Abstract
Gene duplication, expansion, and subsequent diversification are features of the evolutionary process. Duplicated genes can be lost, modified, or altered to generate novel functions over evolutionary timescales. These features make gene duplication a powerful engine of evolutionary change. In this study, we explore these features in the MADF-BESS family of transcriptional regulators. In Drosophila melanogaster, the family contains 16 similar members, each containing an N-terminal, DNA-binding MADF domain and a C-terminal, protein-interacting, BESS domain. Phylogenetic analysis shows that members of the MADF-BESS family are expanded in the Drosophila lineage. Three members, which we name hinge1, hinge2, and hinge3 are required for wing development, with a critical role in the wing hinge. hinge1 is a negative regulator of Winglesss expression and interacts with core wing-hinge patterning genes such as teashirt, homothorax, and jing. Double knockdowns along with heterologous rescue experiments are used to demonstrate that members of the MADF-BESS family retain function in the wing hinge, in spite of expansion and diversification for over 40 million years. The wing hinge connects the blade to the thorax and has critical roles in fluttering during flight. MADF-BESS family genes appear to retain redundant functions to shape and form elements of the wing hinge in a robust and fail-safe manner.
Keywords: MADF, BESS, Wnt/Wg, evolution, development
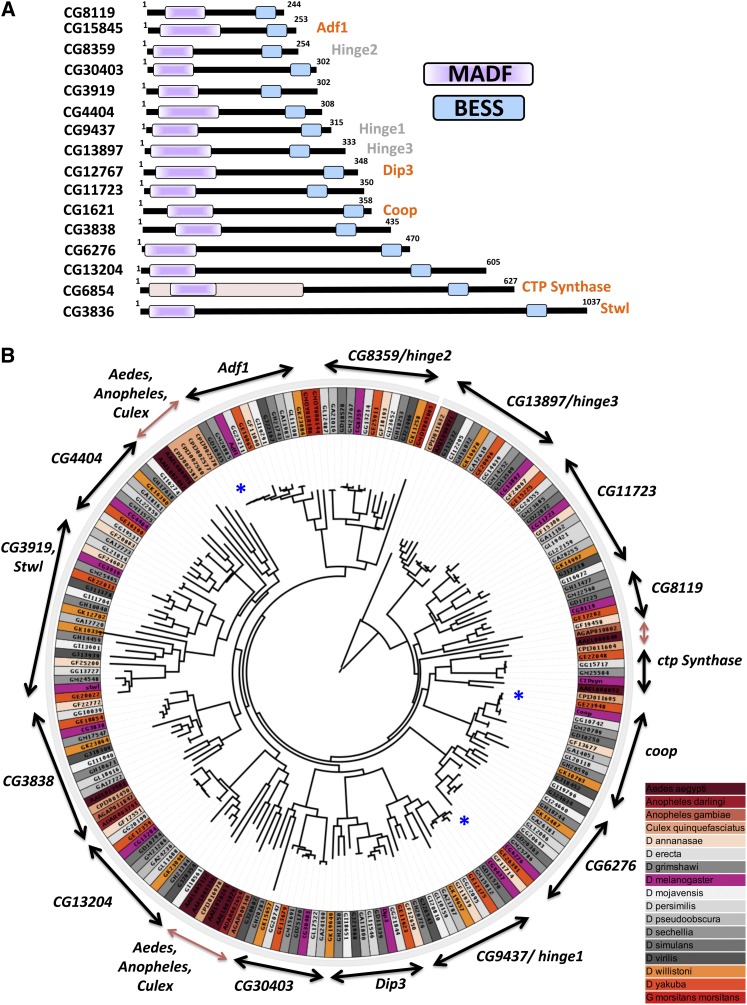
THE MADF-BESS gene family in Drosophila melanogaster consists of 16 transcriptional regulators (Figure 1A), coded by 16 discrete genes. Proteins coded by all 16 members contain an N-terminal Myb-SANT like in Adf (MADF) followed by a C-terminal BEAF, Su-Var(3–7), Stonewall like (BESS) domain (Bhaskar and Courey 2002). Of the 16, most proteins range in size from 200 to 500 amino acids, with three larger-than-600 amino acids. Of the three larger members, CTP-synthase (CG6854; Liu 2010) has gained novel domains that impart additional functions to the encoded protein. The CTP-synthase gene codes for three splice variants of which two remain in the cytoplasm, while the other, isoform A, is nuclear (Azzam and Liu 2013). Excluding CTP-synthase, the 16 genes appear to have only two (MADF, BESS) functional domains. The MADF-BESS family is also in a broader sense a subgroup of the individual, independent MADF and BESS family genes, where both MADF and BESS domains are coded by a single continuous polypeptide.
Figure 1.
The D. melanogaster MADF-BESS family codes for 16 members that are a consequence of gene duplication. (A) MADF-BESS family in D. melanogaster consists of 16 proteins coded by individual genes. The 16 proteins, represented in the figure, contain an N-terminal DNA-binding Myb/SANT like in Adf (MADF) domain and a C-terminal BEAF-32, Stonewall, Su (var) 3-7 homology (BESS) domain. MADF and BESS domains tend to be found together in a single polypeptide chain. Proteins labeled in orange have been characterized, while those in gray are the subject of this study and been named hinge genes based on their loss-of-function phenotype. (B) Maximum-likelihood phylogenetic tree of all MADF-BESS genes in dipterans. Branch lengths are proportional to mean substitutions per site. Orthologs of the 16 MADF-BESS genes cluster separately with the corresponding D. melanogaster gene (labeled on two-sided arrows), indicating that the duplication in the family occurred before the divergence of drosophilids. MADF-BESS genes from Culex, Anopheles, and Glossina morsitans have fewer orthologs and cluster separately (brown arrow) for the most part, indicating that, in their case, expansion in MADF-BESS was independent from that in drosophilids. The genes for different sequenced dipterans are color-coded to bring out this feature. The blue asterisk marks the genes that show short branch lengths and thus minimal sequence divergence.
The architecture of the domains, sequence identity, and the lack of additional defined domains in the polypeptide sequences suggest that the MADF-BESS family members may have similar or identical function. Adf1, a well-studied gene in the family, has been implicated as a transcriptional activator that contains a TAF-like binding motif (England et al. 1992; Cutler et al. 1998). Dorsal interacting protein 3 (Dip3) appears to be a co-activator in NF-κB/Twist function (Bhaskar and Courey 2002; Ratnaparkhi et al. 2008) and also localizes to the heterochromatic chromocenter; while Co-repressor of Pangolin (Coop) is a negative regulator of Wg/Wnt signaling (Song et al. 2010) and stwl is required for germ-cell development (Clark and McKearin 1996) and germ stem-cell maintenance (Maines et al. 2007), acting as a repressor. The MADF domain in Stwl is implicated in chromatin remodeling and histone modification (Boyer et al. 2002). Stonewall is also associated with heterochromatin and colocalizes with HP1 (Yi et al. 2009). The remaining 11 genes have not been named and are predicted to be transcriptional regulators with unknown functions.
The retention of these 16 functional genes in the D. melanogaster genome suggests important roles for this family. If the family members do indeed have similar functions, then functionally they may be completely or partially redundant. Since most genetic studies or even large-scale loss-of-function screens knock down only one gene at a time, functional roles for multiple, redundant genes supporting a single function tend to go undiscovered.
In this study we examine the MADF-BESS family using tools of phylogeny and genetics. By inferring the phylogeny of all MADF-BESS genes in Diptera, we show that these family members probably arose from a common ancestral gene. We have dissected out roles for members of this family to understand if the family members represent a redundant, partially redundant, or a divergent set of genes. Using RNA interference with multiple simultaneous knockdowns of genes, enhancer/suppressor genetics, epistasis, and heterologous transgenic rescue experiments were used to test for functional redundancy and overlapping functions.
We discovered that the MADF-BESS family contains a subset of genes that are involved in wing-hinge patterning that show significant functional redundancy. At least four genes in the family have common roles in patterning the wing hinge by modulating Wingless (Wg)/Homothorax (Hth) expression, while other genes in the family either play supporting roles or retain some functional aspects of the core, hinge-patterning function. The retention of multiple members for tens of million of years for robust development of the wing hinge and thereby the ability of the animal to fly underscores the importance of the MADF-BESS family.
Materials and Methods
Bioinformatics analysis
The number of MADF, BESS, and MADF-BESS domains were counted in different species in the animal kingdom based on information available in databases such as InterPro (Hunter et al. 2012), UniProt (Magrane and Consortium 2011), Flybase (Marygold et al. 2013), Flymine (Lyne et al. 2007), SMART (Schultz et al. 1998), 12 Drosophila genomes (Clark et al. 2007), and ORTHODB (Waterhouse et al. 2013). MADF-BESS domain protein sequences were downloaded from UniProt. The MADF domain was predicted using signatures by ProSite, SMART, and Pfam in InterPro. BESS domains were predicted using Prosite (Sigrist et al. 2002) and Pfam in InterPro.
Using FlyBase for Drosophila proteins and Vectorbase (Megy et al. 2012) for proteins from Aedes, Anopheles, and Culex species, we obtained coding sequences corresponding to each protein. We then removed multiple isoforms corresponding to the same gene, retaining a single isoform. Multiple sequence alignment of the genes was performed using MUSCLE (Edgar 2004). Phylogenetic Tree was constructed using RaxML 7.2.7 (Pfeiffer and Stamatakis 2010) at the CipRES Science Gateway (Miller et al. 2011) with default parameters for DNA. A total of 400 bootstrap iterations were performed and the best scoring maximum-likelihood tree was obtained. Similarly, a tree for the 16 genes corresponding to D. melanogaster was obtained by performing multiple sequence alignment with MUSCLE (Edgar 2004), and a maximum-likelihood tree with RaxML was obtained with the same parameters as for the dipteran MADF-BESS gene tree.
