Abstract
Background
Amoebae and bacteria interact within predator/prey and host/pathogen relationships, but the general response of amoeba to bacteria is not well understood. The amoeba Dictyostelium discoideum feeds on, and is colonized by diverse bacterial species including Gram-positive [Gram(+)] and Gram-negative [Gram(−)] bacteria, two major groups of bacteria that differ in structure and macromolecular composition.
Results
Transcriptional profiling of D. discoideum revealed sets of genes whose expression is enriched in amoebae interacting with different species of bacteria, including sets that appear specific to amoebae interacting with Gram(+), or with Gram(−) bacteria. In a genetic screen utilizing the growth of mutant amoebae on a variety of bacteria as a phenotypic readout, we identified amoebal genes that are only required for growth on Gram(+) bacteria, including one that encodes the cell surface protein gp130, as well as several genes that are only required for growth on Gram(−) bacteria including one that encodes a putative lysozyme, AlyL. These genes are required for parts of the transcriptional response of wild-type amoebae, and this allowed their classification into potential response pathways.
Conclusions
We have defined genes that are critical for amoebal survival during feeding on Gram(+), or Gram(−), bacteria which we propose form part of a regulatory network that allows D. discoideum to elicit specific cellular responses to different species of bacteria in order to optimize survival.
Introduction
The social amoeba D. discoideum inhabits the forest soil and feeds on diverse species of bacteria [1, 2]. As a model eukaryote and proficient phagocyte D. discoideum has proven to be useful for studying aspects of host-pathogen interactions [3–5] and has been used to identify and study bacterial virulence factors [5–8]. It has also been suggested that amoebae serve as environmental reservoir for certain human pathogens [9]. Recent studies have focused on specific amoeba-bacterium interactions, but Dictyostelium amoebae reside in soil environments that are inhabited by thousands of bacterial species [10]. It should be informative to investigate how the amoebae cope with such diversity and to determine how they elaborate physiological responses to different bacteria for feeding and defense. A detailed understanding of the amoebal response should enrich our understanding of the interactions between amoebae and bacteria and may reveal novel antibacterial strategies in eukaryotes.
Anti-bacterial responses in plants and animals have a number of similarities, especially in the recognition of microbial-associated molecular patterns, or MAMPs [11]. For example, the TIR (Toll/Interleukin-1 receptor) domain is often present in MAMP receptors in plants and animals that are involved in microbial recognition. The globular TIR protein domain is an adaptor that signals through protein-protein interactions and is thought to play a role in the specificity of anti-microbial responses [12]. The recent discovery of the function of the TIR domain protein TirA in D. discoideum raises the general question of whether amoebae discriminate between different bacteria as well [13, 14]. Transcriptional profiling of D. discoideum exposed to a variety of bacterial species has revealed the differential accumulation of specific sets of gene transcripts suggesting that the amoebae discriminate between different bacteria [15–17].
One way in which D. discoideum amoebae might handle the diversity of bacterial species in the soil would be to activate specific response pathways for different classes of bacteria. The largest natural grouping of bacteria, the Gram(+) and Gram(−) species, is based on physiological differences that are of particular relevance to bacterial discrimination systems. The cytoplasmic membrane of Gram(+) bacteria is surrounded by a thick outer cell wall of peptidoglycan strands that are cross-linked by short peptides and containing teichoic acid, which is absent in the Gram(−) bacteria [18, 19]. Gram(−) bacteria have a thin peptidoglycan layer that is surrounded by an outer membrane containing lipoproteins and lipopolysaccharide (LPS) in the outer leaflet, which are absent in the Gram(+) bacteria. There is also some genetic evidence that D. discoideum amoebae discriminate between Gram(+) and Gram(−) bacteria. Mutations in several uncharacterized D. discoideum genes preclude growth on Bacillus subtilis, but allow normal growth on K. pneumoniae [20, 21], whereas mutations in the phg1a gene impair growth on K. pneumoniae, but not on B. subtilis [22].
We have undertaken a general approach to investigate the response of amoebae to bacteria by exploring the genetic control of D. discoideum growth on different species of bacteria. Our results suggest that D. discoideum amoebae respond in a highly specific manner to different species of bacteria, and also deploy general response systems for dealing with Gram(+) bacteria that are distinct from those deployed to deal with Gram(−) bacteria.
Results
Distinct D. discoideum transcriptional responses to different bacteria
Transcriptional profiling is a reliable method for detecting differential physiological responses in D. discoideum [23]. To test whether these amoebae can respond differentially to different bacteria, we grew them on two species of Gram(−) bacteria, K. pneumoniae and Pseudomonas aeruginosa, and two species of Gram(+) bacteria, Staphylococcus aureus and B. subtilis. We then measured the steady-state levels of mRNAs within the amoebae by RNA-sequencing (RNA-seq) and obtained transcriptional profiles that include data on ~10,000 genes (supplemental Table S1). We chose to examine the physiological status of amoebae growing exponentially on bacteria to avoid confounding dynamic changes in gene expression resulting from the initial contact with bacteria so that we could rely on the transcriptional profiles as robust indicators of diverse gene regulatory events. Hierarchical clustering of the profiles of growth on the two Gram(+) bacterial species tested are most similar to each other compared with the profiles on the two Gram(−) bacteria (Figure 1a). These results support the hypothesis that the amoebae have differential responses to the two types of bacteria, although, transcriptomes for amoebae growing on several more Gram(+) and Gram(−) bacterial species would have to be compared in order to determine whether this is a consistent trend.
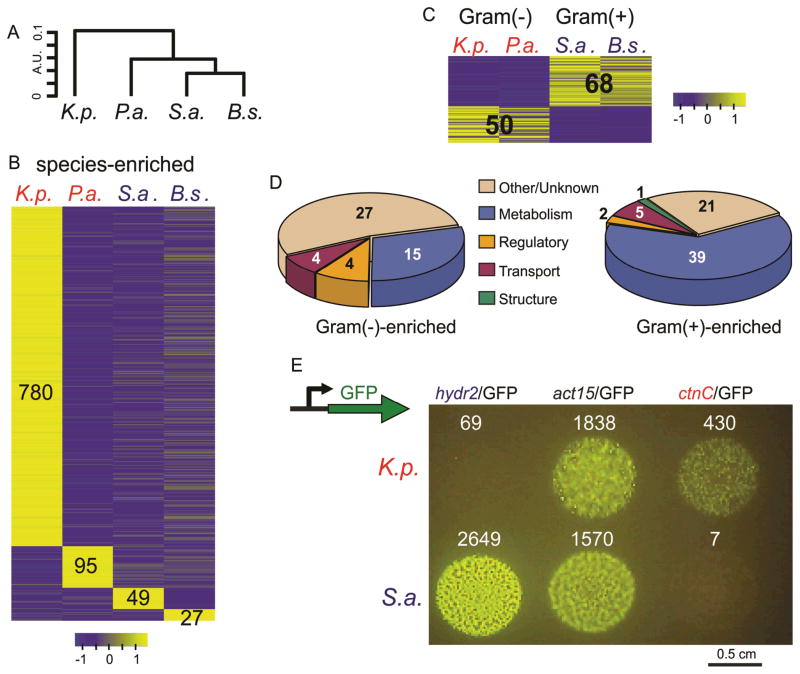
Figure 1. Changes in the physiological response in D. discoideum when feeding on different bacteria.
