Abstract
Cytochrome P450 (P450) enzymes play a critical role in the activation and detoxication of many neurotoxic chemicals. Although research has largely focused on P450-mediated metabolism in the liver, emerging evidence suggests that brain P450s influence neurotoxicity by modulating local metabolite levels. As a first step toward better understanding the relative role of brain P450s in determining neurotoxic outcome, we characterized mRNA expression of specific P450 isoforms in the rodent brain. Adult mice (male and female) and rats (male) were treated with vehicle, phenobarbital, or dexamethasone. Transcripts for CYP2B, CYP3A, CYP1A2, and the orphan CYP4X1 and CYP2S1 were quantified in the liver, hippocampus, cortex, and cerebellum by quantitative (real-time) polymerase chain reaction. These P450s were all detected in the liver with the exception of CYP4X1, which was detected in rat but not mouse liver. P450 expression profiles in the brain varied regionally. With the exception of the hippocampus, there were no sex differences in regional brain P450 expression profiles in mice; however, there were marked species differences. In the liver, phenobarbital induced CYP2B expression in both species. Dexamethasone induced hepatic CYP2B and CYP3A in mice but not rats. In contrast, brain P450s did not respond to these classic hepatic P450 inducers. Our findings demonstrate that P450 mRNA expression in the brain varies by region, regional brain P450 profiles vary between species, and their induction varies from that of hepatic P450s. These novel data will be useful for designing mechanistic studies to examine the relative role of P450-mediated brain metabolism in neurotoxicity.
Introduction
The cytochrome P450 (P450) superfamily is a diverse group of enzymes that catalyze the oxidative metabolism of not only endogenous substrates but also xenobiotics, including environmental contaminants of significant public health concern that target the nervous system, including polychlorinated biphenyls, polybrominated diphenyl ethers, and organophosphorus pesticides (Ariyoshi et al., 1995; Foxenberg et al., 2007; Erratico et al., 2013; Feo et al., 2013). Biotransformation of these compounds by P450s can result in bioactivation or detoxication, and the balance between these activities influences the bioeffective dose, and thus the neurotoxic outcome, following environmental exposures, as shown in studies of humans and animal models (Foxenberg et al., 2007; Curran et al., 2011; Kim et al., 2011; Crane et al., 2012; Khokhar and Tyndale, 2012).
Much of the research effort to characterize P450-mediated metabolism of neurotoxic compounds has focused on the liver. However, it is now evident that P450s are expressed in a number of extrahepatic tissues, including brain (Ding and Kaminsky, 2003; Ferguson and Tyndale, 2011). Although total P450 content in the human and rodent brain is generally significantly lower than that in the liver (Warner et al., 1988; Bhamre et al., 1992; Volk et al., 1995), recent evidence from rat studies demonstrates that P450-mediated metabolism in the brain can contribute significantly to neurotoxicity (Khokhar and Tyndale, 2012; Zhou et al., 2013). These data coupled with reports that P450 expression in the brain may vary between anatomic regions of the brain (Warner et al., 1988; Dutheil et al., 2009) have led to growing interest in the putative role of brain P450s in determining sensitivity and response to neurotoxic compounds via modulation of local metabolite levels (Meyer et al., 2007; Ferguson and Tyndale, 2011; Ravindranath and Strobel, 2013).
