Abstract
The major function of eukaryotic RNA polymerase III is to transcribe transfer RNA, 5S ribosomal RNA, and other small non-protein-coding RNA molecules. Assembly of the RNA polymerase III complex on chromosomal DNA requires the sequential binding of transcription factor complexes TFIIIC and TFIIIB. Recent evidence has suggested that in addition to producing RNA transcripts, chromatin-assembled RNA polymerase III complexes may mediate additional nuclear functions that include chromatin boundary, nucleosome phasing, and general genome organization activities. This study provides evidence of another such “extratranscriptional” activity of assembled RNA polymerase III complexes, which is the ability to block progression of intergenic RNA polymerase II transcription. We demonstrate that the RNA polymerase III complex bound to the tRNA gene upstream of the Saccharomyces cerevisiae ATG31 gene protects the ATG31 promoter against readthrough transcriptional interference from the upstream noncoding intergenic SUT467 transcription unit. This protection is predominately mediated by binding of the TFIIIB complex. When TFIIIB binding to this tRNA gene is weakened, an extended SUT467–ATG31 readthrough transcript is produced, resulting in compromised ATG31 translation. Since the ATG31 gene product is required for autophagy, strains expressing the readthrough transcript exhibit defective autophagy induction and reduced fitness under autophagy-inducing nitrogen starvation conditions. Given the recent discovery of widespread pervasive transcription in all forms of life, protection of neighboring genes from intergenic transcriptional interference may be a key extratranscriptional function of assembled RNA polymerase III complexes and possibly other DNA binding proteins.
Keywords: RNA polymerase III, TFIIIB, TFIIIC, extratranscriptional effects, noncoding transcription
IN eukaryotes, the process of transcription is divided among three RNA polymerases—RNA polymerase I, II, and III. In the yeast Saccharomyces cerevisiae, RNA polymerase III (Pol III) transcribes a variety of small RNAs, including transfer RNA (tRNA), 5S ribosomal RNA (5S rRNA), U6 spliceosomal RNA, snR52 small nucleolar RNA, 7SL RNA, and the RNA component of RNase P. Assembly of the transcription machinery on Pol III genes is mainly determined by the structure of the promoter. A unique feature of most Pol III promoters is the presence of internal control regions (ICRs) that are composed of conserved sequences separated by more variable regions. The most common promoter arrangement used by Pol III is the class II promoter, found mainly in the tRNA genes (tDNAs). Class II promoters consist of the conserved intragenic A-box and B-box sequences that are bound by the transcription factor complex TFIIIC (Pascali and Teichmann 2012; Acker et al. 2013).
In yeast, the entire Pol III transcription machinery bound to tDNAs consists of three multimeric protein complexes: the transcription factors TFIIIC (6 subunits) and TFIIIB (3 subunits), which are required for promoter recognition and preinitiation complex formation, and the 17 subunit Pol III enzyme (Geiduschek and Kassavetis 2001; Huang and Maraia 2001; Acker et al. 2013). The initial step in the transcription of tDNAs in yeast is the binding of the TFIIIC complex to the A- and the B-boxes. The 6 subunits of TFIIIC are organized into two globular domains, τA (Tfc1p, Tfc4p, and Tfc7p) and τB (Tfc3p, Tfc6p, and Tfc8p). τB specifically binds to the B-box with high affinity and favors A-box binding by τA (Geiduschek and Kassavetis 2001). The most currently refined B-box consensus sequence, GWTCRANNC (Marck et al. 2006; Orioli et al. 2012) contains a highly conserved cytosine residue (italicized), and mutation of this cytosine compromises TFIIIC binding (reviewed in Donze 2012). TFIIIC binding is required to recruit TFIIIB at most Pol III promoters. TFIIIB is composed of three proteins, TATA-binding protein (TBP), TFIIB-related factor (Brf1p) and B″ (B-double prime, Bdp1p). Binding of TFIIIB forms an exceptionally kinetically stable TFIIIB–DNA complex (Cloutier et al. 2001), which then recruits the Pol III enzymatic complex and helps maintain it for multiple transcription cycles in a process called facilitated recycling (Dieci and Sentenac 1996; Ferrari et al. 2004).
While Pol III and its transcription factors are generally thought to be dedicated to transcription of Pol III target genes, emerging studies have shown that either partial or complete DNA-bound Pol III transcription complexes can have effects on transcription, chromatin state, and genome organization of neighboring Pol II genes. These so-called “extratranscriptional” (Donze 2012) or “product independent” (Clelland and Schultz 2010) effects of Pol III complexes, mostly demonstrated in S. cerevisiae, include the following activities: targeting integration of Ty retroelements (Chalker and Sandmeyer 1990; Ji et al. 1993; Devine and Boeke 1996), displacement of nucleosomes (Morse et al. 1992), phasing of adjacent nucleosomes (Nagarajavel et al. 2013), position effect repression of adjacent Pol II promoters (Hull et al. 1994), chromatin boundary/insulator functions (Donze 2012), and pausing of replication forks (Deshpande and Newlon 1996; Sekedat et al. 2010). In some instances, the TFIIIC complex alone can mediate extratranscriptional functions, as Extra TFIIIC (ETC) sites (Moqtaderi and Struhl 2004), chromosomal loci that bind only TFIIIC without recruiting TFIIIB or Pol III, can act as insulators (Simms et al. 2008), can directly regulate Pol II promoters (Kleinschmidt et al. 2011), and can tether chromosomal regions to the nuclear periphery (Hiraga et al. 2012).
Using the S. cerevisiae model system, we have previously described multiple types of extratranscriptional functions of the TRT2 tDNA at the STE6–CBT1 locus. In MATα cells, TRT2 serves as a barrier to prevent repression of the neighboring Pol II-transcribed CBT1 gene, whereas in MATa cells TRT2 exerts an apparent tRNA position effect, as deletion of TRT2 results in an increase in CBT1 gene transcription (Simms et al. 2004). This modest position effect (approximately threefold increase in CBT1 mRNA levels) was shown to be due in part to the tDNA acting as an insulator, as it prevents inappropriate activation of the CBT1 promoter by the Mcm1p transcription factor that binds to the nearby STE6 upstream activation sequence (UAS) (Simms et al. 2008).
Manual inspection of the S. cerevisiae genome reveals that about one-quarter of all tDNAs lie between divergently transcribed genes in the yeast genome and could potentially show a similar insulator effect. Given the modest insulator effect observed at the CBT1 locus, we investigated the ATG31–tV(UAC)D–SES1 locus anticipating a more robust effect, as genome-wide expression data indicate that SES1 is transcribed at considerably higher levels (∼70-fold) than is ATG31 in rich media (Holstege et al. 1998; Xu et al. 2009). Our reasoning was that transcription factors responsible for the high level activation of SES1 would more strongly activate ATG31 upon deletion of the tDNA. Surprisingly, when we performed Northern blot analysis on RNA from wild-type and tv(uac)dΔ (referred to hereafter as tdnaΔ) strains, we found that ATG31 mRNA levels were not only increased, but that a longer transcript with an extended 5′-UTR (5′-untranslated region) replaced the normal transcript. We show here that this longer transcript is due to readthrough of the noncoding stable unannotated transcript SUT467 (Xu et al. 2009), and mutations that inhibit TFIIIB complex assembly or stability allow readthrough. Progression of transcription from the upstream SUT467 start site prevents normal ATG31 transcriptional initiation, and the extended 5′-UTR inhibits translation of the ATG31 coding sequence. Since Atg31p is required for autophagy, reduced translation results in compromised autophagy and fitness under nitrogen starvation conditions in strains exclusively expressing the extended transcript. This work identifies another novel extratranscriptional function of tDNAs, the ability to block progression of cryptic intergenic transcription, preventing subsequent deleterious transcriptional interference of an adjacent promoter.
Materials and Methods
Yeast cultures were grown in nutrient-rich YPD media (1% yeast extract, 2% peptone, and 2% dextrose) at 30° on a rotary shaker unless otherwise noted. For induction of autophagy, cells were grown to mid-log phase (A600 = 0.7) in YPD, collected by centrifugation (3000 rpm × 5 min), washed with water, then resuspended in nitrogen starvation media (1.7 g/liter yeast nitrogen base without amino acids and without ammonium sulfate, plus 2% dextrose). For Northern blot analysis of temperature-sensitive mutants, cultures were grown at 30° to an OD600 of 0.7 and then incubated at 37° for 1 hr before RNA extraction.
