Abstract
The essential zinc finger protein ASCIZ (also known as ATMIN, ZNF822) plays critical roles during lung organogenesis and B cell development in mice, where it regulates the expression of dynein light chain (DYNLL1/LC8), but its functions in other species including invertebrates are largely unknown. Here we report the identification of the Drosophila ortholog of ASCIZ (dASCIZ) and show that loss of dASCIZ function leads to pronounced mitotic delays with centrosome and spindle positioning defects during development, reminiscent of impaired dynein motor functions. Interestingly, similar mitotic and developmental defects were observed upon knockdown of the DYNLL/LC8-type dynein light chain Cutup (Ctp), and dASCIZ loss-of-function phenotypes could be suppressed by ectopic Ctp expression. Consistent with a genetic function of dASCIZ upstream of Ctp, we show that loss of dASCIZ led to reduced endogenous Ctp mRNA and protein levels and dramatically reduced Ctp–LacZ reporter gene activity in vivo, indicating that dASCIZ regulates development and mitosis as a Ctp transcription factor. We speculate that the more severe mitotic defects in the absence of ASCIZ in flies compared to mice may be due to redundancy with a second, ASCIZ-independent, Dynll2 gene in mammals in contrast to a single Ctp gene in Drosophila. Altogether, our data demonstrate that ASCIZ is an evolutionary highly conserved transcriptional regulator of dynein light-chain levels and a novel regulator of mitosis in flies.
Keywords: ATM substrate Chk2-interacting Zn2+ finger protein, dynein, mitosis, development, Drosophila
THE ATM substrate Chk2-interacting Zn2+ finger (ZnF) protein (ASCIZ; also known as ATMIN, ZNF822) was identified as a protein that forms nuclear DNA damage-induced foci specifically in response to DNA alkylating or oxidating agents (McNees et al. 2005; Rapali et al. 2011a), and absence of ASCIZ increases sensitivity to these base lesions (McNees et al. 2005; Oka et al. 2008; Jurado et al. 2010; Kanu et al. 2010). ASCIZ contains four N-terminal ZnFs and a C-terminal SQ/TQ-cluster domain (SCD) enriched in phosphorylation sites for ATM-like DNA damage response kinases (Traven and Heierhorst 2005; Matsuoka et al. 2007). However, the SCD also functions as a potent transcription activation domain, and Asciz-deficient mouse models revealed major DNA damage-independent developmental functions as a transcription factor (Jurado et al. 2010). Germline knockout of Asciz results in late embryonic lethality with a range of developmental abnormalities (Jurado et al. 2010; Kanu et al. 2010), including severe foregut separation defects with complete absence of lungs (Jurado et al. 2010; Heierhorst et al. 2011). The mRNA for dynein light-chain DYNLL1 was the most strongly downregulated transcript (∼10-fold) in mouse Asciz knockout cells, and similar DYNLL1 reductions were observed in ASCIZ-deficient human and chicken cells (Jurado et al. 2012a). ASCIZ binds to the Dynll1 promoter in primary mouse cells and activates its transcription in a ZnF-dependent manner, consistent with a function as a ZnF transcription factor.
DYNLL1 (LC8) was first identified as a light chain of the dynein motor complex (King and Patel-King 1995), where it may facilitate the association of dynein intermediate chains with the heavy chains (Williams et al. 2007), but has since emerged as a regulator of possibly more than 100 diverse proteins (King 2008; Barbar 2008; Rapali et al. 2011b). DYNLL1 is structurally highly conserved throughout evolution; for example, human DYNLL1 and its Drosophila ortholog Cutup (Ctp) differ by just four conservative substitutions within the 89-amino-acid proteins (Dick et al. 1996; Phillis et al. 1996). While null mutations of Ctp are lethal in Drosophila, and even hypomorphic mutations lead to severe morphogenesis defects (Dick et al. 1996; Phillis et al. 1996; Batlevi et al. 2010), Dynll1 loss-of-function mutations have not been reported in vertebrates, and it remains unclear if its regulation is as highly conserved as its structure and functions.
Interestingly, beyond acting as a transcriptional activator of Dynll1 gene expression, ASCIZ itself is also a major DYNLL1-binding protein. The ASCIZ SCD contains 11 TQT motifs and 10 of these are functional DYNLL1 binding sites (Rapali et al. 2011a; Jurado et al. 2012a). Importantly, DYNLL1 binding to these sites represses transcriptional activity of ASCIZ in a concentration-dependent manner, and the dual ability of ASCIZ to activate the Dynll1 promoter and to “sense” the concentration of its gene product, therefore, generates a feedback loop to maintain stable, free DYNLL1 protein levels (Jurado et al. 2012a). The extent to which impaired DYNLL1 regulation contributes to organogenesis defects in Asciz KO mice remains to be determined, but severe B cell development defects in conditional Asciz deleters can be rescued by ectopic Dynll1 expression, demonstrating that this phenotype is due to reduced DYNLL1 (Jurado et al. 2012b).
While its transcriptional functions as a Dynll1 regulator and its DNA base damage response functions seem to be conserved from birds to humans (Oka et al. 2008; Jurado et al. 2012a), no ASCIZ orthologs have previously been identified in invertebrates. Here we report the identification of the fly open reading frame CG14962 as the Drosophila melanogaster ortholog of ASCIZ (dASCIZ) and provide the first evidence that ASCIZ is essential for development in Drosophila. Strikingly, although Drosophila ASCIZ is a conserved transcriptional regulator of Dynll1/Ctp expression, we show that its absence leads to severe mitotic defects extending beyond the more restricted and relatively specific organogenesis and B-lymphopoiesis defects in ASCIZ KO mice (Jurado et al. 2010, 2012b), probably as a result of reduced DYNLL redundancy in flies.
Materials and Methods
Fly strains
Unless otherwise stated, Drosophila strains were obtained from the Bloomington Stock Centre, including P{w[+mC]=lacW}ctp[G0371] (Peter et al. 2002) and P{w[+mC]=UASp-GFP-Cnn1}26-1. UAS–Ctp (Batlevi et al. 2010) was a gift from E. Baehrecke, UAS–dASCIZ was generated by cloning full length dASCIZ cDNA into pUAST (as described Quinn et al. 2001), and transgenics were generated by BestGene. The dASCIZ RNAi (v100118), ctp RNAi (v43115 and v43116), and dynein heavy-chain (DHC64C) RNAi (v28054) lines were from the Vienna Drosophila RNAi Center (Dietzl et al. 2007). The alternate, nonoverlapping dASCIZ RNAi line was from the Bloomington stock center (26772 TRiP.JF02336).
Protein expression, antibody generation, and in vitro binding assays
Ctp and the dASCIZ SCD (residues 241–388) were amplified from cDNAs (LD24056 and AT25633, Drosophila Genomics Resource center) and cloned into pQE30 and pGEX4T1, respectively. His6–Ctp and GST–dSQ/TQ were expressed in Escherichea coli BL21[pREP4] or Star-Rosetta cells and purified over Ni–NTA (Qiagen) or GSH–Sepharose (GE Healthcare) columns. Immunization of rabbits was according to standard procedures approved by the St. Vincent’s Hospital Melbourne animal ethics committee. dASCIZ antiserum and affinity-purified Ctp antibodies (using NHS-activated Ctp-coupled Sepharose columns, GE Healthcare) were used for immunofluorescence.