Drosophila husbandry
All flies were raised at 25° in standard corn meal agar. Crosses were set up at 25°. The females of the F1 progeny were screened for the phenotypes in all cases.
RNA interference lines and mutants
MADF-BESS RNAi lines were procured from the Vienna Drosophila RNAi Center (VDRC), and the Transgenic RNAi Project (TRiP) lines were procured from Bloomington Drosophila Stock Center (BDSC) and the National Institute of Genetics (NIG) in Japan. The RNA interference (RNAi) lines used for this study are listed below in the following format: gene name(s) {line #} genotype. Line numbers 4278, 30141, 3204R-2, 8119R-2, and 39733 had 19, 10, 3, 2, and 1 off-targets based on parameters defined in NEXT-RNAi (Horn et al. 2010). Lines that do not show off-target effects were used for our primary experiments.
VDRC:
The lines obtained from the VDRC were the following: CG8359(hng2) {#105177} P{KK103584}VIE-260B; CG13897(hng3) 108487 P{KK111648}VIE-260B; CG13897 {#39733} W[1118];{GD7674}v39733; stwl {#102848}W[1118];P{KK105453}VIE-260B; coop {#14692} w[1118];P{GD6554}v14692; CG3838 {#106551} P{KK112405}VIE-260B; adf-1 {#4278} w[1118]; P{GD1358}v4278; dip3 {#107803} P{KK111529}VIE-260B; CTP synthase {#12762} w[1118];P{GD4740}v12762; CG11723 {#39723} w[1118]; {GD7373}v39723. CG6276 {#30141} w[1118];P{GD11876}v30141; CG4404 {#48183} w[1118];P{GD16746}v4813; CG3919 {#10978} w[1118];P{GD11108}v3895/TM3; CG3919 {#109787} w[1118];P{KK106860}VIE-260B; and CG9437 (hng1) {#100101} P{KK103642}VIE-260B.
NIG:
The lines obtained from the NIG were the following: CG13204 {#13204R-2}W1118; 13204R-2 and CG8119 {#8119R-2} W1118. 8119R-2.
BDSC:
The lines obtained from the BDSC were the following: CG3919 {#33355} y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS00226}attP2; mirror {#31907} y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02196}attP2; CG11723 {#29349} y[1] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.JF02511}attP2; teashirt {#35030} W[1118];P{KK106860}VIE-260B; extradenticle {#34897} y1 sc v1; P{TRiP}attP2; jing {#27024} y1 sc v1; P{TRiP}attP2. tiptop {#35812} y1 sc v1; P{TRiP}attP2. dpp {#33618} y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8]=TRiP.HMS01527}attP2; and UAS mCherry {#35787} y[1] sc[*] v[1]; P{y[+t7.7] [+t1.8]=UAS-mCherry.VALIUM10}attP2.
Deficiencies for hng1 [Df(BSC484); BDSC 24988] and hng2 [Df(BSC306); BDSC 25010] and mutants for vestigial (vg1; BDSC 432), pangolin (UAS-pan-dTCFΔN; BDSC 4784), and nubbin [nub(E37); BDSC 8856] were obtained from the BDSC. UAS-rotund, UAS-jing, and UAS-hth were obtained from the BDSC, the National Centre for Biological Sciences (Bangalore, India) (NCBS) stock center, and the Shashidhara Lab (Indian Institute of Science Education and Research, Pune, India), respectively. UAS-mCherry lines were used to test dilution effects of UAS in the crosses.
Transgenic lines generated
CG9437 cDNA subcloned from the Gold cDNA collection was cloned into pUASp vector between KpnI and NotI sites. pUASt clones for CG13204, CG11723, CG3838, CG6854, exd, and Dip3 were obtained from the Drosophila Genomics Resource Center. These were part of the proteomics collection and expressed proteins tagged with HA and FLAG at the C terminus. Fly injections were done at the National Centre for Biological Sciences (NCBS), Bangalore, Transgenic Fly Facility to generate multiple stable lines on chromosomes II and III for all these constructs. Expression of transgenes was confirmed by expressing these lines using patched-Gal4 (ptc-Gal4) and staining wing imaginal discs with anti-HA antibody. CG3838 and CG6854 did not express and were not used for further experiments.
Gal4 drivers
MS1096 Gal4, which expresses strongly in the dorsal region of the wing disc and weakly in the ventral regions, was used for most of the experiments. MS1096 is a Gal4 P-element insertion in the Beadex/dLMO Enhancer (P{GawB}BxMS1096) (Guillen et al. 1995; Milán et al. 2004). In addition, we utilized da-Gal4, vg-Gal4, ap-Gal4, Sd-Gal4, omb-Gal4, MS209-Gal4, and ptc-Gal4 that express in the wing disc for characterization of our lines.
Flight assay
Flight tests were performed using modified cylinder drop assay (Banerjee et al. 2004). Three-day-old flies were dropped into a 1-m long cylinder, one fly at a time. Flies that fell through the cylinder were scored as “nonfliers” and those that flew and sat on the walls of the cylinder were scored as “fliers.”
Wing measurements and statistical analyses
The wings were detached from the flies and mounted in clove oil. The images were captured using Leica Microsystems Light Microscope. Five wings for every genotype were used to mark the alula and wing boundaries and subsequently measured using Image J software. Wing area includes the proximal and distal wing together. For wing area measurements, wings with folds were avoided, and, when unavoidable, the area of the folded section was added to the total. All graphs were made in Sigma Plot, and statistical analysis was done using Student’s t-test in GraphPad.
Immunostaining and in situ hybridization
Wing discs were dissected in PBS and fixed with 4% paraformaldehyde in 1× PBS for 20 min at room temperature. They were blocked in 1× PBS, 2% BSA, and 0.1% Triton for 1 hr; incubated with the primary antibody (anti-Wg 1:1000, anti-Hth 1:500 and anti-GFP 1:1000) overnight at 4°; washed 4× for 10 min in 1× Phosphate Buffer Saline containing 0.1% Triton × and incubated with the appropriate fluorescent secondary antibody for 1 hr at room temperature in the dark. The wing discs were then washed and mounted in Antifade. Anti-Hth was kindly provided by L. S. Shashidhara. Anti-Wg was purchased from the Developmental Studies Hybridoma Center. Anti-GFP (A11122) was obtained from Invitrogen. Images were taken on Zeiss 710 LSM confocal microscope at 20× and subsequently processed using Image J software. In situ hybridization in larvae was carried out as described in Kraut et al. (2001). Digoxigenin-labeled sense and antisense probes for hng1 and hng2 were generated against 300- to 524-bp and 350- to 570-bp genomic regions, respectively. Anti-Dig was obtained from Roche and used at a dilution of 1:1000. Detection was done using nitroblue tetrazolium salt/5-bromo-4-chloro-3-indolyl phosphate stock solution.
Results
Gene duplication and expansion
A striking feature of the MADF-BESS family of proteins is their conserved protein architecture: all members have an N-terminal MADF domain and a C-terminal BESS domain (Figure 1A). The proteins also show sequence similarity and/or identity for the two domains (Supporting Information, Figure S1A), indicating that the proteins have common evolutionary origins. In the D. melanogaster genome, with the exception of stwl and CG3919, all other members of this family are present in distant locations on multiple chromosomes, pointing to a mechanism for duplication and expansion that is not via a localized duplication as seen for the bithorax complex (Lewis 1978), but instead a possible RNA or transposon-mediated duplication event (Casola et al. 2007). Between the MADF and BESS domains is a “linker” region that ranges from 100 to 600 amino acids. With the exception of CTP synthase, this linker region lacks any recognized functional domain. The roles of the linker polypeptide sequences, if any, are unknown. A phylogenetic tree analysis with all members of the MADF-BESS family in D. melanogaster showed that these genes are related (Figure S1B), suggesting a gene duplication and/or expansion event followed by maintenance of these seemingly redundant genes in the animal.
An expanded phylogenetic tree of all MADF-BESS-containing genes in dipterans showed that the orthologs of the MADF-BESS genes in drosophilids cluster together (Figure 1B) as would be expected if the gene duplication had happened before Drosophila speciation and the genes had diverged from each other. In contrast, the MADF-BESS genes in Culex and Anopheles did not always have equivalent counterparts in drosophilids: their genes clustered separately with each other and were distant from the MADF-BESS genes in drosophilids. Three major clusters for MADF-BESS genes were observed in Culex and Anopheles: The first cluster was close to CG8119; the second was near CG4404, and the third was close to CG13204 (Figure 1B). A small cluster was seen close to CG3838. Thus, the genes in Culex and Anopheles were closer to each other than to genes in other drosophilids. This indicates that gene duplication and expansion in drosophilids was separate from that in other dipterans.
Phylogenetic analyses also showed that the MADF-BESS-containing genes are highly conserved as evident from the distances on the phylogenetic tree within the “melanogaster group” [the melanogaster group contains Drosophila simulans (GD), Drosophila yakuba (GE), Drosophila ananassae (GF), Drosophila erecta (GG), Drosophila sechellia (GM), and Drosophila melanogaster (CG)]. For example, in Figure 1B, the blue asterisk marks strongly conserved sequences for Adf1, hinge1, and Coop where the branches are short and cluster near each other. In comparison, CG8119 and CG4404 DNA sequences do not cluster and are not as strongly conserved (Figure 1B).