a, A dendrogram depicting the distances between the transcriptomes of D. discoideum growing on K. pneumoniae (K.p.), P. aeruginosa (P.a.), S. aureus (S.a.), and B. subtilis (B.s.). The dendrogram was constructed by hierarchical clustering (R function hclust) on the average normalized expression vectors of the two biological replicates from each growth condition consisting of all the genes from RNAseq experiments. We used Spearman’s correlation (SC) to calculate the distances (D = 1-SC) and complete linkage as the clustering criterion. Two objects (individual transcriptomes or joints) are joined by means of a horizontal line if these objects are more similar to one another than any other object in the data. The vertical distance between objects is inversely proportional to the similarity between them, but the horizontal distances are meaningless. b, c, The heat maps represent the patterns of change in standardized mRNA abundance for genes that were differentially expressed in D. discoideum when grown on different bacterial species. Each row represents a gene and each column, a bacterial growth condition. The colors represent relative mRNA abundances. To allow for comparisons between gene expression profiles with different abundances, we normalized the measurements on each gene to have a mean of 0 and a standard deviation of 1, and the scale indicates the number of standard deviations that a measurement is above or below the mean. b, Heat maps representing genes that are differentially expressed between the 4 bacterial species (species-enriched genes). c, genes differentially expressed between D. discoideum cells grown on Gram(+) and Gram(−) bacteria. d, pie charts showing the proportion of group-enriched D. discoideum genes categorized by Gene Ontology annotation (biological process). e, Wild-type amoebae transformed with a green fluorescent protein (GFP) coding sequence placed under the control of D. discoideum bacterial responsive gene promoters, mixed with different bacteria and spotted on buffered agar. A D. discoideum strain expressing GFP under the control of the hydr2 promoter fluoresces specifically when exposed to S.a., a Gram(+) bacterium, but not on K.p., a Gram(−) bacterium, and D. discoideum strain expressing GFP under the control of the ctnC promoter fluoresces when grown on K.p., but not on S.a.. A D. discoideum strain expressing GFP under the control of the actin15 promoter fluoresces under both growth conditions. Numbers indicate averaged normalized read counts from the RNA-seq data (supplemental Table S1).
Differences in the complete profiles were due to specific transcriptional changes observed during growth on individual species of bacteria. For each growth condition we were able to classify a subset of genes whose mRNA levels were elevated, compared to growth on the other three species tested, suggesting that each of the bacteria elicit a specific transcriptional response (Figure 1b and supplemental Table S1). The cohort of 780 genes that were differentially expressed during D. discoideum growth on K. pneumoniae was particularly striking. Although many of these genes likely encode enzymes needed to utilize K. pneumoniae as a food source and represent metabolic adaptation, some may encode specific signaling proteins involved in the detection of K. pneumoniae, or antimicrobial proteins. Indeed, three genes within this cohort, aplM, aplN, and aplQ, encode amoebapore proteins that are known to destabilize bacterial cytoplasmic membranes (supplemental Table S1) [24]. Additional transcriptional responses observed in D. discoideum growing on K. pneumoniae, P. aeruginosa, S. aureus and B. subtilis are described in detail in supplemental Table S1.
We also explored whether we could define transcriptional responses in D. discoideum that were specific to Gram(−) or Gram(+) bacteria. We identified 50 genes that were preferentially expressed on both Gram(−) species tested and 68 genes that were preferentially expressed on both Gram(+) species (Figure 1c and supplemental Table S1). We used Gene Ontology (GO) annotation of the genes that express the Gram(+)- and Gram(−)-enriched transcripts to provide insight into the physiology of the amoebae growing on different groups of bacteria (Figure 1d) [25]. The Gram(+)-enriched gene transcripts were most commonly annotated under the ‘metabolism’ descriptor and included a number of putative hydrolases that are likely to be involved in peptidoglycan cell wall degradation – the amoeba lysozymes (alyA, alyB, alyC and alyD), glycosidases and endopeptidases (supplemental Table S1). On the other hand, the Gram(−)-enriched expression of alyL, a gene encoding an amoeba lysozyme-like protein AlyL [26], suggests that it may be involved in the degradation of the Gram(−) cell walls (see below). The Gram(−)-enriched set also includes the cysteine protease gene, cprF, and a polyketide synthase gene, stlA (supplemental Table S1)[27, 28]. Genes of unknown function represent a substantial proportion of the Gram(+) and the Gram(−) profiles and include sets of genes predicted to encode proteins with signal-peptides that may represent novel secreted antimicrobial peptides. These analyses support the idea that D. discoideum responds differently to Gram(+) and Gram(−) bacteria.
To begin to confirm the regulatory responses underlying the transcriptional profiles, we constructed expression vectors consisting of the green fluorescent protein (GFP) coding sequence placed under the control of species-specific or group-specific gene promoters and introduced them into wild-type D. discoideum cells. We found that D. discoideum cells expressing GFP under the control of the hydr1 or hydr2 promoters fluoresced when exposed to Gram(+) bacteria, but not on any of the Gram(−) bacteria we have tested (Figure 1e, and supplemental Figure S1a). Amoebae expressing GFP under the control of the ctnC promoter fluoresced when feeding on bacteria of the Enterobacteriaceae family, such as K. pneumoniae, but not on P. aeruginosa, or any Gram(+) bacterium that we tested (Figure 1e, and supplemental Figure S1b). The actin15 gene is not differentially expressed during growth on bacteria and, accordingly, the act15/GFP construct was expressed on all bacteria tested (Figure 1e). These results indicate that the increased abundance of mRNAs quantified by RNA-seq are the result of differential promoter regulation, at least for these promoters, suggesting the involvement of a signal transduction cascades in their induction.
Genetic evidence for discrimination between Gram(+) and Gram(−) bacteria
The existence of mutations that restrict amoebal growth on one type of bacteria but not on the other type would provide independent support in favor of bacterial discrimination. We isolated such mutants by screening the growth of 10,000 mutated D. discoideum strains on Gram(−) K. pneumoniae bacteria and on Gram(+) B. subtilis bacteria (supplemental Table S2). D. discoideum mutants that grew well on one bacterial species, but poorly on the other bacterial species were the most interesting to us because we expected they might reveal key elements of bacterial discrimination (Table 1). We first examined the growth of these two classes of mutants on additional species of bacteria to test the generality of their phenotype. Mutations in nagB1, gpi, or swp1 exhibited growth defects on each of the five Gram(+) bacterial species we tested, yet grew well on each of the seven Gram(−) species we tested (Figure 2a, and supplemental Figure S2a). Dictyostelium strains carrying mutations in clkB, spc3, alyL, or the insertion at Ω1334 grew poorly on the seven Gram(−) bacterial species, but grew well on the five Gram(+) bacterial species tested (Figure 2, and supplemental Figure S2a). We also examined a previously characterized mutant of gp130 which we hypothesized might be a receptor for Gram(+) bacteria[29, 30]. Interestingly, the gp130-null mutant exhibited a growth defect on lawns of Gram(+) but not on Gram(−) bacteria (Figure 2a, and supplemental Figure S2a). Thus, we found several genes that are required for growth on a every Gram(+) bacterial species we tested, but are dispensable for growth on every Gram(−) species we tested, and we found several genes with the inverse phenotype. These findings suggest the existence of distinct pathways that allow D. discoideum to grow on each type of bacteria.
Table 1.
Mutant strains used in this study1.
| Mutant | Relevant Gene | GrowthPhenotype | Annotation2 | Reference |
|---|---|---|---|---|
| AX4 | - | wild-type | parental strain | [40] |
| 2F5 | gp130 | Gram(+)-defective | gp130 adhesion protein | [29] |
| AK1321 | swp1 | Gram(+)-defective | oligosaccharide transferase subunit | This work |
| AK1333 | gpi | Gram(+)-defective | glucose phosphate isomerase | This work |
| AK1353 | gpi | Gram(+)-defective | glucose phosphate isomerase | This work |
| AK1372 | nagB1 | Gram(+)-defective | glucosamine-6-phosphate deaminase | This work |
| TirA-KO | tirA | Gram(−)-defective | TIR-domain-containing protein | [13] |
| AK1338 | clkB | Gram(−)-defective | CDC7-related protein kinase, DDB_G0278487 | This work |
| AK1346 | spc3 | Gram(−)-defective | signal peptidase complex subunit 3 | This work |
| AK1350 | alyL | Gram(−)-defective | amoeba lysozyme-related protein | This work |
| AK1334 | unknown | Gram(−)-defective | insertion between two genes, DDB_G0295477 DDB_G0271574 | This work |
All strains are available from the Dictyostelium Stock center (dictyBase.org).
Adapted from DictyBase.org [37].
