Abstract
The central nervous system of Drosophila melanogaster consists of fused segmental units (neuromeres), each generated by a characteristic number of neural stem cells (neuroblasts). In the embryo, thoracic and anterior abdominal neuromeres are almost equally sized and formed by repetitive sets of neuroblasts, whereas the terminal abdominal neuromeres are generated by significantly smaller populations of progenitor cells. Here we investigated the role of the Hox gene Abdominal-B in shaping the terminal neuromeres. We show that the regulatory isoform of Abdominal-B (Abd-B.r) not only confers abdominal fate to specific neuroblasts (e.g. NB6-4) and regulates programmed cell death of several progeny cells within certain neuroblast lineages (e.g. NB3-3) in parasegment 14, but also inhibits the formation of a specific set of neuroblasts in parasegment 15 (including NB7-3). We further show that Abd-B.r requires cooperation of the ParaHox gene caudal to unfold its full competence concerning neuroblast inhibition and specification. Thus, our findings demonstrate that combined action of Abdominal-B and caudal contributes to the size and composition of the terminal neuromeres by regulating both the number and lineages of specific neuroblasts.
Keywords: CNS development, Segmental patterning, Terminal neuromeres, Neuroblasts, Hox genes, Abdominal-B, Caudal, Drosophila
INTRODUCTION
During the development of the central nervous system (CNS), a huge variety of different cell types has to be generated. For the establishment of functional neural circuits it is a prerequisite that characteristic numbers of each cell type become arranged in a reproducible spatial pattern. This pattern is founded during the formation and specification of neural stem cells. Subsequently, tight temporal control of their proliferation and survival as well as specification, differentiation and survival of their progeny ensures the generation of characteristic cell lineages. Drosophila melanogaster is one of the favoured models used to investigate these processes and shares many fundamental mechanisms in CNS development with vertebrate systems (for reviews see Doe et al., 1998; Thor, 1995).
In the embryonic CNS of Drosophila, neural stem cells (termed neuroblasts, NBs) can be individually identified by their characteristic time and position of delamination from the neuroectoderm (NE), and the expression of a unique combination of molecular markers (Broadus et al., 1995; Doe, 1992; Urbach and Technau, 2003). Their specification occurs already in the NE by Cartesian grid-like expression of patterning genes, providing positional information along the anteroposterior (AP) and dorsoventral (DV) axes (for reviews see Bhat, 1999; Skeath, 1999). Hence, each NB generates a unique and almost invariant cell lineage (Bossing et al., 1996; Schmid et al., 1999; Schmidt et al., 1997).
Along the AP axis the CNS is composed of segmental units (neuromeres). In the thoracic (T1-T3) and most of the abdominal segments (A1-A8) each hemineuromere originates from a repetitive set of ∼30 NBs (Broadus et al., 1995; Doe, 1992). Serially homologous NBs, exposed to similar positional cues, produce similar lineages, although some of these include subsets of cells that specifically differ between segments (reviewed by Technau et al., 2006). Neuromeres in the tail region (A9, A10) exhibit a progressively derived character (compared to the assumed developmental ground state in T2; Lewis, 1978). They are formed by only 23 NBs in A9 and by 11 NBs in A10 per hemineuromere, and thus their size is continuously decreased (Birkholz et al., 2013).
Segmental identity and patterning is under the control of the highly conserved Hox genes, each encoding a transcription factor with a DNA-binding homeodomain (e.g. reviewed by McGinnis and Krumlauf, 1992). During neurogenesis in Drosophila, Hox genes have been shown to act in various developmental stages at the level of NE, NBs and their progeny to control cell-fate specification, proliferation and/or apoptosis in a segment-specific manner (for reviews, see Reichert and Bello, 2010; Rogulja-Ortmann and Technau, 2008). Whereas Hox genes clustered in the Antennapedia complex (ANT-C) (Kaufman et al., 1980) control differentiation of the head, gnathal and anterior thoracic segments, those in the Bithorax complex (BX-C) (Lewis, 1978) shape the posterior thoracic and all of the abdominal segments (reviewed by Peifer et al., 1987). Hox gene expression domains coincide with parasegmental boundaries instead of segmental ones. A parasegment (PS) represents a fundamental genetic unit and consists of the posterior compartment of one segment (expressing the segment polarity gene engrailed (en) (DiNardo et al., 1985; Patel et al., 1989) and the anterior compartment of the next segment (En negative) (reviewed by Deutsch, 2004; Martinez-Arias and Lawrence, 1985) (e.g. PS14 consists of posterior A8 and anterior A9). The BX-C comprises the genes Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B) (Casanova et al., 1987; Sánchez-Herrero et al., 1985; Tiong et al., 1985). Arrangement of these genes within the BX-C is collinear with their expression domains along the AP body axis (reviewed by Duncan, 1987; Lewis, 1978) and Abd-B is expressed most posteriorly (Harding et al., 1985). Abd-B contains two distinct genetic elements, which are active in different domains: the morphogenetic (m) subfunction is necessary to generate the morphological diversity of PS10-13, whereas the regulatory (r) element is needed for the identity of PS14-15 (Casanova et al., 1986).
Here we have investigated the role of the different Abd-B isoforms in shaping the most posterior neuromeres of the ventral nerve cord (VNC). We focused on a subset of four NBs and their lineages (NB2-4, NB3-3, NB6-4 and NB7-3) that express the molecular marker Eagle (Eg) (Dittrich et al., 1997; Higashijima et al., 1996). We demonstrate that the r isoform of Abd-B (Abd-B.r) acts at different levels: It confers abdominal fate to NB6-4 (producing glia exclusively), and it triggers programmed cell death (PCD) of several progeny cells within the NB3-3 lineage. Furthermore, it inhibits the formation of a specific set of NBs. In Abd-B null mutants, which show no expression of BX-C genes in PS14-15, NB3-3 and NB6-4 (producing glia plus neurons) in PS14 assume thoracic fate, and in PS15 additional NBs are formed, including NB7-3, which is never generated in PS15 of wild-type embryos. Ectopic expression of the m isoform of Abd-B (Abd-B.m) leads to a rescue of the Abd-B null mutant phenotypes, demonstrating similar potentials of both isoforms. However, Abd-B.r requires co-expression of the ParaHox gene caudal (cad) to repress the formation of NBs in PS15. The combined action of Abd-B.r and cad is also sufficient to ectopically induce posterior identity in anterior neuromeres. We conclude that Abd-B and cad are required to inhibit the formation of specific NBs and to modify particular NB lineages, in order to adjust proper size and composition of the terminal neuromeres.
MATERIALS AND METHODS
Drosophila strains
The following fly strains were used: wild type (Oregon R); eg-Gal4 (Ito et al., 1995); sca-Gal4 (Klaes et al., 1994); elav-Gal4 (Lin and Goodman, 1994); UAS-cad (Moreno and Morata, 1999) (from Ulrich Schaefer, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany); UAS-cad (Hwang et al., 2002) (from Mi-Ae Yoo); en-lacZ (Hama et al., 1990) (from Alfonso Martinez-Arias, University of Cambridge, UK); Df(3R)C4 (Karch et al., 1985) (from François Karch); Df(3L)H99 (White et al., 1994), UAS-P35 (Hay et al., 1994), cad2 (Macdonald and Struhl, 1986) and UAS-nGFP (all from Bloomington Stock Center); Abd-BM1, Abd-BM3, Abd-BM5 (Sánchez-Herrero et al., 1985), Abd-BD18 (Hopmann et al., 1995) and Abd-BUab1 (Karch et al., 1985) (all from Ernesto Sánchez-Herrero); UbxMX12, abd-AM1, Abd-BM8 triple mutant (Casanova et al., 1987), UAS-Abd-B.r (Rivas et al., 2013) and UAS-Abd-B.m (Castelli-Gair et al., 1994) (all from James Castelli-Gair Hombría). All experiments were performed at 25°C.
Immunohistochemistry and in situ hybridisation
Embryos (staging according to Campos-Ortega and Hartenstein, 1997) were dechorionated, fixed and immunostained following previously published protocols (e.g. Patel, 1994). The following primary antibodies were used: mouse anti-Abdominal-A (1:200) (Kellerman et al., 1990) (from Ian Duncan); mouse anti-Abdominal-B (1:20) (Celniker et al., 1989), mouse anti-Invected (1:2) (Patel et al., 1989) and mouse anti-Ultrabithorax (1:20) (White and Wilcox, 1984) (all from DSHB); chicken anti-β-Galactosidase (1:1000) (Abcam); guinea pig anti-Caudal (1:400) and guinea pig anti-Runt (1:500) (Kosman et al., 1998) (from John Reinitz); rabbit anti-Caudal (1:100) (Macdonald and Struhl, 1986) (from Paul Macdonald); rabbit anti-Deadpan (1:100) (Bier et al., 1992) (from Harald Vaessin); mouse anti-Eagle (1:100) (Karcavich and Doe, 2005) (from Chris Doe); rabbit anti-Eagle (1:500) (Dittrich et al., 1997); rabbit anti-Engrailed (1:100) (Santa Cruz Biotechnology); rabbit anti-Eyeless (1:1000) (Kammermeier et al., 2001) (from Uwe Walldorf); mouse anti-GFP (1:250) (Roche); rat anti-Gooseberry-proximal (1:2) (Zhang et al., 1994) (from Robert Holmgren); rabbit anti-Miranda (1:100) (Betschinger et al., 2006) (from Juergen Knoblich); guinea pig anti-Reversed-polarity (1:10,000) (von Hilchen et al., 2013).