Lineage-specific expansion of MADF and BESS domains
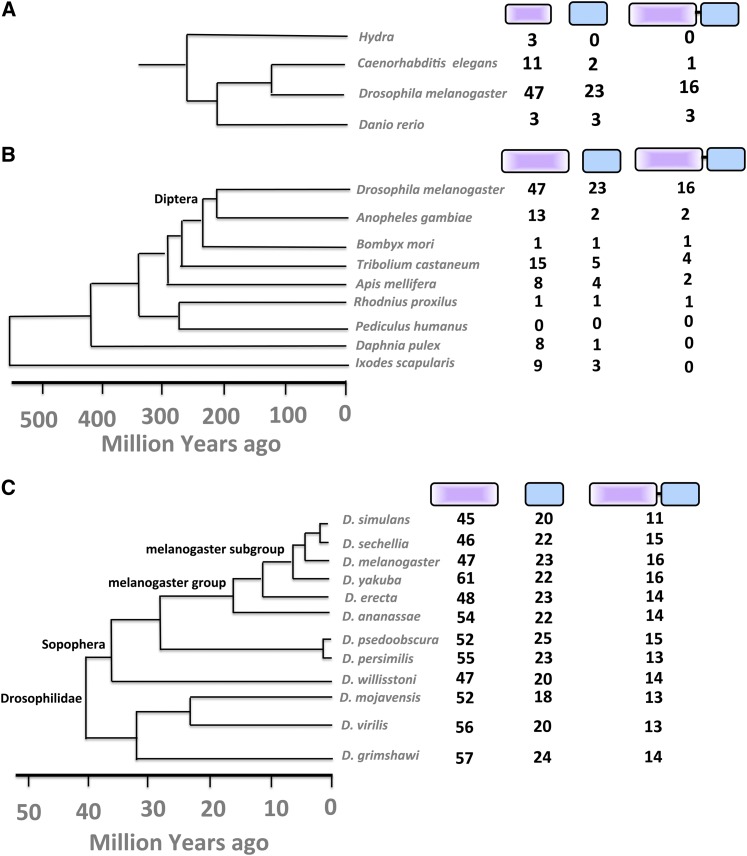
To determine if the MADF and BESS domains have expanded in a specific lineage, the numbers of individual MADF and BESS domains were counted in a few representative organisms where genome sequences were available. A single species was taken as representative for each genera, and the MADF and BESS domains, as defined by the SMART (Schultz et al. 1998), Pfam (Finn et al. 2010), Prosite (Sigrist et al. 2002), and ORTHODB (Waterhouse et al. 2013) databases, were counted. The analysis showed an overrepresentation of both MADF and BESS domains in invertebrates with an unusual increase in numbers for D. melanogaster (Figure 2). Analysis of the phylogeny of dipterans (Figure 1B) and of sequenced Drosophila species indicated that the expansion probably occurred in an ancestor common to the Drosophila lineage, >40 million years ago. From the InterPro database, we found that the MADF domain alone occurs in 908 curated proteins whereas MADF together with a BESS domain occurs in 353 proteins. The most frequent other combination was the presence of multiple MADF domains in the same protein. All other domain combinations such as multiple MADF with a single BESS domain or vice versa were significantly less frequent. The BESS domain was thus found to be most strongly associated with the MADF domain, with 16 of the 20 three BESS-domain-containing genes in D. melanogaster being exclusively associated with the MADF domain, with no other intervening domains in the linker region.
Figure 2.
MADF and BESS domains are expanded specifically in the Drosophila lineage. (A) MADF and BESS protein domains, when counted in representative members of the animal kingdom, indicate expansion in Drosophila lineage. (B) In arthropods, the number of individual MADF and BESS domains coded by the Drosophila genome is higher than in other sequenced species. The dipteran group is marked. (C) The expansion is dramatic in all members of the Drosophila lineage, confirming that that the expansion occurred in a common drosophilid ancestor. The Drosophilidae and the melanogaster group, which are referred to in the text, are marked.
The 353 proteins containing a MADF-BESS domain were also analyzed on the basis of their distribution in different species. We found that 225 are present in dipterans (National Center for Biotechnology Information taxonomy ID 7147) and, furthermore, of these 225 proteins, 197 proteins are in drosophilids. As multiple proteins may arise from a single gene, we also analyzed the distribution of MADF-BESS domains at the gene level. We found that there are 168 MADF-BESS genes across drosophilids and a further 27 identified MADF-BESS genes from other dipterans. Multiple annotated isoforms for almost all genes were found present only in D. melanogaster.
Lineage specific expansion (LSE) is defined as the proliferation of a specific protein family in a genera/species, relative to its sister lineage, with which it is compared (Clark et al. 2007). LSE for the MADF-BESS family (Lespinet et al. 2002) in the Drosophila lineage was confirmed by phylogenetic analysis and by counting the number of MADF and BESS domain family members (InterPro). Of the 1576 MADF domains contained in the databases, 828 were found in Diptera. Furthermore, 755 of these were found in Drosophilidae. At the gene level, we found 626 genes in Drosophilidae containing a MADF domain. By doing a similar analysis for the BESS domain, we found that, of the 644 proteins containing a BESS domain, 388 are in Diptera and 356 of these are in Drosophilidae. At the gene level, we found 311 genes in Drosophilidae with a BESS domain. As mentioned above, among these, 225 proteins in Diptera and 197 proteins in Drosophilidae have a N-terminal MADF and a C-terminal BESS domain with no other domains in the intervening sequence. The number of MADF-BESS genes in the 12 sequenced Drosophila species ranges from 13 to 16 except in the case of D. simulans, which has only 11 MADF-BESS genes. The smaller number of genes in D. simulans may reflect genes that have lost the MADF or BESS domain or incomplete annotation that would have eliminated them from this study (Figure 2C).
Aedes aegypti and Culex quinquefasciatus contain 10 and 9 MADF-BESS genes, respectively, and Anopheles gambiae and darlingi contain only 2 genes each. Glossina morsitans (tsetse fly) contains three MADF-BESS genes. From the phylogenetic tree we can see that despite Culex and Aedes (Culicinae) having a comparable number of genes to Drosophila species, the genes from Culicinae are more closely related between themselves than to genes in Drosophilidae. This indicates that duplication/expansion of genes in Culicinae was separate from that in Drosophilidae.
The MADF and BESS domains, by themselves, appear to be a result of LSE. It follows that the MADF-BESS family of 16 genes in D. melanogaster may be consequence of gene duplication and subsequent expansion in the Drosophila lineage.
Members of the MADF-BESS family pattern the wing hinge
To understand the functions of the MADF-BESS family in Drosophila, we decided on a loss-of-function approach, using the UAS-Gal4 system (Brand and Perrimon 1993; Duffy 2002) to reduce the transcript levels of every member of this family using double-stranded RNAi (Kennerdell and Carthew 2000; Zamore et al. 2000; Dietzl et al. 2007). Drosophila lines available in public stock centers were procured (see Materials and Methods), and a screen was conducted using eye- and wing-specific drivers. The primary result of this screen was that knockdown of three of the unknown MADF-BESS domain genes produced a phenotype in the wing hinge (Figure 3) with multiple wing-specific Gal4 Driver lines. The genes were CG9437, CG8359, and CG13897. Based on the phenotype, we have named these genes hinge1 (hng1), hinge2 (hng2), and hinge3 (hng3), respectively, and will refer to them as such in the subsequent text. A fourth gene from the family, stonewall (stwl), also produced a hinge phenotype (Brun et al. 2006). For the remainder of the text, we also abbreviate a RNAi line specific for the gene by adding an “i” at the end of the gene name. For example, a UAS-hinge1 RNAi line will be abbreviated as hinge1i or hng1i.
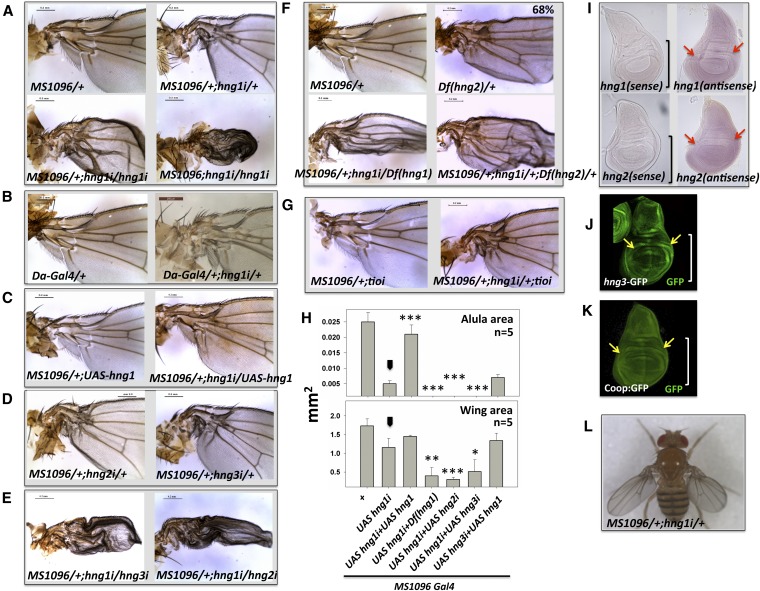
Figure 3.