Rodents are important models for studying the relative influence of brain versus liver P450s on neurotoxicity; however, most of our knowledge of P450 expression in the rodent brain is derived from studies of whole brain homogenates, and there is a paucity of data on regional P450 expression in the rodent brain. Additional questions include whether the well known sex- and species-specific differences in hepatic P450 expression extend to the brain, and whether P450s in the brain respond to classic inducers of hepatic P450 expression. Here, we address these questions by comparing P450 transcript levels in three distinct regions of the rodent brain relative to expression levels in the liver under basal conditions and following treatment with phenobarbital and dexamethasone, which are classic inducers of hepatic CYP2B and CYP3A expression (reviewed by Corcos and Berthou, 2008; Greenblatt et al., 2008). We also assessed the influence of sex and species on P450 expression profiles in the brain using the male mouse as the reference. We studied CYP2B (mouse CYP2B10/rat CYP2B1/2), CYP3A (mouse CYP3A11/rat CYP3A2), and CYP1A2 because these isoforms have been implicated in the metabolism of polychlorinated biphenyls (Kania-Korwel et al., 2008, 2012; Curran et al., 2011), polybrominated diphenyl ethers (Erratico et al., 2013; Feo et al., 2013), and organophosphorus pesticides (Tang et al., 2001; Foxenberg et al., 2007). Two orphan P450s, CYP4X1 and CYP2S1 (Guengerich et al., 2010), were also included in this study because CYP2S1 is abundantly expressed in many extrahepatic tissues (Choudhary et al., 2003) and CYP4X1 is predominantly expressed in the rodent brain (Bylund et al., 2002; Al-Anizy et al., 2006). The data reported herein demonstrate brain region–specific expression of P450s in the rodent brain that is sex- and species-dependent and generally not altered by the classic inducers phenobarbital and dexamethasone under conditions that significantly induce P450 expression in the liver.
Materials and Methods
Animals and Treatments.
Experiments involving animals were approved by the Institutional Animal Care and Use Committee at the University of Iowa. Male and female C57BL/6 mice (7–8 weeks) were obtained from the Jackson Laboratory (Bar Harbor, ME) and randomly assigned to one of four groups: 1) phenobarbital (PB; Sigma-Aldrich, St. Louis, MO) at 102 mg/kg/day in saline or 2) saline at 20 ml/kg/day, i.p., for 3 consecutive days; 3) dexamethasone (DEX; Sigma-Aldrich) at 50 mg/kg/day in corn oil (CO) or 4) CO (Fisher Scientific, Pittsburg, PA) at 10 ml/kg/day, i.p., for 4 consecutive days (Kania-Korwel et al., 2008). Male Sprague-Dawley rats (8 weeks) were purchased from Harlan, Inc. (Indianapolis, IN), acclimated for 1 week, then randomly assigned to one of four groups: 1) PB at 102 mg/kg/day in saline or 2) saline at 5 ml/kg/day, i.p., for 3 consecutive days; 3) DEX at 50 mg/kg/day in CO or 4) CO vehicle control at 5 ml/kg/day, i.p., for 4 consecutive days (Kania-Korwel et al., 2008). Animals were euthanized 24 hours after the last treatment by CO2 asphyxiation followed by cervical dislocation. Brain regions and livers were immediately collected on ice, weighed, placed in RNALater (Qiagen, Valencia, CA) overnight, and then stored at −80°C. The effects of treatments on liver and total body weight are summarized in Supplemental Tables 1–3.
Assessment of mRNA Levels by Quantitative Polymerase Chain Reaction.
Tissue levels of isoform-specific P450 transcripts were quantified using a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). P450 mRNA levels were normalized to the reference gene phosphoglycerate kinase 1, and relative expression ratios between treated and vehicle control animals were calculated by the Pfaffl method (Pfaffl, 2001) using REST 2009 software (Qiagen, Valencia, CA). Statistical analysis was performed using the built-in randomization techniques of REST 2009 (detailed descriptions of RNA isolation and mRNA quantification and analyses are provided in the Supplemental Material; Supplemental Tables 6 and 7 list primer sequences and amplification efficiencies of primers sets, respectively).
