Abstract
The proper dissection of the molecular mechanisms governing the specification and differentiation of specific cell types requires isolation of pure cell populations from heterogeneous tissues and whole organisms. Here, we describe a method for purification of nuclei from defined cell or tissue types in vertebrate embryos using INTACT (isolation of nuclei tagged in specific cell types). This method, previously developed in plants, flies and worms, utilizes in vivo tagging of the nuclear envelope with biotin and the subsequent affinity purification of the labeled nuclei. In this study we successfully purified nuclei of cardiac and skeletal muscle from Xenopus using this strategy. We went on to demonstrate the utility of this approach by coupling the INTACT approach with liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomic methodologies to profile proteins expressed in the nuclei of developing hearts. From these studies we have identified the Xenopus orthologs of 12 human proteins encoded by genes, which when mutated in human lead to congenital heart disease. Thus, by combining these technologies we are able to identify tissue-specific proteins that are expressed and required for normal vertebrate organ development.
Keywords: INTACT, Xenopus, Cardiac muscle, Proteomics, Skeletal muscle
INTRODUCTION
The study of organs and tissues is often confounded by the inability to isolate relatively pure populations of a defined cell type or cell lineage. These experimental limitations reduce the potential of emerging highly sensitive technologies, such as high-throughput sequencing and proteomics. Such technologies are sensitive to contaminating cells or tissue types, which can lead to an escalated noise-to-signal ratio. A number of methods to circumvent these problems have been developed. For example, laser microdissection of cells can be used to isolate relatively pure populations of cells from heterogeneous embryonic or adult tissue types for RNA transcript and proteomics profiling (Agostini et al., 2011; Bonnet et al., 2011; Erickson et al., 2009; Golubeva et al., 2013; Imamichi et al., 2001; Longobardi et al., 2012; Rabien, 2010; Shapiro et al., 2012; Strasser et al., 2010; Zivraj et al., 2010). Alternatively, pure cell populations can be isolated on the basis of the expression of a tissue- or cell-specific surface antigen using, for example, panning or fluorescence-activated cell sorting (FACS) (Barker et al., 1975; Barres et al., 1988; Mage et al., 1977; Wysocki and Sato, 1978, Dorrell et al., 2008; Flores-Langarica et al., 2005; Hoi et al., 2010; Lawson et al., 2007; Osorio et al., 2008; Pruszak et al., 2009; Sugiyama et al., 2007; Shen et al., 2003, Claycomb et al., 1998). However, these approaches require either that the cell population of interest expresses a unique surface antigen or a reporter construct(s) that is exclusive to a specific cell or tissue type (Aubert et al., 2003; Bonn et al., 2012a; Bonn et al., 2012b; Fox et al., 2005; Jesty et al., 2012; Pruszak et al., 2007; Von Stetina et al., 2007). A major shortcoming to these types of approaches is availability of antibodies against cell surface receptors and/or promoters with high fidelity to the cell type(s) of interest.
Current methodologies have made possible a surrogate approach through the induction and differentiation of multipotent or totipotent cells, i.e. embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) (Ahmed et al., 2011; Caiazzo et al., 2011; Kim et al., 2009; Kim et al., 2008; Kubo et al., 2004; Laflamme et al., 2007; Laflamme et al., 2005; Lamba et al., 2006; Lee et al., 2000; Maekawa et al., 2011; Okita et al., 2007; Pang et al., 2011; Park et al., 2008; Qian et al., 2012; Szabo et al., 2010; Wernig et al., 2007; Zhou et al., 2008). The in vitro differentiation of ESCs or iPSCs offers the additional advantage that precursor or intermediary cell states can be isolated and assessed (Chen et al., 2013; Paige et al., 2012; Wamstad et al., 2012). However, these methods are compromised in that the induced differentiated cells are most often not a pure population, nor are they often entirely reflective of an endogenous cell type (e.g. Croft and Przyborski, 2006; Pan et al., 2009).
To bypass these technical issues an alternative approach has been pioneered in Arabidopsis thaliana whereby pure populations of nuclei can be isolated from a defined cell or tissue type using nuclei tagged in specific cell types (INTACT) (Deal and Henikoff, 2010). This technology is based on a binary transgene-mediated approach to specifically label and tag nuclei of a specific cell type(s). When the two transgenes are co-expressed nuclei can be labeled in a spatial and temporal manner. The methodology has been combined with a variety of approaches including RNA, protein and chromatin profiling assays in plants and invertebrates (Deal and Henikoff, 2010; Deal and Henikoff, 2011; Henry et al., 2012; Steiner et al., 2012). However to date, the INTACT method has not been applied to a vertebrate model system.
Here, we describe the application of the INTACT method in the study of the frog, Xenopus, an established model for vertebrate development. We report that nuclei derived from Xenopus heart and skeletal muscle can be labeled and affinity purified using the INTACT method. We further demonstrate the utility of coupling INTACT with a mass spectrometry-based proteomics approach to profile proteins expressed in the nuclei of developing Xenopus hearts. Our optimized bioinformatics workflow identified 502 protein clusters with high confidence, 12 of which represent the Xenopus orthologs of human proteins encoded by genes that when mutated lead to congenital heart disease (CHD). Collectively, these studies present a set of general technologies for the effective enrichment of proteins in a cell- or tissue-specific manner in vertebrates.
RESULTS
Expression of INTACT components in Xenopus embryos
The Xenopus model system has historically been used to study a wide array of developmental processes and has the benefit of producing vast quantities of relatively large, externally developing eggs. Crucially, many of the processes and genes involved in development are highly conserved between Xenopus and human and have been characterized in detail (Angelo et al., 2000; Bartlett et al., 2007; Bartlett and Weeks, 2008; Brown et al., 2003; Burdine and Schier, 2000; Gormley and Nascone-Yoder, 2003; Heimeier et al., 2010; Hellsten et al., 2010; Hirsch and Harris, 1997; Horb and Thomsen, 1999; Jiang and Evans, 1996; Kaltenbrun et al., 2011; Mohun et al., 1987; Pearl et al., 2009; Showell et al., 2006; Steelman et al., 1997; Wallingford, 2006; Zon et al., 1991). Xenopus has the additional advantages that embryos can be manipulated by overexpression through microinjection of mRNA or transgenesis, protein depletion with morpholino oligonucleotides, and targeted mutagenesis by zinc finger nucleases and transcription activator-like effector nucleases (TALENs) (Amaya et al., 1991; Hopwood and Gurdon, 1990; Lei et al., 2012; Nakajima et al., 2012; Sakuma et al., 2013; Tandon et al., 2012; Young et al., 2011). Recent annotation of the X. tropicalis genome (Hellsten et al., 2010), a draft sequence of the X. laevis genome, and the advent of new transgenic lines has reinforced this model as ideal for studying vertebrate development.
The INTACT system is a binary trangenic system. The first transgene, referred to as the nuclear targeting fusion construct (NTF) consists of three parts: (1) a nuclear envelope protein to target the NTF to the nuclear membrane; (2) an enhanced green fluorescent protein (eGFP) cassette to visualize the NTF; and (3) a biotin ligase receptor protein (BLRP) that provides a substrate for the addition of biotin. Crucial to this methodology is the BLRP, a peptide with a sequence that has not been identified in any invertebrate or vertebrate genome, and therefore, labeling of BLRP is highly specific. Moreover, exposure of the BLRP-tagged protein to the BirA enzyme, introduced through a second transgene, results in quick (45 seconds or less) stable conjugation of biotin to the protein of interest (Beckett et al., 1999; Wong et al., 1999). The tagged protein can in turn be identified or isolated by streptavidin (Maine et al., 2010; Roesli et al., 2006; van Werven and Timmers, 2006; Wang et al., 2006).
Previous studies have utilized a species-specific nuclear envelope protein in plants, flies and worms (Deal and Henikoff, 2010; Deal and Henikoff, 2011; Steiner et al., 2012). In this regard, we cloned the nuclear envelope protein nucleoporin (Nup35) from Xenopus and fused it in frame with an eGFP::BLRP cassette to generate the Xenopus nuclear targeting construct, xNTF (Fig. 1A). To visualize the second transgene, which biotinylates xNTF in vivo, we tagged BirA with mCherry (Fig. 1B) (Schäffer et al., 2010).
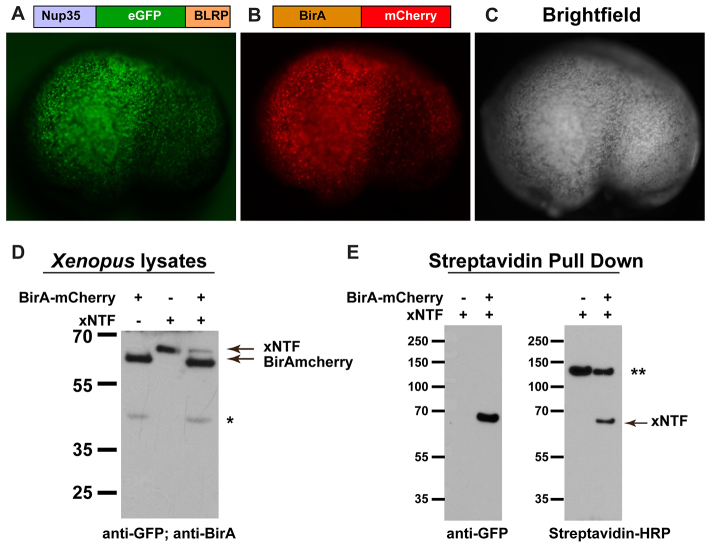
Fig. 1.
Affinity isolation of biotin-tagged proteins from Xenopus embryos. (A-C) Schematic of Nup35::eGFP::BLRP(xNTF) and BirA::mCherry fusion proteins, and corresponding images of a Xenopus embryo co-injected at the one-cell stage with capped mRNA coding for each fusion. Lateral view of a stage 23 embryo with anterior to the left and dorsal up. (D) Western blot analysis of embryos injected individually and together with xNTF and BirA::mCherry shows that the proteins are of the correct size. xNTF was detected with anti-GFP antibodies, and BirA was detected by anti-BirA antibodies. (E) Immunoprecipitation of xNTF with streptavidin-conjugated beads, followed by western blot analysis shows that xNTF is efficiently isolated from embryos by probing with anti-GFP or streptavidin-HRP. *A potential translation product of BirA::mCherry corresponding to the size of BirA protein alone. **Endogenously biotinylated proteins isolated by streptavidin.
