Abstract
Objective
A comprehensive and machine-understandable cancer drug–side effect (drug–SE) relationship knowledge base is important for in silico cancer drug target discovery, drug repurposing, and toxicity predication, and for personalized risk–benefit decisions by cancer patients. While US Food and Drug Administration (FDA) drug labels capture well-known cancer drug SE information, much cancer drug SE knowledge remains buried the published biomedical literature. We present a relationship extraction approach to extract cancer drug–SE pairs from the literature.
Data and methods
We used 21 354 075 MEDLINE records as the text corpus. We extracted drug–SE co-occurrence pairs using a cancer drug lexicon and a clean SE lexicon that we created. We then developed two filtering approaches to remove drug–disease treatment pairs and subsequently a ranking scheme to further prioritize filtered pairs. Finally, we analyzed relationships among SEs, gene targets, and indications.
Results
We extracted 56 602 cancer drug–SE pairs. The filtering algorithms improved the precision of extracted pairs from 0.252 at baseline to 0.426, representing a 69% improvement in precision with no decrease in recall. The ranking algorithm further prioritized filtered pairs and achieved a precision of 0.778 for top-ranked pairs. We showed that cancer drugs that share SEs tend to have overlapping gene targets and overlapping indications.
Conclusions
The relationship extraction approach is effective in extracting many cancer drug–SE pairs from the literature. This unique knowledge base, when combined with existing cancer drug SE knowledge, can facilitate drug target discovery, drug repurposing, and toxicity prediction.
Keywords: Information Extraction, Text Mining, Cancer Drug Toxicity, Natural Language Processing, Drug Target Discovery, Drug Repurposing
Background
Cancer drugs can have potentially severe or even fatal side effects (SEs). The effectiveness of many cancer drugs is greatly limited by their level of toxicity.1 The majority of cytotoxic cancer drugs are not cancer cell-specific, which leads to many common severe SEs such as neutropenia, anemia, and thrombocytopenia. Newer classes of biological agents affect tumors selectively and can cause distinctive drug-specific SEs. Accurate knowledge of the possible SEs of cancer drugs plays a major role in clinician and patient decisions regarding treatment options.2 For example, cancer patients treated with cisplatin may experience serious adverse events affecting hearing, the nervous system, and the kidneys, but such toxicities may be acceptable if the benefit–risk ratio is high and if a given patient can tolerate such severe SEs. The biological cancer drug cetuximab for the treatment of metastatic colorectal cancer produces an acne-like rash, which may be tolerable for many patients. In contrast, the biological agent rituximab (for treating chemotherapy-refractory B-cell non-Hodgkin lymphomas) can cause death, cardiac arrest, and acute kidney failure, a fact that would make this treatment unappealing to all but the most advanced cancer patients. Due to the nature of most cancer drug therapies, drug SEs are often inevitable and cancer treatments frequently involve maintaining a delicate balance between therapeutic benefits and risks. Therefore, the development of a comprehensive and accurate cancer drug–SE relationship knowledge base is important for both patients and physicians because it will make informed and personalized medical decision-making clearer, easier, and more accurate.3
A comprehensive, accurate, and machine-understandable cancer drug–SE relationship knowledge base is also important for computational approaches for cancer drug target discovery, drug repurposing, and toxicity prediction. Using text-mining methods, Kuhn et al4 have compiled from US Food and Drug Administration (FDA) package inserts, a drug SE resource called SIDER (Side Effect Resource) which contains information on FDA-approved drugs only. This computable drug SE information integrated with drug chemical and biological data has been used in both drug target discovery and repurposing5 and drug adverse event prediction.6 In addition to drug SE information on FDA drug labels, a large amount of drug–SE relationship information is also available in other data sources such as FDA spontaneous post-marketing drug safety reporting systems, patient electronic health records (EHRs), and the large body of published biomedical literature. These drug SE knowledge sources overlap with, as well as complement, each other. While the FDA drug labels, spontaneous post-marketing drug safety reporting systems, and EHRs mainly contain SE information on FDA-approved drugs, the published biomedical literature contains SE information on investigational, approved, and even failed drugs. For systems approaches to studying phenotypic relationships among drugs, drug–SE association information from all these sources is necessary to mitigate the data incompleteness problems inherent in many biomedical networks.7