Results and Discussion
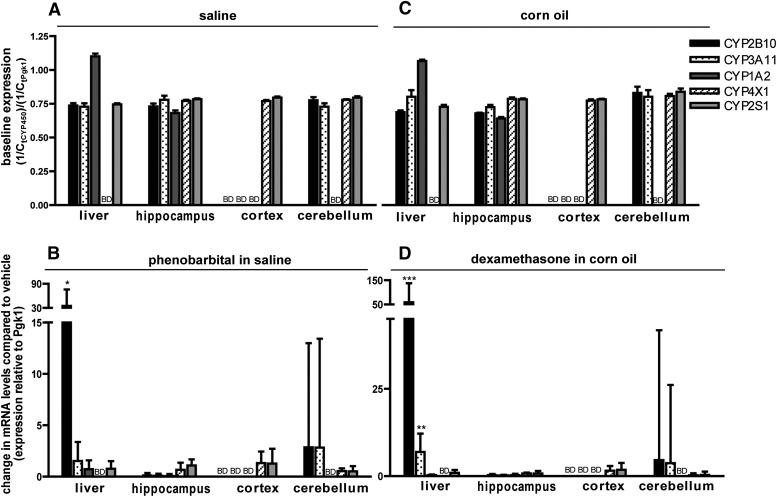
Adult male C57BL/6 mice were used as the reference for comparison of sex- and species-dependent differences in P450 gene expression. In male mice, tissue-specific P450 expression patterns were similar between saline (Fig. 1A) and CO (Fig. 1C) vehicle controls. The liver expressed CYP2B10, 3A11, 1A2, and 2S1, but not CYP4X1, which is consistent with previous reports of brain-specific CYP4X1 expression in mice (Al-Anizy et al., 2006; Renaud et al., 2011). P450 expression profiles in the brain were region-specific: all five P450 isoforms were expressed in the hippocampus; the cerebellum expressed transcripts for all but CYP1A2, and the cortex expressed only CYP2S1 and CYP4X1 mRNA (Fig. 1, A and C). Treatment with either PB or DEX induced P450 expression in the liver (Fig. 1, B and D; see also Supplemental Tables 4 and 5): hepatic CYP2B10 was induced by PB [mean of 35.1; 68% confidence interval (CI): 14.8–75.7] and by DEX (mean of 58.8; 68% CI: 30.6–137.5). DEX also induced hepatic CYP3A11 (mean of 6.9; 68% CI: 3.8–12.2). However, neither PB nor DEX induced expression of any target P450 in the hippocampus, cerebellum, or cortex (Fig. 1, B and D).
Fig. 1.
P450 expression profiles in the brains of adult male C57BL/6 mice. Mice were treated for 3 consecutive days with either saline (20 ml/kg/day, i.p.) (A) or an equal volume of PB in saline (102 mg/kg/day, i.p.) (B), or for 4 consecutive days with either CO (10 ml/kg/day, i.p.) (C) or an equal volume of DEX in CO (50 mg/kg/day, i.p.) (D). Tissues were harvested 24 hours after the last injection and P450 mRNA quantified by quantitative (real-time) polymerase chain reaction. (A and C) Baseline P450 expression determined by normalizing fractional amplification (cycle number at which fluorescence exceeds a user-defined threshold) (Ct) values for P450 transcripts in control samples to Ct values for the reference gene [phosphoglycerate kinase 1 (Pgk1)] in the same sample. (B and D) Change in expression of the target gene in PB- or DEX-treated animals relative to vehicle control (saline for PB and CO for DEX). All data are expressed as the mean relative expression ± S.E. (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001, significantly different from vehicle control as determined by automated randomization and bootstrapping tests (REST 2009 software). BD, below detection limit.
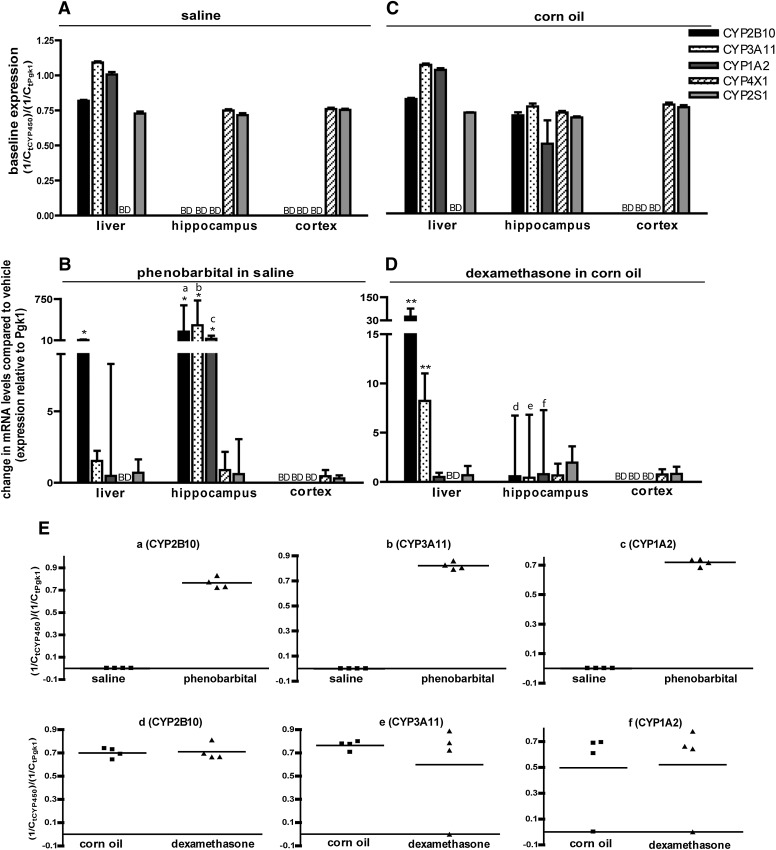