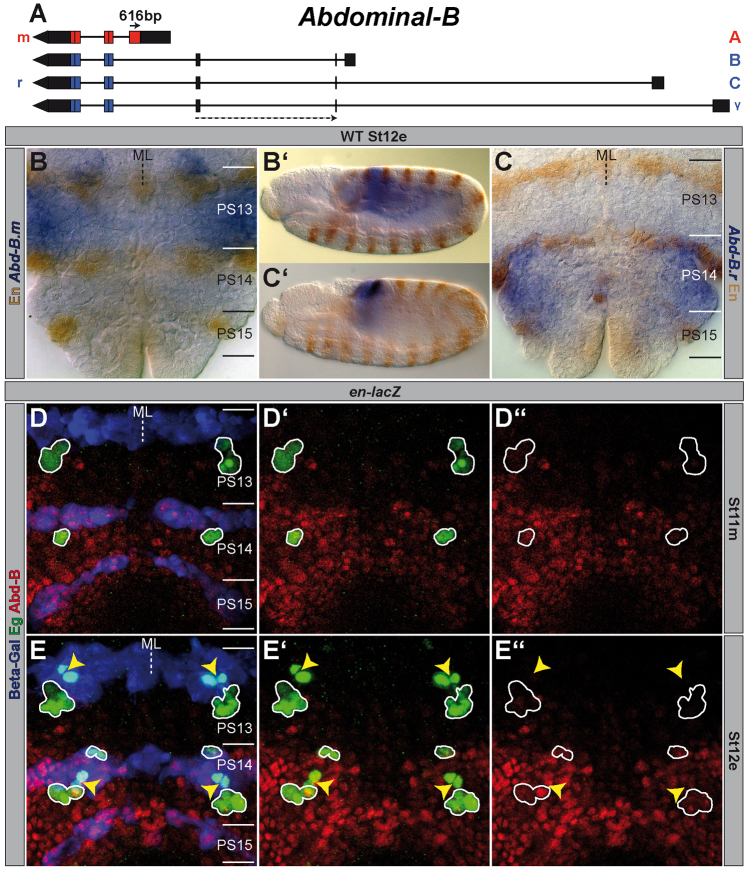
For in situ hybridisation we generated an RNA probe for Abd-B.m (616bp) targeting its unique N-terminal protein coding sequence (CDS). The Abd-B.r RNA probe (220 bp) is directed against two exons, which are exclusively present in all described r-specific transcripts (Fig. 3A). The probes were obtained by amplification from cDNA pAB713 (Abd-B.m) and pAB728 (Abd-B.r) (Kuziora and McGinnis, 1988) (from Ernesto Sánchez-Herrero) and cloned into pDrive (Qiagen). The following primers were used for amplification: for Abd-B.m 5′-CACTGGAGGGAGAAACACTCGC-3′ and 5′-CAACAGCAGCAGCAGCAGCAG-3′; for Abd-B.r 5′-CTGTGGGATGGGAACTGACGC-3′ and 5′-GACCAAAAGCACTACCCAAATAACTG-3′.
Fig. 3.
Expression of different Abdominal-B elements in wild type. (A) The Abd-B gene (drawn to scale) according to FlyBase (McQuilton et al., 2012). Lines depict intron sequences; boxes mark exons (black ones show UTRs, coloured ones CDSs). The localisation of the exon probes are illustrated by arrows (solid one=Abd-B.m (616 bp); stippled one=Abd-B.r (220 bp) spanning over two exons). (B,C) In situ hybridisation of Abd-B.m (B) or Abd-B.r (C) in a wild-type St12e VNC (flat preparation). PSs are depicted on the right and their borders are illustrated by a solid line. (B′,C′) Same stainings in whole mount. (D,E) Flat preparations (maximum projections) of St11m and St12e en-lacZ embryos triple-stained against Abd-B (both isoforms), Eg and beta-galactosidase (Beta-Gal). (E) NB7-3 of PS13 and PS14, which underwent their first division, are marked by yellow arrowheads. All other wild-type Eg-positive cells are encircled. Please note the intensive expression of Abd-B.r in the En stripe of PS15, where the formation of NB7-3 is inhibited. (D′,E′) Abd-B and Eg staining. (D′,E′) Abd-B expression. ML, midline; WT, wild type.
pDrive was linearised and probes were synthesised by a T7 RNA polymerase using FITC (Abd-B.m) or DIG (Abd-B.r) RNA labelling Mix (Roche). The hybridisation was carried out as described before (Plickert et al., 1997; Tautz and Pfeifle, 1989) and the probes were diluted 1:5000 (Abd-B.m) or 1:1000 (Abd-B.r).
As fluorescent secondary antibodies we used the DyLight (Jackson ImmunoResearch) and Alexa (Life Technologies) series. Non-fluorescent secondary antibodies were either biotinylated or alkaline phosphatase-conjugated (Jackson ImmunoResearch). All secondary antibodies were used according to the manufacturer’s protocols.
Non-fluorescent stainings were documented on a Zeiss Axioplan microscope; fluorescent confocal images were acquired on a Leica TCS SP2 or SP5 microscope. All images were processed by Adobe Photoshop and Illustrator.
A two-tailed t-test was performed for statistical significance (see Fig. 1K, Fig. 8G).
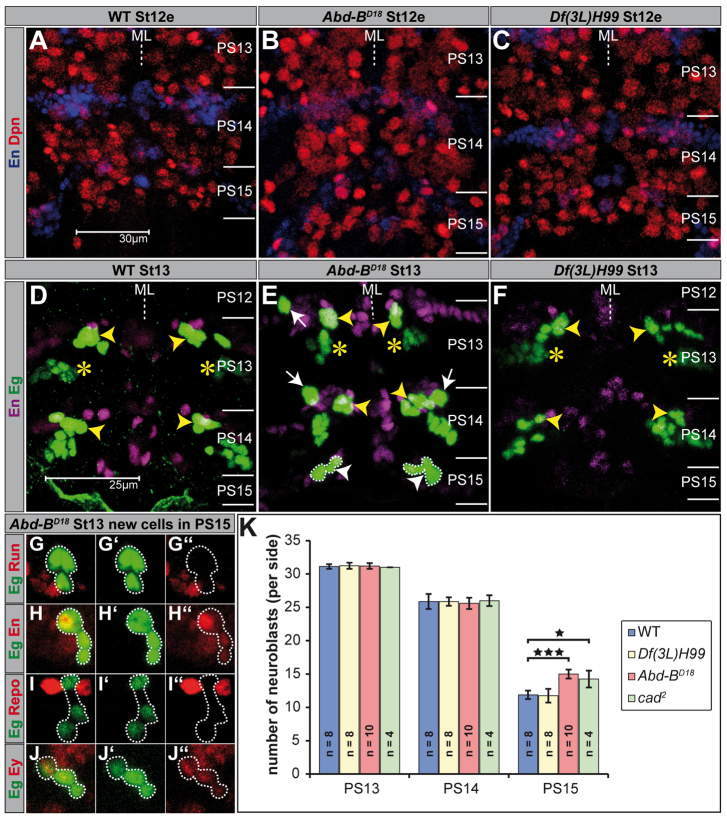
Fig. 1.
Abdominal-B suppresses the formation of neuroblasts in parasegment 15. (A-F) Flat preparations (maximum projections) of St12e and St13 embryos of the indicated genotype double-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. (A-C) Dpn serves as a universal NB marker. (D-F) NB2-4 (dorsal location) and NB3-3 clones (ventral location) are En negative and indistinguishable in maximum projections of PS13 (yellow asterisks). The typical medial location of the NB7-3 cluster (En positive) is marked by yellow arrowheads. (D,F) The abdominal NB6-4 clones (consisting of two glia) already downregulated Eg expression. (E) Transformation of abdominal NB6-4 into thoracic fate is indicated by appearance of the neuronal subclone, showing constant Eg expression (white arrows). Ectopic NB7-3 cells in PS15, which are not found in wild type or in Df(3L)H99 mutant embryos, are surrounded by broken lines and indicated by white arrowheads. (G-J′) The ectopic NB7-3 cells in PS15 of Abd-BD18 mutant embryos were identified by double-staining against Eg (green) and various molecular markers as indicated (red). (K) Statistics for the total number of Dpn-positive NBs (per side) of the indicated genotypes in PS13, PS14 and PS15, respectively. *P<0.05; ***P<0.001. Error bars represent s.d. ML, midline; WT, wild type.
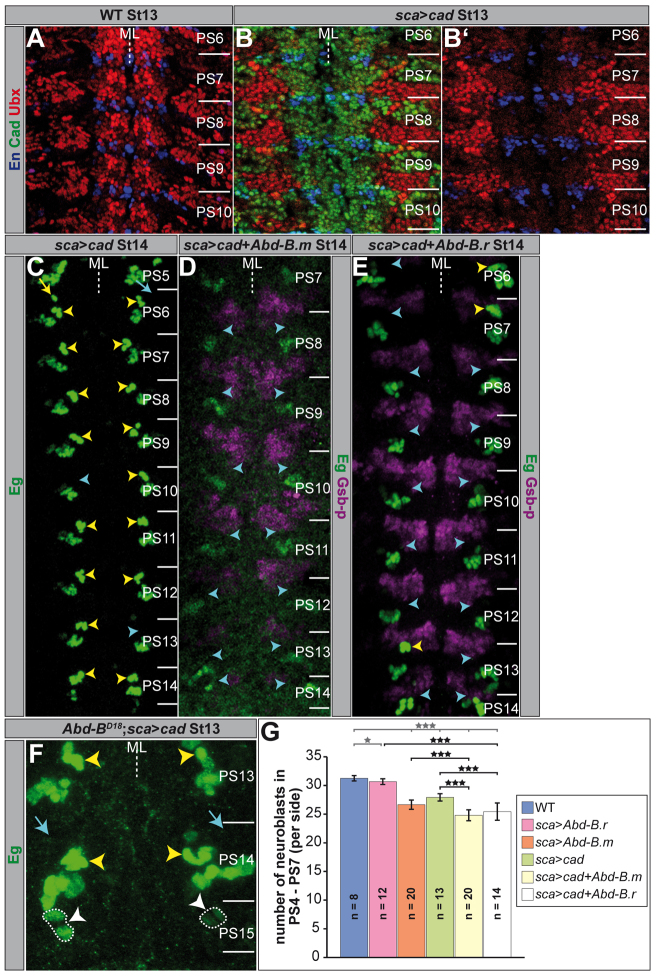
Fig. 8.
