Abstract
Purpose of review
Bacterial colonization of the infant intestinal tract begins at birth. We are at the forefront of understanding complex relationships between bacteria and multiple parameters of health of the developing infant. Moreover, the establishment of the microbiome in the critical neonatal period is potentially foundational for lifelong health and disease susceptibility. Recent studies utilizing state-of-the-art culture-independent technologies have begun to increase our knowledge about the gut microbiome in infancy, the impact of multiple exposures, and its effects on immune response and clinical outcomes such as allergy and infection.
Recent findings
Postnatal exposures play a central role in the complex interactions between the nearly blank canvas of the neonatal intestine, whereas genetic factors do not appear to be a major factor. Infant microbial colonization is affected by delivery mode, dietary exposures, antibiotic exposure, and environmental toxicants. Successive microbiome acquisition in infancy is likely a determinant of early immune programming, subsequent infection, and allergy risk.
Summary
The novel investigation of the neonatal microbiome is beginning to unearth substantial information, with a focus on immune programming that coevolves with the developing microbiome early in life. Several exposures common to neonatal and infant populations could exert pressure on the development of the microbiome and major diseases including allergy and infection in large populations.
Keywords: allergy, infection, microbiome, neonate
INTRODUCTION
The intestinal microbiome has evolved with humans, and is described as creating with its host a metabolic ‘superorganism’, comprised of millions of microbial genes [1]. The complex symbiotic relationship between microbiome and host fills critical physiological roles, and growing evidence suggests a role in immune maturation, as well as diverse metabolic functions [1,2▪]. The microbiome also may induce disorder, and a lengthening list of diseases are now thought to derive from, or be exacerbated by, host–microbiome interactions, including obesity, inflammatory bowel disease, and circulatory diseases. Critical to the pediatric population, the microbiome also may be linked to infection and allergy risk [2▪], as we begin to uncover the multifaceted relationship between bacteria and various parameters of health of the developing infant. Indeed, accumulating data point to establishment of the microbiome during this vulnerable developmental period as fundamentally influencing later disease risk [3]. Potential implications of detailed microbiome data in the neonatal period include informing newborn delivery decision-making, further information regarding the physiology behind the lifelong health benefits of breast-milk exposure in infancy, limiting or altering antibiotic regimens for common infectious diseases, targeted use of specific probiotics to treat and prevent diseases, and ultimately individualization of medication regimens for young children based upon microbial profiles.
Until recently, investigation of serial intestinal colonization patterns and their relationship with exposures in larger human cohorts have focused primarily on adults, whose microbiota is considered to be relatively stable [4▪▪, 5,6▪,7,8▪▪]. In contrast, neonatal intestinal microbial acquisition patterns have traditionally been examined by culture-based or targeted molecular studies, which incompletely characterize the microbiome [9,10,11▪▪,12▪▪,13,14]. Many of these studies focused on premature infant populations, and may reflect specific exposures to this population, including antibiotic treatment, dietary factors, and pathogen-laden hospital environments [13–16]. In healthy neonatal populations, bacterial colonization begins during the process of delivery [3,9,11▪▪,17] and is primarily determined by mode of delivery, gestational age, infant feeding, hospitalization, and antibiotic exposure [10,18,19]. Recent work exploring micro-biome content in relation to age in 326 individuals ages 0–17 found that, regardless of cultural or geographic environment, children evolve an ‘adult-like’ microbiome within the first 3 years of life, but this time period also marks the greatest intrapersonal and interpersonal variation within these microbial communities, possibly reflecting the differential development of the microbiome in relation to environmental factors [8▪▪].
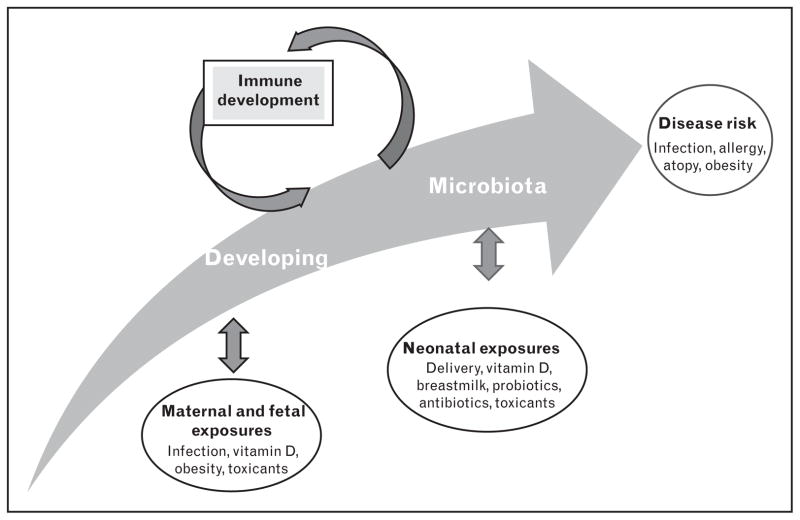
Complete investigation of the developing neonatal microbiome using massively parallel deep sequencing permits an unbiased analysis of acquisition patterns [15,20–22]. Using this technology has helped us to understand how the establishment of symbiotic bacteria can act as a central stimulus for maturation of the immune system and may alter risk of developing immune-mediated diseases [3,23–27] (Fig. 1). External factors, as opposed to genetics, drive the development of the human microbiome; thus, elucidation of these factors will present opportunities to inform decisions that could potentially impact health throughout a child’s lifetime.