In this study, we aimed to build a cancer drug–SE relationship knowledge base from the published biomedical literature. Currently, more than 22 million biomedical records are available on MEDLINE, making it a rich knowledge source of cancer drug–SE relationships. However, the richness of this source, arising from the sheer volume of published articles, limits its usability because much of the important knowledge it contains is buried in free text with limited machine-understandability. Automatic extraction of biomedical relationships from MEDLINE is a highly active area. Common approaches for relationship extraction use rule-based, statistical approaches, machine learning, or natural language processing techniques. Many of these efforts have focused on extracting relationships among drugs, proteins, and genes.8–13 Compared to other biomedical relationship extraction tasks, extracting drug–SE relationships from MEDLINE has been less explored. Recently, Shetty et al14 prototyped a process for applying information mining to discover major drug adverse events associations among 38 drugs and 55 SEs from MEDLINE. The researchers first developed a statistical document classifier (using MeSH index terms) to identify relevant articles and used disproportionality analysis to find signals of disproportionate reporting. Similarly, Wang et al15 reported on a prototype study in which a statistical text classifier trained on MeSH terms was developed in order to automatically determine drug–neutropenia associations. The classifiers in those two studies were constructed based on a set of manually selected ontological and textual features and MeSH terms. Avillach et al16 used MeSH terms (ie, ‘chemically-induced,’ ‘adverse events’) to automate the MEDLINE search to determine if a give drug–SE association was already reported in the literature. There are two perceived limitations in using MeSH terms alone in cancer drug SE extraction: first, cancer drug SE information often appears together with drug–disease treatment information in the same articles. Using MeSH subheadings such as ‘adverse effects’ may retrieve articles containing drug SE information, however, these articles also contain drug treatment information. Since one of the key issues in cancer drug–SE relationship extraction is to differentiate drug–SE pairs from drug–treatment pairs, using MeSH subheadings alone may not be sufficient for such task. Second, it is not clear how sensitive the MeSH terms are in capturing articles that reported any drug adverse events. It is possible that free text abstracts contain drug–SE associations but are not assigned the relevant MeSH terms. Recently, Gurulingappa et al17 developed a machine learning approach trained on a manually annotated corpus to automatically identify adverse drug event assertive sentences in case reports. The system then identified the co-occurring drugs and conditions from positively classified sentences over pre-selected drugs. All of the above mentioned approaches depend on training statistical classifiers in order to remove articles not related to adverse events.
There are three main challenges in automatically extracting cancer drug–SE relationships from the published literature: (1) we need to differentiate cancer drug-specific pairs from non-cancer drug-related pairs; (2) we need to differentiate drug–disease ‘TREAT’ pairs from drug–SE ‘CAUSE’ pairs; and (3) we need to differentiate cancer drug–SE semantic pairs from pure co-occurrence (non-semantic) pairs. To address these challenges, we first built a cancer drug list leveraging known cancer drug treatments and on biomedical ontologies, and identified drug–SE co-occurrence pairs that were cancer drug-related. We then developed a filtering approach to remove drug–disease ‘TREAT’ pairs from extracted cancer drug–SE co-occurrence pairs. Finally, we developed a ranking scheme to further improve the precision of filtered pairs by ranking semantic pairs high and spurious (pure co-occurrence) pairs low. To demonstrate the potential of the constructed knowledge base in cancer drug target discovery and drug repurposing, we analyzed the associations of SEs with drug gene targets and indications. With the knowledge base we created and integrated with existing cancer drug SE knowledge sources, it will be possible to develop phenotype-driven network-based approaches by systematically studying drug SE profile similarities to identify drugs with significant overlap in SEs but different indications or gene targets.
Data and methods
The entire process is depicted in figure 1 and consists of the following steps: (1) obtaining MEDLINE data; (2) building a cancer-specific drug lexicon; (3) building a clean SE lexicon; (4) extracting cancer drug–SE co-occurrence pairs from MEDLINE; (5) filtering extracted drug–SE pairs by automatically removing drug–disease treatment pairs; (6) ranking filtered pairs based on MEDLINE frequency to differentiate true drug–SE pairs from potentially pure co-occurrence pairs; and (7) analyzing the association of SEs with drug gene targets and indications.
Figure 1.
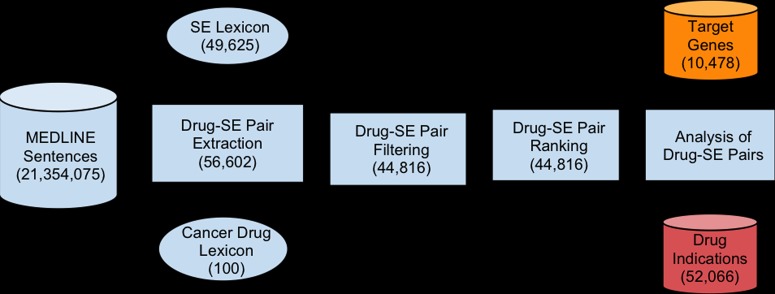
Flow chart depicting the process of cancer drug–SE relationship extraction, filtering, ranking, and analysis. SE, side effect.
MEDLINE data
We used 21 354 075 MEDLINE records (119 085 682 sentences) published between 1965 and 2012 as the text corpus. The 2012 MEDLINE/PubMed XML files were downloaded from NLM's anonymous FTP server at ftp://ftp.nlm.nih.gov/nlmdata/. The MEDLINE XML files were then parsed, and the abstracts (including titles) and PMIDs were extracted from the XML files. Abstracts were then split into sentences.