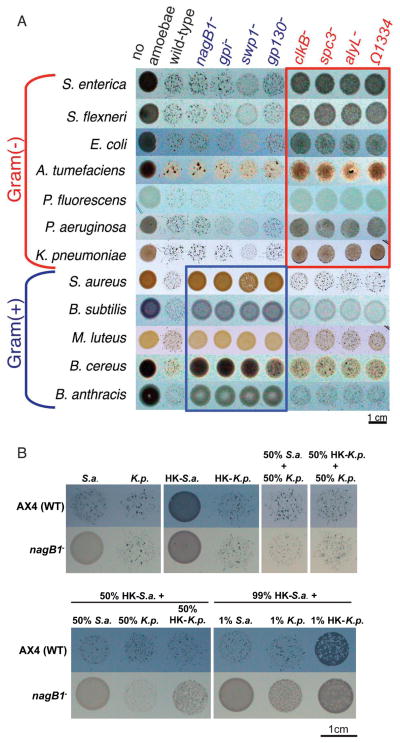
Figure 2. D. discoideum mutants with growth defects on Gram(+) or Gram(−) bacteria.
a, Each spot represents a co-culture of about 500 D. discoideum cells and a thick bacterial culture spotted on buffered agar, imaged after 4 days of incubation. The image (top view) was assembled from 12 images of 12 experiments conducted under similar conditions. All D. discoideum strains were tested on the same day with the same culture of a given bacterium. Rows represent different Gram(−) or Gram(+) bacterial species as indicated on the left, and columns represent different wild-type or mutant amoebae as indicated on the top. The left-most column contains no amoebae. Wild-type amoebae (second column) grew on all the bacterial species (the dark speckles within each spot are D. discoideum fruiting bodies that formed after all the bacteria had been consumed). The nagB1−, gpi−, swp1−, and gp130− mutants exhibit severe growth defects on Gram(+) bacteria compared to the wild type (blue frame). The clkB−, spc3−, alyL−, and Ω1334 (insertion site within the mutant AK1334) mutants exhibit severe growth defect on Gram(−) bacteria (red frame). b, D. discoideum growth on heat-killed S. aureus. Rows represent wild type or nagB1− mutant amoeba as indicated on left, and columns represent different bacterial species or a mixture of bacterial species (live or heat-killed) in different mass ratios as indicated (HK, heat-killed; S.a., S. aureus; K.p., K. pneumoniae). Each spot is a co-culture of about 500 D. discoideum cells and a thick bacterial culture spotted on buffered agar and imaged after 4 days of incubation. The bacteria appear as tan areas within the spots. Intermediate levels of feeding are indicated by partial clearing of the tan bacterial lawn. Similar results were obtained for the other Gram(+)-growth-defective mutants gpi− and swp1− (unpublished observations).
For an amoeba any bacterium can be food, a dangerous pathogen, or both. To begin to test whether the genes we identified are required for feeding on bacteria or for defense against them, we measured the ability of the mutant strains to grow on dead bacteria. We found that each of the Gram(−)-growth defective mutants grew well on heat-killed Gram(−) bacteria (supplemental Figure S2b), and on Gram(−) bacteria treated with ampicillin (unpublished observations). Since these genes are not required to meet a nutritional requirement of amoebae growing on Gram(−) bacteria, we propose that they are required for defense against Gram(−) bacteria. A case in point is the alyL gene which encodes a putative amoeba lysozyme and which is expressed during, and required for, growth on Gram(−) bacteria (Figure 3, supplemental Figure S3, and Table S1). These results suggest that the amoebae respond to Gram(−) bacteria by expressing at least some genes that are essential to their survival on live Gram(−) bacteria.
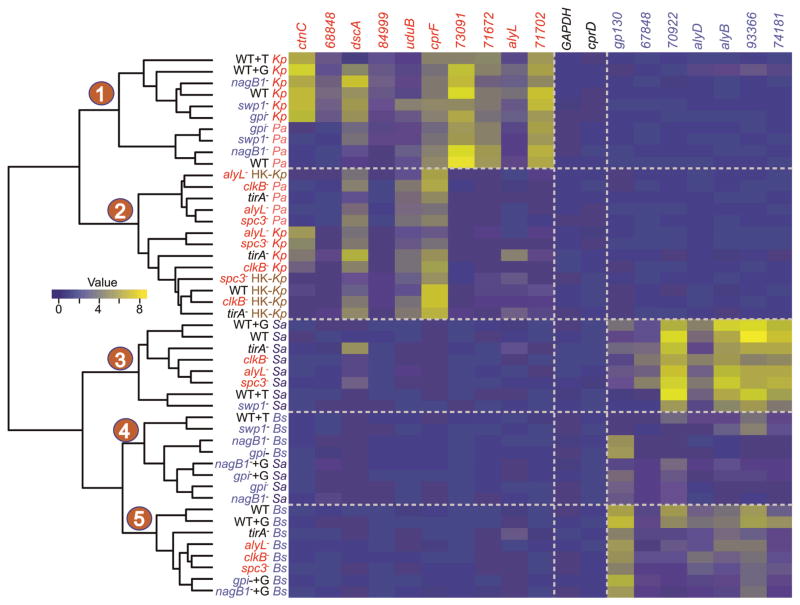
Figure 3. Changes in the transcriptional landscape in mutants with defective growth phenotypes.
The heat map represents the patterns of change in normalized mRNA levels (qRT-PCR) of selected genes in different D. discoideum strains (yellow, higher relative abundance; blue, lower relative abundance). The mRNA levels of each gene (log2 scale) were normalized to the mRNA levels determined for the histone H3a gene. Each row represents a wild type or mutant strain in one bacterial growth condition. Each column represents the relative abundance of a given gene; ctnC and 68848 are K. pneumoniae-specific genes, dscA to 71702 (DDB_G0271702) are Gram(−)-enriched genes, GAPDH and cprD are control genes, and gp130 to 74181 (DDB_G0274181) are Gram(+)-enriched genes [37]. The dendrogram depicts the Euclidean distances between the different D. disoideum strains (wild-type or mutant) grown on different bacterial species: wild-type AX4 (WT); K. pneumoniae (K.p.); Heat-killed K. pneumoniae (HK-K.p.); P. aeruginosa (P.a.); S. aureus (S.a.); B. subtilis (B.s.); G, glucose; and T, tunicamycin. The major divide in the clustering is evident between strains grown on Gram(+) and Gram(−) bacteria. D. discoideum mutants that are defective in growth on Gram(−) bacteria and the tirA− mutant cluster together when grown on live Gram(−) bacteria, or on Heat-killed K.p., along with wild-type grown on heat-killed K.p. (clade 2). D. discoideum mutants that are defective in growth on Gram(−) bacteria and the tirA− mutant cluster together with wild-type D. discoideum when grown on Gram(+) bacteria (clade 3 and 5). Mutant strains nagB1− and gpi that are defective in growth on Gram(+) bacteria cluster together when mixed with Gram(+) bacteria (clade 4), but cluster with the wild type when grown on Gram(−) bacteria (clade 1).
Neither wild-type D. discoideum amoebae, or the Gram(+)-growth defective mutants were able to feed on any dead Gram(+) bacteria that we tested. An example of this type of experiment is shown in Figure 2b using S. aureus, but we obtained similar results with other Gram(+) bacteria. Interestingly, we were able to induce the feeding of amoebae on dead Gram(+) bacteria by mixing in as little as 1% live Gram(+) bacteria, or 1% live Gram(−) bacteria (Figure 2b). This indicates that dead Gram(+) bacteria can be utilized as a food source and suggests that the amoebae must be exposed to live bacteria in order to feed on the dead Gram(+) bacteria. Mixing live Gram(+) bacteria did not induce feeding on dead Gram(+) bacteria by the Gram(+)-growth defective mutants. However, we were able to induce the Gram(+)-growth-defective mutants to feed on live, or heat-killed, Gram(+) bacteria by adding 50%, or even as little as 1%, live K. pneumoniae (Figure 2b). The use of Gram(−) bacteria to stimulate these mutants to feed on Gram(+) bacteria indicates that the Gram(+)-growth defective mutants are not nutritional auxotrophs for feeding on Gram(+) bacteria and suggests that the recognition of live bacteria by the amoebae is required for efficient feeding.