To explore the influence of sex, we measured P450 transcript levels in female C57BL/6 mice. Baseline P450 expression in the liver and cortex was similar between the saline (Fig. 2A) and CO (Fig. 2C) vehicle controls, and comparable to profiles observed in male vehicle controls (Fig. 1, A and C). Similar to males, all five P450 isoforms were expressed in the hippocampus of CO-treated females (Fig. 2C); however, in contrast to males, only CYP4X1 and CYP2S1 were detected in the hippocampus of saline-treated females (Fig. 2A). CO has been previously reported to influence P450 expression in the rat liver (Yoo et al., 1990), but it is unclear whether our findings reflect CO-mediated increase in P450 levels in the female mouse hippocampus. Although differences between saline- and CO-treated female mice could be experimental artifact, this seems unlikely because hippocampal expression levels of CYP4X1 and CYP2S1 were comparable between the two vehicle treatments and between sexes.
Fig. 2.
P450 expression profiles in the brains of adult female C57BL/6 mice. Mice were treated for 3 consecutive days with either saline (20 ml/kg/day, i.p.) (A) or an equal volume of PB in saline (102 mg/kg/dat, i.p.) (B), or for 4 consecutive days with either CO (10 ml/kg/day, i.p.) (C) or an equal volume of DEX in CO (50 mg/kg/day, i.p.) (D). Tissues were harvested 24 hours after the last injection and P450 mRNA measured by quantitative (real-time) polymerase chain reaction. (A and C) Baseline P450 expression determined by normalizing fractional amplification (Ct) values for P450 transcripts in control samples to Ct values for phosphoglycerate kinase 1 (Pgk1) in the same sample. (B and D) Change in expression of the target gene in PB- or DEX-treated animals relative to vehicle control (saline for PB and CO for DEX). (E) Ct values for hippocampal CYP2B10, CYP3A11, and CYP1A2 in the PB group (a,b,c) and the DEX group (d,e,f) of individual mice. Data in (A–D) are expressed as the mean ± S.E. (n = 4–5). *P < 0.05; **P < 0.01; ***P < 0.001, significantly different from vehicle controls as determined using the REST 2009 software. BD, below detection limit.
As observed in male mice, PB induced hepatic CYP2B10 expression in female mice by a mean factor of 13.2 (68% CI: 6.6–23.7), whereas DEX induced hepatic expression of CYP2B10 and CYP3A11 by 48.6 (68% CI: 30.6–91.1) and 8.21 (68% CI: 5.6–11.0), respectively (Fig. 2, B and D). Also consistent with male mice, PB or DEX did not change P450 expression in the cortex of female mice. Similarly, DEX had no effect on P450 expression in the female hippocampus. However, in contrast to males, PB significantly induced expression of CYP2B10 (mean of 164; 68% CI: 56–529), CYP3A11 (mean of 279; 68% CI: 113–724), and CYP1A2 (mean of 36; 68% CI: 14–94) in the hippocampus of females relative to saline vehicle controls (Fig. 2B). Data for these three transcripts are shown for individual female mice in Fig. 2E.