Cancer drug lexicon
We built a cancer drug list based on drug–disease treatment pairs from ClinicalTrials.gov, the registry of federally and privately supported clinical trials conducted in the USA and around the world. Each trial listed at ClinicalTrials.gov is associated with corresponding medical conditions and drug treatments. We downloaded a total of 115 026 clinical trial XML files from Clinicaltrials.gov.18 A total of 196 002 drug–disease pairs were extracted from the downloaded XML files. After drug and disease named entity recognition, mapping, cleaning and normalization, we obtained a total of 52 066 unique drug–disease pairs. Of these, 17 386 pairs were related to cancers as determined by the semantic type (‘Neoplastic Process’) of the disease terms derived from the Unified Medical Language System.19 Not all drug–disease pairs extracted from ClinicalTrials.gov proved to be valid pairs. For example, if m drugs and n diseases were listed in one single clinical report, we extracted a total of m × n drug–disease pairs, some of which did not have valid semantic relationships. We then filtered these drug–cancer pairs by their MEDLINE co-occurrence in order to remove spurious associations. Since our task was to extract cancer drug–SE pairs from MEDLINE, this filtering step was important in creating a MEDLINE-specific cancer drug list. After this MEDLINE-based filtering step, we obtained a list of 358 potential cancer drug names. We then ranked these potential cancer drugs based on their frequencies in ClinicalTrial.gov and manually selected 100 cancer drugs from the top-ranked drugs.
SE lexicon
For drug–SE relationship extraction tasks, the two critical inputs are the drug lexicon and SE lexicon. We have built a cancer-specific drug lexicon as shown above. For the SE lexicon, we manually created a clean SE lexicon based on MedDRA (the Medical Dictionary for Regulatory Activities). MedDRA is a medical terminology used to classify adverse event information by health authorities and the biopharmaceutical industry.20 However, many terms in MedDRA by themselves are not SE terms. For instance, the MedDRA lexicon contains thousands of medical procedure or laboratory test terms such as ‘abdomen scan,’ ‘abdominal cat,’ ‘vasectomy,’ ‘acupuncture,’ and ‘allergy test.’ In addition, the names of many chemicals and proteins are included such as ‘ACTH,’ ‘Aldolase,’ ‘aldosterone,’ ‘alphaglobulin,’ and ‘aluminum.’ We manually curated all the terms in MedDRA. After manual curation, the final clean SE lexicon consisted of a total of 49 625 terms, a significant reduction from the original 70 177 terms.
Drug–SE pair extraction
We tagged MEDLINE sentences with the entities from the cancer drug lexicon and the clean SE lexicon. The tagging was based on case-insensitive extract string matching for high precision and efficiency. Drug–SE co-occurrence pairs were then extracted from both sentences and abstracts.
Drug–SE pair filtering
We filtered the extracted co-occurrence pairs by removing known drug–disease treatment pairs (derived from ClinicalTrials.gov), and by removing drug–SE pairs that contained cancer terms as determined by semantic type ‘Neoplastic Process.’ We compared the precision, recall, and F1 score of pairs before and after filtering using two complementary evaluation datasets (discussed later).
Drug–SE pair ranking
While the above filtering scheme removed many drug–disease treatment pairs, many non-semantic drug–SE pairs were still left. We then ranked the filtered cancer drug–SE pairs based on their MEDLINE frequency counts. The reasoning behind this ranking is that if a drug–SE pair co-occurs in MEDLINE many times, it is likely to have a valid semantic relationship. We used the 11-point interpolated average precision measures, often used for measuring ranked retrieval results for search engines,21 to evaluate the drug SE ranking algorithms. The interpolated precision was measured at the 11 recall levels of 0.0, 0.1, 0.2, …, 1.0.
Evaluation
We created two evaluation datasets for evaluating the precision, recall, and F1 score at each of the extraction steps. The first one was based on SIDER, a drug SE knowledge base compiled from FDA drug labels.4 We downloaded 96 785 drug–SE pairs from the SIDER database. We filtered these pairs by their MEDLINE occurrences and by the cancer drug list we created. We derived an evaluation dataset consisting of 5556 cancer drug–SE pairs. Since not all cancer drug–SE pairs reported in MEDLINE are also captured on FDA drug labels, we created a second evaluation dataset by manually curating all MEDLINE articles containing the drug name ‘irinotecan,’ a commonly used chemotherapy drug. We first retrieved all sentences (10 044 in total) containing the term ‘irinotecan.’ Three curators then manually extracted SEs in which irinotecan was implicated from these sentences. Only the pairs agreed upon by all three curators were used. The resultant evaluation set consisted of 126 irinotecan–SE pairs. Standard precision, recall, and F1 measures were used.