Plasmid pDD1232 was created by cloning a 1.35-kb XhoI–SpeI cut ATG31–SES1 intergenic fragment (PCR amplified with oligos DDO1281/-1282, which added an artificial XhoI site) into Bluescript SK+ (all oligos used are listed in Supporting Information, Table S2). Two-step PCR mutagenesis was performed using pDD1232 as template and T7 and T3 primers with mutagenic primers (DDO184/-1284; DDO183/-1285, respectively) to amplify the fragment containing the tdna deletion; this fragment was then cloned into Bluescript SK+ as above to create pDD1233. Direct-site-directed mutagenesis was performed on pDD1232 to create pDD1248 (tdna B-box∆), pDD1249 (tdna B-box mutant), pDD1261 (tdna A-box mutant), and pDD1260 (tdnaΔ::EcoRI–BamHI linker), using DDO1391/-1392, DDO1393/-1394, DDO1474/-1475, and DDO1466/-1467 primer sets, respectively. The B-box mutant had the invariant cytosine and following guanine bases changed to GC, and the A-box mutant scrambled the entire consensus. Plasmids pDD1262 (flipped orientation of the tDNA) and pDD1272 (tdnaΔ::ETC9) were created by cloning EcoRI–BamHI-digested PCR-amplified fragments using DDO1468/-1469 (flip), or DDO1534/-1535 (ETC9), respectively, into EcoRI–BamHI-digested pDD1260. Yeast genomic DNA was used as PCR template. Plasmid pDD1263 (tdnaΔ::ETC4) was constructed by directly ligating complementary oligonucleotides (DDO1489/-1490) containing EcoRI–BamHI overhangs into pDD1260. All plasmids were confirmed by Sanger sequencing and are listed in Table S1.
Yeast strains were generated from wild-type S. cerevisiae W303-1a; genotypes of all strains used and generated in this study are given in Table 1. Parent tdnaΔ::URA3 (DDY4605–4607) strains were created by amplifying URA3 with primers DDO1279/-1280 containing homology to the flanking region of tV(UAC)D, and then this DNA was transformed into wild-type DDY3 followed by selection of Ura+ colonies and PCR identification of homologous recombinants. Linearized tdna mutant plasmids were digested with XhoI and SpeI, individually transformed into a tdnaΔ::URA3 strain, and 5- fluoroorotic acid (5-FOA) resistant colonies were isolated. Recombinants were identified by PCR and verified by DNA sequencing of the product. Yeast strains for chromatin immunoprecipitation (ChIP) were created by crossing existing BRF1–3X-FLAG (DDY1495) and TFC1–3X-FLAG (DDY3860) strains to DDY4607, and then FLAG-tagged Ura+ progeny were backcrossed to each tdna mutant.
Table 1. S. cerevisiae strains used and generated in this study.
| Name | Genotype | Source |
|---|---|---|
| DDY3 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 | J. Rine |
| DDY232 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 rpc31-236 hmrΔ | Donze lab |
| DDY246 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 rpc160-Δ1::HIS3 p-rpc160-112 | Donze lab |
| DDY261 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 tfc3-G349E hmrΔI | Donze lab |
| DDY416 | MATa his3-11,15 leu2-3,112 lys2Δ trp1-1 ura3-1 hmrΔ brf1Δ::HIS3 p-brf1 II.9 | Donze lab |
| DDY420 | MATa his3-11,15 leu2-3,112 lys2Δ trp1-1 ura3-1 hmrΔ brf1Δ::HIS3 p-brf1 II.6 | Donze lab |
| DDY947 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 sas2∆::TRP1 | Donze lab |
| DDY1376 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 nhp6a:URA3 nhp6b:HIS3 | Donze lab |
| DDY1495 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 BRF1:3XFLAG:KanMX nhp6b:HIS3 | Donze lab |
| DDY1631 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 rsc2∆::TRP1 | Donze lab |
| DDY1676 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 rpd3::LEU2 | Donze lab |
| DDY2058 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 yta7∆::TRP1 | Donze lab |
| DDY2236 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 HMR-ADE2 htz1∆::KanMX | Donze lab |
| DDY2509 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 bdf1∆::HIS3 | Donze lab |
| DDY3860 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 TFC1:3XFLAG:KanMX | Donze lab |
| DDY4300 | MATa his3-11,15 leu2-3,112 lys2Δ trp1-1 ura3-1 tfc6 promoter mutant 3 | Donze lab |
| DDY4607 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆::URA3 | This study |
| DDY4624 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆ | This study |
| DDY4625 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 | This study |
| DDY4652 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆ | This study |
| DDY4653 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆ | This study |
| DDY4764 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 atg31∆::TRP1 | This study |
| DDY4769 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d B-box∆ | This study |
| DDY4816 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tV(UAC)D flip | This study |
| DDY4817 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d A-box mutant | This study |
| DDY4819 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆::ETC4 | This study |
| DDY4901 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::URA3 BRF1:3XFLAG:KanMX | This study |
| DDY4904 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::URA3 TFC1:3XFLAG:KanMX | This study |
| DDY4908 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 TFC1:3XFLAG:KanMX | This study |
| DDY4917 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::ETC4 TFC1:3XFLAG:KanMX | This study |
| DDY4920 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d B-box point mutant TFC1:3XFLAG:KanMX | This study |
| DDY4925 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d B-box point mutant | This study |
| DDY4935 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 tV(UAC)D flip BRF1:3XFLAG:KanMX | This study |
| DDY4938 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 BRF1:3XFLAG:KanMX | This study |
| DDY4943 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d A-box mutant BRF1:3XFLAG:KanMX | This study |
| DDY4946 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d B-box point mutant BRF1:3XFLAG:KanMX | This study |
| DDY4949 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::ETC4 BRF1:3XFLAG:KanMX | This study |
| DDY4970 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d∆::ETC9 | This study |
| DDY5003 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::ETC9 BRF1:3XFLAG:KanMX | This study |
| DDY5006 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 tv(uac)d∆::ETC9 TFC1:3XFLAG:KanMX | This study |
| DDY5010 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 ydl156∆::KanMX | Research Genetics |
| DDY5012 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 ATG31-9X-myc::TRP1 | This study |
| DDY5014 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)dΔ ATG31-9X-myc::TRP1 | This study |
| DDY5018 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d B-boxΔ ATG31-9X-myc::TRP1 | This study |
| DDY5020 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 tv(uac)d B-box pt. mutant ATG31-9X-myc::TRP1 | This study |
| DDY5044 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 atg8Δ::TRP | This study |
| DDY5046 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 atg31Δ::TRP | This study |
| DDY5051 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 | This study |
| DDY5072 | MATα his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 tv(uac)d∆ | This study |
| DDY5074 | MATa his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 | This study |
| DDY5078 | MATα his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 tv(uac)d B-box∆ | This study |
| DDY5081 | MATα his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 tv(uac)d B-box pt. mut. | This study |
| SG154.2 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 bar1∆ SCC2::scc2-D730V::HYG | J. Gerton |
| ROY1032 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 HMR∆I smc1-2::LEU2 ts | R. Kamakaka |
| ROY1060 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 HMR∆I smc3-1:: LEU2 ts | R. Kamakaka |
| ROY1063 | MATα his3-11,15 leu2-3,112 lys2∆ trp1-1 ura3-1 HMR∆I scc1-73::TRP1 ts | R. Kamakaka |
| ZFY155 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pho13::URA3 pho8∆60::HIS3 | D. Klionsky |
All strains are isogenic to S. cerevisiae W303-1A except DDY5010, which is S288C.
The 9X-Myc epitope tag was amplified from a TRP1 marked cassette (Knop et al. 1999) using DDO1462/-1463 and transformed into wild-type and tdna mutants. ATG31–9X-myc Trp+ homologous recombinants were identified by PCR (DDO1419/-1464) and confirmed by Western blotting. ATG31 and ATG8 knockout strains were constructed by standard yeast homologous recombination. For autophagy induction alkaline phosphatase assays, pho13∆ pho8∆60 strains were created by crossing a pho13Δ::URA3pho8∆60::HIS3 strain (kindly provided by Daniel Klionsky) to wild-type and tdna mutants.
RNA isolation and Northern blotting were performed as described (Simms et al. 2004). Most Northern results were verified with three (but at least two) independently isolated mutant strains; Table 1 lists only the specific strains shown in the figures. Primers used to amplify Northern probes are listed in Table S2. 5′-RACE analysis was performed on RNA isolated from the DDY420 brf1 II.6 mutant using the First Choice RLM-RACE kit (Ambion/Life Technologies, AM1700). Individual clones were sequenced by standard Sanger sequencing and mapped to the S. cerevisiae genome on the Saccharomyces Genome Database at http://www.yeastgenome.org (Cherry et al. 2012).
Quantitative RT–PCR was performed as follows: First-strand cDNA was synthesized from 0.5 μg of total RNA after DNase treatment (RQ1 DNase, Promega M6101). Synthesis was extended from long transcript specific primer DDO1284 using ProtoScript M-MuLV first-strand cDNA synthesis kit (NEB E6300S). Quantitative reverse transcription PCR (qRT–PCR) was performed on 1:4 diluted cDNA using primers DDO1606/-1555 and Sybr Green super mix (Bio-Rad 170-8882) with 60° annealing temperature. Results were normalized to amplicons from ACT1 control primers (DDO402+403). Reactions were run and analyzed using a Bio-Rad MyiQ as described (Kim et al. 2011) and examined by agarose gel electrophoresis to verify that only the predicted PCR products were amplified. The primers were designed to specifically amplify readthrough transcripts; the strategy is described and illustrated in Figure S2.