Introduction of xNTF or BirA::mCherry mRNA into Xenopus embryos led to robust expression of eGFP and mCherry, respectively, throughout the embryos, beginning at the mid-blastula transition, with expression persisting until at least late tadpole stages (stage 43; Fig. 1A-C, data not shown). Western blot analysis confirmed expression of xNTF and BirA proteins of the predicted size, and crucially, injected embryos displayed no overt phenotype throughout development (Fig. 1D). Taken together, these data demonstrate that xNTF and BirA::mCherry can be expressed in Xenopus embryos and that their expression is not deleterious to early development.
Affinity isolation of NTF using streptavidin
Having established the expression of xNTF and BirA::mCherry in Xenopus, we sought to confirm that the xNTF transgene can be biotinylated with endogenous biotin. To this end, xNTF mRNA was injected into Xenopus embryos in the presence or absence of BirA::mCherry mRNA. Lysates were collected at mid-neurula (stage 23) and biotinylated proteins were affinity purified with streptavidin-coated Dynabeads (Invitrogen). Western blot analysis of the resulting pull-downs with an anti-GFP antibody showed a highly efficient and specific isolation of xNTF from embryos co-injected with xNTF and BirA::mCherry, but not in samples derived from embryos injected with xNTF alone (Fig. 1E). In vivo labeling of xNTF was further verified by probing western blots with streptavidin-HRP (Fig. 1E). Collectively, these results demonstrate that xNTF can be efficiently biotinylated in vivo using endogenous biotin, and further show the xNTF construct can be isolated in a highly specific manner from Xenopus embryos.
Cell type-specific expression of xNTF
We next recovered nuclei from Xenopus embryos that expressed xNTF and BirA::mCherry or xNTF alone by streptavidin affinity purification. We found that intact nuclei were successfully obtained from embryos expressing xNTF and BirA::mCherry (Fig. 2E-H), whereas we were unable to isolate any nuclei from embryos expressing xNTF alone (Fig. 2A-D). Thus, the binary transgenic system of xNTF and BirA::mCherry can be used to label and recover intact nuclei in vertebrates.
Fig. 2.
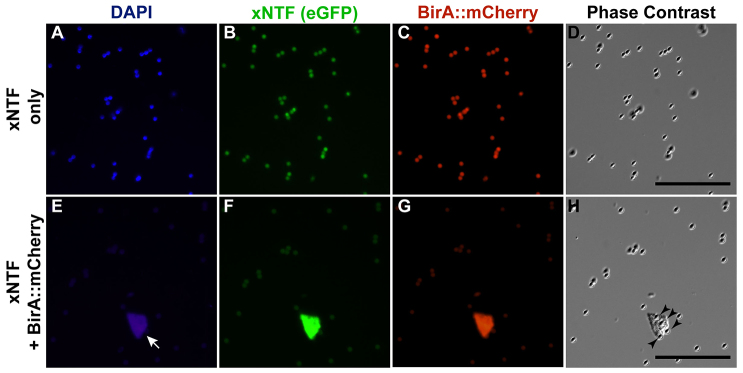
Affinity isolation of xNTF-tagged nuclei. Streptavidin-coated magnetic beads incubated with nuclei from embryos injected with mRNA encoding xNTF with or without BirA::mCherry. (A-D) Fluorescent DAPI (A), xNTF (eGFP; B), BirA::mCherry (Texas Red; C) and phase-contrast (D) images of magnetic beads after nuclear purification of xNTF in the absence of BirA. Autofluorescing beads are detectable throughout the field, but no nuclei. (E-H) Fluorescent DAPI (E), eGFP (F), Texas Red (G) and phase-contrast (H) images of magnetic beads after nuclear purification of xNTF. Note the presence of eGFP and BirA::mCherry-positive nucleus (arrow) and presence of magnetic beads coating nucleus (arrowheads). Scale bars: 50 μm.
To test the tissue specificity of the INTACT system in Xenopus, the xNTF transgene was expressed in a restricted fashion (Fig. 3A,B) using the mlc2p regulatory element, and independently, the cardiac actin regulatory element. mlc2p is expressed in cardiomyocytes from mid-neurula (stage 26) onwards (Latinkić et al., 2004) and the cardiac actin (CA) element, is co-expressed with mlc2p in the cardiomyocytes but in addition is expressed in the developing somitic tissue (Latinkić et al., 2002). In parallel, we generated a transgene that uniformly expresses BirA under control of the CMV promoter.
Fig. 3.
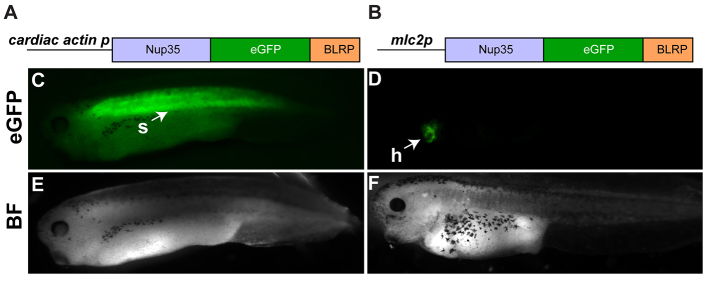
Generation of transgenic embryos expressing xNTF. (A,B) Schematics of constructs used to generate transgenic embryos expressing xNTF in the striated muscle and myocardium (A) and the myocardium only (B). (C-F) Representative images of stage 38 (C,E) and stage 40 (D,F) embryos harboring the respective transgenes. Embryos express xNTF (eGFP) in the expected pattern (C,D), and appear morphologically normal (E,F). eGFP in myocardium is fainter than staining in somites and masked by autofluorescence of the gut in C. Images shown are lateral views with anterior to the left and dorsal up. BF, bright field; h, heart; s, somites.
All transgenes were introduced into Xenopus using restriction enzyme-mediated integration (REMI) (Amaya and Kroll, 1999; Kroll and Amaya, 1996; Mandel et al., 2010). Resulting live embryos were demonstrated to express xNTF in the respective tissue-specific manner (three independent experiments, 40-60% of embryos expressing, n>100 embryos per experiment; Fig. 3C-F). As previously observed for the REMI method in Xenopus (Small and Krieg, 2003), the majority of eggs injected with both xNTF and BirA transgenes co-expressed the two constructs. Integration of the transgenes was verified by genomic PCR of tailclips (Showell and Conlon, 2009) from xNTF, GFP-positive stage 40 embryos (85% co-integration from n=20 animals) and integration of the transgene was further substantiated in juvenile frogs by genomic PCR of toeclips (supplementary material Fig. S1). As predicted, xNTF was localized to the nuclear periphery (visualized by eGFP) in all positive trangenic embryos (Fig. 4A-C).
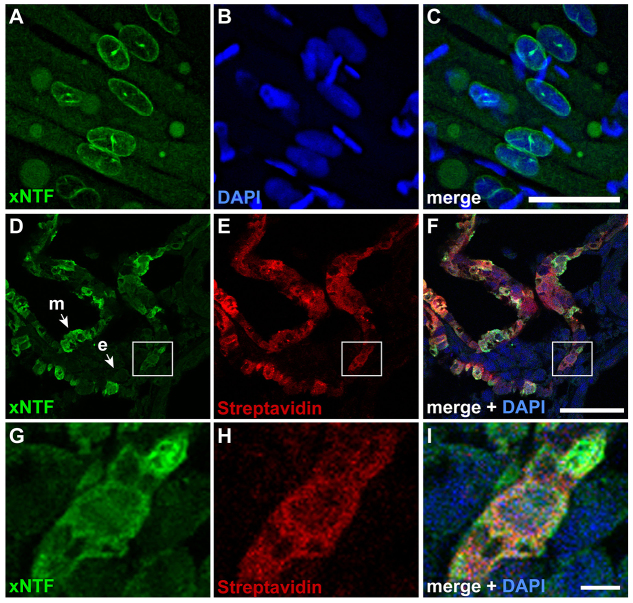
Fig. 4.
Demonstration of xNTF localization and biotin tagging in transgenic animals. (A-C) xNTF localization in the somites as detected by eGFP (A); nuclei were marked with DAPI (B) and the merged image is shown in C. Images are from whole embryos with anterior to the left and dorsal up. (D-F) Transverse sections through the heart (dorsal is up) show expression of xNTF (D) in the myocardium (m) but not the endocardium (e), and corresponding localization of biotinylated xNTF as detected by streptavidin (E). Images were overlaid with DAPI to show the nuclei in F. (G-I) Magnified image of the boxed region in D-F. Scale bars: 200 μm (C); 50 μm (F); 5 μm (I).
To establish that the xNTF transgenes were successfully biotinylated in vivo, we analyzed xNTF in CA::xNTF, CMV::BirA tadpoles by streptavidin staining. Consistent with the expression of CA::reporter constructs, we observed expression of biotinylated xNTF in the myocardium and somites (Fig. 4D-I). Importantly, we did not detect eGFP expression or the presence of biotin in other cell types (Fig. 4D-F). We did not observe xNTF by anti-GFP immunostaining in non-muscle tissue of mlc2p::xNTF; CMV::BirA or CA::xNTF; CMV::BirA embryos, strongly implying that the CMV promoter for the BirA transgene does not interfere with expression of the other two transgenes. Because we did not observe any overt phenotypes or cardiac abnormalities (supplementary material Movie 1), expression of the biotinylated xNTF proteins did not appear to have any adverse effects on muscle or heart development.
Enrichment of muscle-specific nuclei
To confirm that the INTACT method can successfully be used in vertebrates to isolate nuclei in a tissue-specific manner, we examined the expression levels of cardiac- and skeletal muscle-specific genes in nuclei derived from CA::xNTF, CMV::BirA embryos at mid-tadpole stage (stage 40). Indeed, we found a significant enrichment in the expression of the muscle-associated genes acta1, myh6, actc1, tpm2, myoD, mef2a and nkx2-5 isolated from CA::xNTF, CMV::BirA versus non-bead-bound nuclei (Fig. 5A,B). Consistent with the observation that many of these genes are also expressed in non-muscle and non-cardiac tissues (Amin et al., 2013; Tandon et al., 2013; Fu et al., 1998; Hopwood and Gurdon, 1990; Wong et al., 1994; Newman and Krieg, 1998; della Gaspera et al., 2009; Martin and Harland, 2001), we also observe expression of these genes in the non-bound fraction, albeit at significantly lower levels.