These two evaluation datasets overlap with, as well as complement, each other. For example, 133 irinotecan–SE pairs are in SIDER and 126 are in the MEDLINE-based ‘irinotecan–SE’ dataset. However, only 30 pairs overlap, demonstrating that drug SE knowledge on FDA drug labels and in the biomedical literature complement each other. Each of these two evaluation datasets has its own limitations and advantages. The SIDER-based dataset contains pairs for many cancer drugs, but may underestimate the precision of drug SE extraction from MEDLINE, since it does not contain all pairs mentioned in the MEDLINE corpus. The manually curated ‘irinotecan–SE’ dataset captured most of the drug–SE associations in MEDLINE for irinotecan. However, it is limited to one cancer drug due to the intense manual curation required. By using both datasets, we can get a better estimate of both precision and recall than using either one alone.
Analysis of drug–SE pairs
In this study, we investigated whether cancer drugs with the same SEs tend to have overlapping gene targets or indications. Our goals were twofold: first, we aimed to show the effectiveness of our filtering and ranking methods in removing spurious pairs; and second, we aimed to demonstrate that the extracted drug–SE pairs have potential in computational approaches for predicting unknown drug adverse events as well as in drug target discovery and drug repurposing. We extracted 10 478 drug–target gene pairs from DrugBank22 and 52 066 drug–disease pairs from ClinicalTrials.gov. For drug–drug pairs that share SEs at multiple thresholds, we calculated the average numbers of shared gene targets as well as shared indications. More specifically, we calculated the average numbers of shared gene targets and indications for drug–drug pairs sharing at least 0 SEs (all drug–drug combinations), 10, 20, 30, …, or at least 100 SEs.
Results
Filtering out potential drug–disease treatment pairs greatly improves the performance of cancer drug SE extraction from MEDLINE
Using the cancer drug lexicon and the clean SE lexicon we created, we extracted a total of 56 602 drug–SE co-occurrence pairs from sentences and a total of 134 670 pairs from abstracts. Since one of the main complicating factors in drug–SE ‘CAUSE’ relationship extraction is the inclusion of drug–disease ‘TREAT’ pairs, we first filtered the extracted co-occurrence pairs by removing known drug–disease treatment pairs from ClinicalTrials.gov (‘Filtering1’). Then, we removed pairs that have SE terms of the semantic type ‘Neoplastic Process’ (‘Filtering2’), which removed additional drug–disease treatment pairs that were not included in ClinicalTrials.gov. The precision, recall, and F1 score of the extracted drug–SE co-occurrence pairs before and after filtering were evaluated and compared. As shown in table 1, pairs extracted from sentences before any filtering had very low precisions: 0.059 when evaluated using the SIDER dataset and 0.252 when evaluated using the ‘Irinotecan–SE’ dataset. By manual examination, it was possible to see that many of the unfiltered co-occurrence pairs were in fact drug–disease treatment pairs. After the removal of known drug–disease treatment pairs from ClinicalTrials.gov (‘Filtering1’), however, the precision did not improve much. This may be due to the fact that many of the drug–disease treatment pairs in MEDLINE are not included in ClinicalTrials.gov. We then filtered out drug–SE co-occurrence pairs whose SE terms were cancer terms. The precision of the filtered pairs was much improved, while the recall remained the same. When evaluated with the SIDER dataset, the precision of filtered pairs was 0.072, representing a 22% increase from the precision of 0.059 for unfiltered pairs. When evaluated with the MEDLINE-based ‘Irinotecan-SE’ dataset, the precision increased from 0.252 to 0.426, representing a 69% increase; the recall did not decrease. Note that evaluation with the SIDER database may underestimate the actual precision of drug SE extraction from MEDLINE since many true drug–SE pairs reported in MEDLINE are not captured in the SIDER dataset. Similar improvement was observed for abstract-level extraction. Overall, both the precision and F1 score for abstract-level extraction were lower than for sentence-level extraction.
Table 1.
The precision, recall, and F1 score for pairs without filtering (‘No Filtering’), with known drug–disease treatment pairs filtered out (‘Filtering1’), and with pairs containing cancer terms filtered out (‘Filtering2’)
| Document type | Evaluation data | Filtering | p Value | R | F1 |
|---|---|---|---|---|---|
| Sentence | SIDER | No Filtering | 0.059 | 0.599 | 0.107 |
| Filtering1 | 0.059 | 0.563 | 0.107 | ||
| Filtering2 | 0.072 | 0.583 | 0.129 | ||
| Irinotecan–SE | No Filtering | 0.252 | 0.786 | 0.382 | |
| Filtering1 | 0.288 | 0.769 | 0.419 | ||
| Filtering2 | 0.426 | 0.786 | 0.553 | ||
| Abstract | SIDER | No Filtering | 0.038 | 0.927 | 0.073 |
| Filtering1 | 0.038 | 0.890 | 0.073 | ||
| Filtering2 | 0.044 | 0.908 | 0.085 | ||
| Irinotecan–SE | No Filtering | 0.099 | 0.786 | 0.177 | |
| Filtering1 | 0.104 | 0.769 | 0.183 | ||
| Filtering2 | 0.139 | 0.786 | 0.235 |
SE, side effect.