Chromatin immunoprecipitation was performed as described previously (Rusche et al. 2002). Anti-FLAG epitope antibody was purchased from Sigma (F1804). Primers DDO1527/-1555 or DDO1576/-1577 were used to amplify desired regions surrounding tV(UAC)D. For Western analysis, yeast minilysates were prepared by glass bead lysis of log-phase cultures directly in lysis buffer (50 mM Tris pH 7.5, 1% SDS, 5 mM EDTA, 14.3 mM β-mercaptoethanol, 1 mM PMSF, 2 μg/ml leupeptin and pepstatin). Pellets from 5 ml YPD culture at A600 1.0 were resuspended in 200 μl lysis buffer and then vortexed with glass beads at 4° for 10 min. Lysis buffer, 100 μl, was added, and the mixture was boiled for 3 min, cooled on ice, and centrifuged at 4° to remove cell debris. To 100 μl of clarified minilysate, an equal amount of 2× SDS–PAGE loading buffer was added, and after boiling, 15 μl was loaded on 12% acrylamide protein gel. Proteins were transferred to Millipore Immobilon membrane by semidry transfer and incubated in blotto (10× TBS/10% SDS/5% dry milk) for 1 hr. Primary Myc antibody (c-Myc 9E10, Santa Cruz Biotechnology), anti-mouse Ig-horseradish peroxidase secondary antibody (GE healthcare) were used for Western analysis. Immuno-star Western chemiluminescent kit (Bio-Rad) was used to detect the secondary antibody.
The alkaline phosphatase pho8∆60 assay was performed as described (Klionsky 2007). The cell survival assay was adapted from Kabeya et al. (2007). Yeast strains were grown in YPD rich media to A600 = 1.0 (∼107 cells/ml). Cells were harvested by centrifugation, washed once with distilled water, and resuspended at 107 cells/ml in media lacking nitrogen. Cells were incubated for 6 days at 30° on rotatory shaker, and every other day 200 μl of culture dilutions was plated on YPD plates in triplicate. Plates were incubated at 30° for 48 hr and survival rate was obtained by counting resulting colonies.
Results
Mutation of the tDNA upstream of ATG31 results in readthrough of SUT467
Figure 1A depicts the ATG31–SES1 locus, showing the location of the tV(UAC)D tDNA, and the extent of transcripts normally produced from the region. To test the initial hypothesis that tV(UAC)D might act as an insulator by preventing promiscuous activation of ATG31 by regulatory elements associated with the strong promoter of the neighboring divergent SES1 gene, we created tV(UAC)D deleted (tdna∆) mutant strains.
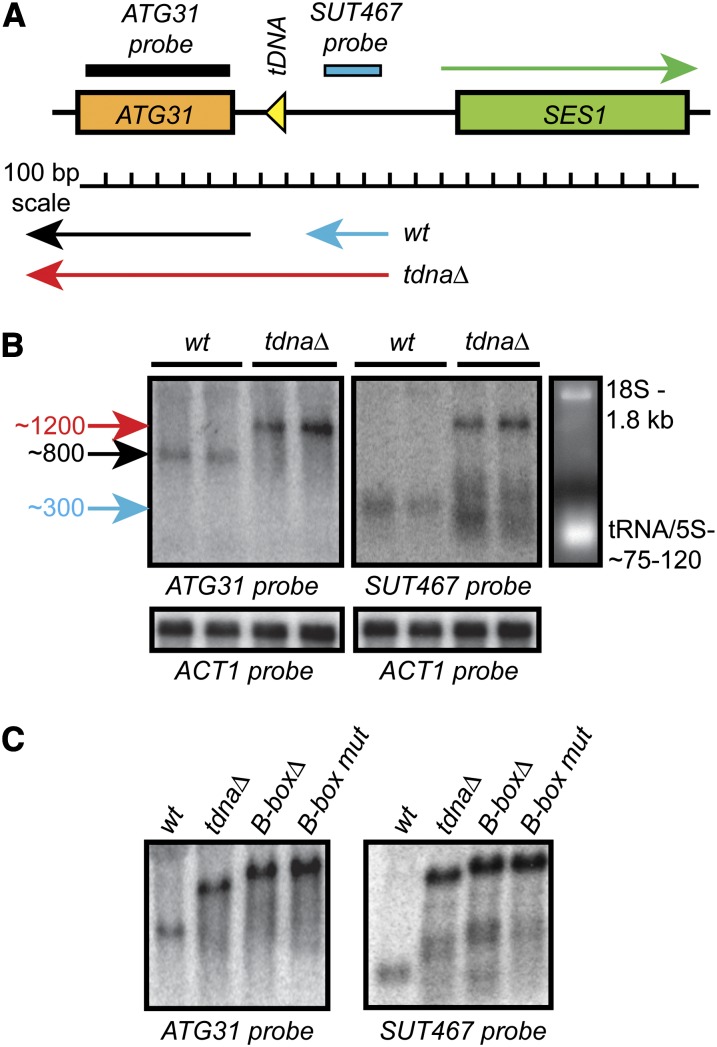
Figure 1.
Mutation of the tV(UAC)D tRNA gene upstream of ATG31 results in readthrough of the intergenic SUT467 transcript. (A) Schematic of the ATG31–SES1 locus on S. cerevisiae chromosome IV. Colored arrows indicate known annotated transcripts: ATG31, black; SUT467, blue. The overlapping red arrow represents the extended readthrough transcript. (B) Northern blot analysis of ATG31 expression in wild-type and tdnaΔ strains reveals the extended transcript. The ATG31 coding sequence probe hybridized to RNA of ∼800 bp in wild-type strains (black arrow) and to RNA of ∼1200 bp after tDNA deletion (red arrow). The SUT467 probe hybridized to the predicted ∼300-bp transcript in wild-type cells (blue arrow) and to the same ∼1200-bp extended transcript in tdnaΔ strains. The normal ATG31 transcript was absent in tdnaΔ strains. Each pair of lanes contained total RNA from independent wild-type and mutant strains. (C) B-box deletion (B-box∆) or mutation of the invariant cytosine in the B-box (B-box mut) also resulted in extended readthrough transcription. Strains used were: (B) DDY4625 and DDY3 (wild-type); DDY4653 and 4624 (tdnaΔ); (C) DDY3 (wt); DDY4652 (tdnaΔ); DDY4769 (B-boxΔ); and DDY4925 (B-box mut).
Northern blot analysis using a probe homologous to the ATG31 coding sequence (Figure 1B, left) showed not only an apparent slight increase in the level of ATG31 mRNA in the tdnaΔ strains (compared to ACT1 controls), but also an increase in the length of the transcript by ∼400 nucleotides (shifting from ∼800 bases in wild-type strains to ∼1200 bases), with apparent absence of the normal length mRNA. Recent tiling array and RNA-seq studies have identified widespread pervasive and intergenic transcription in eukaryotic cells, and in yeast this often appears to occur as bidirectional transcription from strong promoters (Neil et al. 2009; Xu et al. 2009). Inspection of data from these studies indicated that the SUT467 intergenic transcript initiates upstream of the SES1 promoter and terminates near the tDNA (Figure 1A); therefore, we hypothesized that the extended ATG31 transcript in the tdnaΔ strain was a readthrough SUT467 transcript that interferes with the production of the normal ATG31 transcript.
To confirm that the extended transcript in tdnaΔ strains was due to readthrough of SUT467 and not a 3′ extension, we repeated the Northern analysis using a probe specific for the transcribed SUT467 RNA sequence (Figure 1B, right). The results showed that this probe hybridized to RNA of ∼300 bases in wild-type strains, consistent with previous annotations of SUT467. The tdnaΔ strains showed a longer ∼1200-base transcript, the same length as when using the ATG31 coding-sequence probe. We concluded that in the absence of the tDNA at this region, SUT467 readthrough occurs and interferes with normal ATG31 transcription initiation, producing only the observed extended RNA.
Since our gross deletion of the tDNA sequence removed 90 bp of chromosome IV, we confirmed this readthrough effect by creating strains that either had only the B-box sequence of the tDNA deleted or contained a mutation in the invariant cytosine residue in the B-box. Both of these mutations of the tDNA were expected to result in loss of TFIIIC binding and inhibition of Pol III complex assembly. Northern blot analysis with either probe shown in Figure 1C confirmed that in each of these mutant backgrounds, complete readthrough of SUT467 occurred as in the tdnaΔ strains. The slightly shorter transcript observed in the tdnaΔ strain compared to the B-boxΔ and point mutant strains is also consistent with the long transcript being a readthrough from upstream of the tDNA, as this reflects the 90-bp deletion. Also observed were shorter RNAs hybridizing to only the SUT467 probe, which appear to terminate between the tDNA and ATG31.
TFIIIB binding is correlated with blocking of SUT467 readthrough
Our previous studies of heterochromatin barrier and insulator function of tDNAs have shown that assembled TFIIIC alone can block the spread of silencing from the HMR locus and can insulate a UAS from a promoter (Simms et al. 2008). To determine which components of the Pol III complex are required to block SUT467 progression, we analyzed ATG31 transcripts in various temperature-sensitive strains compromised for Pol III complex function and formation when pulsed at the nonpermissive temperature before RNA extraction. Figure 2A shows the results of Northern blot analysis of these strains. Temperature-sensitive mutations in RNA Polymerase III subunit genes rpc31 and rpc160 affect transcription initiation and elongation, respectively (Dieci et al. 1995; Thuillier et al. 1995). These mutants had relatively little effect on the ability of the tDNA to block progression of SUT467 (lanes 2 and 3 compared to wild type in lane 1) as evidenced by a minimal alteration of the ratio of normal to extended transcripts. In contrast, mutations in the TFIIIB subunit encoding BRF1 gene (brf1-II.6 and -II.9) that impair interactions of Brf1p with TBP (Andrau et al. 1999) showed a major shift to the longer extended transcript (lanes 4 and 5), with relatively little normal length RNA. Interestingly, this mutant also showed an intermediate length ATG31 transcript that initiates just upstream of the tDNA coding sequence (see 5′-RACE analysis below).