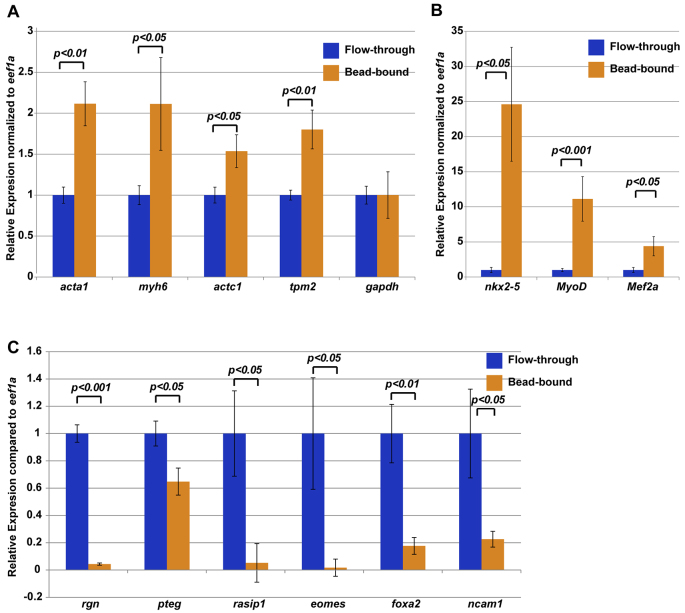
Fig. 5.
INTACT purified nuclei are enriched for cardiac and/or skeletal muscle markers. (A,B) qRT-PCR results show that skeletal muscle structural genes (A) and cardiac or skeletal transcription factors (B) are enriched in bead-bound nuclei compared with flow-through of total nuclei. All expression levels are relative to eef1a and flow-through was normalized to one. Expression of gapdh, another housekeeping gene, was unchanged in bead-bound nuclei compared with flow-through nuclei. (C) qRT-PCR demonstrates that INTACT nuclei show reduced levels of gene expression of non-muscle genes. Error bars represent s.e.m. from a single experiment. Three biological replicates of nuclear purifications were performed. P-values from Student’s t-tests are indicated.
To further validate enrichment of cardiomyocyte and skeletal muscle nuclei, we examined genes not normally detected in muscle or heart including eomes, foxa2, pteg, rgn, rasip1 and ncam1 (Balak et al., 1987; Lee et al., 2010; Misawa and Yamaguchi, 2000; Ryan et al., 1998; Xu et al., 2009; Zorn and Mason, 2001). Crucially, we found a significant reduction in expression of all of these genes in bead-bound nuclei, confirming that the CA::xNTF, CMV::BirA-isolated nuclei are enriched for cardiac and skeletal muscle transcripts (Fig. 5C). Taken together, these results show that the INTACT method successfully enriches for cell-type-specific nuclei of the vertebrate organism Xenopus.
Proteomic profiling of enriched cardiac nuclei
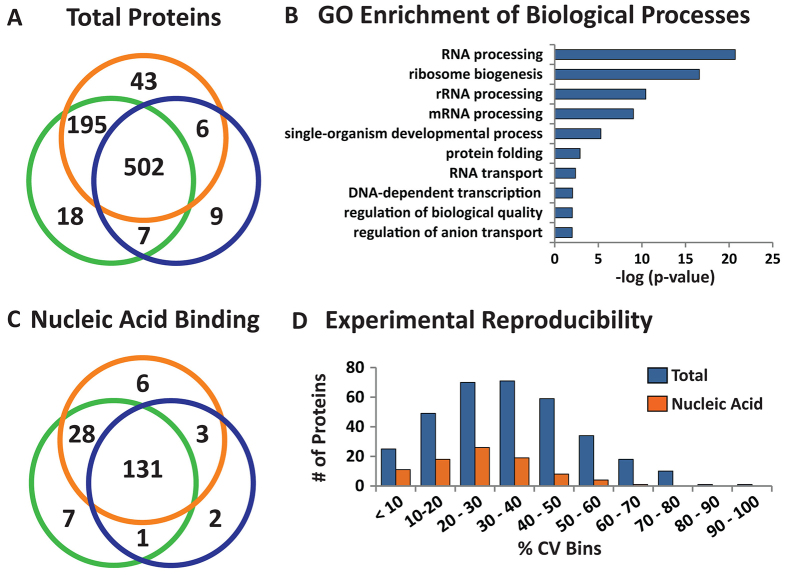
Previous studies have demonstrated the ability to use INTACT-derived nuclei for transcriptional and chromatin profiling (Deal and Henikoff, 2010; Deal and Henikoff, 2011; Steiner et al., 2012). We next set out to test the utility of coupling INTACT with proteomic profiling. Given Xenopus proteomes have not achieved full manual annotation and curation, we used both SwissProt and TrEMBL Xenopus protein sequence databases containing Xenopus laevis and Xenopus tropicalis data. This combination greatly benefits computational peptide-to-spectral assignments by allowing for higher proteome coverage. However, one drawback is the likelihood of obtaining redundant and/or non-unique protein hits. To minimize this effect while retaining relationships between related proteins, we employed both protein grouping and protein clustering, which assembles protein groups based on their shared peptide evidence (see Materials and Methods). From three biological replicates of stage 46 mlc2p::xNTF; CMV::BirAmCherry transgenic embryos, 502 protein clusters containing 909 annotated protein sequences (supplementary material Table S2) were consistently detected in the three replicates with a minimum of two unique peptides (Fig. 6A). Moreover, ten biological process terms were significantly over-represented compared with the total Ensembl Xenopus tropicalis genome, representing predominantly nuclear-related biological functions (Fig. 6B). Specifically, one of the largest molecular function gene ontology terms contained 178 proteins annotated as nucleic acid-binding proteins (supplementary material Table S3), which showed similar detection across biological replicates compared with all proteins (compare Fig. 6C and 6B).
Fig. 6.
Proteomic profiling of INTACT-enriched Xenopus cardiomyocyte nuclei. (A) Venn diagram comparison of the total proteins identified in three biological replicates (n=781). (B) g:profiler biological process gene ontology enrichment analysis. Only terms that were significant versus the annotated Xenopus tropicalis genome are shown (-log(P-value) >2). (C) Venn diagram comparison of proteins identified with ‘nucleic acid binding’ molecular function gene ontology (n=178). (D) Distribution of weighted spectral count percentage coefficient of variation (% CV, n=3). % CV was calculated for shared proteins identified in A and C, corresponding to total proteins (blue bars) and nucleic acid-binding proteins (orange bars) with at least three weighted spectral counts in all biological replicates.
Advancements in proteomic profiling methods also permits simultaneous measurement of abundances in large-scale experiments, often by stable isotope-labeled or ‘label-free’ methods (Baker et al., 2010; Dong et al., 2007; Joshi et al., 2013). To demonstrate the feasibility of quantitative proteomics strategies within enriched cardiac nuclei, we evaluated the peptide-level reproducibility (percentage coefficient of variation; % CV) between biological replicates. Because stable isotope labeling has not yet been demonstrated in Xenopus, we chose a label-free approach as a measure of relative abundance, which can be easily incorporated into existing proteomic workflows. Using weighted spectral counts, the % CV was calculated for each protein cluster with at least three spectral counts detected in all three replicates. For the nucleic acid-binding proteins, more than 94% of proteins had less than 50% spectral count variation (Fig. 6D, orange bars), which was similar to the spectral count variation of total proteins (Fig. 6D, blue bars). These results are consistent with the existing spectral count literature that has shown the 95% confidence range of replicate measurements is often within a relative protein abundance of less than twofold (Old et al., 2005).
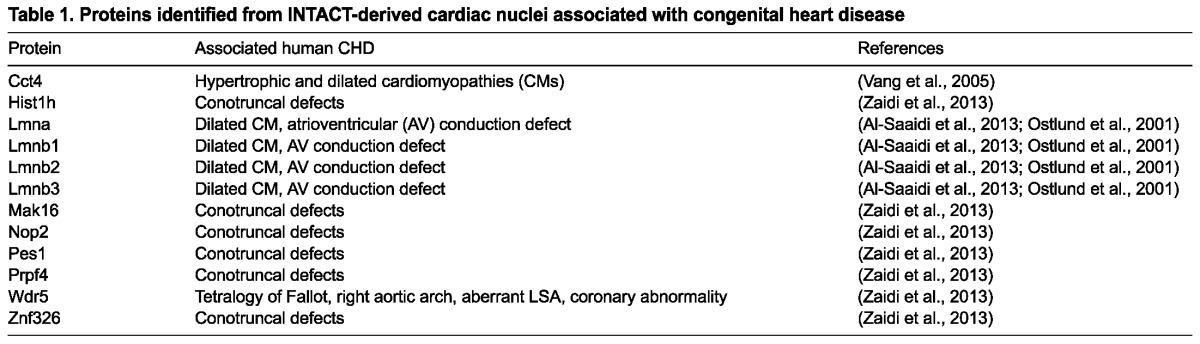
Further data mining of the filtered protein set demonstrated that at this single stage of cardiac development we were able to identify the Xenopus orthologs of 12 human nuclear proteins encoded by genes that when mutated in human are associated with congenital heart disease (CHD; Table 1). Thus, using these technologies, we are able to isolate and profile proteins that are expressed in, and required for, normal cardiac development.
Table 1.