FIGURE 1.
The developing intestinal microbiota beginning at birth. The human intestinal microbiota is shaped by environmental exposures and interacts with the developing immune system.
EXTERNAL FACTORS AND THE IMPACT ON NORMAL NEONATAL GUT MICROBIOME DEVELOPMENT: MODE OF DELIVERY
A healthy human gestation involves a primarily sterile environment, and birth presents the infant’s first encounter with microorganisms that rapidly populate the gut, forming its initial microbiome. Recent studies of infants’ first bacterial gut colonizers after vaginal or cesarean (C-section) delivery suggest that these primary assemblages may be dictated by delivery mode in that vaginally delivered infants more likely become colonized by the organisms comprising the maternal vaginal microbiome, including Lactobacilli and Prevotella, whereas infants delivered by C-section more frequently acquire bacteria present on the mother’s skin and in the surrounding hospital environment, such as Staphylococcus, Propionibacterium, and Corynebacterium [28,29▪▪]. A cross-sectional study of 84 women found that during pregnancy the vaginal microbial community undergoes a decrease in diversity, while becoming simultaneously enriched with Lactobacilli species, which may relate to the vertical transmission that occurs at birth [30▪▪]. Although infants may only retain a portion of the bacteria from the initial colonization, birth can have long-term impacts on the composition of the microbiome [12▪▪,31▪]. In a longitudinal study of 605 infants from five European countries, repeated profiling of the gut microbiome at 6 weeks of age and post-weaning found mode of delivery and preweaning feeding method had persistent effects on microbial composition [31▪]. If early shifts in the development of the microbiota, as may occur with C-section delivery, have lasting health consequences, this would impact a substantial number of children in the United States and elsewhere. An estimated one-third of all births in the United States occur by C-section, many of which are elective [US Centers for Disease Control report – http://www.cdc.gov/nchs/data/databriefs/db35.htm].
There have been some indications within the literature that C-section delivery may be associated with adverse health outcomes and greater susceptibility to infections. For example, babies delivered by C-section appear to have a higher risk of methicillin resistant Staphylococcus aureus (MRSA) infection [32▪,33]. This could be linked to the role of pioneering colonizers in immune development or a lack of protection against pathogenic colonization normally conferred by vaginally transmitted microflora [29▪▪,32▪,33]. However, further studies are warranted, as this has yet to be epidemiologically investigated.
EXTERNAL FACTORS AND THE IMPACT ON NORMAL NEONATAL GUT MICROBIOME DEVELOPMENT: BREASTFEEDING AND DIET
Early life events, such as transitions from breastmilk to formula and the introduction of solid foods, appear to influence bacterial succession in the gut [12▪▪,31▪,34]. In a randomized study, breastfed infants tended to have lower levels of potentially pathogenic Clostridium difficile than their formula-fed counterparts, who also tended to have had higher proportions of Bacteroides and Prevotella [35▪]. Although healthy infants often carry C. difficile asymptomatically in their gut in early infancy, its presence can alter community composition [36▪].
Breastfeeding is associated with a lower risk of childhood and adult-onset obesity (reviewed in [37▪]). This may be due, in part, to the effects of breastfeeding on the development of the microbiome, as early diet guides colonization. Bacteria possess varying abilities to extract nutrients and energy from food; consequently, the microbiome can shift an infant’s energy storage potential [38▪]. Further, oligosaccharides in breastmilk can selectively promote Bifidobacterium growth in the gut, shown by combinatorial genomic and culture approaches with parallel glycoprofiling [39▪▪]. A study of 56 mother–infant pairs found that high maternal BMI during pregnancy is associated with lower levels of key immunomodulators in breast-milk and infant gut Bifidobacterium counts [40▪], which may in turn contribute to long-term health and weight management in breastfed infants [41▪]. A study of 30 children, enrolled in an ongoing longitudinal study, found that at age 10 overweight children had lower levels of gut Bifidobacterium as infants, compared with their normal-weight counterparts [41▪]. However, epidemiological longitudinal studies assessing the microbiome–obesity relation are lacking.