Abdominal-B.r needs caudal to mediate terminal fate. (A-F) Flat preparations (maximum projections) of St13 and St14 embryos of the indicated genotype, stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. (A) In wild type, Ubx is strongly expressed in the VNC and in the periphery. (B) Ectopic cad expression leads to an efficient downregulation of Ubx. (B′) shows only blue and red channel. (C-F) Yellow arrowheads mark regular NB7-3 clusters; cyan ones depict positions, where NB7-3 cells disappeared, and white ones (F) highlight ectopic NB7-3 cells in PS15 (additionally surrounded by broken lines). The yellow arrow marks the wild-type neuronal NB6-4 cluster in the thorax (C); cyan arrows point to locations where the neuronal NB6-4 cells disappeared. (G) Statistics for the total number of Dpn-positive NBs (per side) of the indicated genotypes in PS4-7. All genotypes were compared to wild type (grey stars) and the ectopic expression of single constructs was compared to corresponding ectopic co-expression (black stars). *P<0.05; ***P<0.001; error bars represent s.d. ML, midline; WT, wild type.
RESULTS
Abd-B suppresses formation of specific neuroblasts in PS15
We previously provided a comprehensive map of NBs generated in the tail region and found NB numbers to be significantly reduced in segments A9 (by 28%) and A10 (by 66%), compared with thoracic and anterior abdominal segments (Birkholz et al., 2013). As Hox gene expression domains coincide with parasegmental instead of segmental boundaries, we calculated NB numbers corresponding to terminal PSs. We used the stem cell marker Deadpan (Dpn) (Bier et al., 1992) to label NBs in early stage 12 (St12e) wild-type embryos and found 31.13±0.35 (n=8) Dpn-positive cells per side in PS13, 25.88±1.13 (n=8) in PS14 and 11.88±0.64 (n=8) in PS15 (Fig. 1A,K).
As Abd-B is expressed most posteriorly of all BX-C Hox genes, we investigated its role in shaping terminal neuromeres. Abd-BD18 is a small deficiency, removing the whole Abd-B CDS. In Abd-BD18 mutant embryos, we found the number of Dpn-positive cells in PS15 to be increased (15±0.67, n=10), whereas the more anterior PSs show no significant differences compared with wild type (Fig. 1B,K). Thus, normal Abd-B expression is required to reduce the number of NBs in PS15. In apoptosis-deficient Df(3L)H99 mutant embryos the number of Dpn-positive cells in terminal PSs corresponds to wild type (Fig. 1C,K). This indicates that Abd-B does not reduce NB numbers by inducing PCD, but by suppressing the formation of these NBs.
To identify NBs that are affected by this inhibition, we performed antibody stainings against Eg, which in thoracic and anterior abdominal neuromeres of wild-type embryos is expressed in four NBs (NB2-4, NB3-3, NB6-4 and NB7-3) and all of their progeny cells (Dittrich et al., 1997; Higashijima et al., 1996). According to the NB map of the tail region (Birkholz et al., 2013), NB6-4 is formed in PS14 and PS15 (but rapidly downregulates Eg expression). However, NB7-3 and NB3-3 are not formed in PS15, and NB2-4 is not generated in PS14 and PS15 (see also Fig. 1D). Therefore, these NBs are candidate NBs, which could be suppressed by Abd-B. In St13 Abd-BD18 mutants (Fig. 1E), as well as in Df(3L)H99 mutants (Fig. 1F), we found NB6-4, NB3-3 and NB2-4 to be distributed as in wild type, indicating that Abd-B does not affect formation of these NBs. Yet, their lineages displayed some changes: in Abd-BD18 mutants, the NB6-4 lineage in PS14 (100%, n=28 hemi-PSs) and occasionally in PS13 (14%, n=28) reveals homeotic transformation into thoracic fate, as it consists of neurons in addition to glial cells (Fig. 1E) [normally only glia in abdominal NB6-4 lineages (Schmidt et al., 1997)]. The NB3-3 lineage in PS14 of St16 Abd-BD18 mutants consists of 12 cells, compared to only six cells in wild type (not shown), corresponding to a transformation into thoracic and anterior abdominal fate (Schmidt et al., 1997). As the size of the NB3-3 lineage is also increased in PS14 of Df(3L)H99 mutants (10 cells), Abd-B seems to promote PCD in several progeny cells of NB3-3.
Strikingly, in PS15 of Abd-BD18 mutants we found an ectopic Eg-positive cell cluster (on each side; 100%, n=28; Fig. 1E), that we never observed in wild type (Fig. 1D) or in Df(3L)H99 mutants (Fig. 1F). These cells are negative for Runt (Run) (Fig. 1G), but positive for En (Fig. 1H) and are located within the last En stripe. As they are negative for the glial marker Reversed polarity (Repo) (Fig. 1I), they do not seem to derive from NB6-4. Instead, their Eyeless (Ey) expression unambiguously identifies them as NB7-3 progeny (Fig. 1J). Furthermore, they are located in the same dorsal position as the NB7-3 daughter cells in more anterior neuromeres and their precursor delaminates at St11l-St12e (S5). The cluster consists of up to three cells in St13, five cells in St14, and four cells from St16 on, which is also typical for the NB7-3 cell lineage (Bossing et al., 1996; Karcavich and Doe, 2005; Novotny et al., 2002; Schmid et al., 1999). We therefore conclude that the ectopic Eg-positive cluster generated in Abd-BD18 mutants corresponds to the NB7-3 lineage. Moreover, its absence in Df(3L)H99 mutants indicates that Abd-B does not act to induce PCD, but to repress formation of NB7-3 in PS15 of early wild-type embryos (see Fig. 9A). As the phenotype is also found in an Abd-BD18 heterozygous situation, but with lower penetrance (50%) (supplementary material Fig. S1; Fig. 9A), the effects appear to be dose dependent.
Fig. 9.
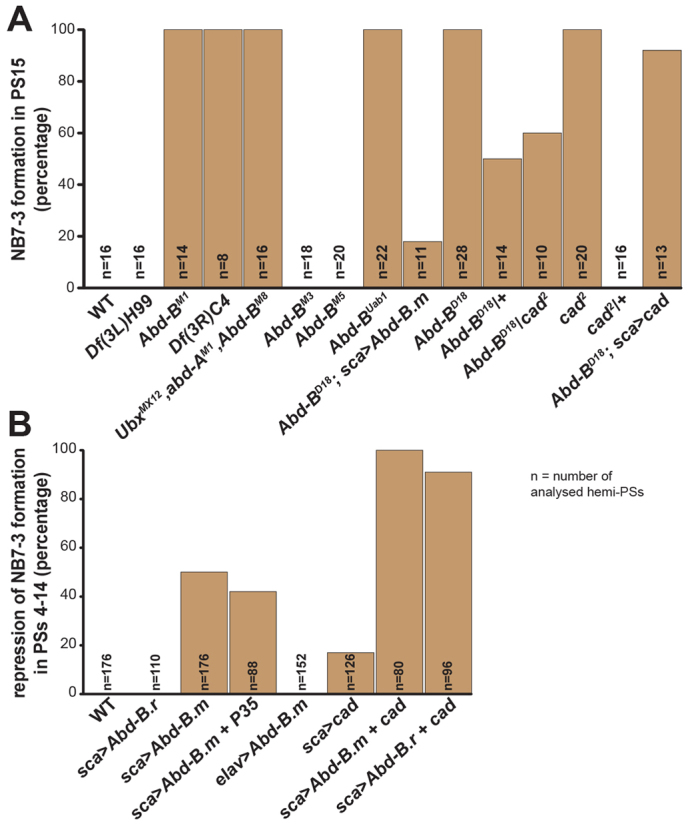
Frequencies of neuroblast 7-3 formation. Diagrams illustrating the relative frequencies of NB7-3 formation in PS15 in different mutant backgrounds (A), and the frequencies of NB7-3 suppression in PSs4-14 upon ectopic expression of different constructs (B), as indicated. WT, wild type.
In the following we use NB7-3 as a model to further elucidate the mechanism controlling restriction of NB formation in the tail region.
The neuroblast 7-3 lineage in PS15 of Abd-B mutants receives no input from the BX-C
Interactions among Hox genes of the BX-C are characterised by ‘posterior prevalence’, which means that Hox genes that are active in posterior PSs tend to repress the expression of more anterior Hox genes (for reviews, see Duboule and Morata, 1994; Morata, 1993). Accordingly, in a St14 wild-type embryo the Abd-A protein domain reaches from PS7 to PS13a, whereas in Abd-B loss-of-function mutants, Abd-A expression is at least expanded to PS14 (Karch et al., 1990; Kellerman et al., 1990; Macias et al., 1990). To examine whether the ectopic Eg-positive cells in Abd-B mutants arise as a result of expansion of the other genes from the BX-C, we investigated their expression in the VNC. In the loss-of-function allele Abd-BM1, and in Df(3R)C4, a chromosomal deletion of Abd-B, we found the anticipated expansion of Abd-A in the VNC, but we never observed Abd-A expression in the ectopic NB7-3 lineage of PS15, or in the NB3-3 and NB6-4 lineages of PS14 (supplementary material Fig. S2A,B). However, in Abd-BD18 mutants we found no obvious expansion of the Abd-A domain within the VNC (compare Fig. 2A,B). Recently it has been postulated that Abd-B is not necessary for the repression of Abd-A in the VNC, but instead the iab-8 non-coding RNA (Gummalla et al., 2012), which is also affected in Abd-BM1 and Df(3R)C4 but not in Abd-BD18. Yet, all mutants displayed the same phenotype concerning the Eg pattern (Fig. 9A). Ubx, the most anterior gene of the BX-C, is normally expressed from PS5 to PS13 in the VNC (White and Wilcox, 1984). In Abd-BD18 mutants we observed no posterior expansion of Ubx expression (Fig. 2C), which is in agreement with a previous report (Struhl and White, 1985). In addition, a triple mutant removing the whole BX-C revealed the same Eg pattern in PS14-15 as the Abd-B single mutants (supplementary material Fig. S3A,B; Fig. 9A).