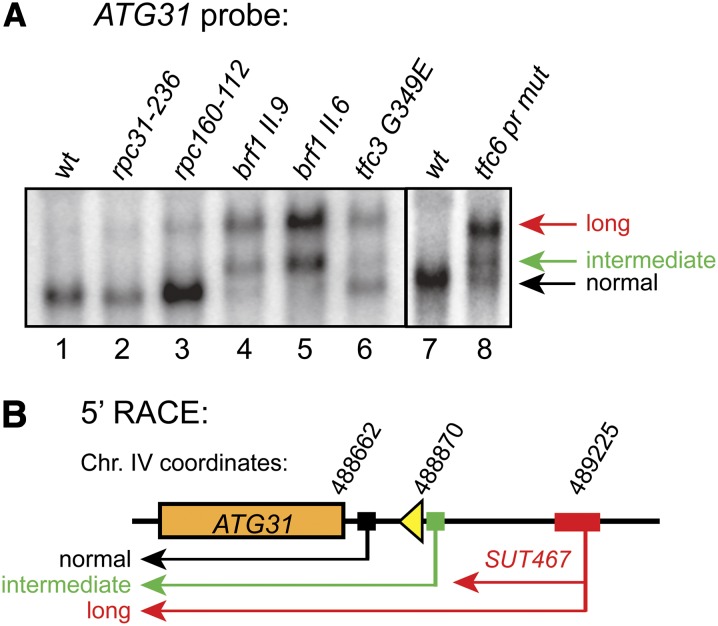
Figure 2.
Pol III transcription factors are required to block SUT467 readthrough transcription. (A) Northern analysis of temperature-sensitive mutants of the Pol III complex was performed as in Figure 1, except that each culture was shifted from 30° to 37° for 1 hr prior to RNA extraction. Extended ATG31 transcripts are most prominent in TFIIIB and TFIIIC subunit mutants, which also express an intermediate length ATG31 transcript. Strains used in lanes 1–8 were DDY3 (wt); DDY232 (rpc31-236); DDY246 (rpc162–112); DDY416 (brf1 II.9); DDY420 (brf1 II.6); DDY261 (tfc3 G349E); DDY3 (wt); and DDY4300 (tfc6 promoter mutant). (B) 5′-RACE analysis of extended and intermediate ATG31 transcripts. 5′-RACE was performed to map transcriptional start sites (TSS) for the various transcripts observed in the Northern blot analysis of the brf1 II.6 mutant. Colored solid boxes represent the range of alternative TSS, which were observed in three distinct clusters. The exact Saccharomyces Genome Database coordinates of all mapped TSS are given in Figure S1.
Mutations involving TFIIIC also resulted in readthrough transcription. RNA isolated from the temperature-sensitive, DNA-binding defective tfc3 G349E mutant strain (Lefebvre et al. 1994) showed apparent equal amounts of both normal and long ATG31 transcripts, with a small relative amount of the intermediate transcript (Figure 2A, lane 6). A strain harboring a mutation in the TFC6 promoter that results in reduced expression of Tfc6p and slow growth (Kleinschmidt et al. 2011) showed a similar pattern (lane 8), shifted a bit more to the long and intermediate transcripts. These results demonstrate that loss of TFIIIC function also results in readthrough SUT467 transcription, but this could be due to loss of TFIIIB assembly in the absence of full TFIIIC activity.
To verify that the transcription start site of our readthrough transcript initiates in the region of the annotated SUT467 transcriptional start site (TSS) and to map the TSS of the observed intermediate transcript, we performed 5′-RACE analysis on RNA isolated from the brf1-II.6 mutant, because it contains all three transcripts as detected by Northern blotting. As shown schematically in Figure 2B, 5′-RACE ends that correspond to the annotated ATG31 mRNA, and within a 94-nucleotide range that overlaps the annotated SUT467 TSS, were mapped. The intermediate transcript was found to begin very close to the beginning of the tRNA coding sequence. The exact Saccharomyces Genome Database chromosome IV coordinates corresponding to each individually mapped 5′-RACE end are listed in Figure S1.
To further assess the mechanistic requirements of each Pol III transcription factor in preventing Pol II readthrough transcription, we constructed yeast strains specifically modified at the ATG31 upstream tDNA locus and analyzed the long vs. short RNA phenotypes. Inverting the orientation of the tDNA had no effect, as no extended ATG31 mRNA was detected (Figure 3A, lane 1). Mutation of the A-box within the tDNA or replacement of the tDNA with the ETC4 site resulted in only the extended transcript being produced (Figure 3A, lanes 2 and 3). Each of these replacements was expected to bind TFIIIC, but not be able to efficiently recruit TFIIIB or Pol III. Interestingly, replacing the tDNA with the tDNA remnant upstream of the TIM21 gene, recently referred to as ETC9 (Nagarajavel et al. 2013), was sufficient to block readthrough transcription (Figure 3A, lane 4). This tDNA remnant has previously been shown to bind both TFIIIC and TFIIIB, but not the Pol III enzymatic complex (Guffanti et al. 2006).
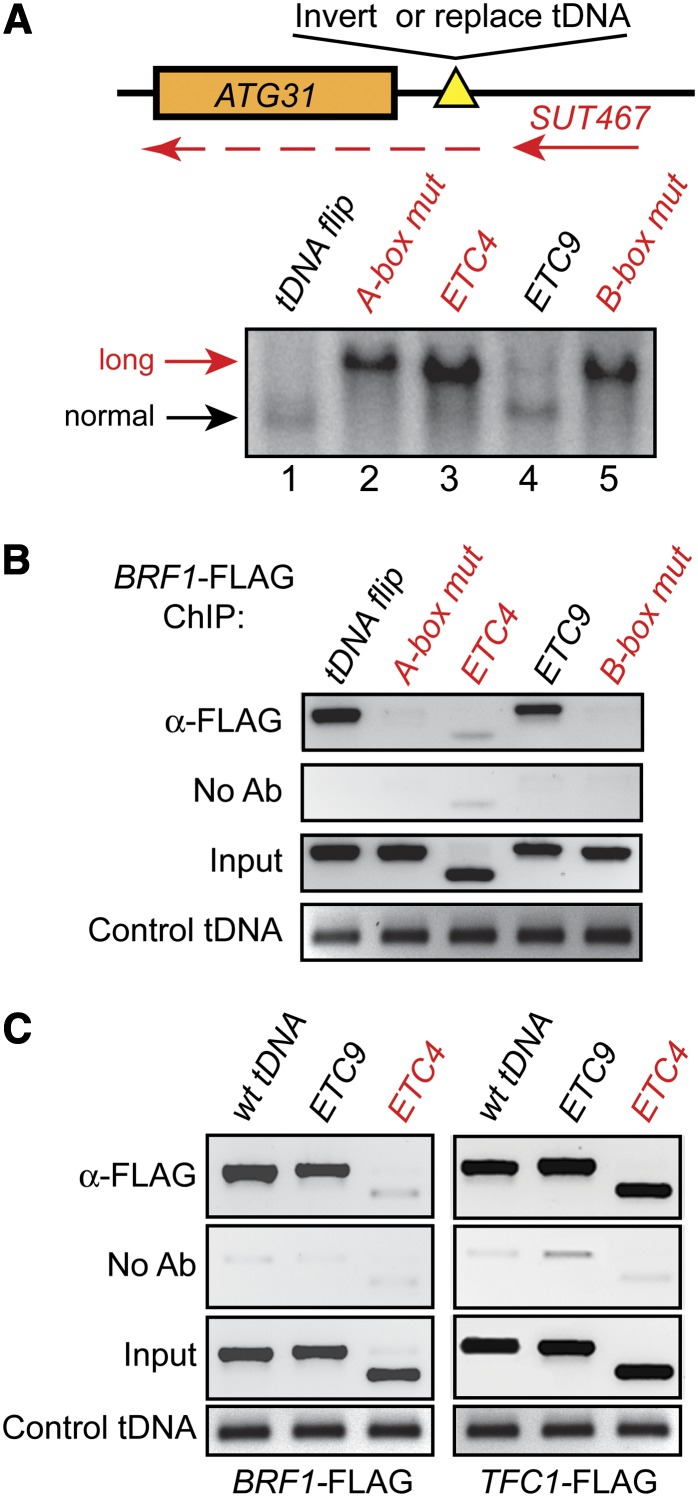
Figure 3.