Transcriptional phenotyping of the Gram(+)- and Gram(−)-growth defective mutants
To explore potential functions of the genes identified in growth-defective mutants, we examined expression profiles of a limited set of differentially expressed genes. We isolated total RNA from mutant amoebae and carried out reverse transcription followed by quantitative PCR (qRT-PCR) analyses with primers against selected Gram(−)-enriched and Gram(+)-enriched transcripts, as well as two K. pneumoniae-specific transcripts. The transcriptional profiles of the Gram(+) growth-defective mutants growing on Gram(−) bacteria were similar to the wild-type D. discoideum profile (Figure 3, clade 1, and supplemental Figure S3). However, when exposed to Gram(+) bacteria, the transcriptional profiles of these mutants were similar to one another and different from the wild type (Figure 3, clade 4, and supplemental Figure S3). Within this group, the nagB1− and gpi− mutants exhibited wild-type levels of gp130 and DDB_G0267848 transcripts, and lower levels of alyB, alyD, DDB_G0293366, DDB_G0274181, and DDB_G0270922. The swp1− mutant on the other hand, expressed near wild-type levels of the putative peptidoglycan-degrading enzyme genes, but had significantly lower expression of gp130 and DDB_G0267848. Hierarchical clustering (Figure 3 and supplemental Figure S4) and multidimensional scaling (MDS, Supplemental Figure S5) indicate that the nagB1 and gpi mutant transcriptional profiles are most similar to each other and distinct from the swp1 mutant profile, suggesting that nagB1 and gpi are components of one pathway that is required for growth on Gram(+) bacteria.
Transcriptional profiles of the Gram(−) growth-defective mutants exposed to Gram(−) bacteria showed lower levels of most of the Gram(−)-enriched genes that we tested (Figure 3, clade 2), whereas their profiles during growth on Gram(+) bacteria were similar to the wild type (Figure 3, clades 3 and 5, and supplemental Figures S3–S5). TirA is a known component of the amoebal response to Gram(−) bacteria [13, 14]. We included the tirA mutant in our analysis and found that its transcriptional profile on Gram(−) bacteria was similar to that of the Gram(−) growth-defective mutants (Figure 3, clade 2, and supplemental Figures S3–5). Two Gram(−)enriched genes showed higher transcript levels, or little change; cprF, a cysteine proteinase gene, and uduB, a gene of unknown function. The spc3 and alyL mutant profiles were the most similar (Figure 3, clade 2). Spc3 is a signal peptidase subunit and AlyL has a signal peptide and likely resides in the lysosome, or is otherwise secreted. The absence of Spc3 may alter signal peptidase function in a way that affects the biogenesis of AlyL, resulting in the similar transcriptional profiles of these two mutants.
To explore the pathway(s) by which the Gram(−) enriched genes are induced, we examined the transcriptional profiles of the mutants after exposure to heat-killed K. pneumoniae. As described in the previous section, growth of the Gram(−)-defective mutants on live K. pneumoniae is defective, but their growth rates on heat-killed K. pneumoniae are comparable to that of the wild type. Hierarchical clustering and MDS analyses indicate that the transcriptional profiles of the Gram(−) growth-defective mutants exposed to live K. pneumoniae are similar to their transcriptional profiles on dead K. pneumoniae, and also similar to the wild type profile on dead K. pneumoniae (Figure 3, clade 2, and supplemental Figure S3–S5). Interestingly, the wild-type amoebae did not induce the expression of 6 out of 10 Gram(−)-enriched gene transcripts when exposed to dead K. pneumoniae, and exhibited higher levels of the cprF and uduB transcript, similar to the expression profile of Gram(−)-defective mutants on live K. pneumoniae (Figure 3, clade 2, and supplemental Figures S3–5). These results suggest that the physiological changes that are specific for growth on live Gram(−) bacteria are not required for feeding on dead Gram(−) bacteria. They also suggest that the predicted lysozyme, AlyL, is not needed to digest Gram(−) bacteria, but instead is needed to kill them, as lysozymes are known to do in other eukaryotes [31]. These data support the idea that the genes identified in our genetic screen are key elements of a bacterial discrimination network in D. discoideum that result in specific responses to either Gram(+), or Gram(−) bacteria.
The role of N-linked glycosylation in the response to Gram(+) bacteria
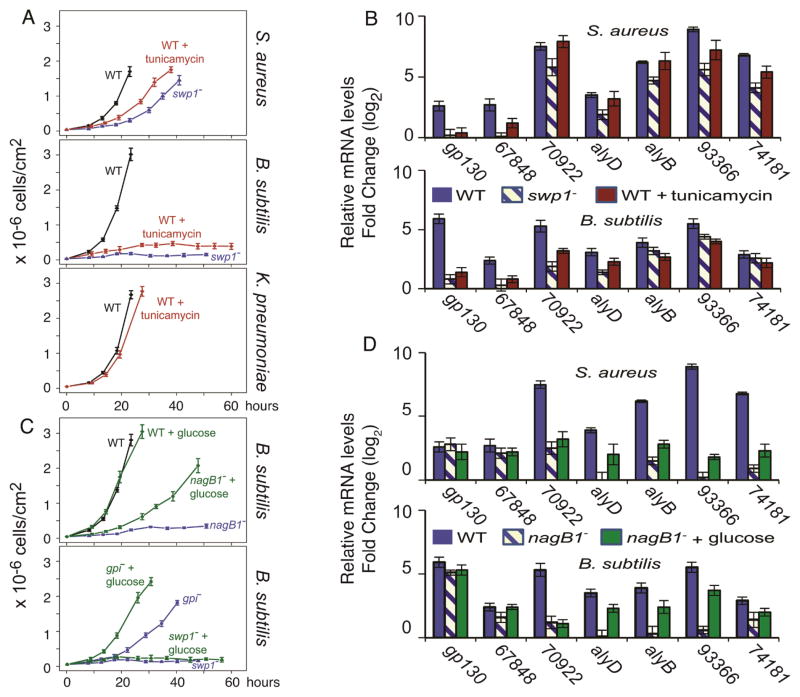
The transcriptional responses of D. discoideum mutants to Gram(+) bacteria suggest a pathway requiring swp1 and a distinct pathway involving gpi and nagB1. The swp1 gene encodes a subunit of the oligosaccharyl transferase protein complex that catalyzes asparagine (N-linked) glycosylation of proteins [32]. We hypothesized that the swp1 gene is important in the biogenesis of glycoproteins that are required for growth on Gram(+) bacteria, such as Gp130 [30]. To test this possibility we treated wild-type D. discoideum with tunicamycin, which blocks N-linked glycosylation in eukaryotes by inhibiting the charging of dolichol phosphate with N-acetyl-glucosamine [33]. Wild-type amoebae treated with tunicamycin displayed growth defects on Gram(+) bacteria, but not on the Gram(−) bacteria K. pneumoniae (Figure 4a), phenocopying the swp1− mutant. Tunicamycin-treated wild-type cells were also similar to the swp1− mutant in their transcriptional profile (Figure 3 – clades 3 and 4). Closer examination revealed that they expressed the Gram(+)-enriched hydrolase genes normally, but had similar low levels of gp130 and DDB_G0267848 transcripts (Figure 4b). This finding suggests that N-linked glycosylation is required for growth on Gram(+) bacteria but dispensable for growth on Gram(−) bacteria.
Figure 4. Two distinct pathways in amoebae for handling Gram(+) bacteria.
a, Growth curves of WT amoebae, WT treated with tunicamycin, and swp1− mutant on S. aureus (top) or B. subtilis (middle), and WT or WT treated with tunicamycin on K. pneumoniae (bottom). b, Relative abundance of mRNA (determined by qRT-PCR) of Gram(+)-enriched genes in WT amoebae, WT treated with tunicamycin, and the swp1− mutant on S. aureus (top), or B. subtilis (bottom) normalized to mRNA levels measured during growth on K. pneumoniae. Legend provided within the panels and the gene names are below the respective bars. c, Growth curves of WT and Gram(+) defective mutants grown on Gram(+) bacteria with or without glucose The axes are as in a. d, relative abundance of mRNA levels (determined by qRT-PCR) of Gram(+)-enriched genes in WT, nagB1− mutants, and nagB1− mutant treated with glucose on S. aureus (top) or B. subtilis (bottom) normalized to mRNA levels measured during growth on K. pneumoniae, as in b.