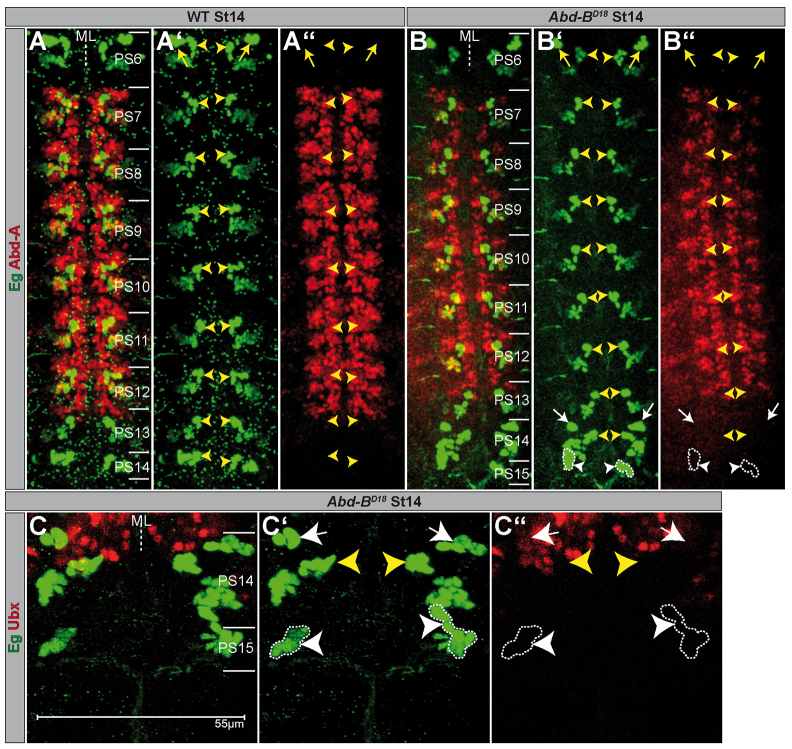
Fig. 2.
Expression of Abdominal-A and Ultrabithorax in the ventral nerve cord of wild type and Abdominal-B mutants. (A-C) Flat preparations (maximum projections) of St14 embryos of the indicated genotype double-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line; (A,B) PS6-15 are shown. There is no obvious expansion of Abd-A in Abd-BD18 mutants compared with wild type. (C) Magnification of PS14-15 is shown. (A′-C′) Eg expression; (A′-C′) Hox gene expression. The wild-type NB7-3 progeny is marked by yellow arrowheads. Ectopic NB7-3 cells are surrounded by broken lines and marked by white arrowheads. Neuronal subclones of thoracic NB6-4 are depicted by yellow arrows; ectopic ones in the abdomen are marked by white arrows. ML, midline; WT, wild type.
Thus, in Abd-B mutant background expression of Abd-A or Ubx is observed neither in the ectopic NB7-3 lineage in PS15, nor in the NB6-4 and NB3-3 clusters in PS14. As a consequence of the missing BX-C input they assume thoracic fate, resembling the ground state (pPS4 + aPS5) (Lewis, 1978).
The r isoform of Abd-B is responsible for the inhibition of neuroblast 7-3 formation in PS15
The Abd-B gene consists of at least four overlapping transcription units (Fig. 3A): Class A [nomenclature according to Zavortink and Sakonju (Zavortink and Sakonju, 1989)] is expressed in PS10-13 and is the only transcript that encodes m function. Class B and C are expressed in PS14-15, γ-transcripts only in PS15, and each of these transcript classes may contribute to r function (Boulet et al., 1991; DeLorenzi et al., 1988; Kuziora and McGinnis, 1988; Sánchez-Herrero and Crosby, 1988; Zavortink and Sakonju, 1989). To recapitulate the localisation of the genetic elements in the VNC by in situ hybridisations, we generated one probe, specific for the A-transcript (Abd-B.m) and a second one, directed against all transcripts encoding r function (Abd-B.r) (Fig. 3A). The stainings confirmed that in St12e Abd-B.m is expressed in PS13 and anterior to it, revealing a sharp posterior border (Fig. 3B), whereas Abd-B.r transcripts are exclusively expressed in PS14-15 (Fig. 3C). cDNA sequences revealed the existence of two different protein isoforms: Abd-B.m (encoded by the A-transcript) and Abd-B.r (encoded by the B, C and γ-transcripts). Abd-B.m is larger (55 kDa) and shares a common C-terminus with Abd-B.r, which harbours the homeodomain but has a unique glutamine-rich N-terminus, comprising 223 additional amino acids that are absent in the truncated r protein (30 kDa) (Fig. 3A) (Celniker et al., 1989; DeLorenzi et al., 1988; Zavortink and Sakonju, 1989). To investigate the wild-type Abd-B protein expression in distinct (Eg-positive) cells of the embryonic VNC, we used an antibody that detects both protein isoforms. As the expression domains of the Abd-B transcripts are spatially non-overlapping, it is reasonable to assume that the antibody detects the Abd-B.m or Abd-B.r protein in the respective PSs. Already at St11m, we detected abundant Abd-B.r expression in PS14, including NB3-3, and in PS15a. The expression of Abd-B.m in PS13 appeared rather weak and was restricted to the posterior, dorsolateral part of this PS (see also Delorenzi and Bienz, 1990). Neither NB2-4 nor NB3-3 in PS13 revealed expression of Abd-B.m (Fig. 3D). From delamination onwards NB6-4 and NB7-3 of PS14 showed very strong expression of Abd-B.r. However, in PS13 neither NB6-4 nor NB7-3 disclosed any Abd-B.m expression before, or directly after, delamination. The weak Abd-B.m expression in PS13 was still restricted to the posterior, dorsolateral part (Fig. 3E) and did not increase or expand in this PS before St12m (not shown) (Delorenzi and Bienz, 1990).
Next, we analysed Abd-B mutants, which specifically disrupt the function of only one isoform. Abd-BM3 and Abd-BM5 both solely disrupt the m function, but possess intact r function (m-r+) (Casanova et al., 1986). In both mutants, we found no obvious differences in the Eg pattern (Fig. 4A; supplementary material Fig. S3C; Fig. 9A) compared to wild type (Fig. 1D). This was expected, considering that Abd-B.m is not expressed posterior to PS13 and that in Abd-B null mutants (m-r-) the Eg pattern is only affected in PS14 and PS15. Consequently, in Abd-BUab1, a mutant specific for the r isoform (m+r-) (Casanova et al., 1986), we obtained almost the same phenotype as in the Abd-B loss-of-function alleles: significantly enlarged NB3-3 clusters and ectopic NB6-4 neuronal subclones in PS14, as well as ectopic NB7-3 lineages (co-expressing Eg, En and Ey) in PS15 (Fig. 4B, Fig. 9A). Thus, the loss of the Abd-B.r isoform is responsible for all described effects. It has been shown before that, in accordance with the ‘posterior prevalence’ phenomenon, Abd-B.r suppresses Abd-B.m (Casanova et al., 1986). Indeed, we observed Abd-B.m expression in PS14 and PS15 of Abd-BUab1 mutants. However, this expression was visible late (St11m) and dorsolaterally, just like in the unaffected PS13. Neither the neuronal NB6-4 clusters in PS14, nor the ectopic NB7-3 clones in PS15 displayed an early ectopic expression of Abd-B.m (Fig. 4C). A broader expression of Abd-B.m in PS14-15 came up in St12m (Fig. 4D), however, at a lower level compared with Abd-B.r expression in wild type (Fig. 3E) (Boulet et al., 1991; Sánchez-Herrero, 1991). Thus, Abd-B.m seems to be expressed too late in PS14 and PS15 of Abd-BUab1 mutants to rescue the phenotype that emerges due to the loss of Abd-B.r.
Fig. 4.
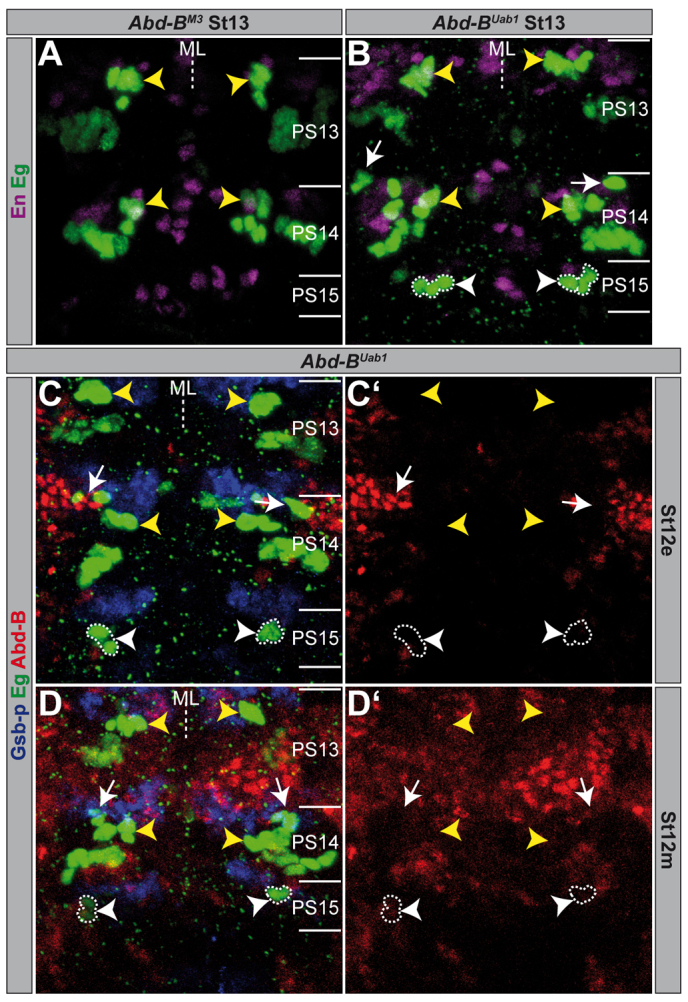
Abdominal-B.r, not Abdominal-B.m is responsible for the Eagle pattern phenotype. (A-D) Flat preparations (maximum projections) of St12e, St12m and St13 embryos of the indicated genotype double- or triple-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. Wild-type NB7-3 cells are in all cases marked by yellow arrowheads. (B-D) Ectopic NB7-3 clones in PS15 are surrounded by broken lines and indicated by white arrowheads. Ectopic NB6-4 neuronal subclones in PS14 are marked by white arrows. (C,D) Gooseberry-proximal (Gsb-p) served as an alternative segmental marker and is expressed anterior to and partially overlapping with En posteriorly (Gutjahr et al., 1993). (C′,D′) Red channel only. ML, midline.