EXTERNAL FACTORS AND THE IMPACT ON NORMAL NEONATAL GUTMICROBIOME DEVELOPMENT: ENVIRONMENTAL TOXICANTS AND THE MICROBIOME
Although microbial transformations may increase bioavailability of some nutrients, these same processes can produce more toxic forms of contaminants. Using an in-vitro model of the human gut microbiome, Diaz-Bone and van de Wiele [42] found that normal human intestinal bacteria metabolize environmental contaminants, turning polycyclic aromatic hydrocarbons into bioactive estrogen-like molecules and transforming metals into volatile, and sometimes toxic, products [43] that can affect the gut’s species balance and function, a condition known as dysbiosis. One study found that dysbiosis can potentially result from bismuth exposure, commonly found in cosmetics and Pepto-Bismol, when Methanobrevibacter smithii, a normal gut inhabitant, transforms bismuth into a form that is toxic to Bacteroides thetaiotaomicron, a beneficial resident that mediates infant dietary transition from breastmilk to starches, and aids the formation of the intestinal mucosal barrier that protects against pathogens [44▪]. Hence, early-life toxicant exposure could shift the microbial balance, potentially affecting both immune and microbiome development.
THE NEONATAL MICROBIOME, IMMUNITY, AND ALLERGY
Several seminal studies in germ-free animals demonstrate that absence of microbial colonization results in altered gut epithelialization, growth, and immune function [45]. Interestingly, specific bacteria have been associated with early-onset allergy (herein defined as any exaggerated immune response to a foreign antigen regardless of mechanism) and atopy (defined as exaggerated immunoglobulin E-mediated immune response or type I hypersensitivity disorders), including C. difficile and Escherichia coli in humans [46,47]. In studies of infants, decreased microbial diversity in the first weeks of life was related to risk of allergy and atopy in infancy and at school age [48,49▪▪]. Correspondingly, in a study of infants exposed to antibiotics, gut microbes appeared to influence the maturation of T helper cells (Th1) immune responses, CD4+ T-cell phenotype, Th1/Th2/Th17 development and activity, and regulatory T-cell function [50].
Targeted investigation of neonatal microbial colonization patterns with Bifidobacterium found associations between enhanced maturation of protective mucosal immunoglobulins and early intense colonization with Bacteroides fragilis might downregulate immune responsiveness in infancy [27]. Moreover, a novel evaluation of diet-dependent interactions within the relationship between the microbiome and host transcriptome identified not only differences in specific bacteriology between breastmilk and formula exposed infants by 3 months, but also metabolic function, immunity, and defense genes, which were more readily upregulated in the breastfed infants [51▪].
THE NEONATAL MICROBIOME, ANTIBIOTIC EXPOSURE, AND INFECTION
Despite advances in sanitation and immunization programs, infectious diseases remain the leading cause of illness in children in the United States, and the primary cause of childhood death in developing countries [52,53]. Public health implications of common infections, for example, otitis media or influenza in the under-5 US population, include antibiotic overuse and resistance, transmission of common infections to pregnant women and their fetuses, or widespread transmission of infectious diseases in unimmunized populations. The average US child is exposed to 10–20 courses of antibiotics before age 18, and this may have far-reaching implications for future disease risk, secondary to the effects on the microbiome [54▪].
Perinatal antibiotic use and antibiotic resistance
Perinatal and early-life antibiotic usage, as well as illnesses themselves, have the potential to influence the establishment of microbial communities and cause large shifts in taxonomic groups, altering overall diversity [12▪▪,31▪,34,55▪]. One study of 31 amoxicillin-treated infants with acute respiratory infection identified complete elimination of Bifido-bacterium adolescentis species, as well as a significant decrease in Bifidobacterium bifidum in the gut, with no change in overall counts of Bifidobacterium but profound shifts in the microbiota at the species level [56]. Indeed, life-threatening complications of empiric antibiotic use in premature neonates, including necrotizing enterocolitis and sepsis, have been observed in a large National Institute of Child Health and Human Development cohort study [57]. However, when early fecal samples from antibiotic-exposed infants were compared with a later post-weaning sample, antibiotic resistance was reduced, and overall diversity had increased [31▪]. This may have been due to the plasticity and rapid rate of change within the gut microbiota in the first year of life, suggesting that effects of early antibiotic usage in infants may be diminished over time [12▪▪,31▪]. Potential long-term effects on the microbiome from early-life antibiotic exposure still may occur, including childhood overweight and obesity associated with antibiotic use [58▪▪]; but these effects remain to be elucidated.
