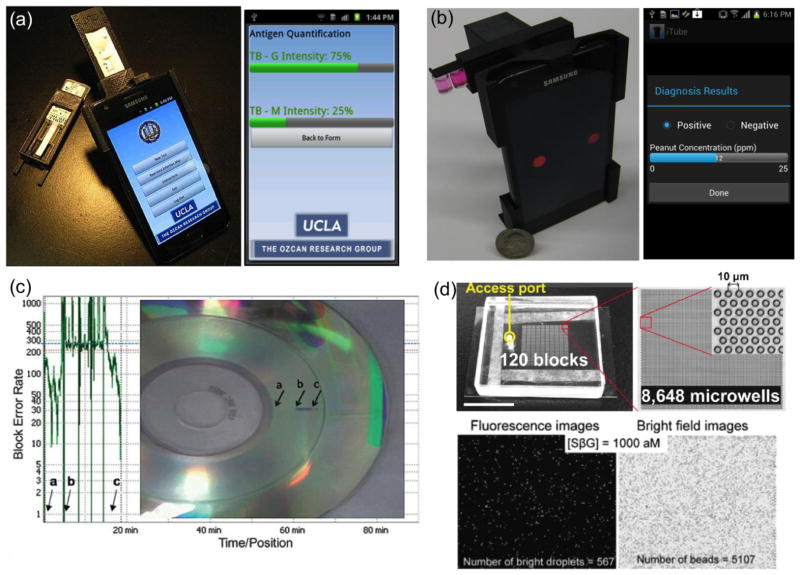
In addition to their microscopic imaging capability, as discussed earlier, cellphones can also be used to build computational sensors to provide cost-effective and field-deployable diagnostic devices for POC applications. As an example of computational sensing on a cellphone, a smart rapid-diagnostic-test (RDT) reader running on Android phones and iPhone [46*] is demonstrated (see Figure 3a), which can automatically image and analyze different lateral flow immunoassays using smart-phones. Through the same interface, test results can be uploaded to central servers, where spatio-temporal maps of various conditions can be visualized in real time making such a cellphone enabled distributed sensing approach quite valuable for e.g., disease surveillance and management as well as epidemiologic studies in general. Another recent emerging computational sensing platform is food allergen testing on a cellphone[47], which is termed as iTube (see Figure 3b). Installed on the existing camera unit of an Android phone, this iTube platform images and automatically analyses colorimetric assays performed in test tubes toward sensitive and specific detection of allergens (e.g., peanut) in food samples. Similar to the cellphone enabled RDT reader platform, the test results acquired using the iTube platform can also be uploaded to secure servers, creating personal and/or public spatio-temporal allergen maps that can be useful for public health in various settings, including e.g., schools, restaurants, etc.
Figure 3.
Computational sensing techniques for telemedicine applications. (a) Integrated rapid diagnostic test reader on a smart-phone. (b) Personalized food allergen testing on a smart-phone. (c) Computational sensing on a standard computer drive. (d) Digital analysis on biochemical assays. (e.g., ELISA or PCR). Reprinted from refs. 44, 45, 49, and 54 with permission from RSC publishing (a–d).
Along the same lines, a computational sensor employing standard computer drives such as compact disk (CD) readers [51] has been recently illustrated (see Figure 3c), where microparticles or live cells loaded into specially designed micro-channels interfere with the laser beam in the optical pickup apparatus of the computer drive, causing an error signal in the reading of the digital data previously written on the CD. The total count of error signals is then used to determine the concentration of the microparticles/cells within the micro-channel, providing a quantitative sensing capability. This CD-based computational sensor has been applied to counting of Chinese Hamster Ovarian (CHO) cells for its proof-of-concept. Such a ubiquitous and cost-effective device could especially be suitable for biosensing applications in remote and resource poor settings.
Another recent direction in computational sensing is to employ digital analysis techniques on biochemical assays that are frequently used to monitor biomolecular interactions or markers for clinical diagnosis of various diseases. Enzyme-linked immunosorbent assay (ELISA) or polymerase chain reaction (PCR) techniques are widely employed for specific and sensitive detection of e.g., protein or nucleic acid bio-markers within large sample volumes (e.g., microliters to milliliters). An emerging digital biochemical analysis method[52*, 53–56] distributes the sample volume of interest into smaller separate regions/volumes such that each droplet of the sample provides “0” or “1” molecules, corresponding to negative or positive reactions occurring within each droplet, respectively (see Figure 3d). Assuming a sufficiently large number of droplets exists per test, the computational analysis of total “0”s and “1”s is expected to follow a Poisson distribution, which in turn can provide a measurement of the target molecule concentration within the sample volume of interest. This “digital assay” method, which can for example be applied to PCR tests, can in general achieve highly sensitive and calibration-free diagnostic measurements, creating a versatile digital sensing approach for especially POC applications.