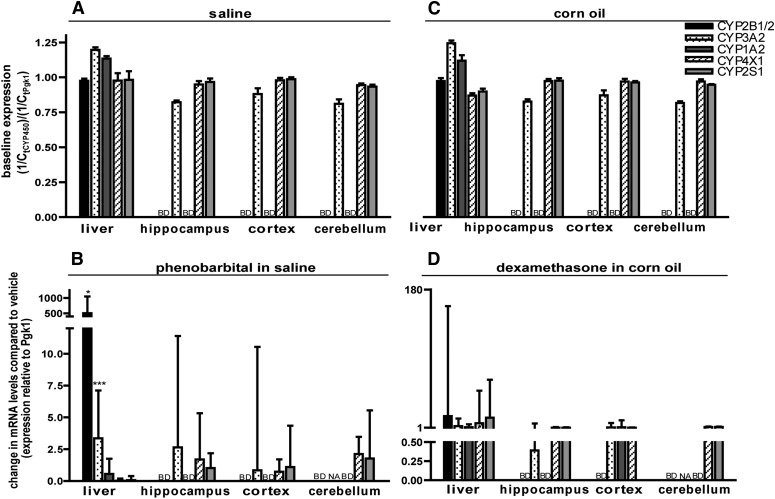
To investigate species-dependent differences, P450 transcripts were quantified in male rats. Tissue-specific P450 expression profiles were similar between saline (Fig. 3A) and CO (Fig. 3C) vehicle controls, but varied from those observed in the comparable male mouse treatment groups (Fig. 1, A and C). Specifically, in the male rat, all five P450 isoforms of interest were expressed in the liver, including CYP4X1 (Fig. 3, A and C). Also in contrast to male mice, rat brain expressed CYP3A2, CYP4X1, and CYP2S1 in the hippocampus, cortex, and cerebellum, but neither CYP2B1/2 nor CYP1A2 was detected in any of these three brain regions (Fig. 3, A and C). These findings are consistent with previous studies of regional P450 expression in rat brain, with the exception that others have reported the presence of CYP2B in rat brain (Schilter and Omiecinski, 1993). Similar to male mice, PB (Fig. 3B) and DEX (Fig. 3D) induced P450 expression in the male rat liver but not in any of the three brain regions. However, the profile of hepatic P450 isoforms induced by these chemical treatments showed species variation. In the rat, PB induced the expression of not only CYP2B1/2 (by a mean factor of 504; 68% CI: 298–1044) but also CYP3A2 (by a mean factor of 3.4; 68% CI: 1.8–7.1). Surprisingly, DEX did not significantly alter hepatic CYP2B1/2 or CYP3A2 expression, but instead significantly upregulated CYP4X1 expression (by a mean factor of 6.5; 68% CI: 1.5–78.8). Although the latter is a novel finding, given the low fold-induction and the lack of protein expression data, the functional significance of this upregulation is not clear.
Fig. 3.
P450 expression profiles in the brain of adult male Sprague-Dawley rats. Rats were treated for 3 consecutive days with either saline (5 ml/kg/day, i.p.) (A) or an equal volume of PB in saline (102 mg/kg/day, i.p.) (B), or for 4 consecutive days with either CO (5 ml/kg/day, i.p.) (C) or an equal volume of DEX in CO (50 mg/kg/day, i.p.) (D). Tissues were harvested 24 hours after the last injection and P450 mRNA quantified by quantitative (real-time) polymerase chain reaction. (A and C) Baseline P450 expression determined by normalizing fractional amplification (Ct) values for CYP transcripts in vehicle control tissues to Ct values for phosphoglycerate kinase 1 (Pgk1) in the same sample. (B and D) Change in expression of the target gene in PB- or DEX-treated animals relative to control (saline for PB and CO for DEX). Data are expressed as the mean ± S.E. (n = 3 except for PB-treated cerebellum, in which n = 2). *P < 0.05; **P < 0.01; ***P < 0.001, significantly different from vehicle controls as determined by automated randomization and bootstrapping tests (REST 2009 software). BD, below detection limit; NA, not available because of low amplification efficiency.