In two studies, a large fraction of healthy, non-antibiotic-treated infants in the first 3 months of life harbored resistant and multiply resistant bacterial strains [59,60], perhaps through maternal transmission [61▪]. Although it has yet to be evaluated epidemiologically, the growing presence of resistant microbes may be due in part to more widespread contaminant exposures from foods and the environment. For instance, several studies demonstrated that individuals exposed to mercury were more likely to possess resistance to multiple antibiotics, suggesting a coselection mechanism [62]. Children are regularly exposed to arsenic, which can be found in well water and foods, such as rice and baby formula [63▪▪,64▪,65]. Metals, such as arsenic, which was used historically as an antibiotic in humans [66] and is currently added to animal feed, have contributed to the emergence of metal/antibiotic coresistant strains arising in livestock, including MRSA isolates [67▪] transmittable to humans via the environment and food supply. These multiresistant pathogens heighten risk of adverse outcomes, especially in young children. Once antibiotic resistance genes are selected for, they may persist within the microbiota for years [68].
Links with infection
A direct link between gut colonization and risk for infection has been described for high-risk neonates [69,70▪]. Unlike full-term infants, many premature infants’ intestines are colonized with pathogenic organisms at birth, likely related to maternal gestational infection and prenatal antibiotic exposure [3,71]. Intestinal pathogenic bacterial predominance and lack of microbial diversity are implicated in neonates with life-threatening infectious diseases (sepsis caused by Enterobacteracea and Coagulase-negative Staphylococcus) [72,73], and in one study with the predominant gut pathogen (Staphylococcus) [74▪]. The potential for healthy term infants, who experience varying environmental exposures from antibiotics, dietary choices, and mode of delivery, among others, to undergo shifts in microbial colonization, altering their underlying risk of infection, clearly warrants epidemiologic investigation.
FUTURE RESEARCH AND IMPLICATIONS FOR INTERVENTION
The neonatal microbiome is an area of emerging interest due to its relative simplicity at its onset at birth, and its subsequent development, which has the potential to dramatically influence lifelong health and disease risk. Culture-independent techniques have become more readily accessible to researchers, in terms of cost and ease of application, along with emerging sophisticated bioinformatics techniques to analyze the high throughput data and these research tools are being applied to neonatal populations. The use of these burgeoning techniques in large prospective epidemiological studies of neonates to define the ‘healthy’ developing microbiome in infancy and the impact of specific exposures in infant life is critical to promoting health. Thus, it will be imperative to study the relationship between maternal, fetal, and the neonatal microbiome as we work to identify preventable causes of premature delivery [30▪▪,75▪], and fetal basis of diseases, which may include fetalimmune programming, as it relates to the microbiome in prenatal and postnatal life. Potential translation to the clinical setting might include: informing newborn delivery decision-making in favor of vaginal deliveries when possible, further reinforcing and illuminating the physiology behind the lifelong health benefits of breastmilk exposure in infancy, limiting or altering antibiotic regimens for common infectious diseases, targeted use of specific probiotics to treat and prevent diseases, and ultimately individualization of medication regimens for young children based upon microbial profiles.
CONCLUSION
Advances in nonculture-based approaches to characterize the microbiome have opened up opportunities to embark on studies of the normal patterns of colonization of neonatal populations and linking specific microbiome patterns to disease risk in pediatric populations, specifically allergy and infection. Certain exposures may have profound effects on the microbiome in early life, including delivery mode, diet, antibiotics, and potentially environmental toxicants, many of which can be eliminated or moderated. Future epidemiologic studies in large populations targeting investigation of the infant microbiome beginning in fetal life will be extremely informative as we strive to define a ‘healthy’ microbiome in childhood to ameliorate disease risk.
KEY POINTS.
The intestinal microbiome, beginning with the nearly sterile newborn, is shaped over time by multiple exposures potentially including delivery mode, diet, antibiotics and toxicants from the environment.
The developing intestinal microbiome, beginning at birth, interacts in a complex interplay with the developing immune system and the immune system, in turn, likely shapes the developing microbiome.
Defining a ‘healthy’ microbiome in the neonatal and infant period is becoming more accessible with culture-independent sequencing technologies to fully identify the microbes that make up the microbiome.
Differences in the neonatal microbiome as they relate to short-term and long-term disease, including infection and allergy/atopy, are being identified, potentially highlighting opportunities for intervention and lifelong disease prevention.