Members of the MADF-BESS family (CG9437/hng1, CG8359/hng2, and CG13897/hng3) have roles in the development of the wing hinge. (A) Reduction of CG9437 transcripts in the wing-imaginal disc, by expressing UAS-CG9437 RNAi line, in the MS1096 expression domain leads to wing-hinge defects in the adult fly. Defects include a reduced/mispatterned alula, a bent hinge, and a disorganized costa region. The phenotype is 100% penetrant and is dose dependent with an increased knockdown leading to a stronger phenotype that also affects the more distal wing blade. (B) Reduction of hng1 transcripts in the more ubiquitous daughterless (Da) expression domain also lead to wing-hinge defects, indicating specific roles for hng1 in the hinge. (C) The defects can be rescued by co-expression of UAS-hng1 in the same expression domain. Expression of UAS-hng1 by itself does not affect normal wing development. (D) Two other genes (CG8359/hng2 and CG13897/hng3) in the family also show a similar phenotype on knockdown. The genes have been named as the hinge genes based on their loss-of-function phenotype. (E) Double knockdowns of hng1+hng2 or hng1+hng3 in a single dose each mimic the phenotypes seen in an increase dose of hng1 knockdowns. (F) Deficiency in the 85B1-85C2 genomic regions removes hng2 completely, and 68% of flies show the wing-hinge phenotype. This, when combined with hng1 (MS1096/+; UAS-hng1-RNAi/+) knockdown, gives enhancement in the Deficiency phenotype. Deficiency in the 57C3-57C7 genomic region removes hng1 completely and also shows enhancement when combined with hng1 (MS1096/+; UAS-hng1-RNAi/+) knockdown. (G) The VDRC KK lines used in this study are inserted into a single site in the regulatory region of the tio locus. A knockdown of tio does not enhance the hng1i phenotype, arguing against a contribution of the tio locus to the observed phenotype. (H) Parameters such as wing size (mm2) and alula size (mm2) are measured to quantify the phenotype in this and subsequent experiments. The phenotype (MS1096/+; UAS-hng1 RNAi/+) is enhanced in the presence of a Deficiency in the CG9437/hng1 locus and is rescued by expressing UAS-hng1. Arrowhead indicates the MS1096/+; UAS-hng1/+ line used as a control for statistical analyses. *P < 0.05, **P < 0.01, and ***P < 0.001. (I) In situ hybridization against hng1 and hng2 transcripts confirms that hng1 and hng2 are expressed in wing imaginal discs. The brackets mark the wing pouch with the red arrows marking part of the hinge-forming region. (J) Expression pattern of hng3 as shown by anti-GFP staining of an enhancer trap line (YB0086DE) in wing imaginal discs. The bracket marks the wing pouch with the yellow arrows marking part of the hinge-forming region. (K) Expression pattern of Coop (CG1621) as shown by anti-GFP staining protein trap line in wing imaginal discs. The bracket marks the wing pouch with the yellow arrows marking part of the hinge-forming region. (L) The hng1i animal has a bent wing hinge, cannot fold its wings over the abdomen normally, and holds out its wings. The animals can flap their wings but cannot fly.
Expression of a UAS-RNAi line driven under the control of MS1096-Gal4 at 25° resulted in a wing-hinge phenotype for hinge1 (hng1i) (Figure 3A). Similar phenotypes were seen with other wing-specific Gal4 drivers such as ptc, sd, vg, and sal (Figure S2A). The primary phenotype with MS1096-Gal4 was a bend in the costa region of the wing hinge and a reduction in the size of the hinge, with a dramatic effect on the patterning and size of the alula with respect to wild type. The phenotype was dose dependent, with drastic reduction in alula and wing-blade size with an increase in UAS or Gal4 dosage. When we used a more ubiquitous driver such as daughterless-Gal4 (da-Gal4), the primary phenotype was still a hinge defect (Figure 3B), indicating specific roles for hng1 in proximal wing development. The hng1i phenotype can be rescued to a significant extent by expressing UAS-hng1 in the same domain (MS1096) where hng1 is knocked down (Figure 3C). The rescue of the phenotype by UAS-hng1 also demonstrates that hng1 can directly affect the hinge phenotype, making it very unlikely that there are significant off-target RNAi effects or effects due to the insertion of the RNAi lines close to some other gene affecting wing development. We checked for the Gal4 dilution effect by crossing MS1096/+; hng1i/+ to UAS-mCherry, but the hng1i phenotype was unchanged. Overexpression of UAS-hng1 by itself did not perturb normal wing patterning (Figure 3C).
Phenotypes similar to hng1i were seen with knockdowns of hng2 and hng3 (Figure 3D) using gene-specific RNAi lines expressed in the MS1096 expression domain. hng1, hng2, and hng3 thus appeared to have critical roles in wing-hinge development. If these three genes are indeed functionally equivalent, as suggested by similar protein sequences in the MADF and BESS domains, it was expected that double knockdowns would enhance the initial phenotype. Simultaneous knockdown of hng2 and hng1 or hng3 and hng1 leads to enhanced wing-hinge phenotypes (Figure 3E). The RNAi lines used for knockdowns in the hng family are specific for their target messenger RNA (mRNA) and do not have off-targets (Materials and Methods) as defined by NEXT-RNAi (Horn et al. 2010), making it unlikely that the observed phenotypes are due to off-target effects. This result is balanced by the observation (Figure S2B) that the hinge phenotype of MS1096/+;UAS-hng3i/+ is not rescued by overexpression of hng1 in the same expression domain. Thus hng1 and hng3 are involved in the same developmental process, but are not functionally equivalent.
One concern in the use of targeted insertions is that the inserted transgenes are all located in same locus. This would give rise to the possibility of the observed phenotypes being an artifact of the insertion rather than a phenotype associated with the hypomorphic, double-stranded RNA-mediated knockdown of the hng genes. In the case of the VDRC KK (phiC31 RNAi Library) lines, the transposons are inserted in the regulatory region of the tiptop (tio) locus. The dose-dependent increase in the hng1 phenotype (Figure 3A) as well as the rescue by a transgene (Figure 3C) indicates that the effect is due to reduction of transcript rather than the insertion at the tio locus. In the absence of other functional knockdown lines for hng genes from the TRiP and NIG collections, we further tested the effect of alternate reagents, namely deficiencies (Df) in the hng1 and hng2 loci, to support our observations. Specific deficiencies in 2R (57C3-57C7) and 3L (85B1-85C2) genomic regions completely remove hng1 and hng2, respectively. Interestingly, heterozygous Df (hng2)/+ flies show a wing-hinge phenotype (Figure 3F), with a penetrance of 68% (38/56 animals), strongly supporting the RNAi loss-of-function phenotype for hng2. The Df (hng2)/+ hinge phenotype closely resembles the hng phenotypes with a disorganized, mis-patterned hinge and a reduced alula (compare Figure 3F to Figure 3D). This result strongly supports the conclusion derived from the RNAi experiments that hng2 is indeed required for normal hinge development. When combined with MS1096;UAS-hng1i, Df (hng1) and Df (hng2) enhanced the wing-hinge phenotype (Figure 3F), again arguing against a general insertion artifact. Further evidence against an insertion-based phenotype is as follows. The KK lines that we use are homozygous viable and do not have a hinge phenotype by themselves either as homozygous (KK-insert/KK-insert) or heterozygous (KK-insert/+). The percentage of all hinge phenotypes is small—∼6% of the total KK lines for ∼65 KK lines tested in a recent screen for interactors of folded gastrulation with MS1096-Gal4 driver (Ratnaparkhi 2013). The phenotype is not an artifact of a combination of any Gal4 line with the KK insert as some of the weaker Gal4 lines that we tested, namely engrailed-Gal4, hth-Gal4, pannier-Gal4, and tsh-Gal4, did not give a wing-hinge phenotype. CG3838, dip3, and CG3919 KK lines did not give a phenotype with MS1096-Gal4, arguing against an artifact that is a combination of any Gal4 with a KK insert. As an additional test, we also reduced tio transcripts using an RNAi line in the hng1i background (Figure 3G). The hng phenotype was unchanged, with the alula size for MS1096/+; hng1i/+; tioi/+ (0.005 ± 0.0005 mm2) similar to that of MS1096/+; hng1i/+ (0.005 ± 0.0009), arguing against a role for tio in hng function. Thus, the KK insertion in the hng1 RNAi line does not appear to contribute to the hng1i phenotype.
We measured the size (Figure 3H) of the alula and wing blade (including the proximal hinge). These measurements allowed us to quantitatively assess the enhancement and suppression of phenotypes in the wing hinge and the wing blade. To confirm expression of the hinge genes in the wing imaginal disc, which gives rise to the adult wing, we visualized transcripts of hng1 and hng2 using in situ hybridization (Figure 3I). hng1 and hng2 are expressed in the third instar larval wing disc, including regions that form the wing hinge (Figure 3I). A GFP enhancer trap line for CG13897 (hng3) shows expression in the hinge (arrows) as well as in the wing pouch (Figure 3J). A GFP protein trap line for Coop also shows expression in similar regions (Figure 3K).