The precision improvements achieved by filtering out potential drug–disease treatment pairs demonstrated that one of the main complicating factors in cancer drug–SE relationship extraction from MEDLINE is the inclusion of drug–disease treatment pairs. By removing potential drug–disease treatment pairs, we greatly improved the precision without causing a decrease in recall. The task for cancer drug–SE relationship extraction is made easier than general drug SE extraction by the fact that we can largely decide whether a cancer drug–SE pair is a ‘TREAT’ or ‘CAUSE’ pair by the semantic type of SE terms alone. This filtering scheme may not be generalized to all drug SE extraction tasks. Even with both filtering steps, the precision of 0.426 for sentence-level extraction is still low. This demonstrates that there are still other complicating factors such as the inclusion of pairs without obvious semantic relationships (pure co-occurrence pairs). Next we developed a ranking algorithm to differentiate true cancer drug–SE pairs from pairs without a strong semantic relationship.
Ranking filtered pairs by frequency further improves precision
We ranked filtered drug–SE pairs (44 816 in sentence-level extraction and 113 649 in abstract-level extraction) by their MEDLINE frequency. Figure 2 shows the ranked precisions at 11 recall values. The evaluation was carried out using the evaluation set ‘Irinotecan–SE’ since the SIDER-based evaluation set tended to underestimate precisions. As shown in figure 2, ranking by MEDLINE frequency effectively ranked true positives highly among all the filtered pairs. For sentence-level extraction, the precision increased from 0.423 for all filtered pairs (at a recall of 1.0) to 0.778 for the top-ranked pairs (at a recall of 0.1). For abstract-level extraction, the precision increased from 0.139 for all pairs (at a recall of 1.0) to 1.000 for the top-ranked pairs (at a recall of 0.1). The precisions of ranked pairs at all recalls (except at 0.1 and 0.2) were higher for sentence-level extraction than for abstract-level extraction.
Figure 2.
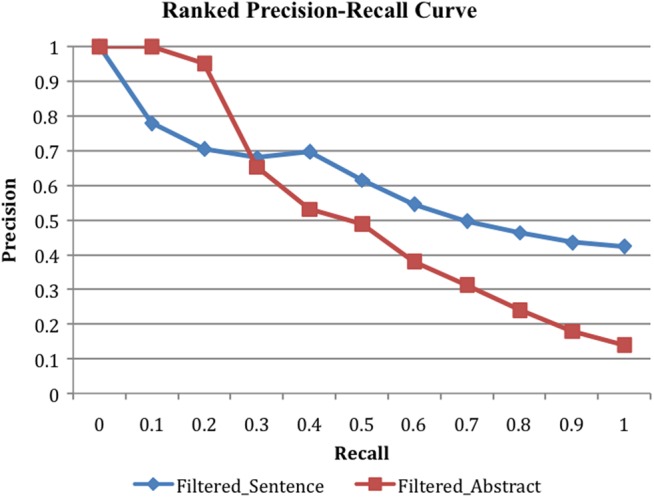
Filtered drug–SE pairs extracted from sentences (‘Filtered_Sentence’) and abstracts (‘Filtered_Abstract’) and ranked by MEDLINE frequency. SE, side effect.
Ranking by MEDLINE frequency to improve precision only worked for filtered pairs. Before filtering, the semantic relationships between a drug and a medical condition can be ‘TREAT,’ ‘CAUSE,’ or other relationships. Ranking by MEDLINE frequency can differentiate pairs with strong semantic relationships from spurious pairs, but not ‘CAUSE’ pairs from ‘TREAT’ pairs. We compared the ranked precisions of unfiltered pairs using two evaluation datasets: the set ‘Irinotecan–SE’ for ‘CAUSE’ relationship evaluation, and drug–disease treatment pairs from ClinicalTrials.gov for ‘TREAT’ relationship evaluation. As shown in figure 3, ranking by MEDLINE frequency is not effective in ranking drug–SE pairs for unfiltered pairs highly (‘Unfiltered_Sentence_Irinotecan_SE’). Instead, it ranked drug–disease treatment pairs highly (‘Unfiltered_Sentence_Drug_Disease_ClinicalTrials’). The same is true for unfiltered pairs extracted from MEDLINE abstracts (data not shown). In summary, before filtering, ranking by MEDLINE frequency ranks drug–disease treatment pairs highly, but not drug–SE causal pairs. After filtering out drug–disease treatment pairs, ranking by MEDLINE frequency was effective in differentiating causal pairs from pairs without strong semantic associations. Therefore, this ranking scheme can assist in prioritizing the filtered cancer drug–SE pairs for further processing.