Future Outlook for Computational Imaging, Sensing and Diagnostics
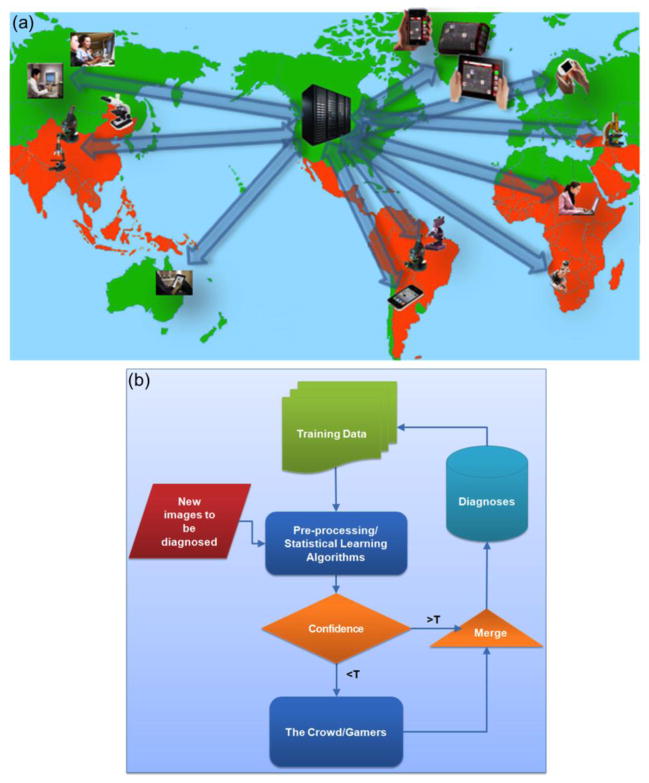
The recent advances in computational imaging and sensing technologies, also outlined in this Review, could potentially flourish a hybrid system/network that integrates unique physical functions of existing tools (e.g., microscopes, cytometers, biosensors etc.) with recently emerging computational measurement/test techniques running on various digital electronics components, including e.g., cellphones or tablet PCs (see Figure 4). Such a hybrid network of micro-analysis, imaging and sensing tools, which are at the same time digitally connected to each other through central servers can output and digitally share massive amounts of high quality and highly relevant biomedical data, forming large scale spatio-temporal databases/maps for various diseases or micro-organisms, among others [57]. This cloud-based micro-analysis and diagnostics network might provide new prospects for e.g., outsourcing medical diagnosis to remote doctors toward building an efficient telemedicine infrastructure, better analysis and management of epidemics and pandemics, as well as aiding epidemiology researchers and decision/policy makers on global health and environmental strategies in general.
Figure 4.
(a) Cloud based micro-analysis and diagnostics devices forming a telemedicine network. (b) A crowd-sourcing method as described in this schematics can be utilized to better handle the large-scale bio-medical imaging and sensing data (the red box) by combining the innate visual recognition and learning capabilities of humans (i.e., medical professionals, diagnosticians, microscopists, etc.) with machine learning based pre-processing computational blocks. Reprinted from ref. 60 with permission from PLoS One (a–b).
On the other hand, we should also emphasize that this massive data to be generated through such widely distributed computational imaging, sensing and diagnostics tools, will also contribute to our emerging “Big Data” challenge[58], which can fundamentally be related to the mismatch between the growth rate of digital electronics devices in general and the availability of human experts (i.e., trained professionals, in the form of e.g., pathologists, medical diagnosticians, microscopists or micro-biologists). To address this big data problem for biomedical image analysis and medical diagnosis, various innovative uses of crowd-sourcing methods [58,59,60*] can be employed to better handle large scale medical imaging and sensing data, and to segment, identify, classify, and reach accurate and sensitive diagnostic conclusions, by e.g., merging machine learning, statistical learning tools with the intrinsic image recognition and learning capabilities of human experts and medical professionals (see Figure 4b).