The hng1i fly tends to keep its wings apart (Figure 3L) and cannot fold its wings over the abdomen. The wings thus remained “held out,” away from the body. The wing hinge functions to connect the wing blade to the thorax and has essential roles in fluttering of the wing during flight and in flexing the wing over the abdomen at rest. Experiments using a high-speed camera indicated that the hng1i females (MS1096/+; UAS-hng1i/+) could flap their wings but not fly, based on experiments using a cylinder drop assay (see Materials and Methods). hng2i and hng3i knockdown flies were also found to be flightless.
hinge1 interacts with genes of wing-hinge gene regulatory network
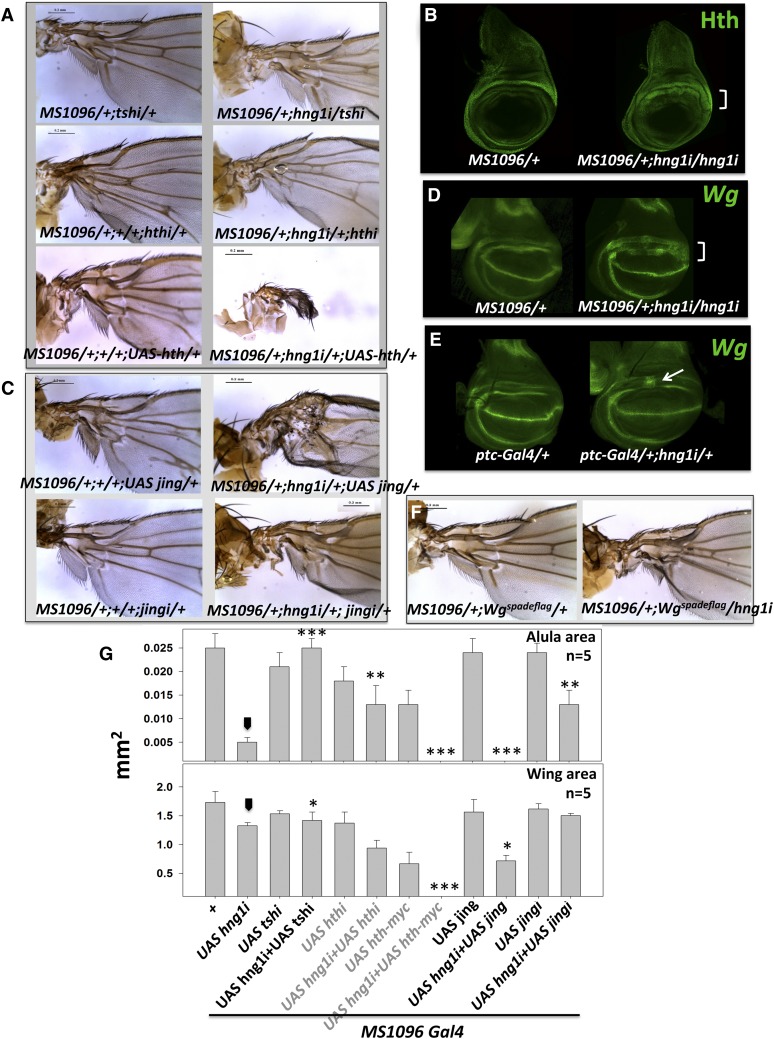
Since flies lacking hng1 have a wing-hinge defect, we tested for genetic interactions between hng1 and genes that play important roles in wing-hinge development. A central pathway involved in wing-hinge development consists of teashirt (tsh), homothorax (hth), and extradenticle (exd) (Mann and Abu-Shaar 1996; Azpiazu and Morata 2000; Casares and Mann 2000; Wu and Cohen 2002). tsh acts like an activator of hth, and binding of Hth is necessary for nuclear localization of Exd. The Hth:Exd complex then activates downstream targets that pattern the wing hinge. A double knockdown of tsh and hng1 rescued the hinge defect and also the size and patterning of alula, indicating that hng1 is a negative regulator of tsh function (Figure 4, A and G). A similar, though a less dramatic rescue, was seen upon simultaneous hth and hng1 knockdown. Based on these results, it was predicted that Hth activity is upregulated in hng1i. Indeeed, an increase in Hth expression (Figure 4B) along with an expansion of the Hth expression domain was observed in hng1i wing discs. Based on this result we predicted that a further increase in Hth expression using UAS-hth in the hng1i background would dramatically enhance the hng1i phenotype, and this was indeed observed (Figure 4A).
Figure 4.
hinge genes are part of the GRN that patterns the wing hinge. (A) A knockdown of tsh and hth in the MS1096/+; hng1i/+ animal rescues the hng1 phenotype, whereas overexpression of hth enhances the hng1 phenotype severely. The RNAi and the UAS lines used to alter transcript levels for hth and tsh, by themselves, have mild hypomorphic effects. (B) Hth is broadened/derepressed in and around the gap region when hng1 expression is reduced in the MS1096-Gal4 expression domain in the wing imaginal discs. (C) A knockdown of jing in the MS1096/+; hng1i/+ animal leads to a rescue of the hng1 phenotype while co-expression of UAS-jing leads to an enhancement of the proximal wing phenotype. (D) Wg is derepressed in and around the gap region between the IR and the OR in the MS1096/+; UAS-hng1i/UAS-hng1i wing imaginal disc. (E) Wg is derepressed in a small stripe in the gap region when hng1 expression is reduced in the ptc-Gal4 expression domain. (F) hng1 knockdown in heterozygous Wgspadeflag background in the MS1096-Gal4 domain mildly rescues the hinge defect. (G) Wing size (mm2) and alula size (mm2) are measured for genetics interactors of hng1 with known wing-hinge GRN genes. Arrowhead indicates the MS1096/+; UAS-hng1/+ line used as a control for statistical analyses. *P < 0.05, **P < 0.01, and ***P < 0.001.
Jing is a zinc-finger transcription factor implicated in repression of tsh and hth in the wing hinge (Culi et al. 2006). We tested if jing and hng1 interact genetically. jing and hng1 double knockdown rescued the hinge defect while Jing overexpression in hng1 knockdown in animals enhanced the hng1i defect (Figure 4, C and G). This indicates that jing negatively regulates hng1.
A major player in wing-hinge development is Wg, which has roles in patterning by restricting the tsh-hth network to the wing hinge. Wg staining in the third instar wing disc marks an outer (wg-OR) and inner ring (wg-IR) (Couso et al. 1993; Neumann and Cohen 1996a,b; Russell 2000; Del Alamo Rodríguez et al. 2002) with a gap in between. The two rings are critical regions for wing-hinge development with the members of the wing-hinge gene regulatory network (GRN) interacting with or regulating Wg or being regulated by expression of Wg. wg-IR, regulated by the Wgspade-flag enhancer (Neumann and Cohen 1996a,b) patterns a major section of the region of the hinge that is affected in the hng1 mutants. wg-IR also drives intercalary proliferation, generating the gap region between the rings (Zirin and Mann 2004). When hng1 is knocked down in the MS1096 expression domain (MS1096/+; UAS-hng1i/UAS-hng1i), a broadening of the Wg expression (Figure 4D) domain with an intrusion into the gap region was observed. A similar broadening was observed for MS1096/+; Df(hng1i)/UAS-hng1i (Figure S2C). Knockdown of hng1 in the patched (ptc-Gal4) expression domain (ptc-Gal4/+; UAS-hng1i/+) also leads to derepression of Wg in the gap region. Interestingly, although ptc-Gal4 also expresses where the anterior/posterior boundary cuts the dorsal/ventral boundary, Wg expression was not derepressed or broadened at the D/V boundary, indicating specific roles for hng1 regulation in the wing hinge. The Wgspadeflag animal lacks expression of wg-IR, resulting in a wing-hinge phenotype (Neumann and Cohen 1996). Since hng1i causes derepression of Wg in the wg-IR, we predicted that a heterozygous combination of hng1i with wgspadeflag would lead to a mild rescue of the hng1i phenotype due to a decrease in Wg in the wg-IR. Indeed, MS1096-Gal4/+; UAS-hng1i/Wgspadeflag animals show a rescue of the wing hinge when compared to hng1i (Figure 4F). Since loss of hng1 in the wing disc showed upregulation of wingless expression, we checked for increased cell proliferation in these animals. We did not, however, detect any significant change in cellular proliferation (Figure S2F).
Wg expression in wg-IR is driven by two independent mechanisms: Nubbin, vg, and rotund are required for wg expression in the wg-IR in the early third instar stages. The second mechanism involves hth function (Del Alamo Rodríguez et al. 2002). We knocked down hng1 in vg and nubbin mutant background (Figure S2D); however, we did not find any significant interaction. Overexpression of rotund in MS1096/+; hng1i background also did not show any change in the phenotype.
Dpp expression in the posterior compartment of the wing disc has been shown to be responsible for the patterning of the proximal wing; the alula and dpp transcription is particularly mirror dependent (Foronda et al. 2009). dpp knockdown in MS1096/+; hng1i/+ background resulted in the enhancement of the hng1 phenotype (Figure S2E). However, we do not see any interaction with mirror.
Testing redundancy using double gene knockdowns
hng1, hng2, and hng3 appear to be genes with similar or equivalent roles in the wing hinge. Other MADF-BESS knockdowns, with the exception of stwl, do not appear to give a hinge phenotype in single knockdown experiments. It is possible that these genes have a partially redundant function in the wing hinge. One method of testing this would be to simultaneously knock down each of theses genes, along with hng1, and check for an enhancement of the hng1 phenotype. Any enhancement would also provide evidence for a functional role for the other MADF-BESS genes, even in the absence of gene expression data.