Binding of the TFIIIB complex is associated with blocking of SUT467 readthrough transcription. Strains were constructed to recruit the entire Pol III complex, TFIIIB and TFIIIC, or TFIIIC alone to the ATG31–SES1 intergenic region. Each construct was tested for the ability to block readthrough and for binding of Pol III transcription factor complexes to the ectopic locations. (A) Schematic of the modified ATG31 loci and Northern blot of each strain using the ATG31 probe. Lane 1, DDY4816 (tDNA flip); lane 2, DDY4817 (A-box mut); lane 3, DDY4819 (ETC4 replacement); lane 4, DDY4970 (ETC9 replacement); and lane 5, DDY4925 (B-box mut). Replacement of the tDNA by ETC4, or mutating the A-box or B-box, resulted in the presence of the extended transcript (red labels). However, inversion of the tDNA sequence or replacement with the ETC9 sequence still blocked readthrough (black labels). (B and C) Confirmation of expected Pol III transcription factor binding in the above mutants by chromatin immunoprecipitation. Each tDNA mutant strain was crossed to strains containing either BRF1–3X-FLAG or TFC1–3X-FLAG alleles, and then subjected to ChIP analysis using anti-FLAG antibody. (B) The absence of TFIIIB upstream of ATG31 in the A-box mutant, ETC4 replacement, and B-box mutant correlates with the presence of the extended transcript, suggesting that TFIIIB binding is required to block readthrough. Strains used (left to right) were DDY4935, -4943, -4949, -5003, and -4946. (C) ChIP analysis of BRF1–3XFLAG and TFC1–3XFLAG strains demonstrates that TFIIIC but not TFIIIB is bound in ETC4 replacement strains, indicating that TFIIIC binding alone cannot block readthrough transcription. Strains used (left to right) were DDY4938, -5003, -4949, -3860, -5006, and -4917.
These results suggest that recruitment of TFIIIB is the critical step that prevents readthrough transcription of SUT467, as binding of TFIIIC alone at the ETC4 site is not sufficient to block Pol II progression. Mutation of the A-box has been demonstrated to impair TFIIIB assembly (Huibregtse and Engelke 1989), and this mutation also allows readthrough. These interpretations assume that each of these sequences used to replace the tDNA have the same in vivo binding characteristics at the ATG31 locus as they do in their native chromosomal locations. To confirm such assumptions regarding the presence or absence of each transcription factor complex at these sequences when moved to the ATG31 locus, we crossed a 3X-FLAG-epitope-tagged BRF1 allele into each of these mutants. ChIP results using anti-FLAG antibody shown in Figure 3B demonstrate that the tDNA flip and ETC9 alleles are strongly enriched for TFIIIB at levels comparable to wild-type tDNAs, while insertions unable to block readthrough transcription (A-box mut, ETC4, and B-box mut) had significantly reduced TFIIIB ChIP signals, comparable to background signals observed in the no antibody control panels. Primers amplifying a separate control tDNA on chromosome III showed similar enrichment in each of the samples, indicating that equivalent amounts of ChIP DNA were added to each PCR reaction.
We also created strains containing a 3X-FLAG-epitope-tagged TFC1 allele to assess the binding of TFIIIC at modified ATG31 loci. The results in Figure 3C show that TFIIIC but not TFIIIB is associated with the ETC4 insertion, and both TFIIIC and TFIIIB are bound at the ETC9 insertion and at the wild-type tDNA locus. The control tDNA again showed equivalent levels of enrichment in the ChIP samples. These results demonstrate an association of TFIIIB binding with blocking of SUT467 transcription. Importantly, and contrary to results seen in our earlier tDNA heterochromatin blocking studies (Simms et al. 2008), TFIIIC binding alone to ETC4 is not sufficient to block cryptic transcript readthrough.
Mutations in genes affecting tDNA heterochromatin barrier function have minimal impact on transcript blocking
Previous studies on the heterochromatin barrier activity of tDNAs revealed the involvement of other chromatin-associated proteins in this extratranscriptional function (Donze et al. 1999; Donze and Kamakaka 2001; Jambunathan et al. 2005; Braglia et al. 2007). To assess the potential role of these tDNA associated proteins in blocking readthrough transcription, we performed Northern blot analysis (using the ATG31 coding sequence probe) on RNA isolated from a number of these mutants. The results in Figure 4A showed that each of these mutants contain mostly normal-length ATG31 transcripts; however, low levels of readthrough are apparent in some strains, most obvious in nhp6 (lane 3) and smc3 (lane 12) mutants in the particular blot shown. However, the intensity of these signals was relatively weak and was often difficult to consistently distinguish from background in different blots.
Figure 4.
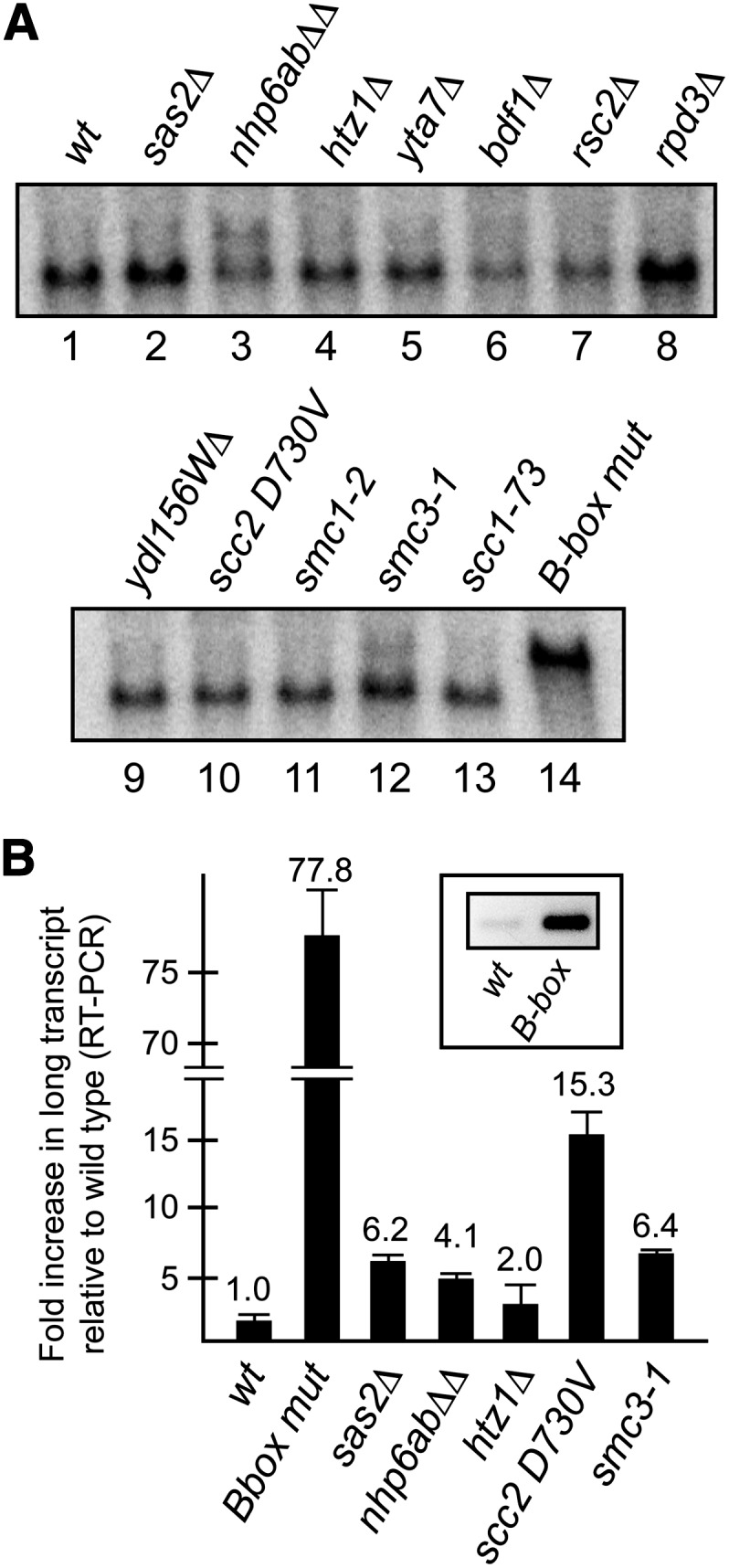
Genetic factors involved in tDNA chromatin boundary function have minimal effects on blocking of Pol II progression through tV(UAC)D. (A) RNA from strains containing mutations that weaken tDNA boundary function were analyzed by Northern blotting using the ATG31 probe. Strains in lanes 1–9 were DDY3, -947, -1376, -2236, -2058, -2509, -1631, -1676, and -5010; lane 10, SG154.2; lanes 11–13, ROY1032, -1060, and -1063; lane 14, DDY4925. (B) RT–PCR analysis of readthrough transcription also shows only minimal effects.
To confirm these apparent low levels of readthrough, we used readthrough transcript-specific primers to develop an RT–PCR assay to measure differences in the relative levels of the long transcript compared to a wild-type strain. The inset in Figure 4B shows an inverted ethidium-stained gel image that verifies that the primers specifically amplified the readthrough cDNA, as the B-box mutant strain showed significantly higher levels of RT–PCR product than the wild-type strain. There also appears to be a low level of readthrough in the wild-type strain, which is consistent with a genome-wide transcriptome analysis that identified a single readthrough clone overlapping this locus (Miura et al. 2006). Quantitative RT–PCR was performed on the same RNA samples shown in the Northern blot in Figure 4A, and those that showed a significant increase in the long ATG31 transcript relative to the wild-type parent are shown in Figure 4B. The B-box mutant strain measured ∼80-fold more readthrough transcript than wild type in this assay, while other mutants were confirmed to have modest (ranging from ∼2- to 15-fold) yet detectable increased levels of the long transcript as suggested by the Northern analysis.