For pulldown assays, 5 µg of each protein was bound to 25 µl GSH–Sepharose beads in PBST (2 hr at 4°) and washed 3× PBS before elution into 25 µl 10 mM reduced glutathione, 50 mM Tris pH 8. A total of 7.5 µl of each supernatant was loaded per lane on 18% SDS–PAGE gels and detected with anti-GST (GE Healthcare; 1/4000) or anti-DYNLL1 antibodies (Santa Cruz; 1/1000).
DNA damage assay
Full-length dASCIZ, its N-terminal ZnF domain (residues 1–170) or C-terminal region (residues 171–388) were fused to N-terminal EGFP-tags and transfected into HeLa cells. Cells were laser irradiated at 405 nm as described (Lan et al. 2004; Yoshimura et al. 2006; Prasad et al. 2007) using conditions that produce DNA damage.
Immunohistochemistry
Third-instar larval discs were fixed (30 min, 4% PFA), blocked (5% BSA/PBST), and incubated overnight at 4° with primary antibody prior to detection with fluorescent secondary antibody. Primary antibodies included β-gal (Sigma) (1:500), Peanut 4C9H4 and β-tubulin (Developmental Studies Hybridoma Bank, 1:10), phospho-histone-H3 (PH3, Millipore, 1:1000), Geminin antiserum (Quinn et al. 2001) (1:500), and caspase 3 (Sigma, 1:500). Wing imaginal discs were counterstained with DAPI and imaged (Zeiss Imager Z with Zen Meta software). Images were processed using Image J 1.43u, and Adobe Photoshop CS4 Version 11.0.2. Mitoses were quantified by measuring the ratio of average PH3-positive cells per fixed area in posterior compartment (PC) compared with the anterior compartment (AC) over at least seven discs. All statistical tests were performed with Prism 4.0c using unpaired two-tailed t-test with 95% confidence interval.
Bromodeoxyuridine labeling
Discs were incubated in 100 μg/ml bromodeoxyuridine (BrdU) in Schneider’s media with 10% FCS (30 min), fixed in 4% PFA, washed in PBST, and treated with DNAse (37° for 1.5 hr) prior to immunostaining with anti-BrdU (Becton Dickinson, 1:100). Ratio of average BrdU-positive cells per fixed area in the PC compared with AC was calculated over at least seven discs.
Adult wing preparation and imaging
Wings of adult male flies were mounted in Canada Balsam in Xylene and imaged under an Olympus SZ microscope at 4.5× magnification. Quantification was performed by measuring the area of PC (posterior to vein L4), the area of AC (anterior to vein L4), and determining the ratio.
cDNA synthesis and qPCR
To overcome embryonic lethality, global UAS–transgene expression in larvae was achieved using tub–gal80ts;tub–GAL4, whereby progeny were raised at 18° for 5–6 days and subsequently shifted to 29° for 72 hr prior to collection of third-instar imaginal tissues for RNA extraction and cDNA synthesis (Bioline kit). For each genotype, qPCR was carried out in biological triplicate using SYBR Green and standard cycles on the 7900HT Fast Real-Time PCR system (Applied Bioscience). Primers were dASCIZ forward, 5′ TGCGACAGCACTACCAGAAG 3′; dASCIZ reverse, 5′ GGGTATGGGAACCTTGTCGG 3′; Ctp forward, 5′ TTGTGCGACACAGGCCCTCG 3′; Ctp reverse, 5′ TGGCGCGTCTCGTGTGTGAC3′; actin forward, 5′ GGCGCAGAGCAAGCGTGGTA 3′; actin reverse, 5′ GGGTGCCACACGCAGCTCAT 3′.
Results
Identification of CG14962 as the Drosophila ortholog of ASCIZ
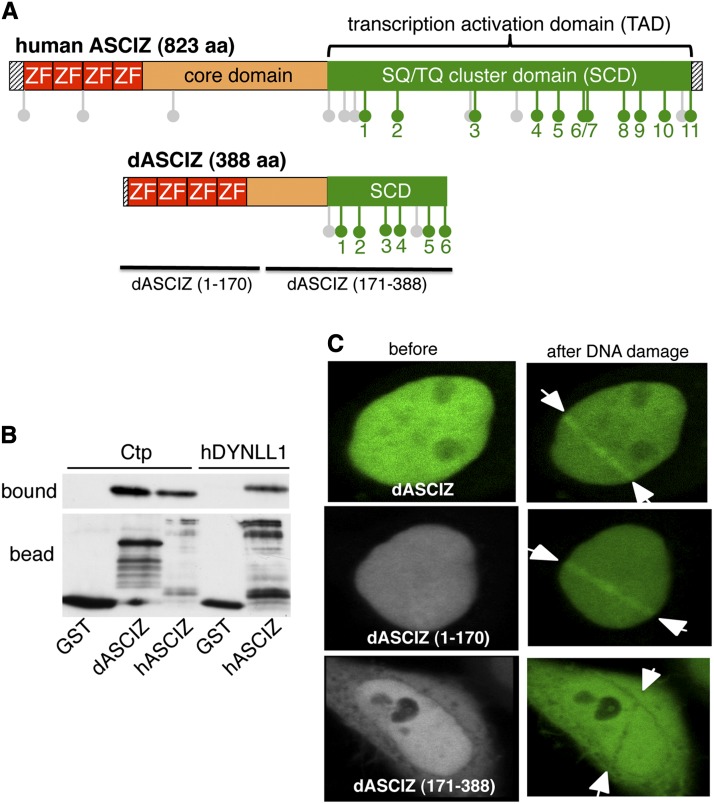
To identify a Drosophila ortholog for ASCIZ, we aligned full-length human ASCIZ against the release-5 genome. The closest match was the 388-amino-acid protein-encoding open reading frame CG14962, which we have therefore named dASCIZ (Figure 1A). The similarity between the human and fly proteins is highest within the N-terminal 4-ZnF domain (Supporting Information, Figure S1A), and dASCIZ also contains a characteristic C-terminal SQ/TQ cluster domain (SCD) with 6 TQT motifs (Figure S1B). Interestingly, similar to mammalian ASCIZ, two independent genome-wide yeast two-hybrid screens had previously detected interaction between dASCIZ and the Drosophila DYNLL1-ortholog Ctp (Giot et al. 2003) (Finley lab two-hybrid screen http://www.droidb.org/Index.jsp). We confirmed this interaction via in vitro pulldown assays, where recombinant Ctp bound to an immobilized TQT motif-containing GST–dASCIZ–SCD fragment, but not GST alone, in a manner comparable to the interaction between recombinant human DYNLL1 and GST–ASCIZ–SCD (Figure 1B). In addition, EGFP-tagged full-length dASCIZ or the isolated N-terminal ZnF-domain, but not the C-terminal region, accumulated within 5 min along tracks of laser-induced oxidative DNA damage (Figure 1C), reminiscent of the association of human ASCIZ with oxidative DNA damage-induced nuclear foci (McNees et al. 2005).
Figure 1.