Proteins identified from INTACT-derived cardiac nuclei associated with congenital heart disease
DISCUSSION
Here, we report the development and application of the INTACT method in Xenopus. We have gone on to demonstrate the feasibility of combining the INTACT methodologies with a liquid chromatography-tandem mass spectrometry (LC-MS/MS) proteomic approach to profile proteins expressed in the nuclei of developing hearts. As part of these studies we generated a vertebrate NTF transgene by cloning the nuclear pore protein nup35 from Xenopus in frame with the GFP-BLRP cassette used previously to visualize and isolate tag nuclei (Deal and Henikoff, 2010; Deal and Henikoff, 2011; Steiner et al., 2012). Nup35 encodes a nucleoporin protein that is highly conserved (71%/84% identical/similar to mouse and 72%/85% to human) and has a crucial role in nuclear envelope formation and function in vertebrates (Hawryluk-Gara et al., 2008). Nup35 attaches to the nuclear lamina through its interaction with lamin B and offers the advantage that it is relatively small and its function and cellular localization are unaffected by fusion to enhanced green fluorescent protein (eGFP) (Hawryluk-Gara et al., 2005). Thus, the vertebrate version of NTF that we have generated from Xenopus is most likely to be capable of functioning similarly in other vertebrate model systems. Thus these methods, protocols, reagents and workflow should be generally applicable to a wide range of vertebrate model systems and tissue types.
Cardiac profiling
CHD occurs in an estimated 1% of live births and an estimated 5% of stillbirths are attributed to human CHD, making it one of the most common diseases among children (Hoffman, 1995; Hoffman and Kaplan, 2002). Moreover, prognosis and outcomes have stagnated over the last few decades and remain exceptionally poor (Bruneau, 2013; Chin et al., 2012). To gain a better understanding of heart development and disease, it has become essential to gain a basic knowledge of proteins that are expressed in the embryonic and the adult heart. To this end, recent advances in proteomic profiling have been applied to cardiac tissue but, to date, have mainly focused on the adult heart and the generation of biomarkers associated with specific cardiac abnormalities (Buscemi et al., 2005; Chen et al., 2008; Cieniewski-Bernard et al., 2008; Gramolini et al., 2008; Kim et al., 2006; Koomen et al., 2006; Peronnet et al., 2006; Weekes et al., 1999; Wei et al., 2009).
Xenopus has historically been used to study a wide array of developmental processes and is particularly amenable to examining the cardiac system because, together with the benefit of producing vast quantities of relatively large externally developing eggs, many of the developmental processes and genes involved in cardiac specification, differentiation and growth are conserved between Xenopus and human and have been characterized in detail (reviewed by Kaltenbrun et al., 2011; Mohun et al., 2003; Warkman and Krieg, 2007). Furthermore, compared with other higher vertebrate models, Xenopus embryos can survive for longer without a properly functioning heart or circulatory system, enabling the observation of later consequences of cardiac malfunction. Moreover, the Xenopus model allows for tissue explantation for study in isolation (Afouda and Hoppler, 2009; Langdon et al., 2007; Raffin et al., 2000), as well as microinjection of overexpression constructs and antisense morpholino oligonucleotides to manipulate gene function (Brown et al., 2007; Brown et al., 2005; Christine and Conlon, 2008; Sive et al., 2000; Tada et al., 1997; Tandon et al., 2012). Crucially, studies have demonstrated a direct correlation between depleting cardiac proteins in Xenopus to that of genetic mutations in mouse or in human CHD patients (Kaltenbrun et al., 2011).
In this study, we examined a compendium of cardiac proteins from late tadpole stage Xenopus (stage 46), a stage equivalent to E12.5 in mouse and day 40 (Carnegie stage 10) in human. Our results identified 502 protein clusters from purified cardiomyocyte nuclei. We note that this number of proteins is less than reported for cardiac protein profiling in the adult fly or mouse heart (Bousette et al., 2009; Cammarato et al., 2011). However, there are several possible reasons for this discrepancy. First, these studies were conducted with whole heart tissue that examined expression in all cellular compartments. By contrast, our study was conducted exclusively on the nuclear fraction and restricted solely to the cardiomyocyte cell population, which constitutes a minority of the total cardiac cell number. Second, the current incompleteness in the sequence and gene ontology annotation of the X. laevis and X. tropicalis proteomes and genomes may limit overall coverage. And third, the reported 502 protein identifications reflect the number of protein clusters, which represent as an upper limit of 909 potential protein products annotated within the X. laevis and X. tropicalis UniProt databases (supplementary material Table S2). In other words, the number of gene products at the cluster level is an underestimate of the true number, as protein clustering can group related but genetically distinct entries. For example, we identified a five member cluster of Ybx1 (Y-box-binding) proteins, which contains the distinct genes ybx1, ybx2-a, ybx2-b, and nsep1 and ybx3. Because this hierarchical relationship is retained, this provides an advantage for comparative proteomic studies, as individual cluster members, from the subset of functionally over-represented or quantitatively regulated protein clusters, can be subjected to a second round of interrogation to reveal isoform-specific regulation. An equally important aspect of protein clustering is the ability to address sequence database redundancy. Although inclusion of manually and automatically annotated protein sequences in the analysis increased the identification of non-shared peptide sequences, this does not necessarily mean the individual cluster members are ‘unique’ gene products. Identifications of this type often have a high rate of sequence identity and could correspond to laevis and tropicalis paralogs or protein fragment database entries within the same species. For these reasons, our bioinformatics analysis was performed on the conservative estimate of the total number of protein clusters identified.
Additionally, the ability to perform data redundancy reduction has the advantage of permitting rapid identification of previously unidentified or uncharacterized protein products that were not assigned to a protein cluster. For example, of the 502 proteins, four protein accessions have not been mapped to Xenopus genes, 17 have been mapped, but to genes of uncertain functions (LOC symbols), and seven were annotated with Mammalian Gene Collection (MGC) symbols. Potential gene products that have only been predicted at the transcript level represent high-value candidates that merit more careful study. Overall, these aspects of protein clustering analysis demonstrate several distinct advantages when performing ‘-omic’ studies with organisms that are not completely annotated.
Cardiac profiling and CHD
Recently, a comprehensive study using high throughout sequencing to identify the incidence of de novo mutations in adult heart tissue from CHD patients found a strong association between CHD and mutations in chromatin-modifying proteins (Zaidi et al., 2013). In our present study we identified the Xenopus ortholog of seven of these proteins in all replicates of cardiac nuclei. Data mining of our samples revealed another five proteins with a known association with CHD. Collectively, these findings emphasize the utility of coupling an INTACT-based approach with LC-MS/MS to identify and characterize those proteins expressed during embryonic development, and further emphasize the utility of using Xenopus to model human heart development (Kaltenbrun et al., 2011). We envisage that changes in absolute levels of these CHD-associated proteins can be studied at different stages of development using targeted mass spectrometry approaches. Our results form an important baseline for such future studies, providing information necessary for selecting signature peptides for targeted quantification by methods such as selected reaction monitoring (Picotti et al., 2010). The use of Xenopus as a model system is further emphasized by the observation that most proteomic profiling studies of embryonic and adult tissue are compromised by the availability of endogenous tissue. Given that Xenopus can provide a virtual limitless supply of tissue, and given the INTACT method is not restricted by sample size, we anticipate that as the annotations of Xenopus near completion, coupling the INTACT method with protein profiling will in the future produce a compendium of proteins in Xenopus that will at the least equal that of studies in fly, mouse or other model systems.
MATERIALS AND METHODS
Generation of INTACT plasmids for Xenopus transgenics and capped mRNA production
Full-length nup35 was cloned from X. laevis cDNA (stage 40 embryos) by PCR; primers: 5′-gcgcccgggATGATGGCTGCTGCTTTCTC-3′ and 5′-cgcgctagcCCAGCCAAACATATACTCCA-3′ with engineered XmaI and NheI sites, respectively. Nup35 PCR product was cloned in frame with pADF8p-eGFP-BLRP (kindly provided by Roger Deal and Steven Henikoff, Fred Hutchinson Cancer Center, Seattle, WA, USA), removing the pADF8p promoter to generate the plasmid UNC208 (Nup35::eGFP::BLRP). BirA::mCherry was cloned from pBY2982 (Addgene) using XhoI and NotI, then cloned into pSP64TxB (Krieg and Melton, 1984) to generate plasmid UNC328. UNC208 and UNC328 were linearized with XhoI or XbaI, respectively, and used to generate capped mRNA with mMESSAGE mMACHINE T3 and SP6 Kits (Ambion), respectively.
NTF was cloned downstream of the tissue-specific promoters mlc2p (Latinkić et al., 2004) and cardiac_actin_p (Latinkić et al., 2002) by amplification from UNC208 with 5′-TAATACGACTCACTATAGG-3′ and 5′-cgcagatctATGATGGCTGCTGCTTTCTC-3′, engineered with a BglII site. Amplified product was cloned in place of GFP using BglII and XhoI, resulting in mlc2p::NTF (UNC256) and cardiac_actin_p::NTF (UNC255). BirA and BirA::mCherry were cloned downstream of the CMV promoter into pBluescript-II KS from pBY2982 (UNC215 and UNC330, respectively). All transgenic constructs were linearized with XbaI for transgenesis by restriction enzyme mediated integration (REMI) (Mandel et al., 2010).
In vitro fertilization and transgenesis of Xenopus embryos
Embryos were obtained and cultured as previously described (Mandel et al., 2010). One nangram of capped NTF and 1 ng of capped BirA-mCherry mRNA were injected into one-cell stage embryos. One nanogram NTF only was injected as a negative control for immunoprecipitation and nuclear isolation expreriments. Injected embryos were cultured in 4% Ficoll in 1× MBS for 2-3 hours. Embryos were collected at stage 23 for imaging, western blotting and immunoprecipitation. Transgenic animals were generated by REMI (Amaya and Kroll, 1999; Kroll and Amaya, 1996; Mandel et al., 2010) and genotyped from tailclips as previously reported (Showell and Conlon, 2009). Fertilized eggs with proper cleavage patterns were sorted 3 hours post-injection, cultured in 0.1× MMR and collected at stage 40 (for nuclear enrichment and qPCR analyses) and stage 46 (for proteome profiling).