Taken together, the isoform Abd-B.r is responsible for inhibition of NB7-3 formation in PS15, for specification of abdominal NB6-4 in PS14 and for NB3-3-fate in PS14.
Ectopic Abd-B.m, but not Abd-B.r, is able to inhibit the formation of neuroblast 7-3 in anterior PSs
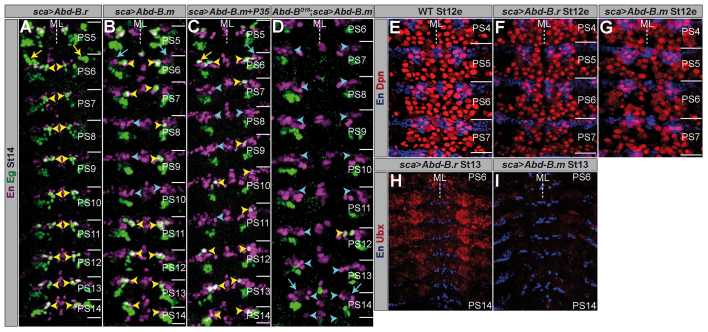
As Abd-B.r suppresses the formation of NB7-3 in PS15, we tested its ability to inhibit the formation of this NB in anterior PSs, when expressed ectopically. To drive the expression of UAS-Abd-B.r we used scabrous (sca)-Gal4, which from St8 onwards reveals a broad and strong expression in the NE and in the NBs that delaminate from it. Surprisingly, ectopic Abd-B.r does not remove NB7-3 (0%, n=110 thoracic and abdominal hemi-PSs) (Fig. 9B) or any other Eg-positive cell cluster in more anterior PSs (Fig. 5A). To investigate if the total number of NBs is altered in this situation, we counted Dpn-positive cells in PS4-7. We noticed only a marginal decrease in the number of NBs per hemi-PS (30.67±0.49, n=12; Fig. 5F) compared with wild type (31.25±0.46, n=8; Fig. 5E, Fig. 8G). In a few cases the ectopic expression of Abd-B.r resulted in a homeotic transformation of the thoracic NB6-4 into an abdominal fate, which is accompanied by the loss of its neuronal sublineage (13%, n=36 thoracic hemi-PSs). However, the frequency of this homeotic transformation was strongly increased when we ectopically expressed Abd-B.m (81%, n=48 thoracic hemi-PSs). Furthermore, we observed a reduction in the total number of NBs per hemi-PS (26.65±0.81, n=20; Fig. 5G, Fig. 8G), which clearly exceeds the effect of ectopic Abd-B.r. This includes efficient removal of thoracic and abdominal NB7-3 cell clusters (50%, n=176 thoracic and abdominal hemi-PSs) (Fig. 5B, Fig. 9B). Upon ectopic expression of Abd-B.m together with P35, a known suppressor of apoptosis, the NB7-3 clusters in anterior PSs could not be rescued (42%; Fig. 9B), which confirms that they do not undergo PCD (Fig. 5C). Additionally, we ectopically expressed Abd-B.m by using an embryonic lethal abnormal vision (elav) driver line, which directs expression in all neuronal tissues from St12l on, but is not expressed in the NE. This late expression of Abd-B.m was not able to remove the NB7-3 cells in thorax or abdomen (0%, n=152) (supplementary material Fig. S4; Fig. 9B). These results emphasise that a strong ectopic expression of Abd-B.m in the NE (as achieved by the sca-Gal4 driver) is required to repress the formation of NB7-3 in more anterior PSs, whereas late expression does not lead to a removal of NB7-3 cells (as in PS13 of wild type).
Fig. 5.
Ectopically expressed Abdominal-B.m, but not Abdominal-B.r has the potential to suppress the formation of neuroblast 7-3. (A-I) Flat preparations (maximum projections) of St12e, St13 and St14e embryos of the indicated genotypes, double-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. (A-D) Wild-type NB7-3 cells are marked by yellow arrowheads. Positions where NB7-3 clusters were removed are marked by cyan arrowheads. Neuronal NB6-4 clusters in the thorax are marked by yellow arrows; positions where they are missing are marked by cyan arrows. ML, midline; WT, wild type.
We also tried to rescue the Eg expression phenotype of Abd-BD18 mutants by applying ectopic Abd-B.m in the mutant situation (using sca-Gal4). In most cases, we were able to remove the neuronal NB6-4 subclone in PS14 (73%, n=11 hemi-PSs) and the ectopic NB7-3 lineage in PS15 (82%, n=11 hemi-PSs) (Fig. 9A). The efficiency of this rescue strongly correlated with the disappearance of NB7-3 cell clusters in more anterior PSs (Fig. 5D). This suggests that Abd-B.m, if expressed early enough, can completely substitute for Abd-B.r function concerning repression of the neuronal NB6-4 sublineage and inhibition of NB formation. In wild type, Abd-B.m cannot adopt this function as its expression starts too late.
Why does ectopic Abd-B.m possess the potency to suppress formation of specific NBs, whereas ectopic Abd-B.r does not? When we compared the ability of the respective constructs to downregulate Ubx expression, we noticed that Abd-B.m was very efficient (Fig. 5I) (Castelli-Gair et al., 1994), whereas Abd-B.r was not (Fig. 5H). Perhaps ectopic Abd-B.r does not mediate a homeotic transformation, because it cannot suppress anterior Hox proteins (see Discussion).
cad also suppresses formation of NB7-3 in PS15
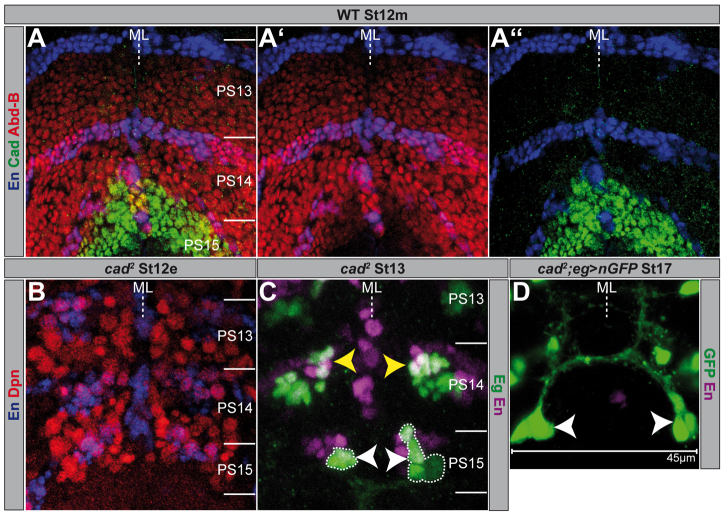
It has been claimed that the ParaHox gene cad determines the character of the most posterior segment. Although it is not a member of the BX-C, ectopic cad expression induces analia development in anterior segments and its loss results in a homeotic transformation into the next anterior segment (Moreno and Morata, 1999). cad reveals maternal and zygotic expression, sharing a common CDS (Mlodzik et al., 1985; Mlodzik and Gehring, 1987). In St12m we observed zygotic Cad expression in the NE of PS15 (Macdonald and Struhl, 1986), where it overlaps with the expression domain of Abd-B.r, but it is also expressed posterior to this (Sánchez-Herrero and Crosby, 1988). Its anterior margin is restricted by the En stripe of PS15, and there is only a minor overlap in the expression of En and Cad (Kuhn et al., 1995) (Fig. 6A). In the VNC we found co-expression of Cad and Abd-B.r in NBs belonging to PS15, although we never observed NBs exclusively displaying Cad expression (supplementary material Fig. S5A).
Fig. 6.
caudal mutants exhibit an ectopic neuroblast 7-3 lineage in parasegment 15. Flat preparations (maximum projections) of St12e, St12m, St13 and St17 embryos of the indicated genotype, double- or triple-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. (A) Domains of Abd-B and Cad expression; (A′) Abd-B and En; (A′) Cad and En. (B) NB pattern in cad2 mutant. (C) Regular NB7-3 progeny cells are marked by yellow arrowheads, ectopic ones are marked by white arrowheads and surrounded by broken lines. (D) Magnification of a completely differentiated NB7-3 lineage in PS15, forming the NB7-3 typical projections. ML, midline; WT, wild type.
Considering that Cad is co-expressed with Abd-B.r in NBs of PS15, we wondered whether Cad might also affect NB formation. In zygotic cad null mutants, cad2, we observed a significant increase of NBs in PS15 (14.25±1.26, n=4; Fig. 6B) compared with wild type (11.88±0.64, n=8; Fig. 1A,K). Similar to Abd-BD18, cad2 mutants show an ectopic cluster of cells on each side of PS15, co-expressing Eg and En (Fig. 6C, Fig. 9A). By combining cad2 with eg-Gal4, UAS-nGFP, we could clearly show that in late stages this cell cluster completely differentiates as a NB7-3 cell lineage, forming its typical contralateral projections (Fig. 6D) (Bossing et al., 1996; Schmid et al., 1999). However, different from Abd-BD18, NBs in PS14 are not affected in cad2 mutants (no ectopic neuronal NB6-4 sublineage or enlarged NB3-3 lineage), as expected from the lack of Cad expression in this PS. The failure to inhibit the formation of NB7-3 in PS15 of cad2 mutants seems to be a recessive effect, as we noticed it only in the homozygous situation, whereas Eg expression in the heterozygotes is indistinguishable from wild type (Fig. 9A). These data show that cad is also involved in the suppression of NB formation in PS15.
cad and Abd-B.r act independently, but need to be co-expressed to efficiently inhibit the formation of NB7-3
As both Abd-B and Cad affect NB7-3 formation in PS15, we attempted to clarify their functional relationship.