Characterization of dASCIZ. (A) Schematic comparison of human and Drosophila ASCIZ; green lollipops indicate TQT motifs, and gray lollipops indicate other SQ or TQ motifs. (B) Pulldown of recombinant His6-tagged Ctp or human DYNLL1 with the GST-fused SCDs of human or Drosophila ASCIZ or GST-only control using GSH beads. Bottom, the immobilized bait proteins probed with a GST antibody; top, the bound ligands after elution from the beads. In the left three samples Ctp was added as ligand; in the two right samples human DYNLL1 was added. (C) Accumulation of EGFP-tagged dASCIZ transfected into HeLa cells along DNA-damaged laser tracks. The images show representative samples before (left) and 5 min after 500 scans of laser irradiation with 405 nm to induce DNA base damage. Arrows indicate the path of the laser. Full-length dASCIZ is shown on top; the range of the other fragments is indicated in C and A.
dASCIZ is required for larval development and wing morphogenesis
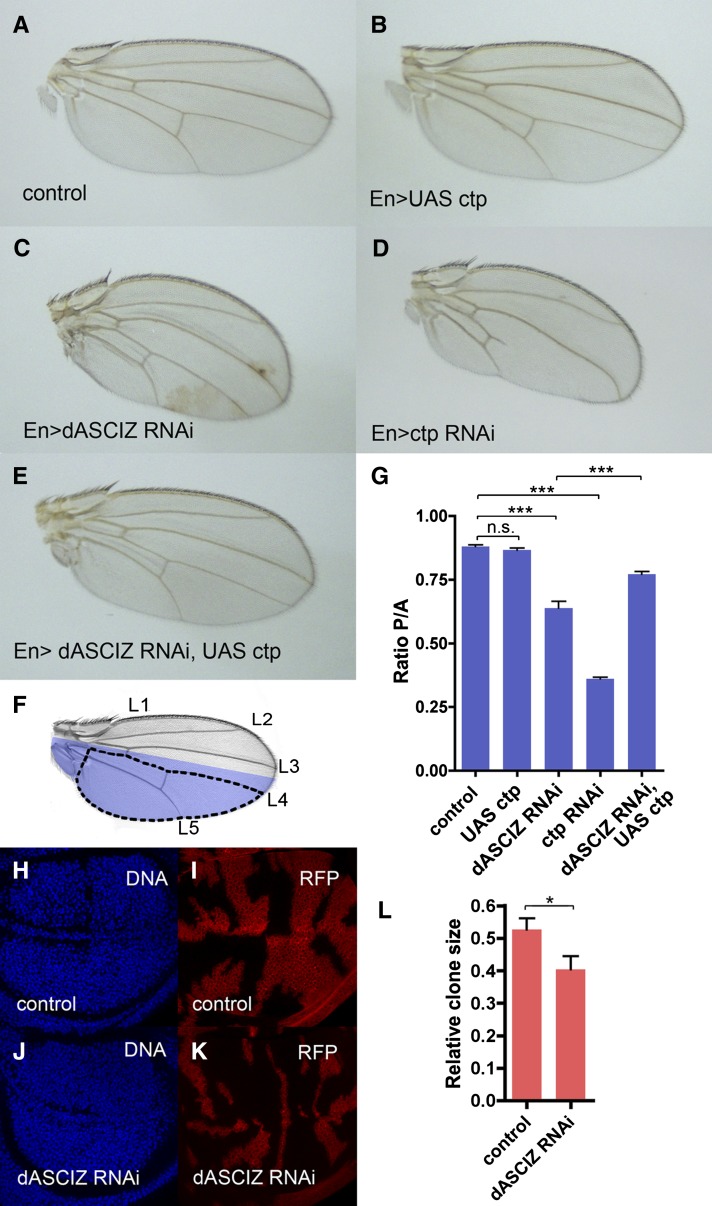
To characterize in vivo functions of dASCIZ, we initially conducted global dASCIZ RNAi knockdowns using tubulin-GAL4 to drive UAS–dASCIZ RNAi transgenes (Dietzl et al. 2007), which led to 100% first-instar larval lethality (data not shown). More localized dASCIZ RNAi knockdown within the PC of the wing imaginal disc using en-GAL4 (Figure 2) resulted in shrunken and curved adult wings (Figure 2C) with a significantly reduced PC relative to the AC (Figure 2G; P < 0.0001). dASCIZ knockdown also caused a “held-out-wings” phenotype (Figure S2), as a likely result of posterior hinge malformation. A qualitatively similar, although more attenuated, PC/AC reduction was also observed in knockdowns using an alternate and nonoverlapping dASCIZ RNAi line (Figure S3A). Further analysis using dASCIZ RNAi “flip-out” clones (Figure 2, H–K) revealed a significant decrease in dASCIZ RNAi clone size compared with the control (Figure 2L). Taken together, these results indicate that dASCIZ is an essential protein and that its locally restricted loss results in impaired tissue growth and developmental defects.
Figure 2.
The dASCIZ RNAi wing phenotype is phenocopied by Ctp knockdown and rescued by Ctp overexpression. (A–E) Representative images of male adult wings with Engrailed–GAL4 (En) driver. (A) Control En–GAL4/+. (B) En > UAS–Ctp overexpression. (C) En > dASCIZ RNAi. (D) En > Ctp RNAi. (E) En > dASCIZ RNAi with En > UAS–Ctp overexpression. (F) Adult wing showing En expression domain in blue and longitudinal veins (L1–5). For quantitation, the area posterior to L4 was used as a measure of the posterior compartment, PC (dotted line), and the area anterior to L4 was used to measure the anterior compartment, AC. (G) Quantification of wing size using the ratio PC/AC as defined in (F). Error bars represent SEM for n > 10. PC/AC ratio is unchanged for UAS–Ctp (P = 0.2446), reduced for dASCIZ RNAi compared to control (P < 0.0001), and reduced for Ctp RNAi (P < 0.0001). Coexpression of dASCIZ RNAi with UAS–Ctp improves the ratio compared to dASCIZ RNAi alone (P = 0.0003). All quantifications were performed using male wings; analogous phenotypes were observed in female wings (data not shown). RFP marked flip-out clones for control (H and I), dASCIZ RNAi (J and K), and quantification of average clone size relative to the total wing disc area (L).