Immunofluorescence and microscopy
Live brightfield and fluorescent images were taken on a Leica dissecting microscope fitted with a QImaging Retiga 4000RV camera. Transgenic embryos were fixed in MEMFA [100 mM MOPS (pH 7.4), 2 mM EGTA, 1 mM MgSO4, 3.7% (v/v) formaldehyde] at room temperature for 30 minutes, washed with 1× PBS + 0.1% Tween 20 (PTw) and incubated with 1 μg/ml DAPI in PTw for 30-40 minutes at room temperature. Embryos were washed with PTw and imaged immediately on a Zeiss LSM5 Pascal confocal microscope. For streptavidin staining, embryos were fixed in 4% paraformaldehyde for 2 hours at room temperature, washed with 1× PBS and incubated in 30% sucrose in PBS overnight at 4°C. Embryos were embedded in OCT and frozen on dry ice. They were then cryosectioned (10 μm thickness) and collected on frosted slides. Slides were stored at -80°C and warmed at 55°C prior to use for immunostaining. Slides were hydrated in 1× PBS for 10 minutes and incubated in blocking buffer (10% heat-inactivated calf serum, 1% Triton X-100, 1× PBS) for 1 hour in a humidity chamber at room temperature. Slides were then incubated with antibody solution containing 1:100 anti-GFP (Molecular Probes, A6455) and 1:100 streptavidin Alexa Fluor 568 (Invitrogen) in wash buffer (1% heat-inactivated calf serum, 0.1% Triton X-100, 1× PBS) overnight at 4°C in a humidity chamber. The next morning they were washed three times in wash buffer, and incubated with 1:500 Alexa-Fluor-488-conjugated goat anti-mouse IgG secondary antibodies (Molecular Probes, A11001) in wash buffer for 2 hours at room temperature in a dark humidity chamber. Slides were washed three times and mounted with Prolong Gold Antifade reagent with DAPI (Invitrogen). Sections were analyzed on a Zeiss 700 confocal microscope with MyZen software.
Purification of nuclei from Xenopus embryos
Nuclei from Xenopus embryos were prepared as previously described (Deal and Henikoff, 2011) with minor modifications. Briefly, 100 embryos were washed in 1× PBS three times and then dropped in groups directly into liquid nitrogen. Frozen embryos were then ground to a powder with a ceramic mortar and pestle. Powder was collected in a tube and resuspended in 6 ml nuclear purification buffer [NPB; 10 mM Tris pH 7.4, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.2 mM dithiothreitol (DTT), 0.5 mM (phenylmethanesulfonyl fluoride (PMSF), 0.5 mM spermine, 0.25 mM spermidine, 1× Roche Complete Protease Inhibitor Cocktail]. DTT, PMSF, spermine, spermidine, and protease inhibitors were all added to NPB immediately prior to use. The powder was thawed in NPB for about 10 minutes on ice and transferred to a polytetrafluoroethylene tissue grinder (VWR) and homogenized with 40 strokes. Lysate was filtered through a 100 μm cell strainer to remove cell clumps and centrifuged at 1000 g for 10 minutes to collect crude nuclei. Nuclei were resuspended in Optiprep (Sigma) diluted to 30% with NPB and pelleted from cell debris at 1000 g for 10 minutes. Nuclei were then washed three times with NPB to remove Optiprep prior to affinity isolation.
Streptavidin immunoaffinity purification of NTF
To determine the efficiency of BirA biotinylation of the nuclear targeting factor protein, nuclear preparations were resuspended in 1 ml HEPES resuspension buffer (20 mM HEPES, 1.2% polyvinylpyrrolidone, pH 7.4, 1× protease inhibitors) and added dropwise to liquid nitrogen. Nuclear pellets were thawed and incubated in 5 ml lysis buffer (200 mM K-HEPES pH 7.4, 1.1 M potassium acetate, 20 mM MgCl2, 1% Tween 20, 10 μM ZnCl2, 10 μM CaCl2, 500 mM NaCl, 1.0% Triton X-100 and 0.5% deoxycholic acid, 1× protease inhibitors). Nuclei were sheared on ice with two 15-second pulses of a Polytron (Thermo Fisher Scientific) at setting 22, with 1 minute recovery between pulses. Lysates were cleared by centrifugation at 7500 g for 10 minutes. Once cleared, lysates were incubated with streptavidin-conjugated magnetic beads (Invitrogen, M-270) for 30 minutes at room temperature, with rotation. Beads were collected on a magnet and washed seven times with lysis buffer at room temperature. Protein was eluted with 30 μl 1× SDS sample buffer (10% glycerol, 100 mM Tris, pH 6.8, 1% SDS, 5% 2-mercaptoethanol, 10 μg/ml Bromophenol Blue) at 95°C for 10 minutes, split in two and run on two lanes of a 10% SDS-PAGE gel. Gels were transferred overnight to PVDF membrane and probed with mouse anti-GFP (JL8, Clontech, 1:10,000) or streptavidin-HRP (Jackson ImmunoResearch, 1:10,000). HRP donkey anti-mouse secondary antibodies (Jackson ImmunoResearch) were used for GFP western blots at 1:10,000. Embryos not injected with BirA mRNA were used as a negative control for the immunoaffinity purification immunoprecipitations.
Affinity isolation of INTACT nuclei
Isolation of tagged nuclei was performed as described previously (Deal and Henikoff, 2011). All steps were performed at 4°C. Briefly, nuclei were prepared as described above, re-suspended in 1 ml NPB and immediately incubated with 50 μl, or 500 μg, of streptavidin-conjugated magnetic beads (Invitrogen, M-270) for 30 minutes, with end-over-end rotation. During incubation, P1000 tips were pre-loaded with 1 ml NPBb (NPB with 0.5% BSA) and laid on their side. After 20 minutes, a tip was inserted into a two-way stopcock and attached to a magnet. The tip was positioned vertically and the stopcock was opened to drain the NPBb. After the 30 minute incubation, the nuclei and bead mixture was mixed with 9 ml NPBt (NPB with 0.1% Triton X-100). This entire mixture was drawn into a 10 ml serological pipette and inserted into the broad opening of the P1000 tip-magnet-stopcock assembly preloaded with 1.2 ml of NPBt. The mixture was drained from the pipette by slowly releasing from stopcock assembly (flow-through, non-bead-bound nuclei). During this process, bead-bound nuclei were collected to the side of the P1000 tip by the magnet. These nuclei were rinsed in 1 ml NPB, diluted to 10 ml with NPBt and purification was repeated using a new tip-magnet assembly. Bead-bound and flow-through samples were centrifuged at 1000 g to pellet the nuclei.
Total RNA isolation and cDNA synthesis
Nuclei were collected in 500 μl Trizol (Invitrogen) for total RNA extraction, and flash frozen in liquid nitrogen. After storage at -80°C, lysates were thawed at room temperature, homogenized in Trizol using a P1000 tip, vortexed briefly, and incubated at room temperature for 5 minutes. Lysate was centrifuged at 22,000 g for 1 minute to precipitate insoluble material. Supernatant was transferred to a new tube containing 100 μl chloroform. The solution was vortexed briefly, incubated at room temperature for 3 minutes, and centrifuged at 12,000 g for 15 minutes at 4°C. The aqueous layer was transferred to a new tube containing 250 μl isopropyl alcohol. After 10 minutes of incubation at room temperature, RNA was precipitated by centrifuging at 12,000 g for 10 minutes at 4°C. RNA was washed twice with 70% ethanol and spun at 8000 g for 5 minutes between washes. The pellet was air-dried, and resuspended in 25 μl DEPC-treated water. Samples were treated with 3 U DNaseI for 30 minutes at 37°C and total RNA was purified using the RNeasy Mini Kit (Qiagen).
Quantitative RT-PCR (qPCR)
cDNA was generated from total RNA with Superscript II (Invitrogen) and random primers according to the manufacturer’s instructions. qPCR was performed on a ABI 7900 Fast HT using SYBRgreen mixed with ROX passive dye (Sigma). To normalize gene expression gapdh and eef1a were used. Relative expression was determined using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Primer sequences for qPCR are summarized in supplementary material Table S1.
Mass spectrometry-based proteome profiling
Streptavidin-bound nuclei from stage 46 mlc2::NTF; CMV::BirA::mCherry transgenic embryos were lysed in RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% deoxycholate, 1% NP-40, 0.1% SDS) and sonicated using a Bioruptor (Diagenode) on high for 15 minutes (cycling 30 seconds on and off) at 4°C. The tube was placed on a magnet and the sample was eluted off the beads. Total protein yield was measured by the BCA assay (ThermoFisher Scientific) and equal protein aliquots (∼30 μg) from three stage 46 biological replicates were analyzed by GeLC-MS/MS analysis as previously described (Guise et al., 2012; Joshi et al., 2013). Briefly, proteins were resolved on 4-12% Bis-Tris polyacrylamide gels. Replicate lanes were excised, cut into 1 mm slices, and grouped into ten fractions per lane. Proteins were digested in parallel with trypsin (12.5 ng/μl in 20 μl of 50 mM ammonium bicarbonate) overnight at 37°C and peptides extracted in 0.5% formic acid/50% ACN. Peptide mixtures were desalted and half (4 μl) of each sample was analyzed by nanoliquid chromatography-tandem mass spectrometry using a Dionex Ultimate 3000 nanoRSLC system coupled online to an LTQ Orbitrap Velos mass spectrometer (ThermoFisher Scientific). Peptides were separated by reverse phase nLC (Acclaim PepMap RSLC C18, 1.8 μm, 75 μm × 25 cm) and selected for MS analysis using a data-dependent ‘Top 15’ acquisition method, as previously described (Greco et al., 2012; Guise et al., 2012). MS/MS spectra were extracted, filtered, and searched using Proteome Discoverer/SEQUEST software (v1.4 ThermoFisher Scientific) against a protein sequence database compiled from UniProt (Swiss-Prot/TrEMBL, 2013-08) containing X. tropicalis and X. laevis sequences appended with common contaminants (39,623 entries). Sequences were automatically reversed and concatenated to the forward sequences to facilitate calculation of peptide and protein false discovery rate (FDR) estimates. SEQUEST peptide-spectrum matches were analyzed using Scaffold (v4.0.5; Proteome Software) and the X!Tandem (Beavis Informatics) refinement search strategy. Probabilities for peptide-spectrum matches were calculated using Scaffold’s Bayesian-based local FDR scoring model. Peptides were assembled into protein groups, i.e. database entries that could not be distinguished by sequence alone. Protein groups were assembled into clusters to maximize sequence coverage while reducing database-dependent redundancy. Weighted peptide probabilities and weighted spectra counts were calculated based on the proportion of unique and shared spectra among related protein clusters. Weighted spectral counts for each protein cluster across replicates were then normalized based on the averaged sum of total weighted spectra between replicates. Data were exported to Excel for processing. Protein ‘clusters’ that did not possess at least two exclusive peptides (weight=1.0) in at least one biological replicate were removed. The coefficient of variation was calculated using weighted spectral counts (at least three in all replicates).