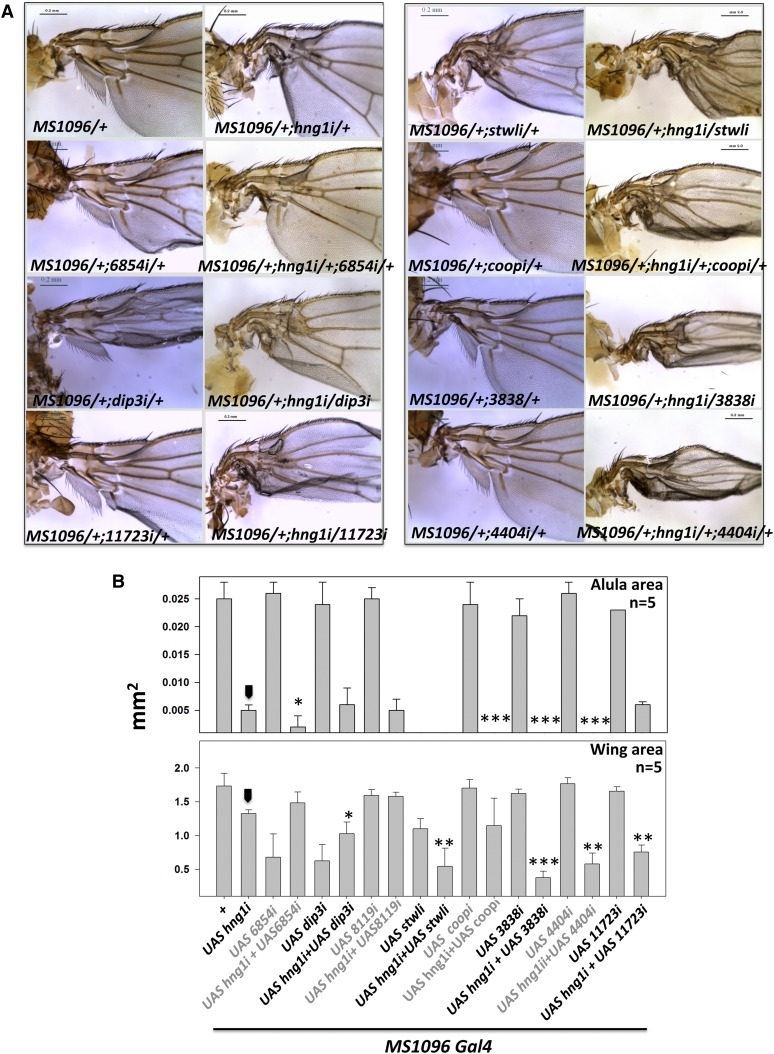
Double knockdowns on all remaining MADF-BESS genes in the background of hng1 RNAi indicated that Dip3, Coop, CG3838, CG11723, and CG4404 (Figure 5, A and B) had roles in wing-hinge patterning as they enhanced the hng1 phenotype, while CG15845, CG8119, CG30403, and CG13204 did not. This indicates that a substantial fraction of the MADF-BESS family (9 of 16) plays a direct or a supporting role in wing-hinge development. The lack of interaction with some of the tested genes may indicate weak RNAi lines that do not reduce transcripts significantly or that these genes do not express in the wing hinge or that these noninteracting genes do not have a role in wing-hinge development. It is also feasible that all members of the family may be involved in wing-hinge patterning.
Figure 5.
hng1 phenotype is enhanced by knockdown of other MADF-BESS genes. (A) CG6854, stwl, coop, CG3838, CG4404, and CG11723 knockdown in the hng1 background enhance the MS1096/+; UAS-hng1i/+ phenotype. The knockdown of these genes by themselves, with the exception of stwl, using RNAi lines does not affect the hinge significantly. The enhancement indicates that these genes are expressed in the cells that pattern the wing hinge and may have partially redundant roles in the hinge-mediated development of the wing hinge. (B) Wing size (mm2) and alula size (mm2) are measured for genetic interactions of hng1 with other MADF-BESS family genes. Arrowhead indicates the MS1096/+; UAS-hng1/+ line used as a control for statistical analyses. *P < 0.05, **P < 0.01, and ***P < 0.001.
Confirmation of redundancy/equivalence by rescue experiments
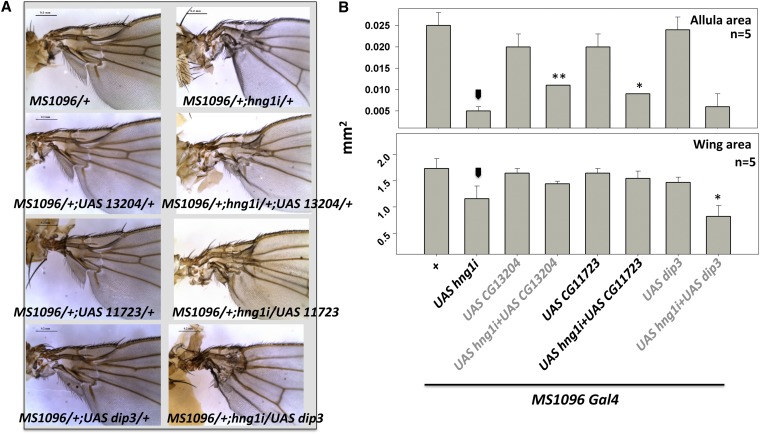
We next tested the ability of the MADF-BESS genes to rescue the hng1 phenotype. As shown in Figure 6, in addition to hng1 itself (Figure 3C), expression of CG11723 and CG13204 could rescue the hng1i phenotype to a significant extent (Figure 6, A and B). Dip3 overexpression, on the other hand, enhanced the phenotype of hng1i. The data for Dip3 is reminiscent of data for Dip3 in Drosophila eye development where both loss and gain of Dip3 function shows similar phenotypes (Duong et al. 2009). hng1 overexpression could not (Figure S2B) rescue the hng3i phenotype, indicating that there are at least some functional differences in the protein products of hng1 and hng3. These data raise the possibility of Hng1 and Hng3 proteins being functionally diverse and also the possibility of the Hng1:Hng3 dimer being the functional entity for hng family function. This possibility is discussed in the next section and incorporated in a model for Hng activity.
Figure 6.
hng1 phenotype is rescued by expression of other MADF-BESS genes. (A) CG13204 and CG11723 expression rescue the MS1096/+;UAS-hng1i/+ phenotype. This indicates that MADF-BESS proteins retain similar biological activity and equivalence in terms of their protein function. (B) Wing size (mm2) and alula size (mm2) are measured for rescue of hng1 phenotype. Arrowhead indicates the MS1096/+; UAS-hng1/+ line used as a control for statistical analyses. *P < 0.05 and **P < 0.01.
Discussion
Fate and consequences of gene duplicates
Gene duplication and subsequent diversification of one or both of the duplicated genes is a well-recognized phenomenon in evolution. Calvin Bridges was one of the first to put forward the idea of genetic units duplicating, specifically in response to symmetric, adjacent banding patterns in polytene chromosomes (Bridges 1936). This idea gained further ground in early studies of the Bar locus (Sturtevant 1925; reviewed in Duncan and Montgomery 2002; Zhang 2003; Taylor and Raes 2004) and took root as a fundamental concept in genetics as a result of studies of the bithorax complex by Ed Lewis (Lewis 1978). The Hox genes are today a textbook example of gene duplication and diversification (Liberles et al. 2010). The idea of gene duplication influencing heredity, evolution, and speciation was enhanced further by Susumu Ohno (Ohno 1970). In recent years, genome sequencing efforts and their resultant analysis has further cemented the reality of gene duplication and its role in invertebrate and vertebrate evolution. Many mechanisms have been proposed to explain the cause and consequence of gene duplication. Gene duplication and expansion have a direct effect on evolution, speciation, and the patterning of new life forms.
The immediate effect of a gene duplication that duplicates both the coding sequence and the regulatory elements is an increase in levels of the mRNA and consequently the level of protein. The duplicated gene at that point can be retained in the genome as a functional copy or lost. If the gene is maintained, it can be a functionally redundant duplicate or the gene sequence may be modified over time. Three possible fates of duplicated genes that are modified have been discussed in the literature (Force et al. 1999; Lynch 2000; Zhang 2003) as pseudogenization, subfunctionalization, and neofunctionalization. Our data seem to agree with a model where the MADF-BESS genes duplicated >40 million years ago and were subfunctionalized. The exception is the CTP synthase gene (Liu 2010), which appears to have neofunctionalized. In Drosophila, CTP synthase codes for a protein where one splice variant (variant C) has a cytoplasmic localization (Azzam and Liu 2013), forms a cellular structure named cytophidia (Liu 2010), and functions to convert UTP to CTP, the last step in pyrimidine nucleotide biosynthesis. This novel function is not seen in any other MADF-BESS family member. CTP synthase, splice variant A, like other MADF-BESS domain proteins, is nuclear (Azzam and Liu 2013). The stwl gene is also an outlier in this family for the following reasons. Stwl is the longest protein in the family with the largest distance between the MADF and BESS domains. Although the protein does not have signatures for additional domains in the intervening sequences, based on current methods, there is a possibility that novel binding motifs exist in that region. Stwl is also the only protein of the family implicated in epigenetic modification of chromatin. If this feature is related to the MADF and BESS domain function, then the other MADF-BESS family members described here may be epigenetic regulators whose functions have not been discovered. Stwl is also the nearest gene to CG3919, another MADF-BESS gene that codes for a small protein (302 amino acids), and appears to be closely related in terms of identity of MADF and BESS domains to Stwl.
The subfunctionalization of a family can happen at three levels. Post duplication, the coding region of a gene can gain mutations that may perturb function at the level of a single polypeptide, but in combination the protein products of a family will retain the ability to perform the original function. This is best explained as the duplication-complementation model (DDC) (Force et al. 1999; Hahn 2009). The DDC model emphasizes that mutations facilitate rather than hinder gene duplication. The second element of subfunctionalization is the change in spatiotemporal expression of the duplicate genes, as compared to the parent, allowing division and diversification of expression domains. In our example, hng1, hng2, hng3, and stwl appear to have overlapping expression patterns in the developing wing hinge, and enhancer genetics indicate that at least five of the other MADF-BESS family genes are expressed and involved in wing-hinge development. The third element of subfunctionalization is the dose of the active species (Hahn 2009). Since members of the MADF-BESS family have the potential to heterodimerize via the BESS domain, common functionality of the family may be dependent on the formation of a heterodimer that may be the functional transcriptional regulator. Differential expression of genes may regulate formation and concentrations of hetero- and homodimers and therefore regulate function.
Are the hinge genes redundant?
The MADF-BESS gene family appears to retain functional redundancy and has also retained similar amino acid sequence in the two “functional” domains. The MADF and BESS domain families independently have ∼50 and ∼25 members, respectively, being in the same ballpark with the largest family in flies, the trypsin gene family, with 111 members (Zhang et al. 2003). It is interesting to note that, in spite of tens of millions of years of evolution, the 16 MADF-BESS genes have retained the N-terminal/C-terminal architectural positioning of the two domains as well as have conserved the domain sequences. The data collected in our study indicate that at least 9 of 16 members participate in the development of the wing hinge. The hinge genes, including stwl, appear to have hinge development as a primary function while five other genes appear to retain at least partial function in the hinge.