In Abd-BD18 (and in Abd-BUab1) mutants, Cad is still expressed in the ectopically formed NB7-3 in PS15 (Fig. 7A). Conversely, in cad2 mutants we noticed Abd-B expression in cells of the ectopic NB7-3 lineage of PS15 (Fig. 7B), although it seems slightly delayed. Furthermore, neither ectopic Abd-B.r nor ectopic Abd-B.m was able to activate or repress Cad expression in the VNC, when driven by sca-Gal4. Moreover, ectopic cad does not change the expression domain and intensity of Abd-B (not shown). These data suggest that Abd-B and Cad do not regulate each other within the VNC, and that they act independently to suppress NB7-3 formation. In cad2/Abd-BD18 transheterozygotes (supplementary material Fig. S5B), we observed the same phenotype as in the Abd-BD18 heterozygous situation (supplementary material Fig. S1; Fig. 9A).
Fig. 7.
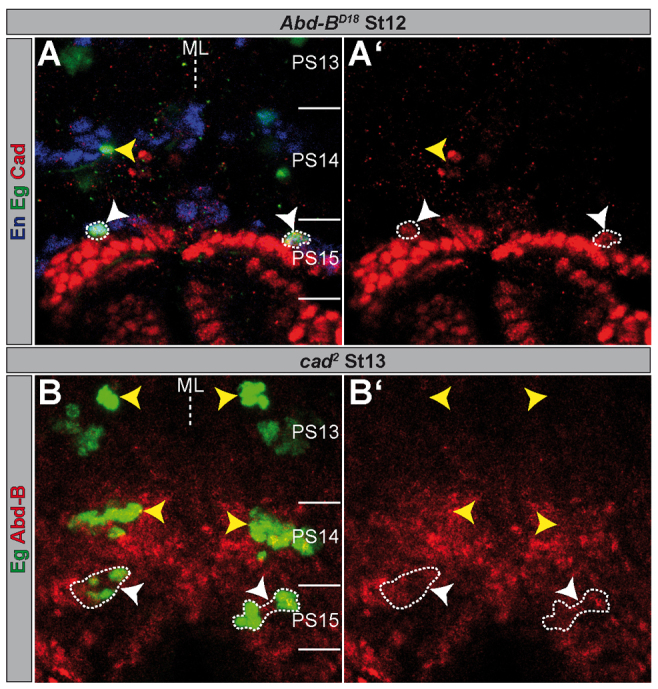
Ectopic neuroblast 7-3 cells in parasegment 15 reveal expression of Abdominal-B.r or Caudal. (A,B) Flat preparations of St12 and St13 embryos of the indicated genotype double- or triple-stained as illustrated. PSs are depicted on the right and their borders are illustrated by a solid line. Regular NB7-3 cells are marked by yellow arrowheads; ectopic ones are marked by white arrowheads and surrounded by broken lines. (A) Single layer; (B) several layers. (A′,B′) Red channel only. ML, midline.
Although it is not able to reduce the expression of Abd-B, ectopic cad strongly downregulates Ubx (Fig. 8B) and Abd-A (supplementary material Fig. S6A), in this respect fulfilling the ‘posterior prevalence’ criterion for Hox genes. Ectopic cad expression (driven by sca-Gal4) was weakly able to inhibit the formation of NB7-3 in anterior PSs (17%, n=126 thoracic and abdominal hemi-PSs) (Fig. 8C, Fig. 9B). In agreement with this, the ectopic NB7-3 lineage in PS15 of Abd-BD18 mutants was just missing in one case when we ectopically expressed cad (8%, n=13 hemi-PSs) (Fig. 8F, Fig. 9A). Additionally, ectopic cad was also able to transform thoracic NB6-4 into abdominal identity (50%, n=30 thoracic hemi-PSs) (Fig. 8C) and rescued the abdominal NB6-4 fate in PS14 of Abd-BD18 mutants (Fig. 8F). Furthermore, like Abd-B.m, cad has the potential to inhibit the formation of further NBs (in addition to NB7-3) in more anterior PSs (27.92±0.64, n=13; supplementary material Fig. S6B; Fig. 8G), although with lower efficiency. However, its ability increases significantly when it is ectopically expressed together with Abd-B.m (24.8±0.95, n=20; supplementary material Fig. S6C; Fig. 8G). This is also reflected by repression of NB7-3 in all investigated PSs (100%, n=80 abdominal and thoracic hemi-PSs; Fig. 8D, Fig. 9B). Upon ectopic expression of Abd-B.r together with cad, which reflects their native co-expression in PS15, we observed almost the same efficiency in suppression of NB7-3 (91%, n=96 hemi-PSs; Fig. 8E, Fig. 9B) and of other NBs (25.43±1.50, n=14; supplementary material Fig. S6D; Fig. 8G), as opposed to the inefficient suppression of NB7-3 upon misexpression of Abd-B.r alone.
Taken together, although Abd-B and Cad act independently of each other, their combined expression is needed to unfold the full potential concerning inhibition of NB formation.
DISCUSSION
A novel role for Abd-B and cad
During the generation of the Drosophila VNC, Hox genes of the BX-C act along the AP axis during different stages of development at the level of NE, NBs and their progeny cells to control segmental patterning (reviewed by Rogulja-Ortmann and Technau, 2008). In the NE they confer segment-specific intrinsic properties to NBs before their delamination (Prokop and Technau, 1994). For example, abd-A and Abd-B specify NB6-4 to produce the abdominal variant of its lineage (only glial cells) as opposed to its thoracic variant (glia plus neurons), which represents the ground state (no input of homeotic genes) (Berger et al., 2005; this study). Accordingly, ectopic expression of Abd-B.m leads to transformation of thoracic NB6-4 into abdominal fate (loss of neuronal sublineage). Early function of Hox genes at NE and NB level also includes the determination of segment-specific subpopulations of NBs that persist at the end of embryogenesis to become postembryonic NBs and (after a period of quiescence) resume proliferation in the larva (Prokop et al., 1998; Tsuji et al., 2008). During later stages Hox genes have been shown to control the specification of particular cell types by segment-specific gene regulation and integration of temporal cues on the level of NBs and their progeny (Estacio-Gómez et al., 2013; Karlsson et al., 2010).
A further mechanism by which Hox genes modulate segmental pattern in the CNS is through regulation of PCD. At the level of differentiated progeny cells, segment-specific control of PCD occurs as a late function of Hox genes in the embryonic VNC (Miguel-Aliaga and Thor, 2004; Rogulja-Ortmann et al., 2008; Suska et al., 2011). At the level of postembryonic NBs, the end of their proliferation is scheduled by Hox genes via PCD during the third instar larval stage (Bello et al., 2003). Furthermore, a set of sex-specific NBs in the terminal region of the VNC undergoes PCD during late embryonic/early larval stages only in females (Birkholz et al., 2013). Thus, it appeared likely that PCD also contributes to the reduction of the total number of embryonic NBs in terminal neuromeres (PS14-15) compared with more anterior PSs. Indeed, localised PCD has been shown to occur at segment boundaries in the ectoderm of gnathal and terminal abdominal segments (Lohmann et al., 2002; Nassif et al., 1998), which is controlled by Hox genes (Deformed and Abd-B) via activation of pro-apoptotic genes (Hueber et al., 2007; Lohmann et al., 2002; Zhai et al., 2010). However, in terminal neuromeres of Df(3L)H99 mutant embryos (St12e) we did not observe significant differences in numbers of NBs compared to wild type, indicating that PCD does not account for the diminished number of NBs generated in the tail region. Instead, sizes of metameric units in the blastodermal tail anlagen are significantly reduced, as revealed by fate-mapping data (Hartenstein et al., 1985; Juergens, 1987; Technau and Campos-Ortega, 1985). Specification of these units is under the control of a number of genes, which affect the identity of terminal segments, including Abd-B, cad, spalt (salm - FlyBase) and forkhead (Jürgens, 1988; Jürgens and Weigel, 1988; Moreno and Morata, 1999; Sánchez-Herrero et al., 1985). One function of Abd-B and probably cad involves suppression of ventral structures in terminal segments (Kuhn et al., 1995). If this also affects the NE, the degree of its rudimentation may account for the reduced numbers of NBs that can be formed in these segments.
Our data show that Abd-B and cad are required in PS15 to suppress the formation of a specific subset of about three to four NBs per side. This could be a consequence of the contribution of these genes to restrict the size of the NE. In this case they would prevent the establishment of only those parts of the NE, which specifically give rise to these NBs. For example, as NB7-3 in more anterior PSs normally derives from the intermediate (along the DV axis) and En-positive NE, one would expect this NE domain to be suppressed by Abd-B and cad in PS15. Alternatively, this domain may be established, but Abd-B and cad may suppress the formation of NB7-3 within this region. They could do so by regulating the expression of proneural genes, which define proneural cell clusters in the NE, each singling out an individual NB (reviewed by Skeath and Carroll, 1994). Indeed, a genetic screen for Hox downstream targets revealed that ectopic expression of Abd-B results in a downregulation of lethal of scute [l(1)sc] (Hueber et al., 2007), one of the four members of the Achaete-Scute complex (Martín-Bermudo et al., 1991). In wild type, L(1)sc is expressed in the proneural cluster that gives rise to NB7-3 and for a short period in the NB itself (S. R. Myneni, G.M.T. and A.R.-O., unpublished results). However, the ectopic co-expression of cad and Abd-B.r reduces the number of NBs from ∼31 to ∼25 in anterior segments, which is significantly different from the amount observed in wild-type PS15 (∼12). This suggests that additional (so far unidentified) posterior patterning cues are required to suppress the formation of further NBs.
