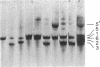
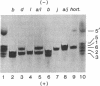
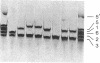
Abstract
The field of biochemical genetics relies heavily upon the detection by electrophoresis of genetically determined variants of proteins. Most of these variants differ by substitutions that involve charged amino acids. Genetic variants of another large class, ones that involve substitutions among neutral amino acids, are not easily detected and are often ignored. Ampholyte isoelectric focusing in some cases can separate proteins indistinguishable by standard electrophoresis, including genetic variants of mouse hemoglobins that differ only by neutral amino acid substitutions. A revolutionary variation of isoelectric focusing, in which gradients covering a small pH range are fixed into place in a polyacrylamide gel, provides greater resolution of these nearly identical proteins. Mouse hemoglobin tetramers that differ only by the substitution of alanine for glycine in the alpha-globin chains are resolved by several millimeters with the new technique; by comparison, these tetramers are imperfectly resolved on a standard pH 7-9 isoelectric focusing gel. This improved technique of isoelectric focusing was used to identify a variety of previously unreported genetic variants of mouse hemoglobin alpha chains. Immobilized gradients tailored to the requirements of the proteins being analyzed will extend greatly the ranges of protein variations that can be easily recognized for diverse applications, including genetic quality-control analyses and in studies of genetics, mutagenesis, and evolution.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjellqvist B., Ek K., Righetti P. G., Gianazza E., Görg A., Westermeier R., Postel W. Isoelectric focusing in immobilized pH gradients: principle, methodology and some applications. J Biochem Biophys Methods. 1982 Sep;6(4):317–339. doi: 10.1016/0165-022x(82)90013-6. [DOI] [PubMed] [Google Scholar]
- Gilman J. G. Hemoglobin Beta chain structural variation in mice: evolutionary and functional implications. Science. 1972 Nov 24;178(4063):873–874. doi: 10.1126/science.178.4063.873. [DOI] [PubMed] [Google Scholar]
- Gilman J. G. Rodent hemoglobin structure: a comparison of several species of mice. Ann N Y Acad Sci. 1974 Nov 29;241(0):416–433. doi: 10.1111/j.1749-6632.1974.tb21897.x. [DOI] [PubMed] [Google Scholar]
- HUTTON J. J., BISHOP J., SCHWEET R., RUSSELL E. S. Hemoglobin inheritance in inbred mouse strains. II. Genetic studies. Proc Natl Acad Sci U S A. 1962 Oct 15;48:1718–1724. doi: 10.1073/pnas.48.10.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUTTON J. J., SCHWEET R. S., WOLFE H. G., RUSSELL E. S. HEMOGLOBIN SOLUBILITY AND ALPHA-CHAIN STRUCTURE IN CROSSES BETWEEN TWO INBREAD MOUSE STRAINS. Science. 1964 Jan 17;143(3603):252–253. doi: 10.1126/science.143.3603.252. [DOI] [PubMed] [Google Scholar]
- Hilse K., Popp R. A. Gene duplication as the basis for amino acid ambiguity in the alpha-chain polypeptides of mouse hemoglobins. Proc Natl Acad Sci U S A. 1968 Nov;61(3):930–936. doi: 10.1073/pnas.61.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel D. A., Maizel J. V., Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- Modiano G., Battistuzzi G., Motulsky A. G. Nonrandom patterns of codon usage and of nucleotide substitutions in human alpha- and beta-globin genes: an evolutionary strategy reducing the rate of mutations with drastic effects? Proc Natl Acad Sci U S A. 1981 Feb;78(2):1110–1114. doi: 10.1073/pnas.78.2.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- POPP R. A. Hemoglobin loci: mice classified for their Hb and Sol alleles. Science. 1963 May 24;140(3569):893–894. doi: 10.1126/science.140.3569.893. [DOI] [PubMed] [Google Scholar]