Knockdown experiments where hng1, hng2, and hng3 transcripts were reduced singly and in combination indicate that the genes are not genetically redundant in the classical sense of where a single knockdown of a gene has no effect, but a double knockdown has a drastic effect. In our example of the MADF-BESS domain proteins, a subset of these proteins can be said to be partially redundant at the genetic level, but since they all affect the same function and many seem to be equivalent at the protein level, we might consider them as redundant at the protein network level. Another point to note is that our experiments are done using an inbred population grown under a single set of laboratory conditions. It is quite possible that, under different conditions of temperature and diet or in a different genetic background, the genes may be demonstrated to be completely redundant genetically. Based on a lack of a phenotype on single gene knockdown, four other genes in the family do not show the ability to function as the hinge genes, but do enhance hng1 phenotypes. These genes are probably expressed in wing-hinge development and retain some activity equivalent to the hng genes, but have diverged enough not to be core hinge genes. It is also possible that these are hng genes, but the RNAi lines used are not efficient enough to give phenotypes. In summary, a subset of the members of the family fit a broad definition of redundant genes.
Why are the hinge family sequences and functions retained?
The reasons behind the expansion and retention of similar function are not clear at this point. One hypothetical possibility is that the founding member of the MADF-BESS family was originally a single transcriptional regulatory adaptor protein involved with many critical transcriptional regulatory pathways such as NF-κB signaling, Wnt signaling, or the Hth/Exd module. A gene duplication/expansion event allowed for the slow diversification (subfunctionalization) of roles so that the primary pathway for each duplicate became distinct over evolutionary timescales. However, since the proteins were still transcriptional regulators, they retained their overall sequences and folds and maintained the ability to perform the adaptor/regulator function for other pathways/modules. At this point in evolution, the proteins have diversified and retained their critical MADF and BESS domains, which for function need to be in a single polypeptide in an N-terminal to C-terminal orientation.
Regulation of Wg/Hth expression is critical for normal wing development
Wg/Wnt is a member of a family of secreted molecules with conserved signaling pathways in animals (Cadigan and Nusse 1997; Wodarz and Nusse 1998; Swarup and Verheyen 2012). Wnt/Wg signaling is activated by binding to receptors such as Frizzled/Arrow, which leads to translocation of stabilized β-catenin to the nucleus and subsequent activation of Wnt target genes. In the Drosophila wing, the dose and spatiotemporal expression of Wg is critical for normal patterning and growth of the wing (Couso et al. 1993). Wg is required for proliferation in the first instar and later for patterning in the third instar. During first instar, tsh and hth are expressed throughout the disc (Zirin and Mann 2004). Repression of tsh by Wg and Dpp in second instar from the pouch is required for the proper development of the wing blade. This is followed by repression of hth and its confinement to the hinge region of the disc (Wu and Cohen 2002). hth expression in late third instar is driven by the Wg expression. Combined signals from vestigial, nubbin, and rotund are required for the wg-IR expression (Del Alamo Rodríguez et al. 2002). wg-IR is also driven by an independent mechanism involving a feedback loop with hth in late third instar (Casares and Mann 2000; Del Alamo Rodríguez et al. 2002).
Wg signaling is regulated at multiple levels, and our data point to roles for the MADF-BESS family as fail-safe regulators for maintaining robust Wg expression. Our data indicate that there appears to be increased activation of the Wg/Tsh/Hth pathway in hng1i animals, which in turn leads to the hinge phenotype. In the absence or reduction of Hng1, Wg is derepressed in the gap region and the Wg spatiotemporal domain broadens. Ectopic expression of Wg earlier was shown to lead to a mispatterned proximal wing (Russell 2000), and our phenotype appears to be of a similar nature.
Coop (Song et al. 2010), a Pangolin-interacting protein and a member of the MADF-BESS family, has been shown to be a negative regulator of Wnt/Wg signaling, regulating Distalless at the D/V boundary. Our data on the other hand show that hng1 regulates Wg expression at the presumptive hinge region, with Coop, which is expressed in the wing hinge (Figure 3K) playing a secondary role. Prima facie, hng1 appears to regulate Wg expression in the regions that form the wing hinge. At the late third instar, because Wg and hth expression in the wg-IR are dependent on each other, hng1 may regulate expression of one or both (Figure 7A). hng1 also interacts genetically with a constitutively repressed variant of pan (UAS-TCF-ΔN), weakly rescuing the pangolin phenotype (Figure S2G), indicating similarity to Coop function, and providing evidence for hng1 regulating Wg signaling. In addition to demonstrating Coop as a negative regulator of Wg signaling, Song et al. (2010) tested Adf1 and CG6854 and showed that these proteins also negatively regulate Wg signaling. This lends support to the idea that MADF-BESS family members in general are involved in regulating Wg expression and or signaling.
Figure 7.
Model for MADF-BESS function in the wing-hinge. (A) The GRN for wing-hinge development includes wg, tsh, hth, and exd as major patterning genes. In the hng1 loss of function, our data indicate an increase in activity of Tsh/Hth/Exd. hng1 appears to negatively regulate the Wg/Hth-positive autoregulatory loop. hng1 also negatively regulates tsh, possibly acting downstream of jing. (B) The three hng genes along with stwl appear to be functionally equivalent and are part of the GRN that patterns the wing hinge. Five additional genes retain, at least partially, functions of the hng family of genes and can replace, to an extent, hng function. The four hinge genes code for proteins (blue circles), which we hypothesize may be part of a dimer/tetramer that is the active transcriptional regulator. Function could be regulated by increasing/decreasing the concentration of the Hng proteins, with the concentration of the functional polymer dependent on spatiotemporal expression and also the levels of the hng genes.
Appendage development and Wnt signaling in vertebrates
In vertebrate appendage development, the homolog of Hth, Meis, and the Prep1 proteins are involved in the nuclear localization of the vertebrate Exd homolog (Selleri et al. 2001). The Pbx/Meis proteins appear to act as cofactors to Hox proteins and confer target selectivity (Galant et al. 2002). Our study has implicated the MADF-BESS family as important regulators of the Hth/Exd pathway, raising the possibility of vertebrate MADF-BESS family members or related SANT domain proteins interacting with Pbx/Meis proteins. Our results raise the possibility of the hng gene family regulating Wnt expression in other Drosophila tissues and in other invertebrates and vertebrates.
Hypothetical model for redundancy and partial overlap of function at the protein level
Our data and analysis suggest that the MADF-BESS gene family is a duplicated expanded family retained in multiple functional copies by the evolutionary process. One reason for the retention of the family would be their involvement in critical functions in the cell during development. The hinge genes appear to be critical regulators of wing-hinge patterning part of the GRN that patterns the Drosophila wing hinge (Figure 7, A and B). These proteins are possibly repressors/corepressors regulating expression of Wg/Hth. Five other genes of the family also retain partial function (Figure 7B) while there is no direct evidence for roles for remaining family members. If the genes were completely redundant, a single knockdown would not produce a phenotype, while a double or triple knockdown would give a strong phenotype. To explain our observations where the three hinge genes are somewhat equivalent, but not genetically redundant, we focus on the polypeptide products of the genes. The BESS domain is a protein interaction domain and may lead to homodimerization of the protein. This leads to the possibility that the conserved BESS domains could heterodimerize. If the active MADF-BESS regulator of function (such as negative regulation of Wg/Wnt) is a heterodimer or heterotetramer, then the activity could be regulated by concentration of the monomer ↔ dimer ↔ tetramer and the equilibrium between these protein species. This model allows us to explain our data of the dose-dependent enhancement of phenotype by decrease in concentration of one protein as well as the supplementation of the loss of one protein by another. The active molecule, a tetramer, would be in equilibrium with the dimer/monomer products of the hng genes. Multiple knockdowns of two or more hng genes would lead to a severe disruption of the active heterodimer or heterotetramer and would give stronger phenotypes. Replacing the loss of any one component by a protein with similar activity at equal or higher concentrations would lead to an increase in the functional dimer/tetramer, leading to a partial rescue of the hng phenotype.
In summary, we discover a new developmental function for the LSE MADF-BESS gene family (the hinge family) with multiple gene members having roles in patterning the wing hinge, possibly by negative regulation of Wg/Hth expression. Four members have central roles in patterning the wing hinge, presumably acting as negative regulators/repressors while another five have supporting/overlapping roles. Our findings and the tools used to define redundancy allow us to appreciate and dissect out the mechanisms of subfunctionalization post gene duplication in evolution.
Supplementary Material
Acknowledgments
We thank the Vienna Drosophila RNAi Center, Transgenic RNAi Project (Harvard University), Bloomington Drosophila Stock Center, Drosophila Genomics Research Consortium, National Centre for Biological Sciences (NCBS) (Bangalore, India) stock center, NCBS transgenic injection facility, National Institute of Genetics (Mishima, Japan) stock center, and K. Vijayraghavan for fly stocks and reagents. We also thank Richa Rikhy, Girish Deshpande, Anuradha Ratnaparkhi, L. S. Shashidhara, Veronica Rodrigues, and members of the Ratnaparkhi Lab for helpful discussions; three anonymous reviewers for comments that helped improve the manuscript; and Richa Rikhy and Vijay Vittal for microscopy training and access to the Indian Institute of Science Education and Research (IISER) confocal facility. This project was financially supported by a Research Grant for Young Investigators (2010–2013) from the Department of Biotechnology (DBT), Government of India, as well as from intramural grants from IISER (Pune, India). G.S.R. is a Wellcome Trust/DBT India Alliance Fellow, V.S. is an IISER Graduate Student supported by a University Grants Commission Senior Research Fellowship, and F.H. is a member of the Centre for Excellence (CoE) in epigenetics, supported by a CoE grant by DBT.