Abd-B.r cooperates with cad to mediate terminal fate in the VNC
Abd-B.m is expressed in PS13 (and anterior to this) and Abd-B.r in PS14 and PS15, where it overlaps with the expression of Cad. Our data indicate that Abd-B.r is necessary, but not sufficient to mediate terminal fate in the VNC. Upon ectopic expression of Abd-B.r we observed no obvious transformation of an anterior PS into posterior fate, regarding total NB number, inhibition of NB7-3 formation or frequent change of NB6-4 identity. In addition, we noticed no detectable changes in Ubx expression in the VNC compared to wild type, although ectopic Abd-B.r expression was described to result in a downregulation of Ubx transcripts (Kuziora, 1993). As shown before, Abd-B.r cannot be limited to its regulatory function as a repressor of Abd-B.m, as it also possesses a morphogenetic role in activating gene expression (Ali and Bienz, 1991; Thali et al., 1988). However, a recent publication revealed that ectopic Abd-B.r has only a weak morphogenetic capacity in anterior segments, as it very inefficiently downregulates co-factors of anterior Hox genes, competing and antagonising Abd-B function (Rivas et al., 2013). Our data underline these observations and indicate that, in terms of mediating posterior identity in the VNC, Abd-B.r requires the co-expression of cad to unfold its complete morphogenetic property. This is in line with a previous report, suggesting that both Abd-B and cad are necessary to repress the formation of ventral structures in terminal PSs (Kuhn et al., 1995). Accordingly, the ectopic expression of cad or Abd-B.r alone has only minor effects on more anterior PSs (Mlodzik et al., 1990; Rivas et al., 2013; this study). Upon ectopic co-expression in the VNC they seem to contribute complementary functions: cad is able to downregulate anterior Hox genes (Ubx, abd-A), which may be a prerequisite for Abd-B.r to repress NB formation. In contrast, Abd-B.m unifies both functions. Why it is equipped with the potency to suppress NB formation, although it does not exhibit this function in its normal expression domain, remains an open question. As the Abd-B.m and the truncated Abd-B.r protein share the same C-terminal region, it may harbour the capability to suppress the formation of NBs, while the N-terminal domain of Abd-B.m may be necessary to downregulate anterior Hox genes. Thus, cad might compensate for the missing N-terminal domain of Abd-B.r, by downregulating Hox genes. At the protein level, the N-terminal domain of Abd-B.m comprises a huge amount of glutamine runs, known as M or opa repeats (Wharton et al., 1985). A similar asparagine stretch, which might be functionally equivalent, has been identified in cad (Mlodzik et al., 1985).
Drosophila cad is located on chromosome 2L and belongs to the group of dispersed ParaHox genes, resembling ancient paralogues of the ANT-C and BX-C. cad and Abd-B might have evolved by duplication from an ancestral ProtoHox gene cluster and therefore reflect evolutionary sisters (Brooke et al., 1998; reviewed by Garcia-Fernàndez, 2005). Accordingly, cad shares several features with Hox genes of the BX-C: a Hox typical hexapeptide motif N-terminal to the homeodomain and an intron at the same position as Abd-B (Gehring et al., 1994). As expected for paralogous genes, we observed overlapping domains of Cad and Abd-B.r in the VNC. Ectopic cad expression efficiently downregulates Ubx- and Abd-A expression. However, in contrast to previous observations in genital discs (Moreno and Morata, 1999), we found no evidence for a downregulation of Abd-B in the VNC, which would be an exception from the ‘posterior prevalence’ rule.
There exist three cad homologues in mammals: Cdx1, Cdx2 and Cdx4 (Ferrier et al., 2005). In mouse (Mus musculus) they are expressed collinearly and overlap in the caudal embryonic neural tube (Beck et al., 1995; Gamer and Wright, 1993; Meyer and Gruss, 1993). Mutations result in anterior homeotic transformations and reveal functional redundancy in the control of posterior identity (Chawengsaksophak et al., 2004; Chawengsaksophak et al., 1997; Subramanian et al., 1995; van den Akker et al., 2002; van Nes et al., 2006), suggesting a conserved function between Mus musculus and Drosophila.
Acknowledgments
We thank James Castelli-Gair Hombría and L. S. Shashidhara for helpful comments on the manuscript and Sudha Rani Myneni for sharing unpublished results. We are very grateful to James Castelli-Gair Hombría, Ernesto Sánchez-Herrero, François Karch, Paul Macdonald, Ulrich Schaefer, Mi-Ae Yoo, John Reinitz, Harald Vaessin, Alfonso Martinez-Arias, Ian Duncan, Chris Doe, Uwe Walldorf, Robert Holmgren, Juergen Knoblich, the Bloomington Stock Center, the Developmental Studies Hybridoma Bank (University of Iowa) and FlyBase for providing flies, antibodies, cDNAs and sequence information.
Footnotes
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft to G.M.T. [TE 130/10; GRK 1044-A1]. Deposited in PMC for immediate release.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
O.B. and G.M.T. conceived the project, designed the experiments and wrote the manuscript. O.B. performed all of the experiments. O.V. contributed to the generation of fly strains. A.R.-O. and C.B. contributed to experimental design and provided crucial input.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.096099/-/DC1
References
- Ali N., Bienz M. (1991). Functional dissection of Drosophila abdominal-B protein. Mech. Dev. 35, 55–64 [DOI] [PubMed] [Google Scholar]
- Beck F., Erler T., Russell A., James R. (1995). Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev. Dyn. 204, 219–227 [DOI] [PubMed] [Google Scholar]
- Bello B. C., Hirth F., Gould A. P. (2003). A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron 37, 209–219 [DOI] [PubMed] [Google Scholar]
- Berger C., Pallavi S. K., Prasad M., Shashidhara L. S., Technau G. M. (2005). A critical role for cyclin E in cell fate determination in the central nervous system of Drosophila melanogaster. Nat. Cell Biol. 7, 56–62 [DOI] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., Knoblich J. A. (2006). Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 [DOI] [PubMed] [Google Scholar]
- Bhat K. M. (1999). Segment polarity genes in neuroblast formation and identity specification during Drosophila neurogenesis. BioEssays 21, 472–485 [DOI] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Younger-Shepherd S., Jan L. Y., Jan Y. N. (1992). deadpan, an essential pan-neural gene in Drosophila, encodes a helix-loop-helix protein similar to the hairy gene product. Genes Dev. 6, 2137–2151 [DOI] [PubMed] [Google Scholar]
- Birkholz O., Rickert C., Berger C., Urbach R., Technau G. M. (2013). Neuroblast pattern and identity in the Drosophila tail region and role of doublesex in the survival of sex-specific precursors. Development 140, 1830–1842 [DOI] [PubMed] [Google Scholar]
- Bossing T., Udolph G., Doe C. Q., Technau G. M. (1996). The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 179, 41–64 [DOI] [PubMed] [Google Scholar]
- Boulet A. M., Lloyd A., Sakonju S. (1991). Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111, 393–405 [DOI] [PubMed] [Google Scholar]
- Broadus J., Skeath J. B., Spana E. P., Bossing T., Technau G., Doe C. Q. (1995). New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mech. Dev. 53, 393–402 [DOI] [PubMed] [Google Scholar]
- Brooke N. M., Garcia-Fernàndez J., Holland P. W. (1998). The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922 [DOI] [PubMed] [Google Scholar]
- Campos-Ortega J. A., Hartenstein V. (1997). The Embryonic Development of Drosophila Melanogaster. New York, NY: Springer Verlag; [Google Scholar]
- Casanova J., Sánchez-Herrero E., Morata G. (1986). Identification and characterization of a parasegment specific regulatory element of the abdominal-B gene of Drosophila. Cell 47, 627–636 [DOI] [PubMed] [Google Scholar]
- Casanova J., Sánchez-Herrero E., Busturia A., Morata G. (1987). Double and triple mutant combinations of bithorax complex of Drosophila. EMBO J. 6, 3103–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli-Gair J., Greig S., Micklem G., Akam M. (1994). Dissecting the temporal requirements for homeotic gene function. Development 120, 1983–1995 [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Keelan D. J., Lewis E. B. (1989). The molecular genetics of the bithorax complex of Drosophila: characterization of the products of the Abdominal-B domain. Genes Dev. 3, 1424–1436 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., James R., Hammond V. E., Köntgen F., Beck F. (1997). Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84–87 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorenzi M., Bienz M. (1990). Expression of Abdominal-B homeoproteins in Drosophila embryos. Development 108, 323–329 [DOI] [PubMed] [Google Scholar]
- DeLorenzi M., Ali N., Saari G., Henry C., Wilcox M., Bienz M. (1988). Evidence that the Abdominal-B r element function is conferred by a trans-regulatory homeoprotein. EMBO J. 7, 3223–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch J. S. (2004). Segments and parasegments in arthropods: a functional perspective. BioEssays 26, 1117–1125 [DOI] [PubMed] [Google Scholar]
- DiNardo S., Kuner J. M., Theis J., O’Farrell P. H. (1985). Development of embryonic pattern in D. melanogaster as revealed by accumulation of the nuclear engrailed protein. Cell 43, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich R., Bossing T., Gould A. P., Technau G. M., Urban J. (1997). The differentiation of the serotonergic neurons in the Drosophila ventral nerve cord depends on the combined function of the zinc finger proteins Eagle and Huckebein. Development 124, 2515–2525 [DOI] [PubMed] [Google Scholar]
- Doe C. Q. (1992). Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116, 855–863 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Fuerstenberg S., Peng C. Y. (1998). Neural stem cells: from fly to vertebrates. J. Neurobiol. 36, 111–127 [PubMed] [Google Scholar]
- Duboule D., Morata G. (1994). Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 10, 358–364 [DOI] [PubMed] [Google Scholar]
- Duncan I. (1987). The bithorax complex. Annu. Rev. Genet. 21, 285–319 [DOI] [PubMed] [Google Scholar]