Loss of dASCIZ leads to impaired mitosis and increased apoptotic cell death
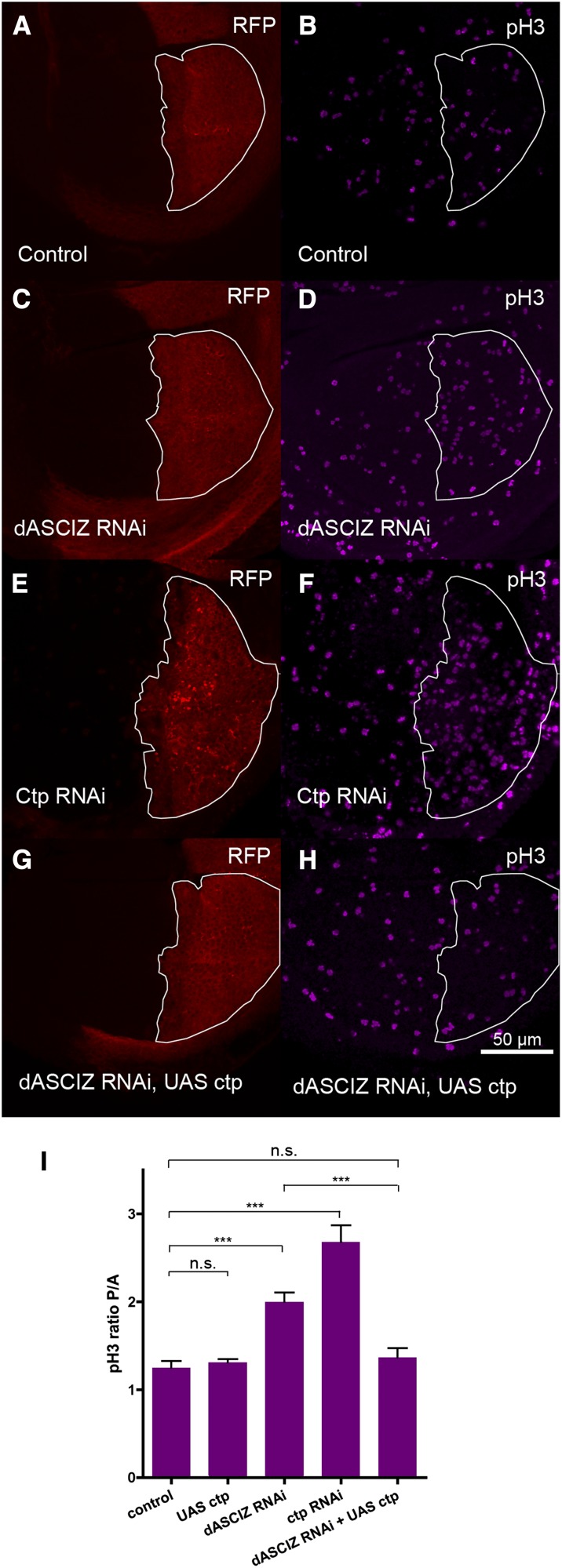
Reduced cell proliferation and/or increased cell death were possible explanations for impaired tissue formation in dASCIZ knockdowns. To assess cell proliferation we stained third-instar wing discs with BrdU as an S-phase marker and phospho-histone H3 (PH3) for mitosis. Interestingly, while BrdU staining was not affected (Figure S4), dASCIZ RNAi significantly increased the ratio of PH3-positive cells in the PC compared with the AC (Figure 3, B, D, and I; P < 0.0001; see also Figure S3B for similar results with the alternative dASCIZ RNAi line), indicating a mitotic delay.
Figure 3.
dASCIZ knockdown results in increased mitotic index, which can be suppressed by Ctp co-overexpression. En–GAL4 expression in the PC of wing imaginal discs is marked with RFP (outlined in white) and mitotic cells are labeled with phospho-histone H3 (PH3) antibody. (A and B) Control, (C and D) dASCIZ RNAi, (E and F) Ctp RNAi, and (G and H) dASCIZ RNAi with UAS–Ctp. (I) Ratios of average mitotic figures in the PC of the wing imaginal disc (RFP positive) to the AC (RFP negative). Error bars represent SEM for n > 11. Ratio is unchanged for UAS–Ctp (P = 0.5582), reduced for dASCIZ RNAi (P < 0.0001) and reduced for Ctp RNAi (P < 0.0001). Coexpression of UAS–Ctp with dASCIZ RNAi improves the ratio compared to dASCIZ RNAi alone (P = 0.0003), to control levels (P = 0.4129).
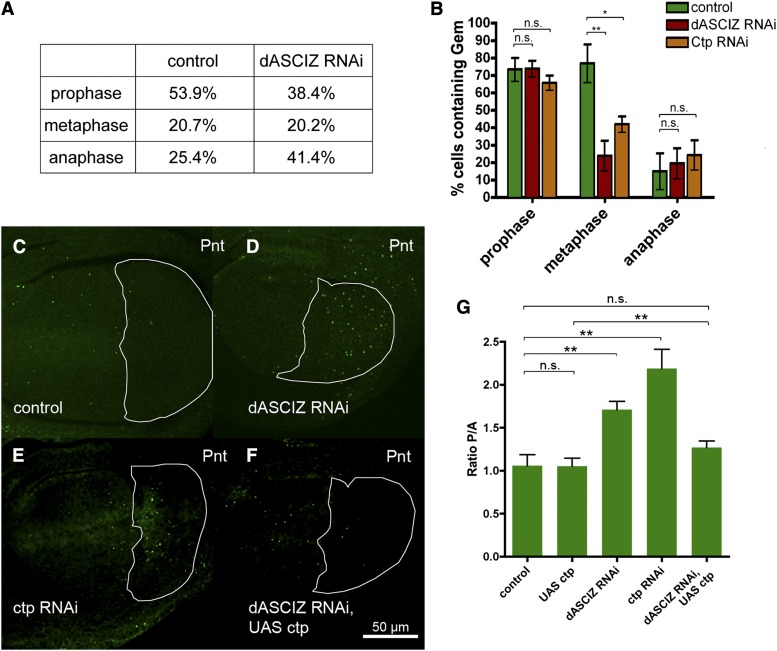
Morphological analysis of mitotic figures from the total population of PH3-positive cells revealed that dASCIZ knockdown was associated with a reduction in prophase and pronounced increase in anaphase (Figure 4A), suggesting impaired completion of anaphase. In addition, although the total proportion of metaphase cells was not altered, the relative proportion of cells containing the replication inhibitor Geminin—that is present at much higher levels in early compared with late metaphase (Quinn et al. 2001)—was considerably reduced in dASCIZ-knockdown metaphase cells (Figure 4B), suggesting that the mitotic delay commences during late metaphase. dASCIZ knockdown also resulted in a higher proportion of cells stained for the septin component Peanut (Pnt), which marks actin-myosin contractile rings (Figure 4, C–G). Pnt first accumulates and becomes concentrated at the spindle interzone in late anaphase, but septin rings are present at all late stages of mitosis (Neufeld and Rubin 1994). This provides additional support for a late mitotic delay, although we cannot exclude a separate role for dASCIZ during cytokinesis, which would also be associated with elevated septin ring levels. Altogether these results indicate that dASCIZ plays a crucial role during late stages of mitosis (metaphase and anaphase; Figure 4, A and B) that may extend into telophase or cytokinesis (Figure 4, C–G).
Figure 4.
dASCIZ and Ctp are required for progression through late stages of mitosis. (A) Proportion of cells in wing imaginal discs at the indicated mitotic stages based on cell morphology and PH3 staining. Compared with the control, wings expressing dASCIZ RNAi contain more cells undergoing anaphase and fewer cells in prophase (n = 449 for control; n = 474 for dASCIZ RNAi). (B) Fraction of PH3-labeled cells at the indicated mitotic stages costained with anti-Geminin antibody. Cells in the PC of the wing disc expressing GAL4 control, dASCIZ RNAi, or Ctp RNAi were sorted into mitotic stages based on morphology and scored for positive staining with Geminin. While control cells degrade Geminin at the metaphase/anaphase transition, staining in cells expressing dASCIZ and Ctp RNAi is reduced during metaphase (P = 0.0036 for dASCIZ RNAi and P = 0.0108 for Ctp RNAi, n > 7), indicating that Geminin is degraded earlier than in control cells and suggesting that these cells spend longer in mitosis. (C–F) Wing imaginal discs stained with Peanut antibody, En–GAL4 expression in the PC is outlined in white. (C) En–GAL4/+ control, (D) En > dASCIZ RNAi, (E) En > Ctp RNAi, and (F) En > dASCIZ RNAi, UAS–Ctp. (G) PC/AC ratio of Peanut staining in the wing imaginal disc. Error bars represent SEM for n > 7. Ratio is unchanged for UAS–Ctp (P = 0.9699), reduced for dASCIZ RNAi (P = 0.0014), and reduced for Ctp RNAi (P = 0.0013). Coexpression of dASCIZ RNAi with UAS–Ctp improves the ratio compared to dASCIZ RNAi alone (P = 0.0064) back to control levels (P = 0.2050).