Footnotes
Communicating editor: M. Wolfner
Literature Cited
- Azpiazu N., Morata G., 2000. Function and regulation of homothorax in the wing imaginal disc of Drosophila. Development 127: 2685–2693. [DOI] [PubMed] [Google Scholar]
- Azzam G., Liu J.-L., 2013. Only one isoform of Drosophila melanogaster CTP synthase forms the cytoophidium. PLoS Genet. 9: e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Lee J., Venkatesh K., Wu C.-F., Hasan G., 2004. Loss of flight and associated neuronal rhythmicity in inositol 1,4,5-trisphosphate receptor mutants of Drosophila. J. Neurosci. 24: 7869–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar V., Courey A. J., 2002. The MADF-BESS domain factor Dip3 potentiates synergistic activation by Dorsal and Twist. Gene 299: 173–184. [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Langer M. R., Crowley K. A., Tan S., Denu J. M., et al. , 2002. Essential role for the SANT domain chromatin remodeling enzymes. Mol. Cell 10: 935–942. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1936. The bar “gene”: a duplication. Science 83: 210–211. [DOI] [PubMed] [Google Scholar]
- Brun S., Rincheval-Arnold A., Colin J., Risler Y., Mignotte B., et al. , 2006. The myb-related gene stonewall induces both hyperplasia and cell death in Drosophila: rescue of fly lethality by coexpression of apoptosis inducers. Cell Death Differ. 13: 1752–1762. [DOI] [PubMed] [Google Scholar]
- Cadigan K. M., Nusse R., 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11: 3286–3305. [DOI] [PubMed] [Google Scholar]
- Casares F., Mann R. S., 2000. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development 127: 1499–1508. [DOI] [PubMed] [Google Scholar]
- Casola C., Lawing A. M., Betrán E., Feschotte C., 2007. PIF-like transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol. 24: 1872–1888. [DOI] [PubMed] [Google Scholar]
- Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., Oliver B., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Clark K. A., McKearin D. M., 1996. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development 122: 937–950. [DOI] [PubMed] [Google Scholar]
- Couso J. P., Bate M., Martinez Arias A., 1993. A wingless-dependent polar coordinate system in Drosophila imaginal discs. Science 259: 484–489. [DOI] [PubMed] [Google Scholar]
- Culi J., Aroca P., Modolell J., Mann R. S., 2006. jing is required for wing development and to establish the proximo-distal axis of the leg in Drosophila melanogaster. Genetics 173: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler G., Perry K. M., Tjian R., 1998. Adf-1 is a nonmodular transcription factor that contains a TAF-binding Myb-like motif. Mol. Cell. Biol. 18: 2252–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Alamo Rodríguez D., Terriente J., Galindo M. I., Couso J. P., Díaz-Benjumea F. J., 2002. Different mechanisms initiate and maintain wingless expression in the Drosophila wing hinge. Development 129: 3995–4004. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila : a fly geneticist’s Swiss Army knife. Genesis 15: 1–15. [DOI] [PubMed] [Google Scholar]
- Duncan I., Montgomery G., 2002. E. B. Lewis and the Bithorax complex: part II. From cis-trans test to the genetic control of development. Genetics 161: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H. A., Nagaraj R., Wang C. W., Ratnaparkhi G., Henry Y., et al. , 2009. Non-cell-autonomous inhibition of photoreceptor development by Dip3. Dev. Biol. 323: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England B. P., Admon A., Tjian R., 1992. Cloning of Drosophila transcription factor Adf-1 reveals homology to Myb oncoproteins. Proc. Natl. Acad. Sci. USA 89: 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Mistry J., Tate J., Coggill P., Heger A., et al. , 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force, A., M. Lynch, F. B. Pickett, A. Amores, Y. L. Yan et al, 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foronda D., Pérez-Garijo A., Martín F. A., 2009. Dpp of posterior origin patterns the proximal region of the wing. Mech. Dev. 126: 99–106. [DOI] [PubMed] [Google Scholar]
- Galant R., Walsh C. M., Carroll S. B., 2002. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development 129: 3115–3126. [DOI] [PubMed] [Google Scholar]
- Guillen I., Mullor J. L., Capdevila J., Sanchez-Herrero E., Morata G., et al. , 1995. The function of engrailed and the specification of Drosophila wing pattern. Development 121: 3447–3456. [DOI] [PubMed] [Google Scholar]
- Hahn M. W., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100: 605–617. [DOI] [PubMed] [Google Scholar]
- Horn T., Sandmann T., Boutros M., 2010. Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol. 11: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., et al. , 2012. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40: D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell J. R., Carthew R. W., 2000. Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18: 896–898. [DOI] [PubMed] [Google Scholar]
- Kraut R., Menon K., Zinn K., 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. [DOI] [PubMed] [Google Scholar]
- Lespinet O., Wolf Y. I., Koonin E. V., Aravind L., 2002. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 12: 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Liberles D. A., Kolesov G., Dittmar K., 2010. Understanding gene duplication through biochemistry and population genetics, pp. 1–21 in Evolution After Gene Duplication. John Wiley & Sons, New York. [Google Scholar]
- Liu J.-L., 2010. Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genomics 37: 281–296. [DOI] [PubMed] [Google Scholar]
- Lynch M., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., et al. , 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics Genome Biol. 8: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M., Consortium U., 2011. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines J. Z., Park J. K., Williams M., McKearin D. M., 2007. Stonewalling Drosophila stem cell differentiation by epigenetic controls. Development 134: 1471–1479. [DOI] [PubMed] [Google Scholar]
- Mann R. S., Abu-Shaar M., 1996. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature 383: 630–633. [DOI] [PubMed] [Google Scholar]
- Marygold S. J., Leyland P. C., Seal R. L., Goodman J. L., Thurmond J., et al. , 2013. FlyBase: improvements to the bibliography. Nucleic Acids Res. 41: D751–DD757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy K., Emrich S. J., Lawson D., Campbell D., Dialynas E., et al. , 2012. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic Acids Res. 40: D729–D734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milán M., Pham T. T., Cohen S. M., 2004. Osa modulates the expression of Apterous target genes in the Drosophila wing. Mech. Dev. 121: 491–497. [DOI] [PubMed] [Google Scholar]
- Miller, M.A., Pfeiffer, W., Schwartz, T., 2010 Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), Nov. 14, 2010, New Orleans, pp. 1–8.
- Neumann Carl J.Cohen S. M., 1996a Sternopleural is a regulatory mutation of wingless with both dominant and recessive effects on larval development of Drosophila melanogaster. Genetics 142: 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann C. J., Cohen S. M., 1996b Distinct mitogenic and cell fate specification functions of wingless in different regions of the wing. Development 122: 1781–1789. [DOI] [PubMed] [Google Scholar]
- Ohno S., 1970. Evolution by Gene Duplication. George Alien & Unwin, London; Springer-Verlag, Berlin; Heidelberg, Germany; New York. [Google Scholar]
- Pfeiffer, W., and A. Stamatakis, 2010 Hybrid MPI/Pthreads parallelization of the RAxML phylogenetics code. Parallel and Distributed Processing, Workshops and Phd Forum, 2010 IEEE International Symposium, pp. 1–8. [Google Scholar]
- Ratnaparkhi A., 2013. Signaling by Folded gastrulation is modulated by mitochondrial fusion and fission. J Cell Sci. 126(Pt 23): 5369–5376. [DOI] [PubMed] [Google Scholar]
- Ratnaparkhi G. S., Duong H. A., Courey A. J., 2008. Dorsal interacting protein 3 potentiates activation by Drosophila Rel homology domain proteins. Dev. Comp. Immunol. 32: 1290–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S., 2000. The Drosophila dominant wing mutation Dichaete results from ectopic expression of a Sox-domain gene. Mol. Gen. Genet. 263: 690–701. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P., 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleri L., Depew M. J., Jacobs Y., Chanda S. K., Tsang K. Y., et al. , 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 128: 3543–3557. [DOI] [PubMed] [Google Scholar]
- Sigrist C. J. A., Cerutti L., Hulo N., Gattiker A., Falquet L., 2002. PROSITE : a documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3: 265–274. [DOI] [PubMed] [Google Scholar]
- Song H., Goetze S., Bischof J., Spichiger-haeusermann C., Kuster M., et al. , 2010. Coop functions as a corepressor of Pangolin and antagonizes Wingless signaling. Genes Dev. 24: 881–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1925. The effects of unequal crossing over at the Bar locus in Drosophila. Genetics 10: 117–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Verheyen E. M., 2012. Wnt/Wingless signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 4: a007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. S., Raes J., 2004. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38: 615–643. [DOI] [PubMed] [Google Scholar]
- Waterhouse R. M., Tegenfeldt F., Li J., Zdobnov E. M., Kriventseva E. V., 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41: D358–D365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz, A., and R. Nusse, 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14: 59–88. [DOI] [PubMed] [Google Scholar]
- Wu J., Cohen S. M., 2002. Repression of Teashirt marks the initiation of wing development. Development 129: 2411–2418. [DOI] [PubMed] [Google Scholar]
- Yi X., De Vries H. I., Siudeja K., Rana A., Lemstra W., et al. , 2009. Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 20: 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P. D., Tuschl T., Sharp P. A., Bartel D. P., 2000. RNAi : double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Zhang J., 2003. Evolution by gene duplication: an update. Trends Ecol. Evol. 18: 292–298. [Google Scholar]
- Zhang P., Gu Z., Li W.-H., 2003. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J. D., Mann R. S., 2004. Differing strategies for the establishment and maintenance of teashirt and homothorax repression in the Drosophila wing. Development 131: 5683–5693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.