Supplementary Material
Acknowledgments
We thank Roger Deal and Steve Henikoff (Fred Hutchinson Cancer Center) for generously providing INTACT plasmids and advice for tagging strategies. Derek Franklin performed the PCR verification of BirA integration in NTF-positive embryos. We thank Jeffrey Kong for his assistance in proteomic sample processing. We also thank members of the Conlon lab for critical discussions and comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
N.M.A. designed and performed the majority of experiments and prepared the manuscript. T.M.G. designed and performed the proteomic profiling experiments and prepared the manuscript. L.M.K., M.M.R. and M.C. performed experiments and edited the manuscript. J.B.W. and I.M.C. contributed reagents and manuscript editing. F.L.C. contributed reagents and manuscript editing and preparation.
Funding
This work is supported by grants from the National Heart, Lung, and Blood Institute [RO1 DE018825, RO1 HL089641 and R21 HD073044 to F.L.C.]; and from the National Institute on Drug Abuse [DP1 DA026192 to I.M.C.]. N.M.A. is supported by an American Heart Association Postdoctoral Fellowship [11POST6320005]. J.B.W. is an Early Career Scientist of the Howard Hughes Medical Institute. Deposited in PMC for release after 6 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098327/-/DC1
References
- Afouda B. A., Hoppler S. (2009). Xenopus explants as an experimental model system for studying heart development. Trends Cardiovasc. Med. 19, 220–226 [DOI] [PubMed] [Google Scholar]
- Agostini M., Tucci P., Killick R., Candi E., Sayan B. S., Rivetti di Val Cervo P., Nicotera P., McKeon F., Knight R. A., Mak T. W., et al. (2011). Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. USA 108, 21093–21098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed R. P., Haider H. K., Buccini S., Li L., Jiang S., Ashraf M. (2011). Reprogramming of skeletal myoblasts for induction of pluripotency for tumor-free cardiomyogenesis in the infarcted heart. Circ. Res. 109, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Saaidi R., Rasmussen T. B., Palmfeldt J., Nissen P. H., Beqqali A., Hansen J., Pinto Y. M., Boesen T., Mogensen J., Bross P. (2013). The LMNA mutation p.Arg321Ter associated with dilated cardiomyopathy leads to reduced expression and a skewed ratio of lamin A and lamin C proteins. Exp. Cell Res. 319, 3010–3019 [DOI] [PubMed] [Google Scholar]
- Amaya E., Kroll K. L. (1999). A method for generating transgenic frog embryos. Methods Mol. Biol. 97, 393–414 [DOI] [PubMed] [Google Scholar]
- Amaya E., Musci T. J., Kirschner M. W. (1991). Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell 66, 257–270 [DOI] [PubMed] [Google Scholar]
- Amin N. M., Tandon P., Osborne Nishimura E., Conlon F. L. (2013). RNA-seq in the tetraploid Xenopus laevis enables genome-wide insight in a classic developmental biology model organism. Methods 13, 00210–00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo S., Lohr J., Lee K. H., Ticho B. S., Breitbart R. E., Hill S., Yost H. J., Srivastava D. (2000). Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech. Dev. 95, 231–237 [DOI] [PubMed] [Google Scholar]
- Aubert J., Stavridis M. P., Tweedie S., O’Reilly M., Vierlinger K., Li M., Ghazal P., Pratt T., Mason J. O., Roy D., et al. (2003). Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc. Natl. Acad. Sci. USA 100 Suppl. 1, 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. A., Reeves G., Hetherington L., Aitken R. J. (2010). Analysis of proteomic changes associated with sperm capacitation through the combined use of IPG-strip pre-fractionation followed by RP chromatography LC-MS/MS analysis. Proteomics 10, 482–495 [DOI] [PubMed] [Google Scholar]
- Balak K., Jacobson M., Sunshine J., Rutishauser U. (1987). Neural cell adhesion molecule expression in Xenopus embryos. Dev. Biol. 119, 540–550 [DOI] [PubMed] [Google Scholar]
- Barker C. R., Worman C. P., Smith J. L. (1975). Purification and quantification of T and B lymphocytes by an affinity method. Immunology 29, 765–777 [PMC free article] [PubMed] [Google Scholar]
- Barres B. A., Silverstein B. E., Corey D. P., Chun L. L. (1988). Immunological, morphological, and electrophysiological variation among retinal ganglion cells purified by panning. Neuron 1, 791–803 [DOI] [PubMed] [Google Scholar]
- Bartlett H. L., Weeks D. L. (2008). Lessons from the lily pad: Using Xenopus to understand heart disease. Drug Discov. Today Dis. Models 5, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett H. L., Sutherland L., Kolker S. J., Welp C., Tajchman U., Desmarais V., Weeks D. L. (2007). Transient early embryonic expression of Nkx2-5 mutations linked to congenital heart defects in human causes heart defects in Xenopus laevis. Dev. Dyn. 236, 2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D., Kovaleva E., Schatz P. J. (1999). A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn S., Zinzen R. P., Girardot C., Gustafson E. H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E. E. (2012a). Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148–156 [DOI] [PubMed] [Google Scholar]
- Bonn S., Zinzen R. P., Perez-Gonzalez A., Riddell A., Gavin A. C., Furlong E. E. (2012b). Cell type-specific chromatin immunoprecipitation from multicellular complex samples using BiTS-ChIP. Nat. Protoc. 7, 978–994 [DOI] [PubMed] [Google Scholar]
- Bonnet A., Bevilacqua C., Benne F., Bodin L., Cotinot C., Liaubet L., Sancristobal M., Sarry J., Terenina E., Martin P., et al. (2011). Transcriptome profiling of sheep granulosa cells and oocytes during early follicular development obtained by laser capture microdissection. BMC Genomics 12, 417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousette N., Kislinger T., Fong V., Isserlin R., Hewel J. A., Emil A., Gramolini A. O. (2009). Large-scale characterization and analysis of the murine cardiac proteome. J. Proteome Res. 8, 1887–1901 [DOI] [PubMed] [Google Scholar]
- Brown D. D., Binder O., Pagratis M., Parr B. A., Conlon F. L. (2003). Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev. Genes Evol. 212, 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Martz S. N., Binder O., Goetz S. C., Price B. M., Smith J. C., Conlon F. L. (2005). Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development 132, 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. D., Christine K. S., Showell C., Conlon F. L. (2007). Small heat shock protein Hsp27 is required for proper heart tube formation. Genesis 45, 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau B. G. (2013). Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb. Perspect. Biol. 5, a008292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine R. D., Schier A. F. (2000). Conserved and divergent mechanisms in left-right axis formation. Genes Dev. 14, 763–776 [PubMed] [Google Scholar]
- Buscemi N., Murray C., Doherty-Kirby A., Lajoie G., Sussman M. A., Van Eyk J. E. (2005). Myocardial subproteomic analysis of a constitutively active Rac1-expressing transgenic mouse with lethal myocardial hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 289, H2325–H2333 [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T. D., Menegon A., Roncaglia P., Colciago G., et al. (2011). Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 [DOI] [PubMed] [Google Scholar]
- Cammarato A., Ahrens C. H., Alayari N. N., Qeli E., Rucker J., Reedy M. C., Zmasek C. M., Gucek M., Cole R. N., Van Eyk J. E., et al. (2011). A mighty small heart: the cardiac proteome of adult Drosophila melanogaster. PLoS ONE 6, e18497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Q., Bi F., Wang S. Q., Xiao S. J., Liu J. N. (2008). Porous silicon affinity chips for biomarker detection by MALDI-TOF-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 875, 502–508 [DOI] [PubMed] [Google Scholar]
- Chen A. E., Borowiak M., Sherwood R. I., Kweudjeu A., Melton D. A. (2013). Functional evaluation of ES cell-derived endodermal populations reveals differences between Nodal and Activin A-guided differentiation. Development 140, 675–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A. J., Saint-Jeannet J. P., Lo C. W. (2012). How insights from cardiovascular developmental biology have impacted the care of infants and children with congenital heart disease. Mech. Dev. 129, 75–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine K. S., Conlon F. L. (2008). Vertebrate CASTOR is required for differentiation of cardiac precursor cells at the ventral midline. Dev. Cell 14, 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieniewski-Bernard C., Mulder P., Henry J. P., Drobecq H., Dubois E., Pottiez G., Thuillez C., Amouyel P., Richard V., Pinet F. (2008). Proteomic analysis of left ventricular remodeling in an experimental model of heart failure. J. Proteome Res. 7, 5004–5016 [DOI] [PubMed] [Google Scholar]
- Claycomb W. C., Lanson N. A., Jr, Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr (1998). HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft A. P., Przyborski S. A. (2006). Formation of neurons by non-neural adult stem cells: potential mechanism implicates an artifact of growth in culture. Stem Cells 24, 1841–1851 [DOI] [PubMed] [Google Scholar]
- Deal R. B., Henikoff S. (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R. B., Henikoff S. (2011). The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 6, 56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- della Gaspera B., Armand A. S., Sequeira I., Lecolle S., Gallien C. L., Charbonnier F., Chanoine C. (2009). The Xenopus MEF2 gene family: evidence of a role for XMEF2C in larval tendon development. Dev. Biol. 328, 392–402 [DOI] [PubMed] [Google Scholar]
- Dong M. Q., Venable J. D., Au N., Xu T., Park S. K., Cociorva D., Johnson J. R., Dillin A., Yates J. R., III (2007). Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science 317, 660–663 [DOI] [PubMed] [Google Scholar]
- Dorrell C., Abraham S. L., Lanxon-Cookson K. M., Canaday P. S., Streeter P. R., Grompe M. (2008). Isolation of major pancreatic cell types and long-term culture-initiating cells using novel human surface markers. Stem Cell Res. 1, 183–194 [DOI] [PubMed] [Google Scholar]