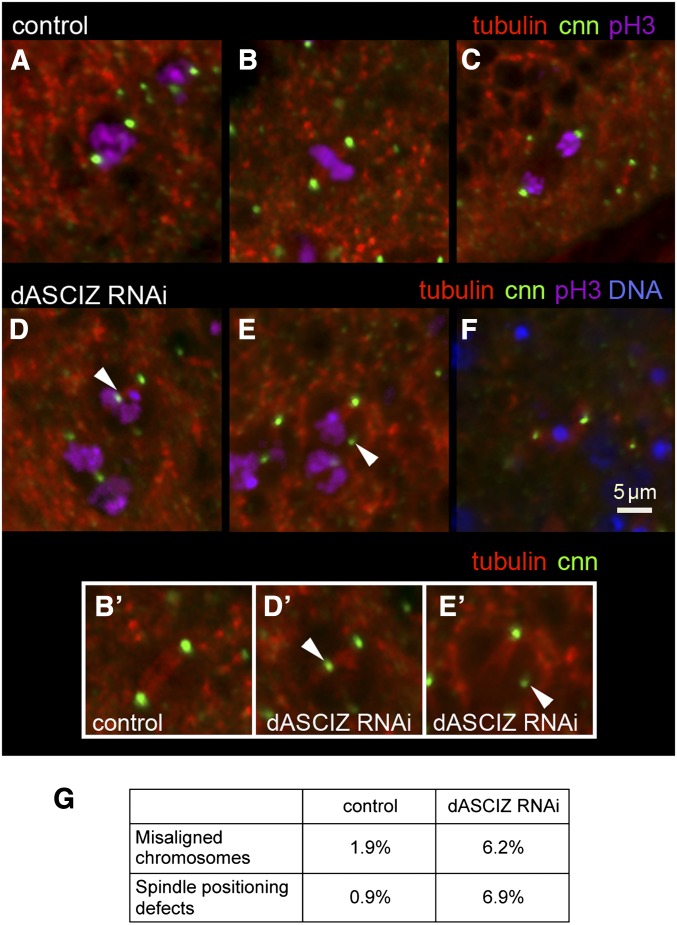
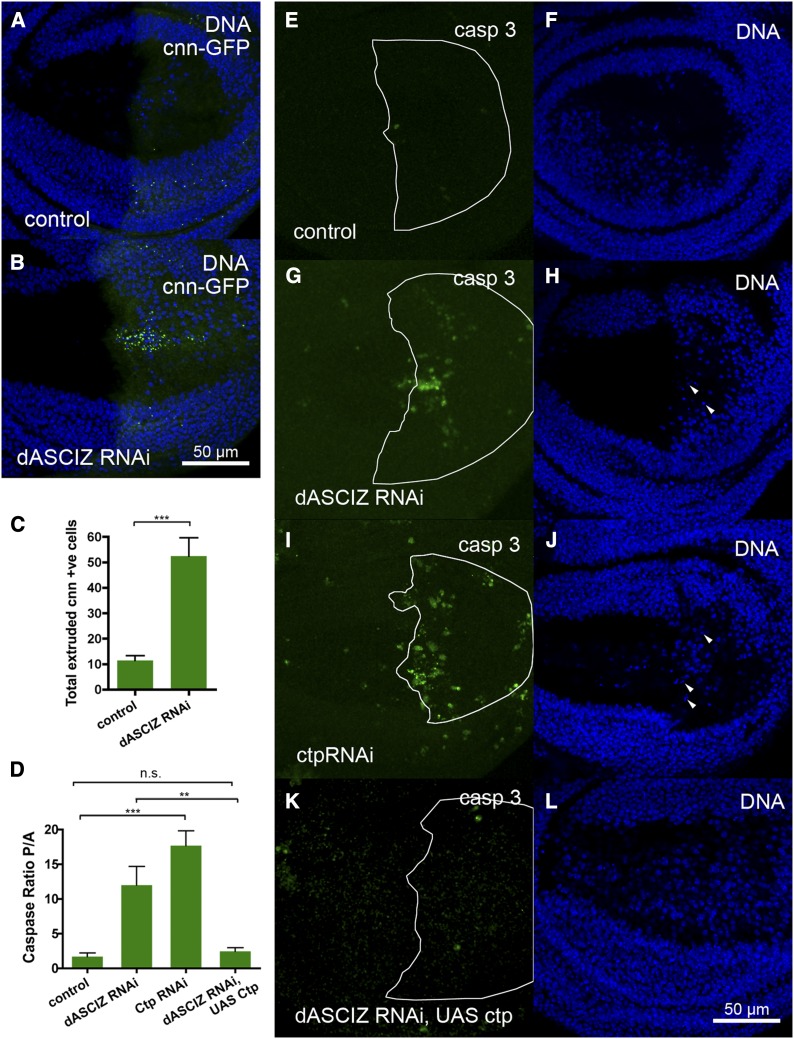
To further analyze the mitotic defects, we monitored centrosomes (using centrosomin–GFP; Megraw et al. 2002), mitotic spindles (using β-tubulin–RFP), and chromosome positioning (using anti-PH3) (Figure 5, A–F). Compared with control prophase, metaphase, and anaphase cells (Figure 5, A–C), dASCIZ RNAi was associated with severe spindle alignment and chromosome alignment defects (Figure 5, D–F). For example, we observed detachment of the spindle from the centrosome (Figure 5, D and D’), mislocalization of the centrosome (Figure 5, E and E’), and spindle misalignments associated with a failure to segregate chromosomes (Figure 5F, quantified in 5G). dASCIZ RNAi also dramatically increased accumulation of centrosomin–GFP-positive cells beneath the wing epithelium (Figure 6, A–C), where dying cells localize after extrusion, suggesting increased cell death due to mitotic catastrophe or irreversible spindle checkpoint activation. Consistently, dASCIZ RNAi also led to increased caspase 3-positive cells in the PC and fragmented DNA below the wing disc basal lamina, indicative of apoptosis (Figure 6, D, G, and H). Thus, impaired completion of mitotic cell divisions and concomitantly increased apoptosis are the likely reason for the decreased PC size in the adult wing following loss of dASCIZ (Figure 2).
Figure 5.
dASCIZ is required for spindle alignment during cell division. (A–C) Control cells undergoing mitosis in prophase (A), metaphase (B), and anaphase (C), expressing tubulin–RFP and centrosomin–GFP and stained for PH3. Inset for control B’. (D and E) Mitotic cells expressing dASCIZ RNAi fail to correctly position the spindle (arrowheads). Insets D’ and E’ lack PH3 staining to show spindles in dASCIZ RNAi cells. (F) Cells expressing dASCIZ RNAi, tubulin–RFP, and centrosomin–GFP, which fail to align the spindle and complete mitosis, are extruded below the epithelial layer. PH3 staining is not observed and the pyknotic DNA is stained with DAPI. (G) Observed proportion of cells undergoing defective late stages of mitosis. Number of cells with chromosome alignment defects and misaligned spindles were recorded on the basis of morphology relative to the total number of cells in late mitosis (n = 207 for control, n = 292 for dASCIZ RNAi).