The extended ATG31 transcript is not efficiently translated
Given the extended 5′-UTR present on the long ATG31 transcript, we next asked to what extent translation of the Atg31 protein was affected by readthrough SUT467 transcription. We created ATG31–9X-myc epitope-tagged strains in wild-type and tDNA mutant backgrounds and then analyzed Atg31 protein expression by Western blotting. In each strain producing the long transcript, we observed a drastic reduction in Atg31p levels (Figure 5A). Atg31p is required for autophagy in yeast. Autophagy is a conserved cellular response that recycles cellular components upon nutrient limitation and during normal regulated molecular turnover and involves the formation of autophagosome vesicles that capture and degrade macromolecules after fusion with other membrane bound vesicles (Reggiori and Klionsky 2013; Stanley et al. 2013).
Figure 5.
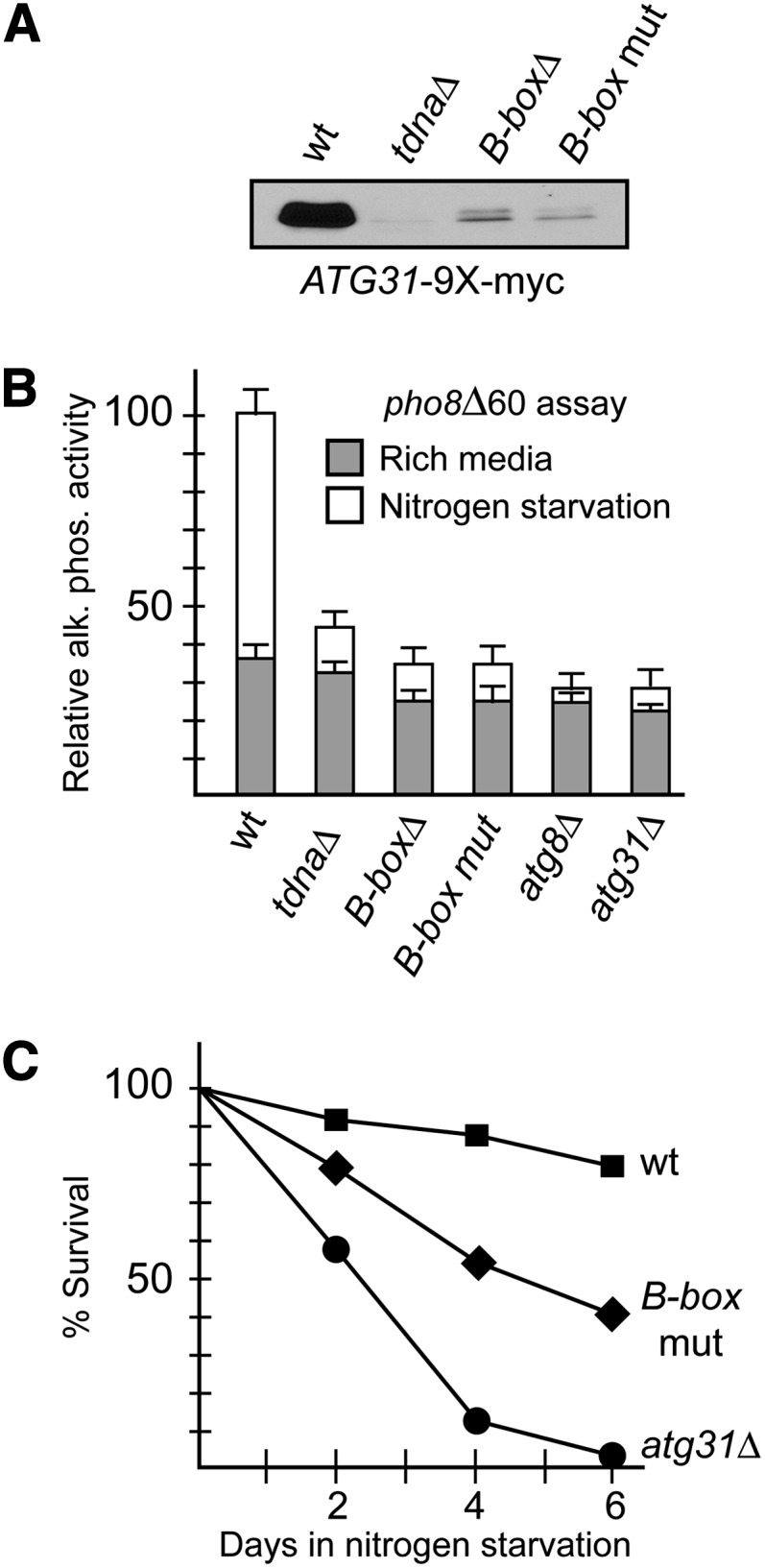
Mutations in tV(UAC)D inhibit expression and function of Atg31 protein. (A) Western blot analysis was performed on wild-type and tDNA mutants containing an ATG31–9X-myc tag allele to determine whether the extended transcript affects translation of Atg31p. Compared to wild type, tDNA mutants showed significantly reduced expression of Atg31p. No other bands were observed on the Western blot, demonstrating that the extended transcript does not lead to production of an extended polypeptide. Strains used were DDY5012 (wt), DDY5014 (tdnaΔ), DDY5018 (B-boxΔ), and DDY5020 (B-box mut). Extracts were prepared from cells grown in YPD media. (B) Inhibition of autophagy induction in yeast expressing the extended transcript as measured by the Pho8Δ60 alkaline phosphatase assay. Strains used were: DDY5051 (wt control), DDY5072 (tdnaΔ), DDY5078 (B-boxΔ), DDY5081 (B-box mut), DDY5044 (atg8Δ), and DDY5046 (atg31Δ). (C) Production of the extended transcript is associated with reduced survival under nitrogen starvation conditions. Strains used were: DDY5012 (wt), DDY5081 (B-box mut), and DDY4764 (atg31Δ).
Since Atg31p is required for the formation of autophagosomes upon nitrogen starvation (Kabeya et al. 2007), we tested the efficiency of this response using two well-characterized assays to measure autophagy induction. We first created a series of strains that produce the readthrough transcript and contain the pho8Δ60 and pho13Δ alleles. These mutations reduce background alkaline phosphatase levels, and the phosphatase activity of the precursor Pho8Δ60 protein can be activated only if it is proteolytically processed during autophagy (Klionsky 2007). Since ATG31 is required for these events, we reasoned that the reduction in Atg31p levels due to the extended 5′-UTR would result in reduced processing of Pho8Δ60p upon induction of autophagy and therefore reduced levels of alkaline phosphatase activity upon shifting cells to nitrogen starvation conditions. Figure 5B shows this to be the case, as strains producing the readthrough transcript showed significantly reduced induction of phosphatase activity compared to a pho13Δ pho8Δ60 strain producing only normal ATG31 mRNA (wt in Figure 5B). Complete deletion of ATG31 or ATG8 in control strains severely reduced starvation-induced phosphatase activity as expected. These results confirm that induction of autophagy is compromised in strains that predominately produce the long ATG31 transcript.
As a second assay for the efficiency of autophagy induction, we tested the viability of yeast cells producing the long transcript when placed under autophagy-inducing conditions. Previous studies have shown that complete deletion of ATG31 results in reduced survival of cells undergoing nitrogen starvation due to inhibition of autophagy (Kabeya et al. 2007). When we tested a strain containing the B-box mutation in the tDNA (producing the long transcript), we found an intermediate level of survival compared with wild-type and atg31Δ strains (Figure 5C). This loss of survival is not due to effects on the neighboring SES1 gene, as wild-type and tdna mutant strains show equivalent expression levels of SES1 when analyzed by Northern blotting (Figure S3). This result suggests that readthrough of the cryptic transcript reduces Atg31p translation to a level that compromises fitness of the cells during nitrogen starvation.
Discussion
The results described in this study demonstrate that stable RNA polymerase III transcription factor complexes containing TFIIIB assembled at tDNAs have the capacity to block the progression of intergenic transcription by RNA polymerase II. High-throughput microarray and sequencing technologies have led to the identification of much more diversity in transcriptomes, from prokaryotes to humans, than was previously appreciated (Core et al. 2008; Dornenburg et al. 2010; Wei et al. 2011). In S. cerevisiae, such pervasive transcripts include the cryptic unstable transcripts (CUTs), stable unannotated transcripts (SUTs), Xrn1-sensitive unstable transcripts (XUTs), and meiotic unannotated transcripts (MUTs) (Wyers et al. 2005; Xu et al. 2009; Lardenois et al. 2011; van Dijk et al. 2011). Additionally, alterations in the prevalence of intergenic transcripts and transcript start and end sites have been observed under different growth and stress conditions (Xu et al. 2009; Waern and Snyder 2013). Since the vast majority of these cryptic transcripts and transcript isoforms have unknown functions, it has been speculated that they may represent inherent sloppiness of the transcriptional process, referred to as “transcriptional noise” (Struhl 2007).