Conclusion
In this Review, we summarized the recent work in computational imaging, sensing and diagnostics techniques, along with some of the emerging non-computational imaging/sensing modalities that can provide significantly improved and yet cost-effective alternatives for global health applications.
Highlights.
Consumer electronics creates new opportunities for imaging and sensing technologies.
We review emerging computational imaging and sensing platforms for global health.
Computational imagers and sensors are in general field-portable and cost-effective.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of this Review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Brady DJ, Choi K, Marks DL, Horisaki R, Lim S. Compressive Holography. Opt Express. 2009;17:13040–13049. doi: 10.1364/oe.17.013040. This work demonstrates a computational 3D tomographic reconstruction method from a single 2D holographic image, where compressive sampling approach achieves multidimensional data recovery from lower dimensional measurements. [DOI] [PubMed] [Google Scholar]
- 3**.Greenbaum A, Luo W, Su T-W, Göröcs Z, Xue L, Isikman SO, Coskun AF, Mudanyali O, Ozcan A. Imaging without lenses: achievements and remaining challenges of wide-field on-chip microscopy. Nature Methods. 2012;9:889–895. doi: 10.1038/nmeth.2114. This work reviews the state-of-the-art performance of various lensfree computational imaging techniques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Xu W, Jericho MH, Meinertzhagen IA, Kreuzer HJ. Digital in-line holography for biological applications. PNAS. 2001;98:11301–11305. doi: 10.1073/pnas.191361398. This digital holographic microscopy platform utilizes an alternative imaging geometry, compared to lensfree on-chip holography presented in this review, creating over-sampled holograms to be used for high-resolution microscopic reconstructions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coskun AF, Sencan I, Su T-W, Ozcan A. Lensless wide-field fluorescent imaging on a chip using compressive decoding of sparse objects. Optics Express. 2010;18:10510. doi: 10.1364/OE.18.010510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Eils R, Athale C. Computational imaging in cell biology. J Cell Biol. 2003;161:477–481. doi: 10.1083/jcb.200302097. This paper highlights the need for computational tools in biomedical sciences, providing a powerful framework combining mathematical models with digital tools for quantitative analysis of biological phenomena. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Yun SH, Tearney GJ, Vakoc BJ, Shishkov M, Oh WY, Desjardins AE, Suter MJ, Chan RC, Evans JA, Jang I-K, et al. Comprehensive volumetric optical microscopy in vivo. Nat Med. 2006;12:1429–1433. doi: 10.1038/nm1450. This work illustrates the use of optical frequency-domain imaging devices for three-dimensional volumetric visualization of mucosal and endothelial tissues. This in-vivo screening tool, combined with computational rendering, could be useful to create virtual histology maps. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Rosen J, Brooker G. Non-scanning motionless fluorescence three-dimensional holographic microscopy. Nat Photon. 2008;2:190–195. This study demonstrates a computational fluorescent microscopy platform that uses an incoherent holographic encoding/decoding scheme, creating three-dimensional microscopic images without the need for mechanical scanning. [Google Scholar]
- 9.Fujimoto JG. Optical coherence tomography for ultrahigh resolution in vivo imaging. Nat Biotech. 2003;21:1361–1367. doi: 10.1038/nbt892. [DOI] [PubMed] [Google Scholar]
- 10.Yelin D, Rizvi I, White WM, Motz JT, Hasan T, Bouma BE, Tearney GJ. Three-dimensional miniature endoscopy. Nature. 2006;443:765–765. doi: 10.1038/443765a. [DOI] [PubMed] [Google Scholar]
- 11.Gehm ME, John R, Brady DJ, Willett RM, Schulz TJ. Single-shot compressive spectral imaging with a dual-disperser architecture. Opt Express. 2007;15:14013–14027. doi: 10.1364/oe.15.014013. [DOI] [PubMed] [Google Scholar]
- 12.Wagadarikar A, John R, Willett R, Brady D. Single disperser design for coded aperture snapshot spectral imaging. Appl Opt. 2008;47:B44–B51. doi: 10.1364/ao.47.000b44. [DOI] [PubMed] [Google Scholar]