The role of a glucose metabolite in the response to Gram(+) bacteria
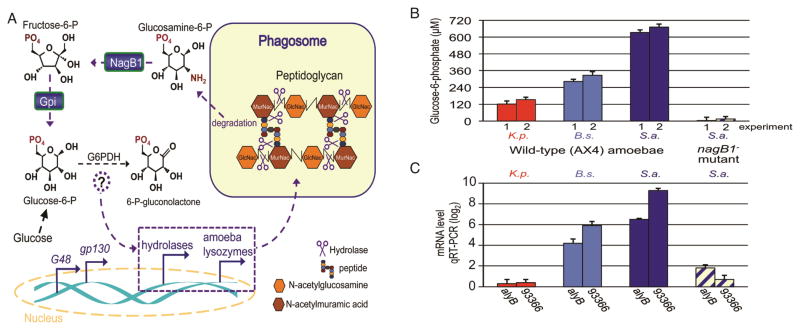
The gpi gene encodes phosphoglucose isomerase, which interconverts glucose-6-phosphate and fructose-6-phosphate, whereas the nagB1 gene product is predicted to catalyze the conversion of glucosamine-6-phosphate to fructose-6-phosphate. These two enzymes are required for the anabolic conversion of glucosamine-6-phosphate into pentose, through glucose-6-phosphate and the pentose phosphate shunt. D. discoideum may rely more on this pathway when growing on Gram(+) bacteria compared to when they are growing on Gram(−) bacteria due to the abundance of hexose monomers produced during the breakdown of thick Gram(+) bacterial cell walls. Supporting this notion, we observed increased expression of nagB1 during amoebal growth on Gram(+) bacteria, compared to Gram(−) bacteria (Figure 1 and supplemental Table S1). Thus, we hypothesize that glucose-6-phosphate, or a metabolite of glucose-6-phosphate, signals the presence of Gram(+) bacteria (Figure 5a). If true, wild-type D. discoideum growing on Gram(+) bacteria would have higher glucose-6-phosphate levels than cells growing on Gram(−) bacteria, and the nagB1− and gpi− mutants would have lower glucose-6-phosphate levels. To test this we grew wild-type (AX4) and nagB1 mutant amoebae on K. pneumoniae, S. aureus and B. subtilis and measured glucose-6-phosphate levels and the expression of Gram(+)-enriched genes. Consistent with our hypothesis, the glucose-6-phosphate levels in the wild-type amoebae did correlate directly with growth on Gram(+) bacteria and with the mRNA levels of the Gram(+)-enriched hydrolase genes, whereas glucose-6-phosphate levels and Gram(+) gene expression were significantly reduced in the nagB1 mutant (e.g., Figure 5b, c).
Figure 5. Correlation between glucose-6-phosphate levels and the induction of a subset of the Gram(+) response genes.
a, During amoebal growth on Gram(+) bacteria, a specific set of lysozymes and hydrolases are expressed that are predicted to aid in the degradation of the thick peptidoglycan cell wall of Gram(+) bacteria. We hypothesize that a glucose metabolite (e.g., glucose-6-phosphate, or 6-phosphoglucolactone) signals the presence of Gram(+) bacteria which results in the induction of these hydrolase gene products. The hypothesis predicts that the nagB1− and the gpi− mutants block the induction of these genes because the enzymatic activities of the NagB1 and Gpi gene products are both required for conversion the major breakdown product of peptidoglycan, glucosamine-6-phosphate, into glucose-6-phosphate. b, Wild-type D. discoideum cells were harvested during exponential growth on K. pneumoniae, S. aureus, and B. subtilis, and the nagB1− mutant mixed with S. aureus. Glucose-6-phosphate levels were determined in cell lysates using the BioVision glucose-6-phosphate assay kit (Materials and Methods). c, In the same samples in b, the mRNA levels of the Gram(+)-enriched hydrolase genes alyB and DDB_G093366 (93366) were determined by qRT-PCR, normalized to the level of the histone H3a gene transcripts.
To test if glucose-6-phosphate (or a metabolite) might serve as an internal cue we added glucose to the growth media to elevate glucose-6-phosphate levels within the amoebae, and tested growth and gene expression. We found that glucose partially rescued the growth of the nagB1− and gpi− mutants on Gram(+) bacteria but had no effect on growth of the wild type (Figure 4c, and supplemental Figure S6). Glucose treatment did not rescue the growth of the swp1− mutant (Figure 4c, and supplemental Figure S6), indicating that glucose rescue is specific to the gpi/nagB1 pathway. The addition of 2-deoxy-glucose, which cannot be processed by the glycolytic enzymes [34], did not rescue the growth of the nagB1− or gpi− mutants on Gram(+) bacteria (supplemental Figure S6). The transcriptional profiles of the nagB1− and gpi− mutants growing on B. subtilis with glucose resembled the wild type profile (Figure 3, clade 5, Figure 4d, and supplemental Figure S3) more than that of the untreated mutants (Figure 3, clade 4). More specifically, the treatment increased the abundance of the hydrolase transcripts to near wild-type levels, and more modest increases in expression were observed in the mutant mixed with S. aureus (Figure 3, clade 4, Figure 4d, and supplemental Figure S3). These data support a model whereby a glucose metabolite signals the presence of Gram(+) bacteria.
Discussion
Amoebae feed on a variety of bacteria and are subject to numerous pathogenic threats, so a regulated response to varying microbiota would be critical for amoebal defense and optimal feeding [3, 35]. It is known that D. discoideum amoeba induce characteristic physiologic changes when engulfing and degrading food bacteria, particularly when the bacteria have pathogenic potential [3, 35]. The transcriptional profiles that we report here expand earlier studies demonstrating differential transcriptional responses to bacteria [3, 15–17], reveal the specificity and extent of the physiological differences in amoebae growing on different bacteria, and are suggestive of adaptive responses by the amoebae that are highly regulated.
Our overarching hypothesis is that a bacterial response network exists in D. discoideum that begins with detection of bacterial elicitors and ends with a differentiated amoebal response that is critical for survival. Included in our hypothesis is the notion that the amoebae discriminate between Gram(+) and Gram(−) bacteria. Our evidence for this is based on the differential regulation of genes critical to the survival of the amoebae on these groups of bacteria, and our genetic results with mutants that displayed inverse growth defects with no intermediate phenotypes. Each of the mutants that could not grow on one species of Gram(+) bacteria were unable to grow on any Gram(+) species tested, but grew normally on all Gram(−) species tested. We observed the inverse of these results with each of the Gram(−)-growth defective mutants. Two of these genes indicate that the transcriptional regulation we have described is directly related to amoebal survival; gp130 is expressed in amoebae growing on Gram(+) bacteria and is also required growth of amoebae on Gram(+) bacterial, and alyL is expressed of in amoebae growing on Gram(−) bacteria and is also required for growth of amoebae on Gram(−) bacteria.
The growth defective mutants also display transcriptional profiles that are entirely consistent with the idea of an amoebal response network. The profiles of the mutants are relatively unaltered under conditions where they grow well, but specific changes in the profiles are apparent under conditions where the mutants cannot grow. Those specific changes can be used to classify the mutants into groups that potentially represent separable functions or pathways. Hierarchical clustering of the mutant profiles consistently link gpi and nagB1, alyL and spc3, clkB and tirA, and swp1 with tunicamycin treatment. These data suggest that during growth on Gram(−) bacteria, spc3 is important for the biogenesis of the AlyL protein and that tirA and clkB function in the same pathway, while, during growth on Gram(+) bacteria, swp1 is required for the asparagine glycosylation of one or more glycoproteins, and that gpi and nagB1 function in the same pathway.