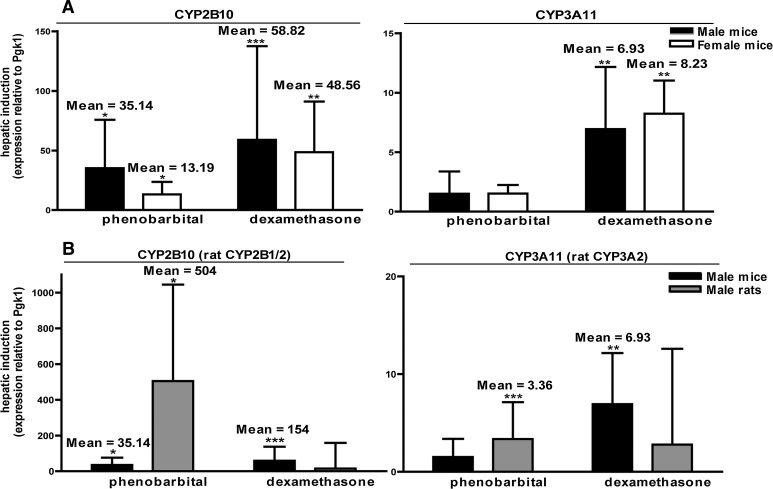
To further investigate sex- and species-specific differences in hepatic P450 induction, we compared relative CYP2B and CYP3A induction in the liver of male versus female mice and between male mice and male rats. We found no significant sex differences in P450 induction patterns in mice (Fig. 4A). Conversely, there were significant differences in hepatic P450 induction between mice and rats (Fig. 4B). Hepatic CYP2B expression was induced by DEX in mice but not rats, and CYP3A expression was induced by PB in mice but not rats and by DEX in rats but not mice.
Fig. 4.
P450 induction in liver is species-dependent but not sex-dependent. (A) In both male and female mice, hepatic CYP2B10 is significantly induced by both PB and DEX, whereas hepatic CYP3A11 is upregulated by DEX but not PB. Differences in induction between sexes are not statistically significant as determined by Student’s t test (P < 0.05). (B) Hepatic CYP2B10 is induced by both PB and DEX in male mice, but the rat ortholog, CYP2B1/2, is induced only by PB in male rats. CYP3A11 is upregulated in male mice by DEX but not by PB. Conversely, the rat ortholog, CYP3A2, is induced by PB but not by DEX. P450 transcript levels in animals treated with either PB or DEX are presented relative to species-specific vehicle controls. *P < 0.05; **P < 0.01; ***P < 0.001, significantly different from vehicle controls as determined by automated randomization and bootstrapping tests (REST 2009 software).
In summary, these data suggest that P450 mRNA expression in the brain 1) differs significantly from hepatic P450 transcript profiles in rodent models; 2) varies between brain regions; 3) exhibits subtle sex-dependent differences in the C57BL/6 mouse, but significant species-specific differences between mouse and rat; and 4) with the possible exception of P450s in the hippocampus of the female mouse, is not induced by PB or DEX regimens that induce hepatic orthologs. With respect to the last finding, previous studies of whole brain homogenates have reported either no P450 induction by these classic inducers (Schilter et al., 2000; Hedlund et al., 2001; Upadhya et al., 2002; Woodland et al., 2008) or CYP2B induction by PB (Schilter and Omiecinski, 1993; Schilter et al., 2000; Upadhya et al., 2002). Discrepancies between studies likely reflect differences in dose and duration of treatment, species and/or strain, whole brain versus isolated brain regions, primer specificity, and methods of mRNA quantification. Although it will be necessary to confirm protein levels and activity of these P450 isoforms to corroborate the significance of these findings, emerging evidence of brain P450-mediated xenobiotic activation strongly suggests that differences in regional expression of brain P450s may be an important mechanism contributing to region-selective neurotoxicity (Spencer and Lein, 2013).
Supplementary Material
Acknowledgments
The authors thank Rachel Shaffer for her assistance with quantitative (real-time) polymerase chain reaction optimization.
Abbreviations
- CI
confidence interval
- CO
corn oil
- DEX
dexamethasone
- P450
cytochrome P450
- PB
phenobarbital
Authorship Contributions
Participated in research design: Stamou, Wu, Kania-Korwel, Lehmler, Lein.
Conducted experiments: Stamou, Wu, Kania-Korwel.
Performed data analysis: Stamou, Wu.
Wrote or contributed to the writing of the manuscript: Stamou, Wu, Kania-Korwel, Lehmler, Lein.