Where functions of such pervasive transcription have been identified in S. cerevisiae, it appears that it is not necessarily the RNA produced but the act of transcription itself that leads to the observed function. The short noncoding SRG1 transcript inhibits SER3 expression by transcriptional interference and promoter occlusion mechanisms, as the path of the SRG1 transcript overlaps transcription factor binding sites within the SER3 promoter (Martens et al. 2004; Martens et al. 2005). In this case, the SRG1 transcript terminates near the beginning of SER3, while mutation of the tDNA at SUT467 results in uncontrolled readthrough all the way to the end of ATG31. Similar cis-linked mechanisms may be at work at other yeast loci where noncoding transcription appears to block initiation or elongation of ADH1 (Bird et al. 2006), IMD2 (Kuehner and Brow 2008), URA2 (Thiebaut et al. 2008), FLO11 (Bumgarner et al. 2009), PHO84 (Camblong et al. 2007), and IME4 (Hongay et al. 2006; Gelfand et al. 2011). There is evidence that there are also trans-effects of the noncoding RNA product regulating the PHO84 locus (Camblong et al. 2009). Additionally, full repression of yeast GAL genes (Houseley et al. 2008), IME1 (van Werven et al. 2012), and again PHO84 (Camblong et al. 2007) requires chromatin modifications associated with ongoing noncoding transcription. While more instances are likely yet to be identified, this handful of yeast genes has incorporated intergenic transcription into their regulatory programs and generally appears to use it as a means of repression. However, we show in this study that if left unchecked, progression of noncoding transcription can have negative consequences on neighboring gene expression, resulting in reduced fitness of cells. This result demonstrating a cryptic transcript-blocking activity of bound Pol III complexes can be added to the list of extratranscriptional effects of the RNA Polymerase III system.
Our results presented here demonstrate that the tDNA upstream of ATG31 protects against such repressive transcriptional interference effects. Our data are consistent with a model in which TFIIIB, as part of the Pol III complex associated with the tV(UAC)D tDNA, serves as a physical impediment to elongating RNA Pol II initiating at the SUT467 transcriptional start site. In the absence of TFIIIB, nearly complete readthrough by Pol II occurs to produce an extended SUT467–ATG31 RNA transcript. This transcript is not efficiently, if at all, translated into Atg31 protein, as scanning ribosomes (Kozak 2005) attaching at the 5′ end of the extended transcript would encounter start and stop codons before reaching the ATG31 start codon. This readthrough transcript appears to be both capped and polyadenylated, as the 5′-RACE protocol includes a phosphatase treatment before decapping and adaptor ligation, ensuring that only capped 5′ ends are mapped, and the long transcript is enriched in Northern analysis of poly(A)-purified RNA (A. Korde, unpublished data).
The small amount of Atg31p we detect in our Western blots likely results from low levels of normal ATG31 transcripts that are undetectable in Northern blots from cells grown in rich media. Extracts from nitrogen-starved tdna mutants show a slight increase in protein levels, along with detectable normal ATG31 transcripts in Northern blots of RNA isolated from the same cultures (A. Korde, unpublished data). This suggests that under conditions that induce ATG31, limited normal initiation is slightly enhanced, but protein levels are still lower than in wild-type cells.
Previous work from our lab and others has shown that certain extratranscriptional effects associated with tDNAs can be mediated by binding of the TFIIIC complex alone. Propagation of silencing at the HMR mating locus can be blocked by replacing the tDNA downstream of the HMR-I silencer with an ETC site, and insertion of an ETC site between UASG and GAL10 insulates the promoter from Gal4p activation (Simms et al. 2008). Heterochromatin boundary activity of TFIIIC-only containing complexes is also observed in Schizosaccharomyces pombe (Noma et al. 2006; Scott et al. 2006). Additionally, the ETC6 site within the TFC6 promoter may modulate transcription by an insulator-like mechanism (Kleinschmidt et al. 2011). In this case of preventing readthrough of intergenic transcription, the binding of TFIIIC alone is clearly not sufficient. While TFIIIC binds to B-box sequences in vitro with extremely high affinity (Lefebvre et al. 1994; Jourdain et al. 2003), this binding is somehow tempered by passage of the Pol III enzymatic complex during transcription of the internal control element regions.
On the other hand, after recruitment of TFIIIB by TFIIIC, the tightly bound TFIIIB complex appears to be fixed, as in vitro experiments have shown that TFIIIB–DNA complexes are resistant to high salt and heparin treatments (Kassavetis et al. 1990; Kassavetis et al. 1995). The fully assembled TFIIIB complex also is thought to be “kinetically trapped” (Cloutier et al. 2001), with a half-life on the order of a full yeast cell cycle, and fully assembled TFIIIB likely persists at tDNAs until regulated release during mitosis or stationary phase (Fairley et al. 2003; Roberts et al. 2003). Such characteristics of TFIIIB are consistent with our results that suggest that formation of this complex is the major impediment to cryptic transcript readthrough by SUT467. These results are also compatible with earlier in vitro studies that demonstrated the ability of Pol III to transcribe through assembled TFIIIC but not assembled TFIIIB (Bardeleben et al. 1994).
Our results suggest that TFIIIC yields to Pol II in a similar manner as it does to Pol III, since replacing the tDNA with ETC4 allowed readthrough of Pol II even though chromatin immunoprecipitation analysis revealed that TFIIIC was bound to the ectopic ETC4 site (Figure 3C). While we have not mapped the exact 3′ end of the SUT467 transcript, the annotated end mapped by tiling array analysis (Xu et al. 2009) places it within ∼20 bp of the 5′ extent of the expected TFIIIB footprint at this tDNA (schematically depicted in Figure S1). Estimation of this 5′ end of the TFIIIB footprint is based on earlier in vitro footprinting studies (Kassavetis et al. 1989) and a recent global “bootprinting” analysis of in vivo bound Pol III transcription factors (Nagarajavel et al. 2013). The location of the 3′ end of SUT467 is consistent with TFIIIB being a transcriptional roadblock that is resistant to displacement by transcribing Pol II.
A curious sidelight to this study is the appearance of the intermediate length transcript in TFIIIB and TFIIIC mutants, but not in tDNA mutants. This is most likely initiated by Pol II, as in brf1, tfc3, and tfc6 mutants, the tDNA terminator sequence is still present, so it is unlikely that this is a Pol III transcript. We speculate that in these mutants, TFIIIB binding still occurs, but is unstable, and perhaps dissociation of the Brf1p and Bdp1p subunits occurs before loss of TBP at the site. Such a lingering TBP might then recruit factors necessary to then subsequently recruit Pol II immediately upstream of the tDNA. Alternatively, the Pol III complex may mask a cryptic Pol II promoter, which is revealed in a subset of cells containing mutations in the Pol III transcription factors.
There is mounting evidence that a much larger fraction of genomes is transcribed than was previously appreciated. While RNA degradation pathways generally keep most of these transcripts at low levels (Wolin et al. 2012), it has become clear that the act of intergenic transcription can have significant effects on neighboring genes. Due to such observations, one must consider how mutation of a specific genomic locus may affect expression of nearby genes in addition to the targeted gene when assigning the actual cause of observed phenotypes (Wei et al. 2011). To assess the global nature of RNA Pol III extratranscriptional effects, we are conducting RNA-seq analysis of wild-type vs. Tfc6p underexpressing mutant strains. Previous studies have been conducted to determine the global effects of Pol III deficiencies (Conesa et al. 2005), but the RNA was analyzed by coding sequence microarray, which could not detect effects involving intergenic transcription. Inspection of preliminary RNA-seq results suggests that when the Pol III complex is globally compromised, several tDNA proximal genes may be affected as described here, and in other possibly unique ways (Q. Wang, A. Korde, and C. Nowak, unpublished results). This type of result also raises the question of how to interpret phenotypes due to mutations that may globally affect intergenic transcription (which may be relevant in mutants of other DNA and chromatin binding proteins), or as shown here for mutation at a specific locus, as unchecked cryptic transcription can lead to unexpected and even detrimental misexpression of downstream genes. While a subset of pervasive transcription products themselves may be noise, multiple mechanisms must exist to keep secondary effects of their production in check.