Figure 6.
dASCIZ is required for cell survival through Ctp. (A and B). Basal sections of wing imaginal discs, showing extruded centrosomin-positive mitotic cells in the disc expressing En > dASCIZ RNAi (B) but not in the control (A), costained for DNA with DAPI. (C) Quantification of the total number of centrosomin-positive cells extruded below the basal lamina after expression of dASCIZ RNAi in the PC (P = 0.0003). Error bars represent SEM for n = 6. (D) PC/AC ratio of caspase staining in the wing imaginal disc. Error bars represent SEM for n = 8. Ratio is increased for dASCIZ RNAi (P = 0.0195) and Ctp RNAi (P = 0.0001). Coexpression of dASCIZ RNAi with UAS–Ctp improves the ratio compared to dASCIZ RNAi alone (P = 0.0064) back to control levels (P = 0.3439). (E–L) Basal sections of wing imaginal discs stained with the activated caspase 3 antibody to detect apoptosis (left), and DAPI-stained pyknotic cells are marked with arrowheads (right). The En–GAL4 expression area in the PC is outlined in white. (E and F) En–GAL4/+ control. (G and H) En > dASCIZ RNAi. (I and J) En > Ctp RNAi. (K and L) En > dASCIZ RNAi with UAS–Ctp overexpression.
Ctp knockdown mimics and Ctp overexpression rescues dASCIZ RNAi phenotypes
Dynein motors play multiple critical roles in the execution of mitosis, including spindle and centrosome positioning (Nguyen-Ngoc et al. 2007; Couwenbergs et al. 2007; Dunsch et al. 2012; Kotak et al. 2013; Kiyomitsu and Cheeseman 2013; Raaijmakers et al. 2013). The mitotic, cell survival, and developmental defects resulting from dASCIZ knockdown were grossly reminiscent of previously reported phenotypes for Ctp or other dynein subunit mutants in Drosophila (Dick et al. 1996; Mische et al. 2008) or dynein RNAi (Morales-Mulia and Scholey 2005). As ASCIZ plays critical roles in dynein light-chain regulation in mammalian cells (Jurado et al. 2012a), we tested if Ctp might modify the dASCIZ RNAi phenotypes. To compare dASCIZ and Ctp loss-of-function phenotypes, we used en-GAL4 to drive UAS–Ctp RNAi (Dietzl et al. 2007). Interestingly, Ctp knockdown resulted in wing phenotypes very similar to dASCIZ knockdown, with decreased PC size (Figure 2, D and G), increased accumulation of cells in mitosis (Figure 3, F and I, and Figure 4, B, E, and G; see also Figure S5 for similar results with an alternate Ctp RNAi line), and increased apoptosis (Figure 6, D, I, and J), suggesting that dASCIZ and Ctp might act in a common pathway. In addition, knockdown of the dynein heavy-chain DHC64C using en-GAL4 also significantly increased PH3 staining in the PC (Figure S6). Based on these findings, we tested whether ectopic Ctp expression might rescue the dASCIZ RNAi phenotypes. Importantly, while Ctp overexpression using a previously characterized UAS–Ctp line (Batlevi et al. 2010) alone did not result in an observable phenotype (Figure 2B), its overexpression in the en-GAL4 dASCIZ RNAi knockdown background significantly restored the adult PC/AC size ratio (Figure 2E,G; P = 0.0003) and normalized the PC/AC ratio of mitotic (Figure 3, H and I, and Figure 4, F and G) and apoptotic cells (Figure 6, D, K, and L) back to control levels. Thus, taken together, these results indicate that dASCIZ functions upstream of Ctp in the regulation of wing morphogenesis, mitosis, and apoptosis.
dASCIZ functions as a transcriptional regulator of Ctp expression
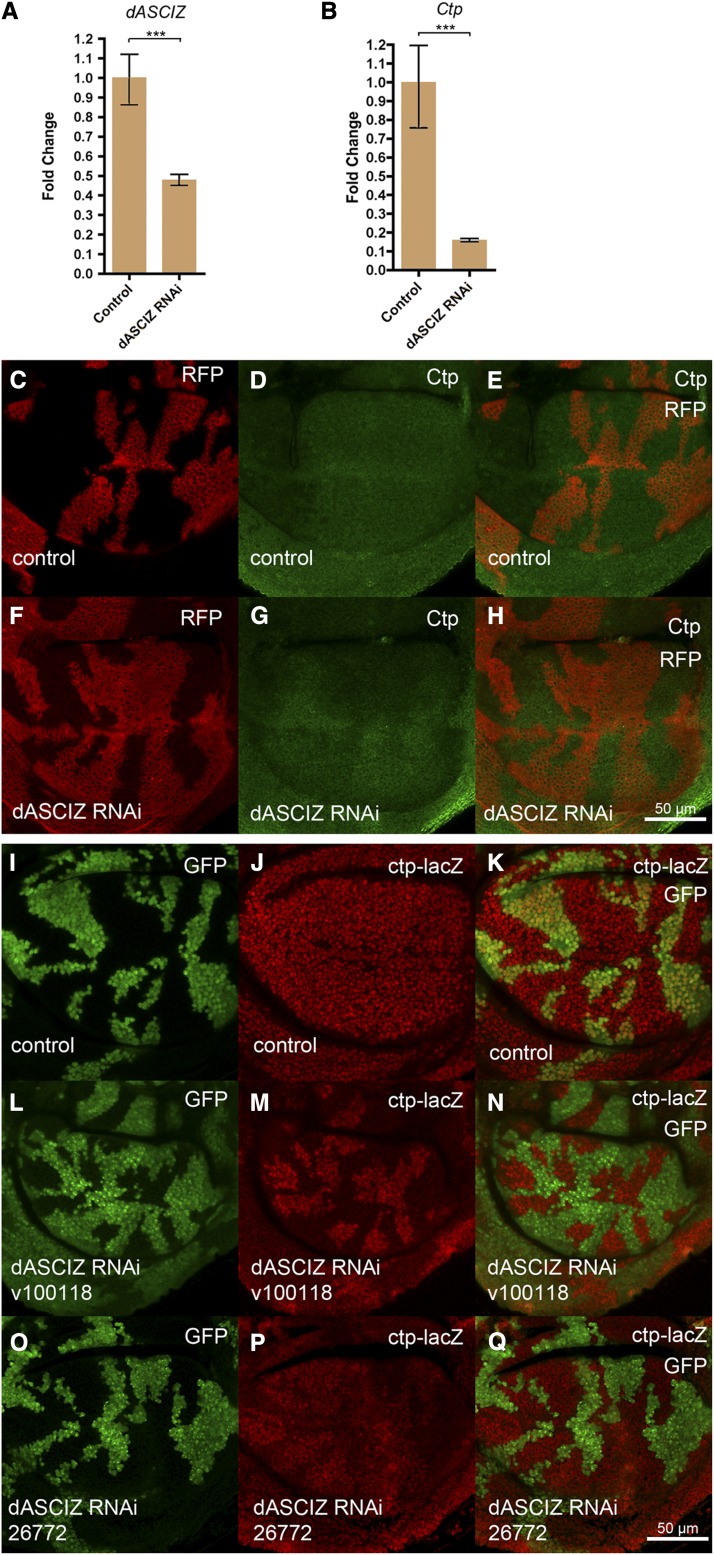
Given that dASCIZ genetically acts upstream of Ctp, we sought to directly determine if dASCIZ regulates Ctp expression. Global dASCIZ RNAi knockdown driven by tubulin–GAL4, but in the presence of the temperature-sensitive GAL4 antagonist GAL80 to prevent early larval lethality (see above), resulted in a significant reduction in dASCIZ mRNA (Figure 7A, P = 0.0008) that was associated with ∼10-fold decrease in Ctp mRNA (Figure 7B, P = 0.0003) 3 days after GAL80 inactivation. We also generated a polyclonal Ctp antibody to monitor Ctp protein levels in dASCIZ RNAi flip-out clones (Pignoni and Zipursky 1997). In control wings, staining for Ctp was even across the entire wing disc (Figure 7, C–E). In contrast, consistent with reduced Ctp mRNA levels after dASCIZ knockdown (Figure 7B), Ctp staining was reduced in dASCIZ RNAi clones (Figure 7, F–H).