- Erickson H. S., Albert P. S., Gillespie J. W., Rodriguez-Canales J., Marston Linehan W., Pinto P. A., Chuaqui R. F., Emmert-Buck M. R. (2009). Quantitative RT-PCR gene expression analysis of laser microdissected tissue samples. Nat. Protoc. 4, 902–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Langarica A., Meza-Perez S., Calderon-Amador J., Estrada-Garcia T., Macpherson G., Lebecque S., Saeland S., Steinman R. M., Flores-Romo L. (2005). Network of dendritic cells within the muscular layer of the mouse intestine. Proc. Natl. Acad. Sci. USA 102, 19039–19044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R. M., Von Stetina S. E., Barlow S. J., Shaffer C., Olszewski K. L., Moore J. H., Dupuy D., Vidal M., Miller D. M., III (2005). A gene expression fingerprint of C. elegans embryonic motor neurons. BMC Genomics 6, 42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Yan W., Mohun T. J., Evans S. M. (1998). Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development 125, 4439–4449 [DOI] [PubMed] [Google Scholar]
- Golubeva Y., Salcedo R., Mueller C., Liotta L. A., Espina V. (2013). Laser capture microdissection for protein and NanoString RNA analysis. Methods Mol. Biol. 931, 213–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gormley J. P., Nascone-Yoder N. M. (2003). Left and right contributions to the Xenopus heart: implications for asymmetric morphogenesis. Dev. Genes Evol. 213, 390–398 [DOI] [PubMed] [Google Scholar]
- Gramolini A. O., Kislinger T., Alikhani-Koopaei R., Fong V., Thompson N. J., Isserlin R., Sharma P., Oudit G. Y., Trivieri M. G., Fagan A., et al. (2008). Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Mol. Cell. Proteomics 7, 519–533 [DOI] [PubMed] [Google Scholar]
- Greco T. M., Miteva Y., Conlon F. L., Cristea I. M. (2012). Complementary proteomic analysis of protein complexes. Methods Mol. Biol. 917, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise A. J., Greco T. M., Zhang I. Y., Yu F., Cristea I. M. (2012). Aurora B-dependent regulation of class IIa histone deacetylases by mitotic nuclear localization signal phosphorylation. Mol. Cell. Proteomics 11, 1220–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara L. A., Shibuya E. K., Wozniak R. W. (2005). Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell 16, 2382–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara L. A., Platani M., Santarella R., Wozniak R. W., Mattaj I. W. (2008). Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol. Biol. Cell 19, 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimeier R. A., Das B., Buchholz D. R., Fiorentino M., Shi Y. B. (2010). Studies on Xenopus laevis intestine reveal biological pathways underlying vertebrate gut adaptation from embryo to adult. Genome Biol. 11, R55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U., Harland R. M., Gilchrist M. J., Hendrix D., Jurka J., Kapitonov V., Ovcharenko I., Putnam N. H., Shu S., Taher L., et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science 328, 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry G. L., Davis F. P., Picard S., Eddy S. R. (2012). Cell type-specific genomics of Drosophila neurons. Nucleic Acids Res. 40, 9691–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch N., Harris W. A. (1997). Xenopus Pax-6 and retinal development. J. Neurobiol. 32, 45–61 [PubMed] [Google Scholar]
- Hoffman J. I. (1995). Incidence of congenital heart disease: I. Postnatal incidence. Pediatr. Cardiol. 16, 103–113 [DOI] [PubMed] [Google Scholar]
- Hoffman J. I., Kaplan S. (2002). The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39, 1890–1900 [DOI] [PubMed] [Google Scholar]
- Hoi C. S., Lee S. E., Lu S. Y., McDermitt D. J., Osorio K. M., Piskun C. M., Peters R. M., Paus R., Tumbar T. (2010). Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol. Cell. Biol. 30, 2518–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood N. D., Gurdon J. B. (1990). Activation of muscle genes without myogenesis by ectopic expression of MyoD in frog embryo cells. Nature 347, 197–200 [DOI] [PubMed] [Google Scholar]
- Horb M. E., Thomsen G. H. (1999). Tbx5 is essential for heart development. Development 126, 1739–1751 [DOI] [PubMed] [Google Scholar]
- Imamichi Y., Lahr G., Wedlich D. (2001). Laser-mediated microdissection of paraffin sections from Xenopus embryos allows detection of tissue-specific expressed mRNAs. Dev. Genes Evol. 211, 361–366 [DOI] [PubMed] [Google Scholar]
- Jesty S. A., Steffey M. A., Lee F. K., Breitbach M., Hesse M., Reining S., Lee J. C., Doran R. M., Nikitin A. Y., Fleischmann B. K., et al. (2012). c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc. Natl. Acad. Sci. USA 109, 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Evans T. (1996). The Xenopus GATA-4/5/6 genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev. Biol. 174, 258–270 [DOI] [PubMed] [Google Scholar]
- Joshi P., Greco T. M., Guise A. J., Luo Y., Yu F., Nesvizhskii A. I., Cristea I. M. (2013). The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 9, 672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbrun E., Tandon P., Amin N. M., Waldron L., Showell C., Conlon F. L. (2011). Xenopus: An emerging model for studying congenital heart disease. Birth Defects Res. A Clin. Mol. Teratol. 91, 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Lee Y., Kim H., Joo H., Youm J. B., Park W. S., Warda M., Cuong D. V., Han J. (2006). Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics 6, 1237–1249 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Zaehres H., Wu G., Gentile L., Ko K., Sebastiano V., Araúzo-Bravo M. J., Ruau D., Han D. W., Zenke M., et al. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454, 646–650 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Greber B., Araúzo-Bravo M. J., Meyer J., Park K. I., Zaehres H., Schöler H. R. (2009). Direct reprogramming of human neural stem cells by OCT4. Nature 461, 649–3 [DOI] [PubMed] [Google Scholar]
- Koomen J. M., Wilson C. R., Guthrie P., Androlewicz M. J., Kobayashi R., Taegtmeyer H. (2006). Proteome analysis of isolated perfused organ effluent as a novel model for protein biomarker discovery. J. Proteome Res. 5, 177–182 [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. (1984). Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 12, 7057–7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll K. L., Amaya E. (1996). Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122, 3173–3183 [DOI] [PubMed] [Google Scholar]
- Kubo A., Shinozaki K., Shannon J. M., Kouskoff V., Kennedy M., Woo S., Fehling H. J., Keller G. (2004). Development of definitive endoderm from embryonic stem cells in culture. Development 131, 1651–1662 [DOI] [PubMed] [Google Scholar]
- Laflamme M. A., Gold J., Xu C., Hassanipour M., Rosler E., Police S., Muskheli V., Murry C. E. (2005). Formation of human myocardium in the rat heart from human embryonic stem cells. Am. J. Pathol. 167, 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M. A., Chen K. Y., Naumova A. V., Muskheli V., Fugate J. A., Dupras S. K., Reinecke H., Xu C., Hassanipour M., Police S., et al. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 25, 1015–1024 [DOI] [PubMed] [Google Scholar]
- Lamba D. A., Karl M. O., Ware C. B., Reh T. A. (2006). Efficient generation of retinal progenitor cells from human embryonic stem cells. Proc. Natl. Acad. Sci. USA 103, 12769–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon Y. G., Goetz S. C., Berg A. E., Swanik J. T., Conlon F. L. (2007). SHP-2 is required for the maintenance of cardiac progenitors. Development 134, 4119–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinkić B. V., Cooper B., Towers N., Sparrow D., Kotecha S., Mohun T. J. (2002). Distinct enhancers regulate skeletal and cardiac muscle-specific expression programs of the cardiac alpha-actin gene in Xenopus embryos. Dev. Biol. 245, 57–70 [DOI] [PubMed] [Google Scholar]
- Latinkić B. V., Cooper B., Smith S., Kotecha S., Towers N., Sparrow D., Mohun T. J. (2004). Transcriptional regulation of the cardiac-specific MLC2 gene during Xenopus embryonic development. Development 131, 669–679 [DOI] [PubMed] [Google Scholar]
- Lawson D. A., Xin L., Lukacs R. U., Cheng D., Witte O. N. (2007). Isolation and functional characterization of murine prostate stem cells. Proc. Natl. Acad. Sci. USA 104, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Lumelsky N., Studer L., Auerbach J. M., McKay R. D. (2000). Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat. Biotechnol. 18, 675–679 [DOI] [PubMed] [Google Scholar]
- Lee S. J., Kim S., Choi S. C., Han J. K. (2010). XPteg (Xenopus proximal tubules-expressed gene) is essential for pronephric mesoderm specification and tubulogenesis. Mech. Dev. 127, 49–61 [DOI] [PubMed] [Google Scholar]
- Lei Y., Guo X., Liu Y., Cao Y., Deng Y., Chen X., Cheng C. H., Dawid I. B., Chen Y., Zhao H. (2012). Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs). Proc. Natl. Acad. Sci. USA 109, 17484–17489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Longobardi L., Li T., Myers T. J., O’Rear L., Ozkan H., Li Y., Contaldo C., Spagnoli A. (2012). TGF-β type II receptor/MCP-5 axis: at the crossroad between joint and growth plate development. Dev. Cell 23, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N., Yamanaka S. (2011). Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature 474, 225–229 [DOI] [PubMed] [Google Scholar]
- Mage M. G., McHugh L. L., Rothstein T. L. (1977). Mouse lymphocytes with and without surface immunoglobulin: preparative scale separation in polystyrene tissue culture dishes coated with specifically purified anti-immunoglobulin. J. Immunol. Methods 15, 47–56 [DOI] [PubMed] [Google Scholar]
- Maine G. N., Li H., Zaidi I. W., Basrur V., Elenitoba-Johnson K. S., Burstein E. (2010). A bimolecular affinity purification method under denaturing conditions for rapid isolation of a ubiquitinated protein for mass spectrometry analysis. Nat. Protoc. 5, 1447–1459 [DOI] [PubMed] [Google Scholar]
- Mandel E. M., Kaltenbrun E., Callis T. E., Zeng X. X., Marques S. R., Yelon D., Wang D. Z., Conlon F. L. (2010). The BMP pathway acts to directly regulate Tbx20 in the developing heart. Development 137, 1919–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. L., Harland R. M. (2001). Hypaxial muscle migration during primary myogenesis in Xenopus laevis. Dev. Biol. 239, 270–280 [DOI] [PubMed] [Google Scholar]
- Misawa H., Yamaguchi M. (2000). The gene of Ca2+-binding protein regucalcin is highly conserved in vertebrate species. Int. J. Mol. Med. 6, 191–196 [DOI] [PubMed] [Google Scholar]