Acknowledgments
Special thanks to Claudia Cornejo for her research assistance. M.R.K. and J.C.M. are members of the formative Children’s Environmental Health and Disease Prevention Research Center at Dartmouth (P20 ES018175 from NIEHS and RD-83459901 from the EPA). S.F.F. is funded by R25CA134286. J.C.M. also received funds from The Hearst Foundation, The Synergy Grant (Dartmouth), The Joshua Burnett Career Development Award through the Hitchcock Foundation (Dartmouth), the Department of Pediatrics, Dartmouth and a pilot grant to J.C.M. from the Cystic Fibrosis Foundation Research Development Program (STANTO07R0) and the CF Foundation Harry Shwachman Clinical Investigator Award. P.L.H. is funded by NIH grant 2K24AT003683.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–818. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2▪.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. Review of recent studies (including metagenomic and metabonomic approaches) outlining the interaction between symbiotic microorganisms and the human gastrointestinal tract and their collective interacting genomes in relationship to health and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mshvildadze M, Neu J, Shuster J, et al. Intestinal microbial ecology in premature infants assessed with nonculture-based techniques. J Pediatr. 2010;156:20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4▪▪.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. A large international study of combined sequenced fecal metagenomes of individuals from four countries identified three robust clusters, which they termed enterotypes, that were not nation specific, they were varied, stratified; and not continuous; enterotypes were driven by species composition, highlighting the potential to respond differently to diet and drug intakes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. This study of 98 individuals linked diet inventories to sequencing data from fecal samples demonstrating that communities clustered into enterotypes that were distinguished by levels of Bacteroides and Prevotella, and associated with long-term diet (particularly protein and animal fat versus carbohydrate) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manson JM, Rauch M, Gilmore MS. The commensal microbiology of the gastrointestinal tract. Adv Exp Med Biol. 2008;635:15–28. doi: 10.1007/978-0-387-09550-9_2. [DOI] [PubMed] [Google Scholar]
- 8▪▪.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. This large cross-cultural study of over 500 individuals ranging in age from newborn to 70 years examined microbiome differences in relation to age, diet, and geographical location. In infants, researchers found the largest interindividual and intraindividual variation in gut microbial communities when compared with all other ages, and these are largely dominated by Bifidobacterium species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 11▪▪.Eggesbo M, Moen B, Peddada S, et al. Development of gut microbiota in infants not exposed to medical interventions. APMIS. 2011;119:17–35. doi: 10.1111/j.1600-0463.2010.02688.x. A large, longitudinal study of the colonization process during the first 4 months after birth in 85 infants who have no major medical or dietary interventions (vaginal delivery, healthy, term infants, with no antibiotics, who were exclusively breastfed for at least 1 month and then at least partially breastfed up to 4 months). Almost all infants harbored gamma-Proteobacteria and Bifidobacterium, with associations between day 4 microbiota and day 120, indicating early microbiota influences later microbiota. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12▪▪.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infantgut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. A longitudinal study of one healthy infant over 2.5 years (case study) of 60 fecal samples and sequencing of the phylogenetic diversity (structure and function) over time in relationship to life events. Diversity increased over time, but major taxonomic groups showed abrupt shifts in abundance with diet or health changes, with profound shifts with the onset of solid food mirroring adult microbiome structure and function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwiertz A, Gruhl B, Lobnitz M, et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatr Res. 2003;54:393–399. doi: 10.1203/01.PDR.0000078274.74607.7A. [DOI] [PubMed] [Google Scholar]
- 14.Magne F, Abely M, Boyer F, et al. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57:128–138. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacquot A, Neveu D, Aujoulat F, et al. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. 2011;158:390–396. doi: 10.1016/j.jpeds.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Rouge C, Goldenberg O, Ferraris L, et al. Investigation of the intestinal micro-biota in preterm infants using different methods. Anaerobe. 2010;16:362–370. doi: 10.1016/j.anaerobe.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 17.Turroni F, Ribbera A, Foroni E, et al. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek. 2008;94:35–50. doi: 10.1007/s10482-008-9232-4. [DOI] [PubMed] [Google Scholar]
- 18.Adlerberth I, Wold AE. Establishment of the gut microbiota in western infants. Acta Paediatr. 2009;98:229–238. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 19.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:13–29. doi: 10.1159/000146245. discussion 29–33. [DOI] [PubMed] [Google Scholar]