Supplementary Material
Acknowledgments
We thank Ro Kamakaka, Jennifer Gerton, and Dan Klionsky for kindly providing yeast strains. We sincerely thank Anne Donaldson and Giorgio Dieci for comments on the manuscript. We also thank members of the Joo Kim lab for assistance with the quantitative PCR analysis. This work was funded by grants MCB-0817823 and MCB-1329446 from the National Science Foundation.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Acker J., Conesa C., Lefebvre O., 2013. Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta 1829: 283–295. [DOI] [PubMed] [Google Scholar]
- Andrau J. C., Sentenac A., Werner M., 1999. Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol. 288: 511–520. [DOI] [PubMed] [Google Scholar]
- Bardeleben C., Kassavetis G. A., Geiduschek E. P., 1994. Encounters of Saccharomyces cerevisiae RNA polymerase III with its transcription factors during RNA chain elongation. J. Mol. Biol. 235: 1193–1205. [DOI] [PubMed] [Google Scholar]
- Bird A. J., Gordon M., Eide D. J., Winge D. R., 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 25: 5726–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P., Dugas S. L., Donze D., Dieci G., 2007. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F., 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717. [DOI] [PubMed] [Google Scholar]
- Camblong J., Beyrouthy N., Guffanti E., Schlaepfer G., Steinmetz L. M., et al. , 2009. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 23: 1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Sandmeyer S. B., 1990. Transfer RNA genes are genomic targets for de novo transposition of the yeast retrotransposon Ty3. Genetics 126: 837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. M., Hong E. L., Amundsen C., Balakrishnan R., Binkley G., et al. , 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40: D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland B. W., Schultz M. C., 2010. Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription 1: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier T. E., Librizzi M. D., Mollah A. K., Brenowitz M., Willis I. M., 2001. Kinetic trapping of DNA by transcription factor IIIB. Proc. Natl. Acad. Sci. USA 98: 9581–9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa C., Ruotolo R., Soularue P., Simms T. A., Donze D., et al. , 2005. Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription. Mol. Cell. Biol. 25: 8631–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J., Lis J. T., 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. M., Newlon C. S., 1996. DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Devine S. E., Boeke J. D., 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Dieci G., Sentenac A., 1996. Facilitated recycling pathway for RNA polymerase III. Cell 84: 245–252. [DOI] [PubMed] [Google Scholar]
- Dieci G., Hermann-Le Denmat S., Lukhtanov E., Thuriaux P., Werner M., et al. , 1995. A universally conserved region of the largest subunit participates in the active site of RNA polymerase III. EMBO J. 14: 3766–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., 2012. Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene 493: 169–175. [DOI] [PubMed] [Google Scholar]
- Donze D., Kamakaka R. T., 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D., Adams C. R., Rine J., Kamakaka R. T., 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornenburg, J. E., A. M. Devita, M. J. Palumbo and J. T. Wade, 2010 Widespread antisense transcription in Escherichia coli. MBio 1. [DOI] [PMC free article] [PubMed]
- Fairley J. A., Scott P. H., White R. J., 2003. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 22: 5841–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R., Rivetti C., Acker J., Dieci G., 2004. Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation. Proc. Natl. Acad. Sci. USA 101: 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek E. P., Kassavetis G. A., 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310: 1–26. [DOI] [PubMed] [Google Scholar]
- Gelfand B., Mead J., Bruning A., Apostolopoulos N., Tadigotla V., et al. , 2011. Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Mol. Cell. Biol. 31: 1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti E., Percudani R., Harismendy O., Soutourina J., Werner M., et al. , 2006. Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J. Biol. Chem. 281: 29155–29164. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Botsios S., Donze D., Donaldson A. D., 2012. TFIIIC localizes budding yeast ETC sites to the nuclear periphery. Mol. Biol. Cell 23: 2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege F. C., Jennings E. G., Wyrick J. J., Lee T. I., Hengartner C. J., et al. , 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Hongay C. F., Grisafi P. L., Galitski T., Fink G. R., 2006. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 127: 735–745. [DOI] [PubMed] [Google Scholar]
- Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M., 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32: 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Maraia R. J., 2001. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 29: 2675–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Engelke D. R., 1989. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5′-flanking region. Mol. Cell. Biol. 9: 3244–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M. W., Erickson J., Johnston M., Engelke D. R., 1994. tRNA genes as transcriptional repressor elements. Mol. Cell. Biol. 14: 1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N., Martinez A. W., Robert E. C., Agochukwu N. B., Ibos M. E., et al. , 2005. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics 171: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., et al. , 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018. [DOI] [PubMed] [Google Scholar]
- Jourdain S., Acker J., Ducrot C., Sentenac A., Lefebvre O., 2003. The tau95 subunit of yeast TFIIIC influences upstream and downstream functions of TFIIIC.DNA complexes. J. Biol. Chem. 278: 10450–10457. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Kawamata T., Suzuki K., Ohsumi Y., 2007. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 356: 405–410. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P., 1989. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 9: 2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Braun B. R., Nguyen L. H., Geiduschek E. P., 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60: 235–245. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Nguyen S. T., Kobayashi R., Kumar A., Geiduschek E. P., et al. , 1995. Cloning, expression, and function of TFC5, the gene encoding the B” component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc. Natl. Acad. Sci. USA 92: 9786–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kang K., Ekram M. B., Roh T. Y., Kim J., 2011. Aebp2 as an epigenetic regulator for neural crest cells. PLoS ONE 6: e25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt R. A., LeBlanc K. E., Donze D., 2011. Autoregulation of an RNA polymerase II promoter by the RNA polymerase III transcription factor III C (TF(III)C) complex. Proc. Natl. Acad. Sci. USA 108: 8385–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., 2007. Monitoring autophagy in yeast: the Pho8Delta60 assay. Methods Mol. Biol. 390: 363–371. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., et al. , 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Kozak M., 2005. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361: 13–37. [DOI] [PubMed] [Google Scholar]
- Kuehner J. N., Brow D. A., 2008. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell 31: 201–211. [DOI] [PubMed] [Google Scholar]
- Lardenois A., Liu Y., Walther T., Chalmel F., Evrard B., et al. , 2011. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. USA 108: 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O., Ruth J., Sentenac A., 1994. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J. Biol. Chem. 269: 23374–23381. [PubMed] [Google Scholar]
- Marck C., Kachouri-Lafond R., Lafontaine I., Westhof E., Dujon B., et al. , 2006. The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 34: 1816–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Wu P. Y., Winston F., 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., et al. , 2006. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA 103: 17846–17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moqtaderi Z., Struhl K., 2004. Genome-wide occupancy profile of the RNA polymerase III machinery in Saccharomyces cerevisiae reveals loci with incomplete transcription complexes. Mol. Cell. Biol. 24: 4118–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Roth S. Y., Simpson R. T., 1992. A transcriptionally active tRNA gene interferes with nucleosome positioning in vivo. Mol. Cell. Biol. 12: 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajavel V., Iben J. R., Howard B. H., Maraia R. J., Clark D. J., 2013. Global ‘bootprinting’ reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 41: 8135–8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H., Malabat C., d’Aubenton-Carafa Y., Xu Z., Steinmetz L. M., et al. , 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. [DOI] [PubMed] [Google Scholar]
- Noma K., Cam H. P., Maraia R. J., Grewal S. I., 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125: 859–872. [DOI] [PubMed] [Google Scholar]
- Orioli A., Pascali C., Pagano A., Teichmann M., Dieci G., 2012. RNA polymerase III transcription control elements: themes and variations. Gene 493: 185–194. [DOI] [PubMed] [Google Scholar]
- Pascali C., Teichmann M., 2012. RNA polymerase III transcription: regulated by chromatin structure and regulator of nuclear chromatin organization. Subcell. Biochem. 61: 261–287. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J., 2013. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194: 341–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D. N., Stewart A. J., Huff J. T., Cairns B. R., 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 100: 14695–14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2002. Ordered nucleation and spreading of silenced chromatin in saccharomyces cerevisiae. Mol. Biol. Cell 13: 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. C., Merrett S. L., Willard H. F., 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16: 119–129. [DOI] [PubMed] [Google Scholar]
- Sekedat M. D., Fenyo D., Rogers R. S., Tackett A. J., Aitchison J. D., et al. , 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol. Syst. Biol. 6: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms T. A., Miller E. C., Buisson N. P., Jambunathan N., Donze D., 2004. The Saccharomyces cerevisiae TRT2 tRNAThr gene upstream of STE6 is a barrier to repression in MATalpha cells and exerts a potential tRNA position effect in MATa cells. Nucleic Acids Res. 32: 5206–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms T. A., Dugas S. L., Gremillion J. C., Ibos M. E., Dandurand M. N., et al. , 2008. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell 7: 2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley R. E., Ragusa M. J., Hurley J. H., 2013. The beginning of the end: how scaffolds nucleate autophagosome biogenesis. Trends Cell Biol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14: 103–105. [DOI] [PubMed] [Google Scholar]
- Thiebaut M., Colin J., Neil H., Jacquier A., Seraphin B., et al. , 2008. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol. Cell 31: 671–682. [DOI] [PubMed] [Google Scholar]
- Thuillier V., Stettler S., Sentenac A., Thuriaux P., Werner M., 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 14: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E. L., Chen C. L., d’Aubenton-Carafa Y., Gourvennec S., Kwapisz M., et al. , 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117. [DOI] [PubMed] [Google Scholar]
- van Werven F. J., Neuert G., Hendrick N., Lardenois A., Buratowski S., et al. , 2012. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 150: 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waern, K., and M. Snyder, 2013 Extensive transcript diversity and novel upstream open reading frame regulation in yeast. G3 Genes Genomes Genet. 3: 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Pelechano V., Jarvelin A. I., Steinmetz L. M., 2011. Functional consequences of bidirectional promoters. Trends Genet. 27: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S. L., Sim S., Chen X., 2012. Nuclear noncoding RNA surveillance: Is the end in sight? Trends Genet. 28: 306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., et al. , 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.