Our observations suggest that D. discoideum uses at least two distinct pathways for handling Gram(+) bacteria. Amoebal growth on Gram(+) bacteria appears to be critically dependent on one or more glycoproteins and Gp130 appears to be one of them, so the swp1 mutant growth phenotype might be entirely attributable to reduced Gp130 function or levels [29, 30]. Pharmacological interruption of the N-linked glycosylation pathway of the amoebae would allow Gram(+) bacteria to evade predation by amoebae, and this appears to be a weakness that the Gram(+) Streptomyces bacteria seem to exploit for their survival by secreting tunicamycin [33, 36]. The glucose metabolite signal appears to influence the expression of hydrolases, such as amoeba lysozymes, that are predicted to degrade peptidoglycan. Glucosamine-6-phosphate and N-acetylglucosamine (a major bacterial cell wall component) link the degradation of peptidoglycan with central metabolism [37] and the NagB1 and Gpi enzymes convert glucosamine-6-phosphate to glucose-6-phosphate, which is needed to fuel the pentose phosphate pathway for the production nucleic acid precursors. Taken together these observations suggest that the metabolic flux of hexose monomers, from the catabolic breakdown of bacterial cell walls to the anabolic production of pentose monomers, is used by D. discoideum to effect appropriate responses to Gram(+) bacteria. We favor the hypothesis that anabolic glucose metabolites induce the expression of Gram(+) specific hydrolases because exogenous glucose specifically rescues expression of those genes in both gpi and nagB1 mutant cells, ruling out glycolytic metabolites (Figure 5a). It would be interesting to determine whether a similar mechanism operates in other eukaryotes, including humans.
During growth on K. pneumoniae, the amoebae appear to receive cues from the live bacteria that lead to the induction of Enterobacteriaceae-specific and Gram(−)-specific genes. D. discoideum mutants unable to grow on live do not induce these genes, but are able to grow on dead Gram(−) bacteria nonetheless, and dead bacteria do not induce the expression of these genes in wild-type cells. Furthermore, we found that live (and not dead) Gram(−) bacteria can induce amoebae to feed on dead Gram(+) bacteria. These findings reinforce the notion that the differential response we observe is at least in part an amoebal defense response to live bacteria and not simply a set of responses that optimize the digestion of bacteria.
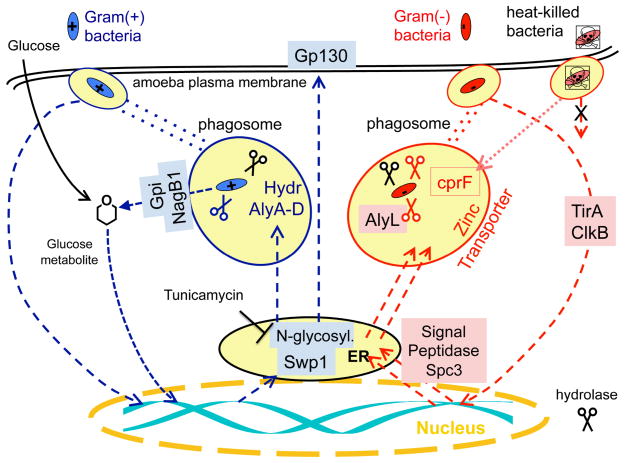
Our work has provided new insights into the interaction of amoebae and bacteria by showing that amoebae use distinct pathways to discriminate between different types of bacteria and by identifying components of the regulatory network that allow amoebae to recognize specific groups of bacteria, including potential receptors and signaling molecules that directly facilitate recognition (Figure 6). Phagocytosis, killing by superoxide radicals, and enzymatic digestion of bacteria are well-known examples of universal eukaryotic defense mechanisms [3, 4, 35, 38], but there are likely others that have yet to be uncovered. The Amoebozoa are a monophyletic group of eukaryotes that arose in evolution soon after the divergence of the plants and animals, about a billion years ago [27, 39]. A closer examination of bacterial recognition by amoebae, and characterization of the responses that recognition engenders, should inform strategies for subverting bacterial pathogenesis and may also provide useful insight into the origin of innate immune function in plants and animals.
Figure 6. Components in the amoebal response to Gram(+) and Gram(−) bacteria.
In this cartoon of the components described in this paper, proteins required for growth of amoebae on Gram(+) bacteria are boxed in light blue and proteins required for growth on Gram(−) bacteria are boxed in light pink (descriptions of each are provided in the main text and in Table 1). Two components are involved in the biogenesis of lysozomal, secreted, or membrane proteins that may affect more than one protein; N-linked glycosylation (Swp1) is critical for proteins required for growth on Gram(+) bacteria, while some aspect of signal peptide cleavage (Spc3) is critical for proteins required for growth on Gram(−) bacteria.
Experimental Procedures
Growth of D. discoideum
We used Dictyostelium discoideum laboratory strain AX4 [40] for all growth experiments. D. discoideum was recovered on K. pneumoniae bacterial lawn on SM medium, then grown axenically on HL5 medium and maintained at mid-log phase (2.5×106 cells/ml) [41] before subsequent experiments.
For the growth of D. discoideum on bacteria, the different bacterial strains were inoculated into Nutrient Media [Per 1 Liter: 6 gm beef extract (Difco, BD Biosciences), 10 gm Protease Peptone (Difco), 7.2 gm dextrose, 2.7 gm KH2PO4, 1.4 gm Na2HPO4, and 0.25 gm sodium chloride] for over-night culture. 1–1.5 ml of the over-night bacterial culture was spread on Nutrient media agar plates [10 cm petri plates with 40 ml, 2% Bacto agar (Difco) in Nutrient Media], and left to dry for 1–2 days at 22°C. The bacterial lawn that formed on the Nutrient agar plates was harvested by scraping it off of the agar plate and re-suspending the bacteria into modified Sorenson buffer (“mSor” is 30 mM phosphate made from a 50X stock solution; 150 gm/L KH2PO4, 21.6 gm/L Na2HPO4, pH 6.0). Each gram of wet bacteria was resuspended in 3 ml of mSor buffer to make a thick paste with a density equivalent to an OD600 light-scattering reading of 130–150 (a 100-fold dilution of the bacterial mixture would have an OD600 reading between 1.3 to 1.5). For heat-killed bacteria, a falcon tube containing the thick bacterial mixture was submerged in hot water bath (70–80 °C) for 20–30 minutes, mixing occasionally.
For the spot assay on buffered agar, we mixed D. discoideum cells (250 to 2500) as described, with 100 μl of the thick bacterial culture, and spotted 20 μL on buffered agar plate [10 cm petri plate made from 2% Nobel Agar (Difco) in mSor buffer], and scored for growth/ no growth phenotype over 5 days while keeping plates in a humid chamber.
For the spot assay on Nutrient media agar, we mixed D. discoideum cells (250 to 2500) as described, with 100 μl of overnight bacterial culture in Nutrient media, and spotted 10 μL on Nutrient media agar plates, and scored for growth phenotype over 5 days while keeping plate in a humid chamber.
For growth curves of D. discoideum on bacteria, we mixed 1×107 D. discoideum cells into 2 ml of the thick bacterial culture, spread the suspension evenly on buffered agar and let it dry on level surface (even distribution of mixture on plate is crucial). We then transferred to a humid chamber to prevent agar from further drying. We counted the amoebae present in one “plug” of agar taken by plunging the wide end of the Pasteur pipette (0.2 cm in diameter) into the agar plate and removing the agar disk along with the surface contents. The plug was transferred to 0.33–1 ml of buffer (volume determined empirically to get a count of 20–200 amoebae in each 0.1-μl aliquot, mixture was diluted further if cell count exceeded 200) in a 2 ml Eppendorf tube, vortexed and shaken vigorously until the amoebae cells were dissociated completely from the agar and the bacteria, and cells were then counted in a hemocytometer.
For growth curve of D. discoideum on HL5 axenic medium, we inoculated 5×104 cells/ml into a shaking culture of HL5 media, and counted cells on a hemocytometer.