- 20.Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huse SM, Huber JA, Morrison HG, et al. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sogin ML. Characterizing microbial population structures through massively parallel sequencing. In: Epstein SS, editor. Uncultivated Microorganisms. Berlin Heidelberg: Springer-Verlag Publishing; 2009. pp. 19–34. [Google Scholar]
- 23.Backhed F. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: the normal gut microbiota in health and disease. Clin Exp Immunol. 2010;160:80–84. doi: 10.1111/j.1365-2249.2010.04123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung H, Kasper DL. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr Opin Immunol. 2010;22:455–460. doi: 10.1016/j.coi.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Neu J, Chen M, Beierle E. Intestinal innate immunity: how does it relate to the pathogenesis of necrotizing enterocolitis. Semin Pediatr Surg. 2005;14:137–144. doi: 10.1053/j.sempedsurg.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Shanahan F. 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: host-microbe interactions in the gut: target for drug therapy, opportunity for drug discovery. Clin Exp Immunol. 2010;160:92–97. doi: 10.1111/j.1365-2249.2010.04135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjogren YM, Tomicic S, Lundberg A, et al. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39:1842–1851. doi: 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29▪▪.Dominguez-Bello MG, Blaser MJ, Ley RE, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–1719. doi: 10.1053/j.gastro.2011.02.011. A focused review highlighting nonculture-based work describing the development of gastrointestinal microbiota and key early exposures that affect colonization, with reference to evolutionary and age-related trends in variation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30▪▪.Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. This study showing decreased vaginal microbial diversity during pregnancy may shed light on the relationship between the maternal and neonatal microbiome and help to identify causes of premature delivery and perinatal infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31▪.Fallani M, Amarri S, Uusijarvi A, et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157 (Pt 5):1385–1392. doi: 10.1099/mic.0.042143-0. This cohort study was among the first to characterize variations in species prevalence in relation to early events, such as weaning and solid food introduction, using repeated infant fecal samples. [DOI] [PubMed] [Google Scholar]
- 32▪.Gregory KE. Microbiome aspects of perinatal and neonatal health. J Perinat Neonatal Nurs. 2011;25:158–162. doi: 10.1097/JPN.0b013e3182169346. quiz 163–164. A review of the role of the microbiome in perinatal and neonatal health, including exposures to potentially pathogenic organisms that can cause necrotizing enterocolitis and MRSA infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson J, Jones RC, Cortes C, et al. Community-Associated Methicillin-Resistant. JAMA. 2006;296:36–38. [Google Scholar]
- 34.Fallani M, Young D, Scott J, et al. Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010;51:77–84. doi: 10.1097/MPG.0b013e3181d1b11e. [DOI] [PubMed] [Google Scholar]
- 35▪.Holscher HD, Faust KL, Czerkies LA, et al. Effects of prebiotic-containing infant formula on gastrointestinal tolerance and fecal microbiota in a randomized controlled trial. JPEN J Parenter Enteral Nutr. 2012;36 (1 Suppl):95S–105S. doi: 10.1177/0148607111430087. This study, which randomized infants to breast or formula feeding, found that formula-fed infants had higher levels of potentially pathogenic C. difficile. [DOI] [PubMed] [Google Scholar]
- 36▪.Rousseau C, Levenez F, Fouqueray C, et al. Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J Clin Microbiol. 2011;49:858–865. doi: 10.1128/JCM.01507-10. A comparison of infant microbiome profiles showed that infants positive for C. difficile had different dominant species, which may affect gut microbial composition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37▪.Thompson AL. Developmental origins of obesity: early feeding environments, infant growth, and the intestinal microbiome. Am J Hum Biol. 2012;24:350–360. doi: 10.1002/ajhb.22254. A comprehensive review examining the evidence linking obesity to early exposures and the development of the microbiome, as well as an analysis of how breastfeeding practices may dually alter metabolism and microflora. [DOI] [PubMed] [Google Scholar]
- 38▪.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58–65. doi: 10.3945/ajcn.110.010132. This study demonstrated that caloric load can induce short-term changes in the gut microbiota and that the balance between microbes corresponds to differences in energy harvest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39▪▪.Zivkovic AM, German JB, Lebrilla CB, et al. Human milk glycobiome and its impact on the infant gastrointestinal microbiota. Proc Natl Acad Sci USA. 2011;108 (Suppl 1):4653–4658. doi: 10.1073/pnas.1000083107. Using bacterial genomic sequencing and culture techniques in parallel with breast-milk glycoprofiling, this group demonstrated how human milk oligosaccharides tend to selectively promote the growth of beneficial intestinal microbes in the infant. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40▪.Collado MC, Laitinen K, Salminen S, et al. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatr Res. 2012;72:77–85. doi: 10.1038/pr.2012.42. This prospective study found that overweight mothers tended to have lower levels of important immunomodulators in breastmilk, which may in turn affect an infant’s microbiome and immune development. [DOI] [PubMed] [Google Scholar]