Footnotes
This work was supported by the National Institutes of Health National Institute of Environmental Health Sciences [Grants R01 ES017425 and P42 ES04699] and the J.B. Johnson Foundation [unrestricted gift].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Al-Anizy M, Horley NJ, Kuo CW, Gillett LC, Laughton CA, Kendall D, Barrett DA, Parker T, Bell DR. (2006) Cytochrome P450 Cyp4x1 is a major P450 protein in mouse brain. FEBS J 273:936–947 [DOI] [PubMed] [Google Scholar]
- Ariyoshi N, Oguri K, Koga N, Yoshimura H, Funae Y. (1995) Metabolism of highly persistent PCB congener, 2,4,5,2′,4′,5′-hexachlorobiphenyl, by human CYP2B6. Biochem Biophys Res Commun 212:455–460 [DOI] [PubMed] [Google Scholar]
- Bhamre S, Anandatheerthavarada HK, Shankar SK, Ravindranath V. (1992) Microsomal cytochrome P450 in human brain regions. Biochem Pharmacol 44:1223–1225 [DOI] [PubMed] [Google Scholar]
- Bylund J, Zhang C, Harder DR. (2002) Identification of a novel cytochrome P450, CYP4X1, with unique localization specific to the brain. Biochem Biophys Res Commun 296:677–684 [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Schenkman JB, Sarfarazi M, Stoilov I. (2003) Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Arch Biochem Biophys 414:91–100 [DOI] [PubMed] [Google Scholar]
- Corcos L, Berthou F. (2008) The CYP2B subfamily, RSC Publishing, Cambridge [Google Scholar]
- Crane AL, Klein K, Olson JR. (2012) Bioactivation of chlorpyrifos by CYP2B6 variants. Xenobiotica 42:1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran CP, Nebert DW, Genter MB, Patel KV, Schaefer TL, Skelton MR, Williams MT, Vorhees CV. (2011) In utero and lactational exposure to PCBs in mice: adult offspring show altered learning and memory depending on Cyp1a2 and Ahr genotypes. Environ Health Perspect 119:1286–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Kaminsky LS. (2003) Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol 43:149–173 [DOI] [PubMed] [Google Scholar]
- Dutheil F, Dauchy S, Diry M, Sazdovitch V, Cloarec O, Mellottée L, Bièche I, Ingelman-Sundberg M, Flinois JP, de Waziers I, et al. (2009) Xenobiotic-metabolizing enzymes and transporters in the normal human brain: regional and cellular mapping as a basis for putative roles in cerebral function. Drug Metab Dispos 37:1528–1538 [DOI] [PubMed] [Google Scholar]
- Erratico CA, Szeitz A, Bandiera SM. (2013) Biotransformation of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) by human liver microsomes: identification of cytochrome P450 2B6 as the major enzyme involved. Chem Res Toxicol 26:721–731 [DOI] [PubMed] [Google Scholar]
- Feo ML, Gross MS, McGarrigle BP, Eljarrat E, Barcelo D, Aga DS, and Olson JR (2013) Biotransformation of BDE-47 to potentially toxic metabolites is predominantly mediated by human CYP2B6. Environ Health Perspect 121:440–446, 446e441–447. [DOI] [PMC free article] [PubMed]
- Ferguson CS, Tyndale RF. (2011) Cytochrome P450 enzymes in the brain: emerging evidence of biological significance. Trends Pharmacol Sci 32:708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foxenberg RJ, McGarrigle BP, Knaak JB, Kostyniak PJ, Olson JR. (2007) Human hepatic cytochrome p450-specific metabolism of parathion and chlorpyrifos. Drug Metab Dispos 35:189–193 [DOI] [PubMed] [Google Scholar]
- Greenblatt DJ, He P, von Moltke LL, Court MH.(2008) The CYP3 family, in Cytochromes P450: Role in the Metabolism and Toxicity of Drugs and Other Xenobiotics, pp 354–383, The Royal Society of Chemistry, London [Google Scholar]
- Guengerich FP, Tang Z, Salamanca-Pinzón SG, Cheng Q. (2010) Characterizing proteins of unknown function: orphan cytochrome p450 enzymes as a paradigm. Mol Interv 10:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedlund E, Gustafsson JA, Warner M. (2001) Cytochrome P450 in the brain; a review. Curr Drug Metab 2:245–263 [DOI] [PubMed] [Google Scholar]