- POPP R. A. Studies on the mouse hemoglobin loci. II. Position of the hemoglobin locus with respect to albinism and shaker-1 loci. J Hered. 1962 Mar-Apr;53:73–80. doi: 10.1093/oxfordjournals.jhered.a107127. [DOI] [PubMed] [Google Scholar]
- POPP R. A. Studies on the mouse hemoglobin loci. J Hered. 1962 Jul-Aug;53:142–151. doi: 10.1093/oxfordjournals.jhered.a107153. [DOI] [PubMed] [Google Scholar]
- Popp R. A., Bailiff E. G. Sequence of amino acids in the major and minor chains of the diffuse hemoglobin from BALB-c mice. Biochim Biophys Acta. 1973 Mar 23;303(1):61–67. doi: 10.1016/0005-2795(73)90148-7. [DOI] [PubMed] [Google Scholar]
- Popp R. A., Bailiff E. G., Skow L. C., Johnson F. M., Lewis S. E. Analysis of a mouse alpha-globin gene mutation induced by ethylnitrosourea. Genetics. 1983 Sep;105(1):157–167. doi: 10.1093/genetics/105.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp R. A., Bailiff E. G., Skow L. C., Whitney J. B., 3rd The primary structure of genetic variants of mouse hemoglobin. Biochem Genet. 1982 Feb;20(1-2):199–208. doi: 10.1007/BF00484946. [DOI] [PubMed] [Google Scholar]
- Popp R. A. Hemoglobins of mice: sequence and possible ambiguity at one position of the alpha chain. J Mol Biol. 1967 Jul 14;27(1):9–16. doi: 10.1016/0022-2836(67)90347-6. [DOI] [PubMed] [Google Scholar]
- Popp R. A. Sequence of amino acids in the chain of single hemoglobins from C57BL, SWR, and NB mice. Biochim Biophys Acta. 1973 Mar 23;303(1):52–60. doi: 10.1016/0005-2795(73)90147-5. [DOI] [PubMed] [Google Scholar]
- Popp R. A. Studies on the mouse hemoglobin loci. IX. A fifth alpha-chain phenotype. J Hered. 1969 May-Jun;60(3):128–131. [PubMed] [Google Scholar]
- Popp R. A. Studies on the mouse hemoglobin loci. X. Linkage of duplicate genes at the alpha-chain locus, Hba. J Hered. 1969 May-Jun;60(3):131–133. [PubMed] [Google Scholar]
- Rifkin D. B., Rifkin M. R., Konigsberg W. The presence of two major hemoglobin components in an inbred strain of mice. Proc Natl Acad Sci U S A. 1966 Mar;55(3):586–592. doi: 10.1073/pnas.55.3.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell E. S., Blake S. L., McFarland E. C. Characterization and strain distribution of four alleles at the hemoglobin -chain structural locus in the mouse. Biochem Genet. 1972 Dec;7(3):313–330. doi: 10.1007/BF00484831. [DOI] [PubMed] [Google Scholar]
- Skow L. C., Burkhart B. A., Johnson F. M., Popp R. A., Popp D. M., Goldberg S. Z., Anderson W. F., Barnett L. B., Lewis S. E. A mouse model for beta-thalassemia. Cell. 1983 Oct;34(3):1043–1052. doi: 10.1016/0092-8674(83)90562-7. [DOI] [PubMed] [Google Scholar]
- WOLFE H. G., RUSSELL E. S., PACKER S. O. HEMOGLOBINS IN MICE. SEGREGATION OF GENETIC FACTORS AFFECTING ELECTROPHORETIC MOBILITY AND SOLUBILITY. J Hered. 1963 May-Jun;54:107–112. doi: 10.1093/oxfordjournals.jhered.a107236. [DOI] [PubMed] [Google Scholar]
- Whitney J. B., 3rd, Copland G. T., Skow L. C., Russell E. S. Resolution of products of the duplicated hemoglobin alpha-chain loci by isoelectric focusing. Proc Natl Acad Sci U S A. 1979 Feb;76(2):867–871. doi: 10.1073/pnas.76.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J. B., 3rd, Martinell J., Popp R. A., Russell L. B., Anderson W. F. Deletions in the alpha-globin gene complex in alpha-thalassemic mice. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7644–7647. doi: 10.1073/pnas.78.12.7644. [DOI] [PMC free article] [PubMed] [Google Scholar]