- 41▪.Luoto R, Kalliomaki M, Laitinen K, et al. Initial dietary and microbiological environments deviate in normal-weight compared to overweight children at 10 years of age. J Pediatr Gastroenterol Nutr. 2011;52:90–95. doi: 10.1097/MPG.0b013e3181f3457f. This case–control study of childhood obesity was one of the first to begin to define a mechanism by which the microbiome, along with contributing enviromental exposures, may affect metabolic programming from an early age. [DOI] [PubMed] [Google Scholar]
- 42.Diaz-Bone RA, van de Wiele TR. Biovolatilization of metal(loid)s by intestinal microorganisms in the simulator of the human intestinal microbial ecosystem. Environ Sci Technol. 2009;43:5249–5256. doi: 10.1021/es900544c. [DOI] [PubMed] [Google Scholar]
- 43.Van de Wiele T, Vanhaecke L, Boeckaert C, et al. Human colon microbiota transform polycyclic aromatic hydrocarbons to estrogenic metabolites. Environ Health Perspect. 2005;113:6–10. doi: 10.1289/ehp.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44▪.Bialek B, Diaz-Bone RA, Pieper D, et al. Toxicity of methylated bismuth compounds produced by intestinal microorganisms to bacteroides thetaiotaomicron, a member of the physiological intestinal microbiota. J Toxicol. 2011;2011:608349. doi: 10.1155/2011/608349. This study showed the potential for the production of methylated organometal(loid) species in the human gut by resident methanoarchaea, which may inhibit beneficial microorganisms that aid digestion and protect against pathogens, in addition to causing systemic toxicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meurens F, Berri M, Siggers RH, et al. Commensal bacteria and expression of two major intestinal chemokines, TECK/CCL25 and MEC/CCL28, and their receptors. PLoS One. 2007;2:e677. doi: 10.1371/journal.pone.0000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sepp E, Julge K, Mikelsaar M, et al. Intestinal microbiota and immunoglobulin E responses in 5-year-old Estonian children. Clin Exp Allergy. 2005;35:1141–1146. doi: 10.1111/j.1365-2222.2005.02315.x. [DOI] [PubMed] [Google Scholar]
- 47.Bjorksten B, Sepp E, Julge K, et al. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 48.Wang M, Karlsson C, Olsson C, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 49▪▪.Bisgaard H, Li N, Bonnelykke K, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652. e1–e5. doi: 10.1016/j.jaci.2011.04.060. The link between atopic diseases and the developing microbiome is studied (by influencing immune maturation potentially) in 411 high-risk children from birth through 6 years as a substudy of the Copenhagen Prospective Study on asthma in Childhood, with analysis of 1-month and 12-month microbiome. Risk of allergy was associated with decreased bacterial diversity at 1 and 12 months. [DOI] [PubMed] [Google Scholar]
- 50.Oyama N, Sudo N, Sogawa H, et al. Antibiotic use during infancy promotes a shift in the T(H)1/T(H)2 balance toward T(H)2-dominant immunity in mice. J Allergy Clin Immunol. 2001;107:153–159. doi: 10.1067/mai.2001.111142. [DOI] [PubMed] [Google Scholar]
- 51▪.Schwartz S, Friedberg I, Ivanov IV, et al. A metagenomic study of diet dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol. 2012;13:r32. doi: 10.1186/gb-2012-13-4-r32. A novel study of the host transcriptome and microbiome simultaneously in infants, highlighting differences in breastfed and formula-fed infants via gut colonization, through host expression of genes associated with the innate immune system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuchat A, Polder JA. [Accessed 5 July 2012];Child Health: Bacterial and other infectious diseases. 2012 [cited 2012 1-31-2012]. http://www.cdc.gov/reproductivehealth/ProductsPubs/DatatoAction/pdf/Chlt1.pdf.
- 53.WHO. [Accessed 5 July 2012];Global Health Observatory, causes of child mortality. 2008 http://www.who.int/gho/child_health/mortality/causes/en/
- 54▪.Blaser M. Antibiotic overuse: stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. This comment published in Nature highlights several concerning facts linking the increase in antibiotic exposure in children in the United States with profound shifts in the microbiome linked to significant disease, including inflammatory bowel disease, and calls for further investigation of antibiotic-free microbiota versus antibiotic-exposed microbiota and more targeted, judicious use of antibiotics. [DOI] [PubMed] [Google Scholar]
- 55▪.Antunes LC, Finlay BB. A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis. Gut Microbes. 2011;2:105–108. doi: 10.4161/gmic.2.2.15610. A review article based on the group’s animal studies highlighting the impact of enteric pathogenic infection and antibiotics on the animal intestinal microbiome and its metabolic function, specifically in the production of steroids and eicosanoids. [DOI] [PubMed] [Google Scholar]
- 56.Mangin I, Suau A, Gotteland M, et al. Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe. 2010;16:433–438. doi: 10.1016/j.anaerobe.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Cotten CM, Taylor S, Stoll B, et al. Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics. 2009;123:58–66. doi: 10.1542/peds.2007-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58▪▪.Ajslev TA, Andersen CS, Gamborg M, et al. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, prepregnancy weight and early administration of antibiotics. Int J Obes (Lond) 2011;35:522–529. doi: 10.1038/ijo.2011.27. In a cohort study of nearly 30 000 mother–child pairs, the researchers found that antibiotic exposure in the first 6 months of life increases a child’s risk of overweight in later childhood, suggesting a role for the microbiome in establishment of long-term metabolism and energy storage early in life. [DOI] [PubMed] [Google Scholar]