- Kania-Korwel I, Barnhart CD, Stamou M, Truong KM, El-Komy MH, Lein PJ, Veng-Pedersen P, Lehmler HJ. (2012) 2,2′,3,5′,6-Pentachlorobiphenyl (PCB 95) and its hydroxylated metabolites are enantiomerically enriched in female mice. Environ Sci Technol 46:11393–11401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania-Korwel I, Hrycay EG, Bandiera SM, Lehmler H-J. (2008) 2,2′,3,3′,6,6′-Hexachlorobiphenyl (PCB 136) atropisomers interact enantioselectively with hepatic microsomal cytochrome P450 enzymes. Chem Res Toxicol 21:1295–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhar JY, Tyndale RF. (2012) Rat brain CYP2B-enzymatic activation of chlorpyrifos to the oxon mediates cholinergic neurotoxicity. Toxicol Sci 126:325–335 [DOI] [PubMed] [Google Scholar]
- Kim KH, Bose DD, Ghogha A, Riehl J, Zhang R, Barnhart CD, Lein PJ, Pessah IN. (2011) Para- and ortho-substitutions are key determinants of polybrominated diphenyl ether activity toward ryanodine receptors and neurotoxicity. Environ Health Perspect 119:519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RP, Gehlhaus M, Knoth R, Volk B. (2007) Expression and function of cytochrome p450 in brain drug metabolism. Curr Drug Metab 8:297–306 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindranath V, Strobel HW. (2013) Cytochrome P450-mediated metabolism in brain: functional roles and their implications. Expert Opin Drug Metab Toxicol 9:551–558 [DOI] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M, Klaassen CD. (2011) Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci 124:261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilter B, Andersen MR, Acharya C, Omiecinski CJ. (2000) Activation of cytochrome P450 gene expression in the rat brain by phenobarbital-like inducers. J Pharmacol Exp Ther 294:916–922 [PubMed] [Google Scholar]
- Schilter B, Omiecinski CJ. (1993) Regional distribution and expression modulation of cytochrome P-450 and epoxide hydrolase mRNAs in the rat brain. Mol Pharmacol 44:990–996 [PubMed] [Google Scholar]
- Spencer PS, Lein PJ.(2013) Neurotoxicity, in Encyclopedia of Toxicology (Wexler P. ed), Elsevier, Oxford, UK [Google Scholar]
- Tang J, Cao Y, Rose RL, Brimfield AA, Dai D, Goldstein JA, Hodgson E. (2001) Metabolism of chlorpyrifos by human cytochrome P450 isoforms and human, mouse, and rat liver microsomes. Drug Metab Dispos 29:1201–1204 [PubMed] [Google Scholar]
- Upadhya SC, Chinta SJ, Pai HV, Boyd MR, Ravindranath V. (2002) Toxicological consequences of differential regulation of cytochrome p450 isoforms in rat brain regions by phenobarbital. Arch Biochem Biophys 399:56–65 [DOI] [PubMed] [Google Scholar]
- Volk B, Meyer RP, von Lintig F, Ibach B, Knoth R. (1995) Localization and characterization of cytochrome P450 in the brain. In vivo and in vitro investigations on phenytoin- and phenobarbital-inducible isoforms. Toxicol Lett 82-83:655–662 [DOI] [PubMed] [Google Scholar]
- Warner M, Köhler C, Hansson T, Gustafsson J-Å. (1988) Regional distribution of cytochrome P-450 in the rat brain: spectral quantitation and contribution of P-450b,e, and P-450c,d. J Neurochem 50:1057–1065 [DOI] [PubMed] [Google Scholar]
- Woodland C, Huang TT, Gryz E, Bendayan R, Fawcett JP. (2008) Expression, activity and regulation of CYP3A in human and rodent brain. Drug Metab Rev 40:149–168 [DOI] [PubMed] [Google Scholar]
- Yoo JSH, Hong JY, Ning SM, Yang CS. (1990) Roles of dietary corn oil in the regulation of cytochromes P450 and glutathione S-transferases in rat liver. J Nutr 120:1718–1726 [DOI] [PubMed] [Google Scholar]
- Zhou K, Khokhar JY, Zhao B, Tyndale RF. (2013) First demonstration that brain CYP2D-mediated opiate metabolic activation alters analgesia in vivo. Biochem Pharmacol 85:1848–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.