RNA extraction for RNA-seq
1–3×107 wild type or mutant D. discoideum cells were mixed with 2 ml of a thick bacterial culture as mentioned above (slow growing mutants were mixed at higher density), and spread evenly on a 10-cm petri plate of buffered agar. After 14–16 hours, amoebae and bacteria were harvested and resuspended in ice-cold mSor buffer to wash away bacteria by low centrifugation (200×g at 4°C). Supernatant with bacteria was discarded. This procedure was repeated 2–4 times until no visible bacteria were detected in supernatant. 2.5×107 washed amoebae cells were immediately lysed in TRIZOL reagent, and total RNA was prepared according to the manufacturer’s protocol (Invitrogen by Life Technologies).
cDNA library preparation, RNA-seq data and mapping
To prepare cDNA libraries, we processed 20 μg of total RNA through one round of poly-A selection, RNA fragmentation, first-strand and second-strand cDNA synthesis, carried out according to Parikh et al. [42]. Two biological replicates were analyzed for each condition. We sequenced the cDNA libraries (read length = 35 bases) on a high-throughput Illumina Genome Analyzer II using the manufacturer’s recommended pipeline (versions 1.2 and 1.3). The resulting FASTQ files were mapped using the short-read alignment software bowtie (version 0.12.7, 64-bit) [43] allowing only for single hits (-m 1) and trimming unmapped reads up to 10bp iteratively by 2bp. The mapping procedure was similar to that explained in Parikh et al [42] and the data is maintained on the PIPAx server (http://pipa.biolab.si). Raw abundance level of a transcript is defined as the sum of all the reads that uniquely map to that transcript. In order to compare transcript abundance between different bacterial growth conditions, we normalized raw abundance values to account for differences in total number of mapable reads obtained with each RNA-seq run and differences in mapable gene length as described in Parikh et al. [42].
Data visualization
We generated the heat maps in Figure 1 using the heatmap.2 function from the gplots package in R [44]. To allow comparisons between gene expression profiles with different abundances, we normalize them to have a mean of 0 and a standard deviation of 1. The resulting z-scores are used to color the heat map. To calculate similarity between transcriptomes of D. discoideum grown on different bacteria (supplemental Figure S1a, we performed hierarchical clustering (R function hclust) on the expression vectors from each growth condition consisting of all the genes and visualized the results as a dendrogram. The expression vectors consist of normalized mRNA abundance levels averaged between the two replicates. We used Spearman’s correlation (SC) to calculate the distance (D = 1-SC) and complete linkage as the clustering criterion. Two objects (individual transcriptomes, or joints) are joined by means of a horizontal line if these objects are more similar to one another than any other object in the data. The vertical distance between objects is inversely proportional to the similarity between them. The horizontal distances in the dendrogram are meaningless. The pie charts were generated using annotation categorization using GO annotations as a reference framework (R function pie).
Differential expression from RNAseq data
baySeq was used to perform differential expression analysis and genes with False Discovery Rates (FDR) of 0.2 or lesser were considered to be differentially expressed [45]. We calculated fold-change of a gene on a bacterial species as the log2 ratio of averaged normalized mRNA abundance on that bacterial species to the maximum of the averaged scaled mRNA abundance on the other bacterial species. Genes that were differentially expressed and up regulated in D. discoideum cells grown on one bacterial species were categorized as species-specific genes. Genes that were differentially expressed between D. discoideum cells grown on Gram(+) and those grown on Gram(−) bacteria were categorized as group-enriched genes. The genes that were present in both group-enriched and species-enriched categories were removed from the set of species-enriched genes.
Genetic Screen
To generate random D. discoideum mutants, we performed restriction enzyme-mediated integration (REMI) on AX4 cells [46]. The following day, each transformation was diluted into two 96-well plates, and kept under blasticidin selection. We monitored clonal growth of D. discoideum mutants in each well (no more than one colony formation per well for clonal growth). Clonally growing mutants were consolidated into 96-well plates, and maintained at mid-log phase density (total HL5 volume in each well was 100 μL). Using a multichannel pipette, we mixed 5 μL from D. discoideum mutant culture from each well with 120 μL of overnight bacterial culture of K. pneumoniae (K.p.) or B. subtilis (B.s) bacteria in Nutrient media, and spotted each mixture on 9 × 9 inch Nutrient agar plate (made from 2% Bacto agar in Nutrient Media broth), each row had 12 different D. discoideum mutants mixed with K.p and next to it another row of the same 12 mutants mixed with B.s. Each spot contained 150–200 amoebae cells. Growth phenotype of each D. discoideum mutant was monitored over 5 days on K.p or B.s (supplemental Figure S7). Selected mutants were tested for their growth phenotype on other Gram-positive or Gram-negative bacterial species with spot assays on buffered and nutrient media agar, and a subset of mutants were assayed for growth rates on different bacteria.
Identification of insertion sites and recreating mutants
To identify plasmid insertion sites we carried out plasmid rescue using ClaI, BclI, KpnI, NcoI, NdeI, and BamHIII restriction enzymes, and sequencing the plasmid’s flanking regions using T7 and Sp6 universal primers, as described previously [46]. Recapitulation of the original genomic insertion was carried out by transforming linearized plasmid (from the plasmid-rescue procedure) into wild-type AX4 cells. Homologous recombination events that re-established the original insertion observed in the primary mutant were verified by junctional PCR and Southern blots. Recapitulated mutants were re-tested for the growth phenotype.
qRT-PCR for transcriptional profile analysis
1–3×107 AX4 or mutant D. discoideum cells, with or without treatment with glucose or tunicamycin, were mixed with thick bacterial culture and spread evenly on buffered agar (refer to growth of amoebae on different bacteria). 14–16 hours post incubation we extracted total RNA populations from amoebae (see total RNA extraction).
1 μg of total RNA was treated with DNAse I according to manufacturer’s recommendation (Invitrogen 18068-015), followed by cDNA synthesis using Bio Rad iScript cDNA Synthesis kit (170-8891). Approximately 17 ng of the original total RNA sample was used for each 50 μL reaction mixture for quantitative polymerase chain reaction (qPCR) using an Opticon2 real-time thermocycler (MJ Research). H3a, gpdA, and cprD mRNA were used to normalize for total mRNA load. MJ Opticon Monitor Analysis Software version 3.1 was used to compare relative mRNA abundance of each gene among different bacterial species growth condition. The difference was expressed as log2 ratio. Data were collected from three biological replicates, with three technical replicates each.
The data were subjected to hierarchical clustering using the R function pvclust [47] and classical multidimensional scaling (MDS) analysis (the R function, cmdscale), both of which depict the dissimilarities of the normalized mRNA levels of differentially expressed genes.
Glucose-6-phosphate determination
5×107 washed wild type or mutant D. discoideum cells mixed with or growing exponentially on K. pneumoniae, B. subtilis, or S. aureus bacteria (14–16 hours, see above for growth of amoebae on bacteria on buffered agar) were dounced homogenized with 2X volume of ice-cold PBS buffer. Samples were centrifuged at 10,000×g for 10 minutes at 4°C. Supernatant was spun down in 10-kDa molecular weight cut off filters to remove enzymes that utilize glucose-6-phosphate as a substrate. Flow through was processed to quantify glucose-6-phosphate concentration using Glucose-6-Phosphate Assay Kit according to manufacturer’s recommended protocol (BioVision; Milpitas, California, Cat# K657-100). We measured OD at 450 nm with ASYS UVM 340 microplate reader (Hitech GmbH, Austria).
Reporter gene plasmid construction
We cloned each gene’s promoter (350–700 bp fragment immediately upstream of gene start codon) into pPT165 plasmid [48] using the Gateway cloning system (Invitrogen). This positioned the promoters immediately upstream of the green fluorescent protein (GFP) coding sequence, allowing for differential control of GFP expression in D. discoideum cells. The ctnC promoter fragment that we used was the 557 bp immediately upstream of the start codon; the DDB_G0293366 (hydr1) promoter that we used was 364 bp; the DDB_G0274181 (hydr2) promoter was 700 bp; and the act15 promoter was 393 bp.
We transformed 10μg of plasmid into 5×106 wild-type AX4 cells using electroporation in H-50 buffer [49]. Transformed cells were maintained in HL5 media supplemented with 20μg/ml concentration of G418 (Geneticin). Transformed amoebae were mixed with different bacterial species and spotted on buffered agar, see above for spot assay on buffered agar. 1–2 days later, fluorescent amoebae were visualized under a fluorescence microscope or under the Dark Reader spot lamp (Clare Chemical Research, Inc., SL9S).
Supplementary Material
Acknowledgments
This work was supported by the Dictyostelium Functional Genomics Program Project Grant from the National institutes of Health (PO1 HD39691). The authors thank Anjana Sankara Narayanan for help with setting up the genetic screen and Moshe Harel for critical comments on the manuscript.