Figure 7.
dASCIZ is required for Ctp expression. (A) qPCR for dASCIZ after global knockdown, fold change relative to control (error bars represent 95% CI, P = 0.0008 based on ΔΔCT value, normalized to actin). (B) Ctp mRNA levels are reduced in response to global dASCIZ knockdown (error bars represent 95% CI, P = 0.0003 based on ΔΔCT value, normalized to actin). (C–E) Control UAS–RFP-marked clones stained with Ctp antibody 3 days after clone induction. (F–H) Ctp protein levels detected using Ctp antibody in dASCIZ RNAi-expressing clones marked with UAS–RFP. (I–K) Control actin–GAL4 flip-out clones positively marked with GFP generated in the Ctp–lacZ enhancer trap background, stained with β-gal antibody 3 days after heatshock. (L–N) GFP-marked clones in the Ctp–lacZ background expressing dASCIZ RNAi stained with β-gal display reduced Ctp promoter activity compared to the surrounding wild-type tissue. (O and Q) dASCIZ knockdown with an alternate nonoverlapping RNAi in clones marked with GFP in the Ctp–lacZ background.
To determine if dASCIZ regulates Ctp expression at the transcriptional level, we generated dASCIZ RNAi flip-out clones in the Ctp–lacZ enhancer trap background. For this purpose, we used the previously generated P{w[+mC]=lacW}ctp[G0371] allele, which among the P-element insertions mapped to the Ctp promoter (Peter et al. 2002) appeared most strongly expressed in larval wing discs (Figure S7). Interestingly, whereas the GAL4 driver alone had no effect on Ctp–lacZ reporter activity (Figure 7, I–K), staining for Ctp–lacZ activity was mutually exclusive with staining for dASCIZ RNAi knockdown clones (marked with UAS–GFP, Figure 7, L–N) and qualitatively similar results were obtained with the alternate nonoverlapping dASCIZ RNAi line (Figure 7, O–Q). Thus, the dramatic reduction of Ctp promoter activity in dASCIZ knockdown cells strongly indicates that dASCIZ regulates Ctp levels through transcriptional activation of the Ctp promoter.
In contrast, dASCIZ overexpression in flip-out clones (shown with dASCIZ antisera in Figure S8B) did not further increase Ctp expression or Ctp–lacZ promoter activity (Figure S8, E–H). This is similar to previous findings in mammalian cells (Jurado et al. 2012a), where high-level retroviral ASCIZ overexpression did not increase DYNLL1 levels, presumably because all available ASCIZ binding sites in the Dynll1 promoter are bound by the endogenous protein.
Discussion
Here we have identified dASCIZ as a new transcription factor with important mitotic and developmental functions in Drosophila. Our data that loss of dASCIZ results in lower Ctp promoter activity and reduced Ctp protein levels, as well as similar phenotypes to loss of Ctp function that can be rescued by ectopic Ctp overexpression, demonstrate that dASCIZ exerts its developmental functions through regulation of Ctp expression. This molecular mechanism is remarkably similar to the recently reported functions of mammalian ASCIZ in the regulation of DYNLL1 expression, indicating that the ASCIZ–DYNLL1/Ctp axis has been strikingly conserved during metazoan evolution.
The mitotic and cell survival defects we have observed here for dASCIZ and Ctp RNAi knockdowns are consistent with previous reports linking Ctp to roles in embryonic cell survival and developmental apoptosis (Dick et al. 1996; Phillis et al. 1996; Batlevi et al. 2010). While here we have analyzed only defects resulting from the loss of these two proteins in the developing wing, we presume that knockdown of dASCIZ would also mimic Ctp loss of function phenotypes in other tissues. The mitotic block/delay, with accumulation of cells from metaphase through telophase (Figure 3 and Figure 4), is very similar to phenotypes reported for other dynein motor subunits in Drosophila (Morales-Mulia and Scholey 2005; Mische et al. 2008; Sitaram et al. 2012) and those observed for dynein heavy-chain knockdowns in our system (Figure S6). Although Ctp and DYNLL1 have also been linked to dynein-independent roles during mitosis in Drosophila (Wang et al. 2011) and mammalian cells (Dunsch et al. 2012; Kotak et al. 2013), the simplest explanation for the phenotypes observed here is that loss of dASCIZ or Ctp impairs cell division as a result of dynein-dependent mitotic spindle-related defects. In this sense, we suspect that the increased incidence of cell death during wing development after dASCIZ or Ctp RNAi knockdown is a consequence of the mitotic block, rather than reflective of a role in cell survival pathways per se, as failure to complete mitosis is a well-known cause of apoptosis (Quinn et al. 2001; Castedo et al. 2004).
A striking difference between the Drosophila RNAi phenotypes reported here and previously reported vertebrate RNAi or knockout phenotypes is that loss of dASCIZ causes pronounced, presumably dynein-dependent, mitotic defects, whereas developmental phenotypes in Asciz KO mice seem to be more restricted to relatively specific organogenesis and B-lymphopoiesis defects without overt mitotic deficits (Jurado et al. 2010; Jurado et al. 2012b). A possible explanation could be that Drosophila contains a single Ctp gene, whereas mammals contain two DYNLL-encoding genes, of which only Dynll1 is regulated by ASCIZ. Thus, DYNLL2, which is normally expressed at much lower levels than DYNLL1 and not affected by loss of ASCIZ (Jurado et al. 2012a), may provide a buffer for the mitotic or other dynein-related functions of DYNLL1 in ASCIZ-deficient mice. Nevertheless, it would be conceivable that loss of DYNLL1 may contribute to aspects of the Asciz KO phenotype through other, more subtle, effects on mitotic spindle functions. For example, proposed roles for the dynein complex in realigning mitotic spindles and cell polarity for asymmetric cell divisions in Drosophila neuroblasts (Wang et al. 2011) or mammalian cells (Dunsch et al. 2012; Kotak et al. 2013; Kiyomitsu and Cheeseman 2013) might provide a paradigm for the lack of lung buds in Asciz KO mice: Maybe respiratory precursors, which are present in these mice (Jurado et al. 2010), need to be rotated in some manner by dynein motors or another DYNLL1-specific effector for lung buds to emerge from the ventral foregut endoderm? While the answer to such questions is beyond the scope of the present study, our findings provide a solid basis for future studies to determine the extent to which ASCIZ via DYNLL1 regulates mitotic and dynein motor functions in mammalian cells, as well as for future investigations into developmental functions of the dASCIZ–DYNLL1/Ctp axis in the experimentally more amenable Drosophila model system.