- 13.Wagadarikar AA, Pitsianis NP, Sun X, Brady DJ. Video rate spectral imaging using a coded aperture snapshot spectral imager. Optics Express. 2009;17:6368. doi: 10.1364/oe.17.006368. [DOI] [PubMed] [Google Scholar]
- 14.Hong S-H, Jang J-S, Javidi B. Three-dimensional volumetric object reconstruction using computational integral imaging. Opt Express. 2004;12:483–491. doi: 10.1364/opex.12.000483. [DOI] [PubMed] [Google Scholar]
- 15.Javidi B, Ponce-Díaz R, Hong S-H. Three-dimensional recognition of occluded objects by using computational integral imaging. Opt Lett. 2006;31:1106–1108. doi: 10.1364/ol.31.001106. [DOI] [PubMed] [Google Scholar]
- 16*.Goda K, Tsia KK, Jalali B. Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature. 2009;458:1145–1149. doi: 10.1038/nature07980. This real-time computational imaging method creates 2D images of biological samples using a single-pixel photo-detector encoded with a serial time-domain data stream, which can monitor very fast dynamical processes that would have been impossible to record with conventional image sensor-arrays (e.g., CCD and CMOS) [DOI] [PubMed] [Google Scholar]
- 17**.Brady DJ, Gehm ME, Stack RA, Marks DL, Kittle DS, Golish DR, Vera EM, Feller SD. Multiscale gigapixel photography. Nature. 2012;486:386–389. doi: 10.1038/nature11150. This work describes an emerging computational camera design that utilizes a parallel array of micro-cameras to digitally create high-pixel-count (e.g., 50 gigapixels) photographs. [DOI] [PubMed] [Google Scholar]
- 18.Isikman SO, Greenbaum A, Luo W, Coskun AF, Ozcan A. Giga-Pixel Lensfree Holographic Microscopy and Tomography Using Color Image Sensors. PLoS ONE. 2012;7:e45044. doi: 10.1371/journal.pone.0045044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossairt OS, Miau D, Nayar SK. Gigapixel Computational Imaging. 2011 IEEE International Conference on Computational Photography (ICCP); 2011. pp. 1–8. [Google Scholar]
- 20*.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine. 2007;58:1182–1195. doi: 10.1002/mrm.21391. This study introduces a compressive sensing based MR imaging technique, improving the performance of MRI platforms in terms of spatial resolution and image acquisition speed. [DOI] [PubMed] [Google Scholar]
- 21.Mudanyali O, Tseng D, Oh C, Isikman SO, Sencan I, Bishara W, Oztoprak C, Seo S, Khademhosseini B, Ozcan A. Compact, light-weight and cost-effective microscope based on lensless incoherent holography for telemedicine applications. Lab Chip. 2010;10 :1417–1428. doi: 10.1039/c000453g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt Lett. 1978;3:27–29. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 23.Park SC, Park MK, Kang MG. Super-resolution image reconstruction: a technical overview. IEEE Signal Processing Magazine. 2003;20:21–36. [Google Scholar]
- 24.Mudanyali O, Oztoprak C, Tseng D, Erlinger A, Ozcan A. Detection of waterborne parasites using field-portable and cost-effective lensfree microscopy. Lab Chip. 2010;10 :2419–2423. doi: 10.1039/c004829a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenbaum A, Sikora U, Ozcan A. Field-portable wide-field microscopy of dense samples using multi-height pixel super-resolution based lensfree imaging. Lab Chip. 2012;12 :1242–1245. doi: 10.1039/c2lc21072j. [DOI] [PubMed] [Google Scholar]
- 26.Greenbaum A, Feizi A, Akbari N, Ozcan A. Wide-field computational color imaging using pixel super-resolved on-chip microscopy. Opt Express. 2013;21:12469–12483. doi: 10.1364/OE.21.012469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27**.Mudanyali O, McLeod E, Luo W, Greenbaum A, Coskun AF, Hennequin Y, Allier CP, Ozcan A. Wide-field optical detection of nanoparticles using on-chip microscopy and self-assembled nanolenses. Nature Photonics. 2013;7:247–254. doi: 10.1038/nphoton.2012.337. This paper demonstrates a high-throughput nano-imaging approach based on lensfree holographic computational imaging, which specifically employs a contrast enhancement mechanism through the use of self-assembled liquid nanolenses covering single nanoparticles or viral particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui X, Lee LM, Heng X, Zhong W, Sternberg PW, Psaltis D, Yang C. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proceedings of the National Academy of Science. 2008;31:10670–75. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishara W, Su T-W, Coskun AF, Ozcan A. Lensfree on-chip microscopy over a wide field-of-view using pixel super-resolution. Opt Express. 2010;18:11181–11191. doi: 10.1364/OE.18.011181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SA, Leitao R, Zheng G, Yang S, Rodriguez A, Yang C. Color Capable Sub-Pixel Resolving Optofluidic Microscope and Its Application to Blood Cell Imaging for Malaria Diagnosis. PLoS ONE. 2011;6:e26127. doi: 10.1371/journal.pone.0026127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishara W, Isikman SO, Ozcan A. Lensfree Optofluidic Microscopy and Tomography. Ann Biomed Eng. 2012;40:251–262. doi: 10.1007/s10439-011-0385-3. [DOI] [PubMed] [Google Scholar]
- 32.Isikman SO, Bishara W, Zhu H, Ozcan A. Optofluidic Tomography on a Chip. Applied Physics Letters. 2011;98 doi: 10.1063/1.3548564. 161109–161109–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isikman SO, Bishara W, Mavandadi S, Yu FW, Feng S, Lau R, Ozcan A. Lens-free optical tomographic microscope with a large imaging volume on a chip. Proceedings of the National Academy of Science. 2011;18:7296–7301. doi: 10.1073/pnas.1015638108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee M, Yaglidere O, Ozcan A. Field-portable reflection and transmission microscopy based on lensless holography. Biomed Opt Express. 2011;2:2721–2730. doi: 10.1364/BOE.2.002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu H, Yaglidere O, Su T-W, Tseng D, Ozcan A. Cost-effective and compact wide-field fluorescent imaging on a cell-phone. Lab Chip. 2011;11:315–322. doi: 10.1039/c0lc00358a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Ramachandraiah H, Amasia M, Cole J, Sheard P, Pickhaver S, Walker C, Wirta V, Lexow P, Lione R, Russom A. Lab-on-DVD: standard DVD drives as a novel laser scanning microscope for image based point of care diagnostics. Lab Chip. 2013;13:1578–1585. doi: 10.1039/c3lc41360h. A computational microscope is implemented on a computer DVD drive. [DOI] [PubMed] [Google Scholar]
- 37.Janzen MC, Ponder JB, Bailey DP, Ingison CK, Suslick KS. Colorimetric Sensor Arrays for Volatile Organic Compounds. Anal Chem. 2006;78:3591–3600. doi: 10.1021/ac052111s. [DOI] [PubMed] [Google Scholar]
- 38.Lapresta-Fernández A, Capitán-Vallvey LF. Scanometric potassium determination with ionophore-based disposable sensors. Sensors and Actuators B: Chemical. 2008;134:694–701. [Google Scholar]
- 39*.Levin-Reisman I, Gefen O, Fridman O, Ronin I, Shwa D, Sheftel H, Balaban NQ. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat Meth. 2010;7:737–739. doi: 10.1038/nmeth.1485. A computational imaging platform is installed on a flatbed scanner that monitors the growth patterns of bacterial phenotypes, revealing rare events in various bacterial communities. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H, Mavandadi S, Coskun AF, Yaglidere O, Ozcan A. Optofluidic Fluorescent Imaging Cytometry on a Cell Phone. Anal Chem. 2011;83:6641–6647. doi: 10.1021/ac201587a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Zhu H, Sencan I, Wong J, Dimitrov S, Tseng D, Nagashima K, Ozcan A. Cost-effective and rapid blood analysis on a cell-phone. Lab Chip. 2013;13:1282–1288. doi: 10.1039/c3lc41408f. This work describes a computational imager on smart-phone, integrating simple sample preparation steps with digital imaging/processing on the same platform, for automated blood analysis in low resource settings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42*.Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, Schnitzer MJ. Miniaturized integration of a fluorescence microscope. Nat Methods. 2011;8:871–878. doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Shin D, Pierce MC, Gillenwater AM, Williams MD, Richards-Kortum RR. A Fiber-Optic Fluorescence Microscope Using a Consumer-Grade Digital Camera for In Vivo Cellular Imaging. PLoS ONE. 2010;5:e11218. doi: 10.1371/journal.pone.0011218. A non-computational fiber-optic fluorescence microscope implemented on a digital camera that could be useful for in-vivo cellular imaging at the point of care. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS ONE. 2009;4:e6320. doi: 10.1371/journal.pone.0006320. This paper illustrates a non-computational cellphone-based bright-field microscopy platform for field diagnostics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith ZJ, Chu K, Espenson AR, Rahimzadeh M, Gryshuk A, Molinaro M, Dwyre DM, Lane S, Matthews D, Wachsmann-Hogiu S. Cell-Phone-Based Platform for Biomedical Device Development and Education Applications. PLoS ONE. 2011;6:e17150. doi: 10.1371/journal.pone.0017150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Mudanyali O, Dimitrov S, Sikora U, Padmanabhan S, Navruz I, Ozcan A. Integrated rapid-diagnostic-test reader platform on a cellphone. Lab Chip. 2012;12:2678–2686. doi: 10.1039/c2lc40235a. This work demonstrates a computational sensing scheme running on a smart phone that can image and analyze various rapid diagnostic tests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coskun AF, Wong J, Khodadadi D, Nagi R, Tey A, Ozcan A. A personalized food allergen testing platform on a cellphone. Lab Chip. 2013;13:636–640. doi: 10.1039/c2lc41152k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kido H, Maquieira A, Hammock BD. Disc-based immunoassay microarrays. Analytica Chimica Acta. 2000;411:1–11. [Google Scholar]
- 49.Lange SA, Roth G, Wittemann S, Lacoste T, Vetter A, Grässle J, Kopta S, Kolleck M, Breitinger B, Wick M, et al. Measuring Biomolecular Binding Events with a Compact Disc Player Device. Angewandte Chemie. 2006;118:276–279. doi: 10.1002/anie.200501243. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Ou LML, Yu H-Z. Digitized molecular diagnostics: reading disk-based bioassays with standard computer drives. Anal Chem. 2008;80:8216–8223. doi: 10.1021/ac8012434. [DOI] [PubMed] [Google Scholar]
- 51.Imaad SM, Lord N, Kulsharova G, Liu GL. Microparticle and cell counting with digital microfluidic compact disc using standard CD drive. Lab Chip. 2011;11:1448–1456. doi: 10.1039/c0lc00451k. [DOI] [PubMed] [Google Scholar]
- 52*.Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Digital PCR on a SlipChip. Lab Chip. 2010;10:2666–2672. doi: 10.1039/c004521g. This study demonstrates a SlipChip that can perform PCR in simple well-format plates. Based on a computational strategy performed on counting of nucleic acids, this smart chip may find applications in various biomedical applications, especially for point of care. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vogelstein B, Kinzler KW. Digital PCR. PNAS. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Q, Gao Y, Yu B, Ren H, Qiu L, Han S, Jin W, Jin Q, Mu Y. Self-priming compartmentalization digital LAMP for point-of-care. Lab Chip. 2012;12:4755–4763. doi: 10.1039/c2lc40774d. [DOI] [PubMed] [Google Scholar]
- 55.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotech. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim SH, Iwai S, Araki S, Sakakihara S, Iino R, Noji H. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip. 2012;12:4986–4991. doi: 10.1039/c2lc40632b. [DOI] [PubMed] [Google Scholar]
- 57.Mavandadi S, Feng S, Yu F, Dimitrov S, Yu R, Ozcan A. BioGames: A Platform for Crowd-Sourced Biomedical Image Analysis and Telediagnosis. Games for Health Journal. 2012;1:373–376. doi: 10.1089/g4h.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mavandadi S, Dimitrov S, Feng S, Yu F, Yu R, Sikora U, Ozcan A. Crowd-sourced BioGames: managing the big data problem for next-generation lab-on-a-chip platforms. Lab Chip. 2012;12:4102–4106. doi: 10.1039/c2lc40614d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Ahn L, Maurer B, McMillen C, Abraham D, Blum M. reCAPTCHA: Human-Based Character Recognition via Web Security Measures. Science. 2008;321:1465–1468. doi: 10.1126/science.1160379. [DOI] [PubMed] [Google Scholar]
- 60*.Mavandadi S, Dimitrov S, Feng S, Yu F, Sikora U, Yaglidere O, Padmanabhan S, Nielsen K, Ozcan A. Distributed Medical Image Analysis and Diagnosis through Crowd-Sourced Games: A Malaria Case Study. PLoS ONE. 2012;7:e37245. doi: 10.1371/journal.pone.0037245. This paper illustrates the use of crowd-sourcing to potentially address the emerging Big Data challenge for biomedical image analysis and related diagnosis, where innate visual recognition capabilities of humans and machine learning tools can be used together to handle large-scale diagnostics data. [DOI] [PMC free article] [PubMed] [Google Scholar]