Figure 3.
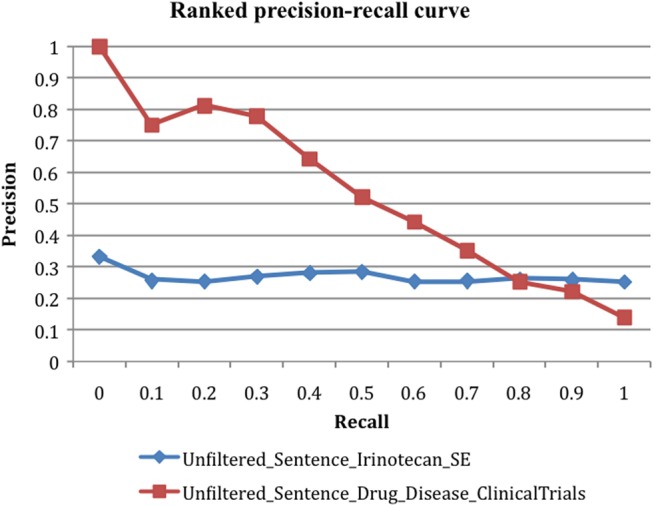
Unfiltered drug–SE pairs ranked by MEDLINE frequency and evaluated using dataset ‘Irinotecan–SE’ pairs (‘Unfiltered_Sentence_Irinotecan_SE’) and drug–disease treatment pairs from ClinicalTrials.gov (‘Unfiltered_Sentence_Drug_Disease_ClinicalTrials’). SE, side effect.
Cancer drugs with the same SEs tended to have overlapping gene targets
We analyzed the relationship between the extracted cancer drug–SE pairs with drug gene targets. A total of 10 478 drug–gene pairs, representing 3 454 drugs and 88 cancer drugs, were parsed from DrugBank. The average number of shared gene targets was 0.041 for all drug–drug combinations and 0.711 for cancer drug–drug combinations. Among 3828 cancer drug–drug pairs that shared any gene targets, 3524 (92%) shared at least 10 SEs. The average shared gene targets for these pairs slightly increased to 0.762 (‘Cancer_Drug_SE’ in figure 4). The number of shared genes increased as the number of shared SEs increased, from 0.711 for all cancer drug–drug pairs to 1.076 for pairs sharing 100 or more SEs. We also showed that the association between drug–SE pairs from the SIDER database and drug gene targets was weaker (‘SIDER_Drug_SE’). The difference diminished as drug–drug pairs shared more SEs. In summary, as the average number of shared SEs increased for drug–drug pairs, so did the number of shared gene targets. The trend was stronger for cancer drug–SE pairs extracted from MEDLINE than for drug–SE pairs from the SIDER database.
Figure 4.
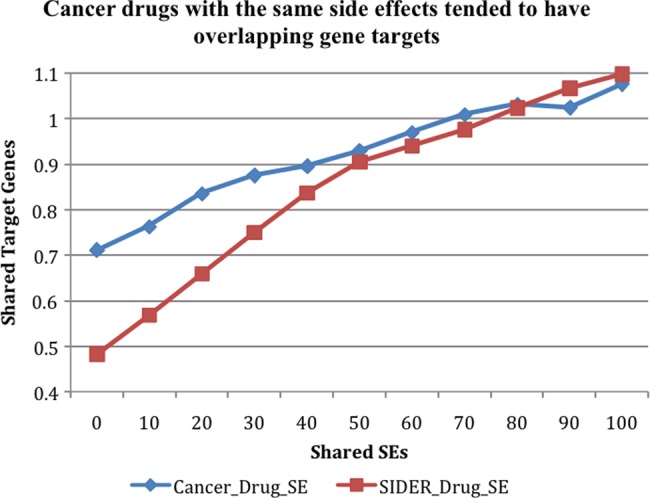
The positive association between drug target genes and drug–SE pairs: cancer specific drug–SE pairs extracted from MEDLINE (‘Cancer_Drug_SE’) and all pairs from the SIDER database (‘SIDER_Drug_SE’). SE, side effect.
Cancer drugs with the same SEs tended to have overlapping indications
We studied the relationship between SEs and drug indications. Drug–disease treatment pairs were removed from the set of drug–SE pairs under consideration during the filtration process. A total of 52 066 drug–disease pairs were extracted from ClinicalTrials.gov, representing 2035 unique drugs and 9591 disease names. As shown in figure 5, the number of shared indications increased from 21.28 for all drug–drug pairs to 37.145 for drug–drug pairs sharing 100 or more SEs (‘Cancer_Drug_SE’). Drug–SE pairs from the SIDER database showed a much weaker upward trend (‘SIDER_Drug_SE’): the number of shared indications increased from 1.022 for all drug–drug pairs to 3.308 for pairs sharing 100 or more SEs.