Supplementary Material
Acknowledgments
We thank Sabine Jurado and Tony Tiganis for reading the manuscript and Eric Baehrecke for the UAS-Ctp strains. We are grateful to the Bloomington and Vienna Drosophila RNAi Center (VDRC) stock centers for Drosophila strains and to the Developmental Studies Hybridoma Bank (DSHB) for antibodies. Funded by Project Grants and a Senior Research Fellowship from the National Health and Medical Research Council of Australia (ID1009763, ID1025125, and ID1022469) and in part by the Victorian State Governments Operational Infrastructure Support Program to J.H.
Footnotes
Communicating editor: I. K Hariharan
Literature Cited
- Barbar E., 2008. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 47: 503–508. [DOI] [PubMed] [Google Scholar]
- Batlevi Y., Martin D. N., Pandey U. B., Simon C. R., Powers C. M., et al. , 2010. Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila. Proc. Natl. Acad. Sci. USA 107: 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M., Perfettini J. L., Roumier T., Andreau K., Medema R., et al. , 2004. Cell death by mitotic catastrophe: a molecular definition. Oncogene 23: 2825–2837. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C., Labbé J.-C., Goulding M., Marty T., Bowerman B., et al. , 2007. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J. Cell Biol. 179: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick T., Ray K., Salz H. K., Chia W., 1996. Cytoplasmic dynein (ddlc1) mutations cause morphogenetic defects and apoptotic cell death in Drosophila melanogaster. Mol. Cell. Biol. 16: 1966–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Dunsch A. K., Hammond D., Lloyd J., Schermelleh L., Gruneberg U., et al. , 2012. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J. Cell Biol. 198: 1039–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., et al. , 2003. A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Heierhorst J., Smyth I., Jurado S., 2011. A breathtaking phenotype: unexpected roles of the DNA base damage response protein ASCIZ as a key regulator of early lung development. Cell Cycle 10: 1222–1224. [DOI] [PubMed] [Google Scholar]
- Jurado S., Smyth I., Van Denderen B., Tenis N., Hammet A., et al. , 2010. Dual functions of ASCIZ in the DNA base damage response and pulmonary organogenesis. PLoS Genet. 6: e1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S., Conlan L. A., Baker E. K., Ng J. L., Tenis N., et al. , 2012a ATM substrate Chk2-interacting Zn2+ finger (ASCIZ) Is a bi-functional transcriptional activator and feedback sensor in the regulation of dynein light chain (DYNLL1) expression. J. Biol. Chem. 287: 3156–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado S., Gleeson K., O’donnell K., Izon D. J., Walkley C. R., et al. , 2012b The zinc-finger protein ASCIZ regulates B cell development via DYNLL1 and Bim. J. Exp. Med. 209: 1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanu N., Penicud K., Hristova M., Barnaby W., Lrvine E., et al. , 2010. The ATM cofactor ATMIN protects against oxidative stress and accumulation of DNA damage in the aging brain. J. Biol. Chem. 285: 38534–38542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. M., 2008. Dynein-independent functions of DYNLL1/LC8: redox state sensing and transcriptional control. Sci. Signal 1: pe51. [DOI] [PubMed] [Google Scholar]
- King S. M., Patel-King R. S., 1995. The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J. Biol. Chem. 270: 11445–11452. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Cheeseman I. M., 2013. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell 154: 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., Gonczy P., 2013. NuMA phosphorylation by CDK1 couples mitotic progression with cortical dynein function. EMBO J. 32: 2517–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L., Nakajima S., Oohata Y., Takao M., Okano S., et al. , 2004. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc. Natl. Acad. Sci. USA 101: 13738–13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B. A., Smogorzewska A., Mcdonald E. R., 3rd, Hurov K. E., et al. , 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166. [DOI] [PubMed] [Google Scholar]
- McNees C. J., Conlan L. A., Tenis N., Heierhorst J., 2005. ASCIZ regulates lesion-specific Rad51 focus formation and apoptosis after methylating DNA damage. EMBO J. 24: 2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T. L., Kilaru S., Turner F. R., Kaufman T. C., 2002. The centrosome is a dynamic structure that ejects PCM flares. J. Cell Sci. 115: 4707–4718. [DOI] [PubMed] [Google Scholar]
- Mische S., He Y., Ma L., Li M., Serr M., et al. , 2008. Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol. Biol. Cell 19: 4918–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Mulia S., Scholey J. M., 2005. Spindle pole organization in Drosophila S2 cells by dynein, abnormal spindle protein (Asp), and KLP10A. Mol. Biol. Cell 16: 3176–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T. P., Rubin G. M., 1994. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77: 371–379. [DOI] [PubMed] [Google Scholar]
- Nguyen-Ngoc T., Afshar K., Gönczy P., 2007. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 9: 1294–1302. [DOI] [PubMed] [Google Scholar]
- Oka H., Sakai W., Sonoda E., Nakamura J., Asagoshi K., et al. , 2008. DNA damage response protein ASCIZ links base excision repair with immunoglobulin gene conversion. Biochem. Biophys. Res. Commun. 371: 225–229. [DOI] [PubMed] [Google Scholar]
- Peter A., Schottler P., Werner M., Beinert N., Dowe G., et al. , 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3: 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillis R., Statton D., Caruccio P., Murphey R. K., 1996. Mutations in the 8 kDa dynein light chain gene disrupt sensory axon projections in the Drosophila imaginal CNS. Development 122: 2955–2963. [DOI] [PubMed] [Google Scholar]
- Pignoni F., Zipursky S. L., 1997. Induction of Drosophila eye development by decapentaplegic. Development 124: 271–278. [DOI] [PubMed] [Google Scholar]
- Prasad R., Liu Y., Deterding L. J., Poltoratsky V. P., Kedar P. S., et al. , 2007. HMGB1 is a cofactor in mammalian base excision repair. Mol. Cell 27: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn L. M., Herr A., Mcgarry T. J., Richardson H., 2001. The Drosophila Geminin homolog: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev. 15: 2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J. A., Tanenbaum M. E., Medema R. H., 2013. Systematic dissection of dynein regulators in mitosis. J. Cell Biol. 201: 201–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P., Garcia-Mayoral M. F., Martinez-Moreno M., Tarnok K., Schlett K., et al. , 2011a LC8 dynein light chain (DYNLL1) binds to the C-terminal domain of ATM-interacting protein (ATMIN/ASCIZ) and regulates its subcellular localization. Biochem. Biophys. Res. Commun. 414: 493–498. [DOI] [PubMed] [Google Scholar]
- Rapali P., Szenes A., Radnai L., Bakos A., Pal G., et al. , 2011b DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278: 2980–2996. [DOI] [PubMed] [Google Scholar]
- Sitaram P., Anderson M. A., Jodoin J. N., Lee E., Lee L. A., 2012. Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development 139: 2945–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Heierhorst J., 2005. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. BioEssays 27: 397–407. [DOI] [PubMed] [Google Scholar]
- Wang C., Li S., Januschke J., Rossi F., Izumi Y., et al. , 2011. An ana2/ctp/mud complex regulates spindle orientation in Drosophila neuroblasts. Dev. Cell 21: 520–533. [DOI] [PubMed] [Google Scholar]
- Williams J. C., Roulhac P. L., Roy A. G., Vallee R. B., Fitzgerald M. C., et al. , 2007. Structural and thermodynamic characterization of a cytoplasmic dynein light chain: intermediate chain complex. Proc. Natl. Acad. Sci. USA 104: 10028–10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura M., Kohzaki M., Nakamura J., Asagoshi K., Sonoda E., et al. , 2006. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell 24: 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.