- 59.Mitsou EK, Kirtzalidou E, Oikonomou I, et al. Fecal microflora of Greek healthy neonates. Anaerobe. 2008;14:94–101. doi: 10.1016/j.anaerobe.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Gueimonde M, Salminen S, Isolauri E. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol Med Microbiol. 2006;48:21–25. doi: 10.1111/j.1574-695X.2006.00112.x. [DOI] [PubMed] [Google Scholar]
- 61▪.de Vries LE, Valles Y, Agerso Y, et al. The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS One. 2011;6:e21644. doi: 10.1371/journal.pone.0021644. This case study provided evidence to support the hypothesis that antibiotic resistance genes can be transmitted from mother to child by comparing fecal microbial signatures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skurnik D, Ruimy R, Ready D, et al. Is exposure to mercury a driving force for the carriage of antibiotic resistance genes? J Med Microbiol. 2010;59 (Pt 7):804–807. doi: 10.1099/jmm.0.017665-0. [DOI] [PubMed] [Google Scholar]
- 63▪▪.Gilbert-Diamond D, Cottingham KL, Gruber JF, et al. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci USA. 2011;108:20656–20660. doi: 10.1073/pnas.1109127108. This study assessing rice consumption in pregnant women found that eating 0.5cups of rice per day was equivalent to drinking a liter of water containing 10 g As/l, the maximum contaminant limit set by the US Environmental Protection Agency. Rice consumption during pregnancy may be a potential source of fetal arsenic exposure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64▪.Jackson BP, Taylor VF, Karagas MR, et al. Arsenic, organic foods, and brown rice syrup. Environ Health Perspect. 2012;120:623–626. doi: 10.1289/ehp.1104619. This study found that many first foods that are fortified with rice, including some infant formulas, contain inorganic arsenic at levels that could result in significant exposures in young children. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.National Research Council. Arsenic in Drinking Water. Washington, DC: National Academy Press; 2001. [Google Scholar]
- 66.Sepkowitz KA. One hundred years of Salvarsan. N Engl J Med. 2011;365:291–293. doi: 10.1056/NEJMp1105345. [DOI] [PubMed] [Google Scholar]
- 67▪.Cavaco LM, Hasman H, Aarestrup FM. Zinc resistance of Staphylococcus aureus of animal origin is strongly associated with methicillin resistance. Vet Microbiol. 2011;150:344–348. doi: 10.1016/j.vetmic.2011.02.014. This study examines the co-occurrence of metal/antibiotic coresistance in over 600 MRSA isolates from livestock; it is the latest from this group to demonstrate that zinc resistance genes may drive the selection of MRSA and that the addition of zinc to animal feed may have contributed to its emergence. [DOI] [PubMed] [Google Scholar]
- 68.Jakobsson HE, Jernberg C, Andersson AF, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut micro-biome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mshvildadze M, Neu J. Probiotics and prevention of necrotizing enterocolitis. Early Hum Dev. 2009;85 (10 Suppl):S71–S74. doi: 10.1016/j.earlhumdev.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 70▪.Wardwell LH, Huttenhower C, Garrett WS. Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr Infect Dis Rep. 2011;13:28–34. doi: 10.1007/s11908-010-0147-7. A review article investigating the relationship between the host microbiota and infection through colonization resistance, immune programming, responses to antibiotics, and interaction with viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.DiGiulio DB, Romero R, Amogan HP, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gewolb IH, Schwalbe RS, Taciak VL, et al. Stool microflora in extremely low birthweight infants. Arch Dis Child Fetal Neonatal Ed. 1999;80:F167–F173. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fanaroff AA, Korones SB, Wright LL, et al. Incidence, presenting features, risk factors and significance of late onset septicemia in very low birth weight infants. The National Institute of Child Health and Human Development Neonatal Research Network Pediatr Infect Dis J. 1998;17:593–598. doi: 10.1097/00006454-199807000-00004. [DOI] [PubMed] [Google Scholar]
- 74▪.Madan JC, Salari RC, Saxena D, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012 doi: 10.1136/fetalneonatal-2011-301373. [Epub ahead of print]. An in-depth study of the relationship between the developing microbiome, beginning at birth, studied weekly in six extremely low birthweight infants in a neonatal intensive care unit setting, highlighting that sepsis risk was associated with a predominance of particular gut pathogens, most notably Staphylococcus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75▪.Lamont RF, Nhan-Chang CL, Sobel JD, et al. Treatment of abnormal vaginal flora in early pregnancy with clindamycin for the prevention of spontaneous preterm birth: a systematic review and metaanalysis. Am J Obstet Gynecol. 2011;205:177–190. doi: 10.1016/j.ajog.2011.03.047. A systematic review and metaanalysis of five trials including 2346 women total, investigating whether the administration of clindamycin to women with abnormal vaginal flora at less than 22 weeks of gestation reduces risk of preterm birth and late miscarriage. The study found no differences with clindamycin in risk of preterm birth at less than 33 weeks; however, a reduction of spontaneous preterm birth at less than 37 weeks was found, highlighting the benefit of future studies most importantly understanding the vaginal microbiome and mucosal immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]