Figure 5.
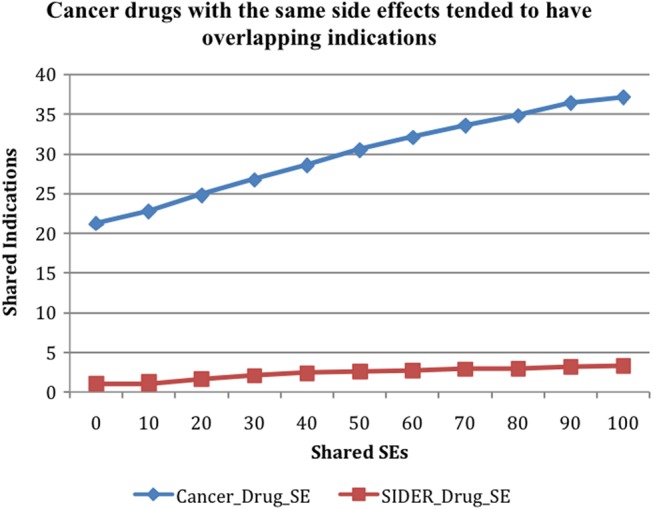
The positive association between drug disease indications and drug–SE pairs: cancer drug–SE pairs extracted from MEDLINE (‘Cancer_Drug_SE’) and all pairs from the SIDER database (‘SIDER_Drug_SE’). SE, side effect.
Discussion
We have presented a multi-step process to extract cancer drug-specific SE association knowledge from the biomedical literature available on MEDLINE. We first built a cancer drug list leveraging known cancer drug treatment pairs. We then extracted cancer drug–SE pairs from MEDLINE using the cancer drug list and a manually created clean SE lexicon. After filtering out potential drug–disease treatment pairs, we greatly improved the precision from 0.252 at baseline to 0.426 without decreasing the recall. We then developed a ranking algorithm to further improve the precision from 0.426 to 0.778 for top-ranked pairs. We demonstrated that as the number of shared SEs between cancer drugs increased, so did the number of shared gene targets and disease indications, showing strong associations between SEs and gene targets, and between SEs and indications.
Many aspects of the current method can be improved. (1) Our approach may only work for cancer drug–SE pair extraction and may not be generalizable to extract other drug–SE pairs. We relied on the semantic type (‘Neoplastic Process’) of SE terms to differentiate ‘CAUSE’ from ‘TREAT’ semantic relationships. In general, we cannot classify drug–SE pairs by the semantic types of SE terms alone. (2) The ranking by MEDLINE co-occurrence frequency ranked drug–disease treatment pairs highly, therefore it only works on filtered drug–SE co-occurrence pairs where the majority of drug–disease treatment pairs have already been filtered out. For general drug–SE relationship extraction from MEDLINE, one of the key issues is to differentiate ‘CAUSE’ from ‘TREAT’ pairs. The most intuitive way is to filter out known drug–disease treatment pairs. However, we often do not have a comprehensive drug–disease treatment relationship knowledge base that contains pairs for both FDA-approved drug indications and pairs that are not approved but are reported in MEDLINE articles. Automatic approaches in extracting drug–disease treatment pairs from MEDLINE can potentially greatly facilitate drug–SE relationship extraction tasks. (3) We only extracted pairs from MEDLINE abstracts, not full-text articles. In one of our ongoing studies, we downloaded all full-text oncology articles from cancer-specific journals such as the Journal of Clinical Oncology and are developing methods to extract cancer drug–SE pairs from these cancer-specific full text articles. We will compare the cancer drug–SE relationships extracted from MEDLINE abstracts to those extracted from cancer-specific full text articles. (4) Most cancer drug therapies are drug combinations consisting of more than one drug. Our study only considered single drugs, not drug combinations. In general, it will be a difficult task to automatically attribute a SE to a specific drug or drug combination. (5) In this study, we only used free text abstracts and ignored the information captured in MeSH terms. Currently, there are no studies comparing the advantages of one method over the other in the context of drug–SE relationship extraction. In our future studies, we will investigate this interesting topic by comparing and combining free text abstracts and MeSH terms as well as publication types to further improve the performance of the drug SE extraction task. (6) Since the overall precision of the extracted cancer drug–SE pairs is still low, the data are not yet useful for patients and clinicians. We are developing additional approaches (both automatic and manual) to further improve the coverage and the precision of the database. However, we believe that this dataset can be used to increase the data completeness of existing cancer drug SE knowledge sources to facilitate systems approaches for cancer drug target discovery and drug repurposing.