References
- 1.Raper KB. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J Agr Res. 1935;50:135–147. [Google Scholar]
- 2.Raper KB. The influence of the bacterial associate and of the medium upon the growth and development of Dictyostelium discoideum. Vol. 22. Cambridge, MA: Harvard University; 1936. [Google Scholar]
- 3.Clarke M. Recent insights into host-pathogen interactions from Dictyostelium. Cell Microbiol. 2010;12:283–291. doi: 10.1111/j.1462-5822.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- 4.Steinert M. Pathogen-host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens. Semin Cell Dev Biol. 2011;22:70–76. doi: 10.1016/j.semcdb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Bozzaro S, Eichinger L. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011;12:942–954. doi: 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solomon JM, Rupper A, Cardelli JA, Isberg RR. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Inf Immun. 2000;68:2939–2947. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, van Delden C, Curty LK, Kohler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bact. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hasselbring BM, Patel MK, Schell MA. Dictyostelium discoideum as a model system for identification of Burkholderia pseudomallei virulence factors. Infect Immun. 2011;79:2079–2088. doi: 10.1128/IAI.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci U S A. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iriti M, Faoro F. Review of innate and specific immunity in plants and animals. Mycopathologia. 2007;164:57–64. doi: 10.1007/s11046-007-9026-7. [DOI] [PubMed] [Google Scholar]
- 12.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walk A, Callahan J, Srisawangvong P, Leuschner J, Samaroo D, Cassilly D, Snyder ML. Lipopolysaccharide enhances bactericidal activity in Dictyostelium discoideum cells. Dev Comp Immunol. 2011;35:850–856. doi: 10.1016/j.dci.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farbrother P, Wagner C, Na J, Tunggal B, Morio T, Urushihara H, Tanaka Y, Schleicher M, Steinert M, Eichinger L. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol. 2006;8:438–456. doi: 10.1111/j.1462-5822.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- 16.Sillo A, Bloomfield G, Balest A, Balbo A, Pergolizzi B, Peracino B, Skelton J, Ivens A, Bozzaro S. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics. 2008;9:291. doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carilla-Latorre S, Calvo-Garrido J, Bloomfield G, Skelton J, Kay RR, Ivens A, Martinez JL, Escalante R. Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 2008;8:109. doi: 10.1186/1471-2180-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salton MR. The anatomy of the bacterial surface. Bacteriol Rev. 1961;25:77–99. doi: 10.1128/br.25.2.77-99.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 20.Newell PC, Henderson RF, Mosses D, Ratner DI. Sensitivity to Bacillus subtilis: A novel system for selection of heterozygous diploids of Dictyostelium discoideum. J Gen Microbiol. 1977;100:207–211. [Google Scholar]
- 21.Morrissey JH, Wheeler S, Loomis WF. New loci in Dictyostelium discoideum determining pigment formation and growth on Bacillus subtilis. Genetics. 1980;96:115–123. doi: 10.1093/genetics/96.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, Lardy B, Klein G, Sansonetti P, Charette SJ, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 23.Van Driessche N, Demsar J, Booth EO, Hill P, Juvan P, Zupan B, Kuspa A, Shaulsky G. Epistasis analysis with global transcriptional phenotypes. Nat Genet. 2005;37:471–477. doi: 10.1038/ng1545. [DOI] [PubMed] [Google Scholar]
- 24.Andra J, Herbst R, Leippe M. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev Comp Immunol. 2003;27:291–304. doi: 10.1016/s0145-305x(02)00106-4. [DOI] [PubMed] [Google Scholar]
- 25.dictybase.org2012
- 26.Muller I, Subert N, Otto H, Herbst R, Ruhling H, Maniak M, Leippe M. A Dictyostelium mutant with reduced lysozyme levels compensates by increased phagocytic activity. J Biol Chem. 2005;280:10435–10443. doi: 10.1074/jbc.M411445200. [DOI] [PubMed] [Google Scholar]
- 27.Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narita TB, Koide K, Morita N, Saito T. Dictyostelium hybrid polyketide synthase, SteelyA, produces 4-methyl-5-pentylbenzene-1,3-diol and induces spore maturation. FEMS Microbiol Lett. 2011;319:82–87. doi: 10.1111/j.1574-6968.2011.02273.x. [DOI] [PubMed] [Google Scholar]
- 29.Chia CP, Gomathinayagam S, Schmaltz RJ, Smoyer LK. Glycoprotein gp130 of dictyostelium discoideum influences macropinocytosis and adhesion. Mol Biol Cell. 2005;16:2681–2693. doi: 10.1091/mbc.E04-06-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feasley CL, Johnson JM, West CM, Chia CP. Glycopeptidome of a heavily N-glycosylated cell surface glycoprotein of Dictyostelium implicated in cell adhesion. J Proteome Res. 2010;9:3495–3510. doi: 10.1021/pr901195c. [DOI] [PubMed] [Google Scholar]
- 31.Masschalck B, Michiels CW. Antimicrobial properties of lysozyme in relation to foodborne vegetative bacteria. Crit Rev Microbiol. 2003;29:191–214. doi: 10.1080/713610448. [DOI] [PubMed] [Google Scholar]
- 32.Lennarz WJ. Studies on oligosaccharyl transferase in yeast. Acta Biochim Pol. 2007;54:673–677. [PubMed] [Google Scholar]
- 33.Takatsuki A, Arima K, Tamura G. Tunicamycin, a new antibiotic. I Isolation and characterization of tunicamycin. J Antibiot (Tokyo) 1971;24:215–223. doi: 10.7164/antibiotics.24.215. [DOI] [PubMed] [Google Scholar]
- 34.Tresse E, Kosta A, Giusti C, Luciani MF, Golstein P. A UDP-glucose derivative is required for vacuolar autophagic cell death. Autophagy. 2008;4:680–691. doi: 10.4161/auto.6084. [DOI] [PubMed] [Google Scholar]
- 35.Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Yamada H, Hirano T, Miyazaki T, Takatsuki A, Tamura G. Effects of tunicamycin on cell adhesion and biosynthesis of glycoproteins in aggregation-competent cells of Dictyostelium discoideum. J Biochem. 1982;92:399–406. doi: 10.1093/oxfordjournals.jbchem.a133946. [DOI] [PubMed] [Google Scholar]
- 37.Komatsuzawa H, Fujiwara T, Nishi H, Yamada S, Ohara M, McCallum N, Berger-Bachi B, Sugai M. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol Microbiol. 2004;53:1221–1231. doi: 10.1111/j.1365-2958.2004.04200.x. [DOI] [PubMed] [Google Scholar]
- 38.Casadevall A, Pirofski LA. Accidental virulence, cryptic pathogenesis, martians, lost hosts, and the pathogenicity of environmental microbes. Eukaryot Cell. 2007;6:2169–2174. doi: 10.1128/EC.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bapteste E, Brinkmann H, Lee JA, Moore DV, Sensen CW, Gordon P, Durufle L, Gaasterland T, Lopez P, Muller M, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci U S A. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knecht DA, Cohen SM, Loomis WF, Lodish HF. Developmental regulation of Dictyostelium discoideum actin gene fusions carried on low-copy and high-copy transformation vectors. Mol Cell Biol. 1986;6:3973–3983. doi: 10.1128/mcb.6.11.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Meth Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- 42.Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, et al. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warnes GR. gplots: Various R programming tools for plotting data. 2008 available at: http://cran.r-project.org/web/packages/gplots/index.html.
- 45.Hardcastle TJ, Kelly KA. baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics. 2010;11:422. doi: 10.1186/1471-2105-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuspa A. Restriction enzyme-mediated integration (REMI) mutagenesis. Methods Mol Biol. 2006;346:201–209. doi: 10.1385/1-59745-144-4:201. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki R, Shimodaira H. pvclust: Hierarchical Clustering with P-Values via Multiscale Bootstrap Resampling. 2006. [Google Scholar]
- 48.Thomason PA, Brazill DT, Cox EC. A series of Dictyostelium expression vectors for recombination cloning. Plasmid. 2006;56:145–152. doi: 10.1016/j.plasmid.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 49.Eichinger L, Rivero F. Dictyostelium discoideum Protocols. Vol. 346. Totowa, NJ: Humana Press; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.