- Mohun T., Garrett N., Treisman R. (1987). Xenopus cytoskeletal actin and human c-fos gene promoters share a conserved protein-binding site. EMBO J. 6, 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohun T., Orford R., Shang C. (2003). The origins of cardiac tissue in the amphibian, Xenopus laevis. Trends Cardiovasc. Med. 13, 244–248 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Nakajima T., Takase M., Yaoita Y. (2012). Generation of albino Xenopus tropicalis using zinc-finger nucleases. Dev. Growth Differ. 54, 777–784 [DOI] [PubMed] [Google Scholar]
- Newman C. S., Krieg P. A. (1998). tinman-related genes expressed during heart development in Xenopus. Dev. Genet. 22, 230–238 [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. (2007). Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005). Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- Osorio K. M., Lee S. E., McDermitt D. J., Waghmare S. K., Zhang Y. V., Woo H. N., Tumbar T. (2008). Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 135, 1059–1068 [DOI] [PubMed] [Google Scholar]
- Ostlund C., Bonne G., Schwartz K., Worman H. J. (2001). Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 114, 4435–4445 [DOI] [PubMed] [Google Scholar]
- Paige S. L., Thomas S., Stoick-Cooper C. L., Wang H., Maves L., Sandstrom R., Pabon L., Reinecke H., Pratt G., Keller G., et al. (2012). A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151, 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Kumar C., Bohl S., Klingmueller U., Mann M. (2009). Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., et al. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- Pearl E. J., Bilogan C. K., Mukhi S., Brown D. D., Horb M. E. (2009). Xenopus pancreas development. Dev. Dyn. 238, 1271–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peronnet E., Becquart L., Poirier F., Cubizolles M., Choquet-Kastylevsky G., Jolivet-Reynaud C. (2006). SELDI-TOF MS analysis of the Cardiac Troponin I forms present in plasma from patients with myocardial infarction. Proteomics 6, 6288–6299 [DOI] [PubMed] [Google Scholar]
- Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010). High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- Pruszak J., Sonntag K. C., Aung M. H., Sanchez-Pernaute R., Isacson O. (2007). Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells 25, 2257–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruszak J., Ludwig W., Blak A., Alavian K., Isacson O. (2009). CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 27, 2928–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Huang Y., Spencer C. I., Foley A., Vedantham V., Liu L., Conway S. J., Fu J. D., Srivastava D. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485, 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabien A. (2010). Laser microdissection. Methods Mol. Biol. 576, 39–47 [DOI] [PubMed] [Google Scholar]
- Raffin M., Leong L. M., Rones M. S., Sparrow D., Mohun T., Mercola M. (2000). Subdivision of the cardiac Nkx2.5 expression domain into myogenic and nonmyogenic compartments. Dev. Biol. 218, 326–340 [DOI] [PubMed] [Google Scholar]
- Roesli C., Neri D., Rybak J. N. (2006). In vivo protein biotinylation and sample preparation for the proteomic identification of organ- and disease-specific antigens accessible from the vasculature. Nat. Protoc. 1, 192–199 [DOI] [PubMed] [Google Scholar]
- Ryan K., Butler K., Bellefroid E., Gurdon J. B. (1998). Xenopus eomesodermin is expressed in neural differentiation. Mech. Dev. 75, 155–158 [DOI] [PubMed] [Google Scholar]
- Sakuma T., Hosoi S., Woltjen K., Suzuki K., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y., et al. (2013). Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18, 315–326 [DOI] [PubMed] [Google Scholar]
- Schäffer U., Schlosser A., Müller K. M., Schäfer A., Katava N., Baumeister R., Schulze E. (2010). SnAvi - a new tandem tag for high-affinity protein-complex purification. Nucleic Acids Res. 38, e91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. P., Biswas S., Merchant A. S., Satoskar A., Taslim C., Lin S., Rovin B. H., Sen C. K., Roy S., Freitas M. A. (2012). A quantitative proteomic workflow for characterization of frozen clinical biopsies: laser capture microdissection coupled with label-free mass spectrometry. J. Proteomics 77, 433–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Collier J. M., Hlaing M., Zhang L., Delshad E. H., Bristow J., Bernstein H. S. (2003). Genome-wide examination of myoblast cell cycle withdrawal during differentiation. Dev. Dyn. 226, 128–138 [DOI] [PubMed] [Google Scholar]
- Showell C., Conlon F. L. (2009) Tissue sampling and genomic DNA purification from the western clawed frog Xenopus tropicalis. Cold Spring Harb. Protoc. 2009, pdb prot5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell C., Christine K. S., Mandel E. M., Conlon F. L. (2006). Developmental expression patterns of Tbx1, Tbx2, Tbx5, and Tbx20 in Xenopus tropicalis. Dev. Dyn. 235, 1623–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H. L., Grainger R. M., Harland R. M. (2000) Early Development of Xenopus Laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Small E. M., Krieg P. A. (2003). Transgenic analysis of the atrialnatriuretic factor (ANF) promoter: Nkx2-5 and GATA-4 binding sites are required for atrial specific expression of ANF. Dev. Biol. 261, 116–131 [DOI] [PubMed] [Google Scholar]
- Steelman S., Moskow J. J., Muzynski K., North C., Druck T., Montgomery J. C., Huebner K., Daar I. O., Buchberg A. M. (1997). Identification of a conserved family of Meis1-related homeobox genes. Genome Res. 7, 142–156 [DOI] [PubMed] [Google Scholar]
- Steiner F. A., Talbert P. B., Kasinathan S., Deal R. B., Henikoff S. (2012). Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 22, 766–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser G. A., Kaminker J. S., Tessier-Lavigne M. (2010). Microarray analysis of retinal endothelial tip cells identifies CXCR4 as a mediator of tip cell morphology and branching. Blood 115, 5102–5110 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Rodriguez R. T., McLean G. W., Kim S. K. (2007). Conserved markers of fetal pancreatic epithelium permit prospective isolation of islet progenitor cells by FACS. Proc. Natl. Acad. Sci. USA 104, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo E., Rampalli S., Risueño R. M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature 468, 521–526 [DOI] [PubMed] [Google Scholar]
- Tada M., O’Reilly M. A., Smith J. C. (1997). Analysis of competence and of Brachyury autoinduction by use of hormone-inducible Xbra. Development 124, 2225–2234 [DOI] [PubMed] [Google Scholar]
- Tandon P., Showell C., Christine K., Conlon F. L. (2012). Morpholino injection in Xenopus. Methods Mol. Biol. 843, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tandon P., Miteva Y. V., Kuchenbrod L. M., Cristea I. M., Conlon F. L. (2013). Tcf21 regulates the specification and maturation of proepicardial cells. Development 140, 2409–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven F. J., Timmers H. T. (2006). The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 34, e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang S., Corydon T. J., Børglum A. D., Scott M. D., Frydman J., Mogensen J., Gregersen N., Bross P. (2005). Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. FEBS J. 272, 2037–2049 [DOI] [PubMed] [Google Scholar]
- Von Stetina S. E., Watson J. D., Fox R. M., Olszewski K. L., Spencer W. C., Roy P. J., Miller D. M., III (2007). Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 8, R135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford J. B. (2006). Planar cell polarity, ciliogenesis and neural tube defects. Hum. Mol. Genet. 15, R227–R234 [DOI] [PubMed] [Google Scholar]
- Wamstad J. A., Alexander J. M., Truty R. M., Shrikumar A., Li F., Eilertson K. E., Ding H., Wylie J. N., Pico A. R., Capra J. A., et al. (2012). Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- Warkman A. S., Krieg P. A. (2007). Xenopus as a model system for vertebrate heart development. Semin. Cell Dev. Biol. 18, 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes J., Wheeler C. H., Yan J. X., Weil J., Eschenhagen T., Scholtysik G., Dunn M. J. (1999). Bovine dilated cardiomyopathy: proteomic analysis of an animal model of human dilated cardiomyopathy. Electrophoresis 20, 898–906 [DOI] [PubMed] [Google Scholar]
- Wei Y. J., Huang Y. X., Shen Y., Cui C. J., Zhang X. L., Zhang H., Hu S. S. (2009). Proteomic analysis reveals significant elevation of heat shock protein 70 in patients with chronic heart failure due to arrhythmogenic right ventricular cardiomyopathy. Mol. Cell. Biochem. 332, 103–111 [DOI] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- Wong M. W., Pisegna M., Lu M. F., Leibham D., Perry M. (1994). Activation of Xenopus MyoD transcription by members of the MEF2 protein family. Dev. Biol. 166, 683–695 [DOI] [PubMed] [Google Scholar]
- Wong J., Chilkoti A., Moy V. T. (1999). Direct force measurements of the streptavidin-biotin interaction. Biomol. Eng. 16, 45–55 [DOI] [PubMed] [Google Scholar]
- Wysocki L. J., Sato V. L. (1978). ‘Panning’ for lymphocytes: a method for cell selection. Proc. Natl. Acad. Sci. USA 75, 2844–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Chong D. C., Rankin S. A., Zorn A. M., Cleaver O. (2009). Rasip1 is required for endothelial cell motility, angiogenesis and vessel formation. Dev. Biol. 329, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. J., Cherone J. M., Doyon Y., Ankoudinova I., Faraji F. M., Lee A. H., Ngo C., Guschin D. Y., Paschon D. E., Miller J. C., et al. (2011). Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 108, 7052–7057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S., Choi M., Wakimoto H., Ma L., Jiang J., Overton J. D., Romano-Adesman A., Bjornson R. D., Breitbart R. E., Brown K. K., et al. (2013). De novo mutations in histone-modifying genes in congenital heart disease. Nature 498, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj K. H., Tung Y. C., Piper M., Gumy L., Fawcett J. W., Yeo G. S., Holt C. E. (2010). Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 30, 15464–15478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L. I., Mather C., Burgess S., Bolce M. E., Harland R. M., Orkin S. H. (1991). Expression of GATA-binding proteins during embryonic development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 88, 10642–10646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn A. M., Mason J. (2001). Gene expression in the embryonic Xenopus liver. Mech. Dev. 103, 153–157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.