Conclusions
We have presented a three-step information extraction approach to extract cancer drug–SE relationships from over 21 million published biomedical abstracts available on MEDLINE. After automatically removing non-cancer drug-related pairs, drug–disease treatment pairs, and pairs without strong semantic relationships, we greatly improved the precision from 0.252 at baseline to 0.778, all without causing a decrease in the recall. We showed that cancer drugs that have the same SEs tend to have overlapping gene targets and overlapping indications, indicating potential value for in silico cancer drug target discovery and drug repurposing. As the precision and coverage of the cancer drug–SE relationship knowledge base we have created continues to improve, it will greatly enhance the ability of practitioners and patients to make more accurate risk–benefit assessments with regard to cancer drug treatment options, and increase the power of SE-based cancer drug target discovery, repurposing, and toxicity prediction models.
Footnotes
Contributors: RX and QW jointly conceived the idea, designed and implemented the algorithms, and wrote the manuscript.
Funding: RX was supported by Case Western Reserve University/Cleveland Clinic CTSA Grant UL1 RR024989, and QW was supported by ThinTek LLC.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement Data are available by contacting rxx@case.edu or qwang@thintek.com.
References
- 1.Richey EA, Lyons EA, Nebeker JR, et al. Accelerated approval of cancer drugs: improved access to therapeutic breakthroughs or early release of unsafe and ineffective drugs? J Clin Oncol 2009;27:4398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladewski LA, Belknap SM, Nebeker JR, et al. Dissemination of information on potentially fatal adverse drug reactions for cancer drugs from 2000 to 2002: first results from the research on adverse drug events and reports project. J Clin Oncol 2003;21:3859–66 [DOI] [PubMed] [Google Scholar]
- 3.Ely JW, Osheroff JA, Ebell MH, et al. Obstacles to answering doctors’ questions about patient care with evidence: qualitative study. BMJ 2002;324:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol 2010;6:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campillos M, Kuhn M, Gavin AC, et al. Drug target identification using side-effect similarity. Science 2008;321:263–6 [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Wu Y, Chen Y, et al. Large-scale prediction of adverse drug reactions using chemical, biological, and phenotypic properties of drugs. J Am Med Inform Assoc 2012;19:e28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mestres J, Gregori-Puigjané E, Valverde S, et al. Data completeness—the Achilles heel of drug-target networks. Nat Biotechnol 2008:26(9):983–4 [DOI] [PubMed] [Google Scholar]
- 8.Xu R, Wang Q. A knowledge-driven conditional approach to extract pharmacogenomics specific drug-gene relationships from free text. J Biomed Inform 2012;45:827–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaschke C, Andrade MA, Ouzounis C, et al. Automatic extraction of biological information from scientific text: protein-protein interactions. Proc Int Conf Intell Syst Mol Biol 1999;7:60–7 [PubMed] [Google Scholar]
- 10.Friedman C, Kra P, Yu H, et al. GENIES: a natural-language processing system for the extraction of molecular pathways from journal articles. Bioinformatics 2001;17(Suppl 1):S74–82 [DOI] [PubMed] [Google Scholar]
- 11.Craven M. Learning to extract relations from MEDLINE. AAAI-99 Workshop on Machine Learning for Information Extraction; July 1999:25–30 [Google Scholar]
- 12.Rindflesch TC, Tanabe L, Weinstein JN, et al. EDGAR: extraction of drugs, genes and relations from the biomedical literature. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing; NIH Public Access, 2000:517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosario B, Hearst MA. Classifying semantic relations in bioscience texts. Proceedings of the 42nd Annual Meeting on Association for Computational Linguistics; Association for Computational Linguistics, July 2004:430 [Google Scholar]
- 14.Shetty KD, Dalal SR. Using information mining of the medical literature to improve drug safety. J Am Med Inform Assoc 2011;18:668–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Haerian K, Salmasian H, et al. A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. AMIA Annual Symposium Proceedings; American Medical Informatics Association, 2011:. 1464–70. [PMC free article] [PubMed] [Google Scholar]
- 16.Avillach P, Dufour JC, Diallo G, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU–ADR project. J Am Med Inform Assoc 2013;20:446–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurulingappa H, Fluck J, Hofmann-Apitius M, et al. Identification of adverse drug event assertive sentences in medical case reports. First International Workshop on Knowledge Discovery and Health Care Management (KD-HCM), European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases (ECML PKDD); 2011 [Google Scholar]
- 18.ClinicalTrials.gov. http://clinicaltrials.gov/ [Google Scholar]
- 19.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32(Suppl 1):D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medical Dictionary for Regulatory Activities (MedDRA). http://www.meddramsso.com/ (accessed May 2012). [DOI] [PubMed]
- 21.Manning CD, Raghavan P, Schütze H. Introduction to information retrieval (Vol. 1). Cambridge: Cambridge University Press, 2008. [Google Scholar]
- 22.Wishart DS, Knox C, Guo AC, et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 2008;36(Suppl 1):D901–6 [DOI] [PMC free article] [PubMed] [Google Scholar]