- Estacio-Gómez A., Moris-Sanz M., Schäfer A. K., Perea D., Herrero P., Díaz-Benjumea F. J. (2013). Bithorax-complex genes sculpt the pattern of leucokinergic neurons in the Drosophila central nervous system. Development 140, 2139–2148 [DOI] [PubMed] [Google Scholar]
- Ferrier D. E., Dewar K., Cook A., Chang J. L., Hill-Force A., Amemiya C. (2005). The chordate ParaHox cluster. Curr. Biol. 15, R820–R822 [DOI] [PubMed] [Google Scholar]
- Gamer L. W., Wright C. V. (1993). Murine Cdx-4 bears striking similarities to the Drosophila caudal gene in its homeodomain sequence and early expression pattern. Mech. Dev. 43, 71–81 [DOI] [PubMed] [Google Scholar]
- Garcia-Fernàndez J. (2005). Hox, ParaHox, ProtoHox: facts and guesses. Heredity (Edinb.) 94, 145–152 [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Affolter M., Bürglin T. (1994). Homeodomain proteins. Annu. Rev. Biochem. 63, 487–526 [DOI] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S., Edwards K. A., Karch F., Bender W. (2012). abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8, e1002720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr T., Patel N. H., Li X., Goodman C. S., Noll M. (1993). Analysis of the gooseberry locus in Drosophila embryos: gooseberry determines the cuticular pattern and activates gooseberry neuro. Development 118, 21–31 [DOI] [PubMed] [Google Scholar]
- Hama C., Ali Z., Kornberg T. B. (1990). Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 4, 1079–1093 [DOI] [PubMed] [Google Scholar]
- Harding K., Wedeen C., McGinnis W., Levine M. (1985). Spatially regulated expression of homeotic genes in Drosophila. Science 229, 1236–1242 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Technau G. M., Campos-Ortega J. A. (1985). Fate-mapping in wild-type Drosophila melanogaster: III. A fate map of the blastoderm. Rouxs Arch. Dev. Biol. 194, 213–216 [Google Scholar]
- Hay B. A., Wolff T., Rubin G. M. (1994). Expression of baculovirus P35 prevents cell death in Drosophila. Development 120, 2121–2129 [DOI] [PubMed] [Google Scholar]
- Higashijima S., Shishido E., Matsuzaki M., Saigo K. (1996). eagle, a member of the steroid receptor gene superfamily, is expressed in a subset of neuroblasts and regulates the fate of their putative progeny in the Drosophila CNS. Development 122, 527–536 [DOI] [PubMed] [Google Scholar]
- Hopmann R., Duncan D., Duncan I. (1995). Transvection in the iab-5,6,7 region of the bithorax complex of Drosophila: homology independent interactions in trans. Genetics 139, 815–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber S. D., Bezdan D., Henz S. R., Blank M., Wu H., Lohmann I. (2007). Comparative analysis of Hox downstream genes in Drosophila. Development 134, 381–392 [DOI] [PubMed] [Google Scholar]
- Hwang M. S., Kim Y. S., Choi N. H., Park J. H., Oh E. J., Kwon E. J., Yamaguchi M., Yoo M. A. (2002). The caudal homeodomain protein activates Drosophila E2F gene expression. Nucleic Acids Res. 30, 5029–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath J. B., Nabeshima Y., Doe C. Q., Matsuzaki F. (1997). Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–629 [DOI] [PubMed] [Google Scholar]
- Ito K., Urban J., Technau G. M. (1995). Distribution, classification, and development of Drosophila glial cells in the late embryonic and early larval ventral nerve cord. Rouxs Arch. Dev. Biol. 204, 284–307 [DOI] [PubMed] [Google Scholar]
- Juergens G. (1987). Segmental organisation of the tail region in the embryo of Drosophila melanogaster. Rouxs Arch. Dev. Biol. 196, 141–157 [DOI] [PubMed] [Google Scholar]
- Juergens G., Weigel D. (1988). Terminal versus segmental development in the Drosophila embryo - the role of the homeotic gene fork head. Rouxs Arch. Dev. Biol. 197, 345–354 [DOI] [PubMed] [Google Scholar]
- Jürgens G. (1988). Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 7, 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammermeier L., Leemans R., Hirth F., Flister S., Wenger U., Walldorf U., Gehring W. J., Reichert H. (2001). Differential expression and function of the Drosophila Pax6 genes eyeless and twin of eyeless in embryonic central nervous system development. Mech. Dev. 103, 71–78 [DOI] [PubMed] [Google Scholar]
- Karcavich R., Doe C. Q. (2005). Drosophila neuroblast 7-3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J. Comp. Neurol. 481, 240–251 [DOI] [PubMed] [Google Scholar]
- Karch F., Weiffenbach B., Peifer M., Bender W., Duncan I., Celniker S., Crosby M., Lewis E. B. (1985). The abdominal region of the bithorax complex. Cell 43, 81–96 [DOI] [PubMed] [Google Scholar]
- Karch F., Bender W., Weiffenbach B. (1990). abdA expression in Drosophila embryos. Genes Dev. 4, 1573–1587 [DOI] [PubMed] [Google Scholar]
- Karlsson D., Baumgardt M., Thor S. (2010). Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol. 8, e1000368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Lewis R., Wakimoto B. (1980). Cytogenetic analysis of chromosome 3 in DROSOPHILA MELANOGASTER: The homoeotic gene complex in polytene chromosome interval 84a-B. Genetics 94, 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellerman K. A., Mattson D. M., Duncan I. (1990). Mutations affecting the stability of the fushi tarazu protein of Drosophila. Genes Dev. 4, 1936–1950 [DOI] [PubMed] [Google Scholar]
- Klaes A., Menne T., Stollewerk A., Scholz H., Klämbt C. (1994). The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell 78, 149–160 [DOI] [PubMed] [Google Scholar]
- Kosman D., Small S., Reinitz J. (1998). Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208, 290–294 [DOI] [PubMed] [Google Scholar]
- Kuhn D. T., Turenchalk G., Mack J. A., Packert G., Kornberg T. B. (1995). Analysis of the genes involved in organizing the tail segments of the Drosophila melanogaster embryo. Mech. Dev. 53, 3–13 [DOI] [PubMed] [Google Scholar]
- Kuziora M. A. (1993). Abdominal-B protein isoforms exhibit distinct cuticular transformations and regulatory activities when ectopically expressed in Drosophila embryos. Mech. Dev. 42, 125–137 [DOI] [PubMed] [Google Scholar]
- Kuziora M. A., McGinnis W. (1988). Different transcripts of the Drosophila Abd-B gene correlate with distinct genetic sub-functions. EMBO J. 7, 3233–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 [DOI] [PubMed] [Google Scholar]
- Lin D. M., Goodman C. S. (1994). Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron 13, 507–523 [DOI] [PubMed] [Google Scholar]
- Lohmann I., McGinnis N., Bodmer M., McGinnis W. (2002). The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110, 457–466 [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. (1986). A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature 324, 537–545 [DOI] [PubMed] [Google Scholar]
- Macias A., Casanova J., Morata G. (1990). Expression and regulation of the abd-A gene of Drosophila. Development 110, 1197–1207 [DOI] [PubMed] [Google Scholar]
- Martín-Bermudo M. D., Martínez C., Rodríguez A., Jiménez F. (1991). Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development 113, 445–454 [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Lawrence P. A. (1985). Parasegments and compartments in the Drosophila embryo. Nature 313, 639–642 [DOI] [PubMed] [Google Scholar]
- McGinnis W., Krumlauf R. (1992). Homeobox genes and axial patterning. Cell 68, 283–302 [DOI] [PubMed] [Google Scholar]
- McQuilton P., St, Pierre S. E., Thurmond J.; FlyBase Consortium (2012). FlyBase 101 - the basics of navigating FlyBase. Nucleic Acids Res. 40, D706–D714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. I., Gruss P. (1993). Mouse Cdx-1 expression during gastrulation. Development 117, 191–203 [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I., Thor S. (2004). Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development 131, 6093–6105 [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Gehring W. J. (1987). Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell 48, 465–478 [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W. J. (1985). Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 4, 2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Gibson G., Gehring W. J. (1990). Effects of ectopic expression of caudal during Drosophila development. Development 109, 271–277 [DOI] [PubMed] [Google Scholar]
- Morata G. (1993). Homeotic genes of Drosophila. Curr. Opin. Genet. Dev. 3, 606–614 [DOI] [PubMed] [Google Scholar]
- Moreno E., Morata G. (1999). Caudal is the Hox gene that specifies the most posterior Drosophile segment. Nature 400, 873–877 [DOI] [PubMed] [Google Scholar]
- Nassif C., Daniel A., Lengyel J. A., Hartenstein V. (1998). The role of morphogenetic cell death during Drosophila embryonic head development. Dev. Biol. 197, 170–186 [DOI] [PubMed] [Google Scholar]
- Novotny T., Eiselt R., Urban J. (2002). Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development 129, 1027–1036 [DOI] [PubMed] [Google Scholar]
- Patel N. H. (1994). Imaging neuronal subsets and other cell types in whole mount Drosophila embryos and larvae using antibody probes. In Methods in Cell Biology. Drosophila melanogaster: Practical Uses in Cell Biology, Vol. 44 (ed. Goldstein L. S., Fyrberg E.), pp. 445–487 New York, NY: Academic Press; [DOI] [PubMed] [Google Scholar]
- Patel N. H., Martin-Blanco E., Coleman K. G., Poole S. J., Ellis M. C., Kornberg T. B., Goodman C. S. (1989). Expression of engrailed proteins in arthropods, annelids, and chordates. Cell 58, 955–968 [DOI] [PubMed] [Google Scholar]
- Peifer M., Karch F., Bender W. (1987). The Bithorax complex - control of segmental identity. Genes Dev. 1, 891–898 [DOI] [PubMed] [Google Scholar]
- Plickert G., Gajewski M., Gehrke G., Gausepohl H., Schlossherr J., Ibrahim H. (1997). Automated in situ detection (AISD) of biomolecules. Dev. Genes Evol. 207, 362–367 [DOI] [PubMed] [Google Scholar]
- Prokop A., Technau G. M. (1994). Early tagma-specific commitment of Drosophila CNS progenitor NB1-1. Development 120, 2567–2578 [DOI] [PubMed] [Google Scholar]
- Prokop A., Bray S., Harrison E., Technau G. M. (1998). Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech. Dev. 74, 99–110 [DOI] [PubMed] [Google Scholar]
- Reichert H., Bello B. (2010). Hox genes and brain development in Drosophila. Adv. Exp. Med. Biol. 689, 145–153 [DOI] [PubMed] [Google Scholar]
- Rivas M. L., Espinosa-Vázquez J. M., Sambrani N., Greig S., Merabet S., Graba Y., Hombría J. C. (2013). Antagonism versus cooperativity with TALE cofactors at the base of the functional diversification of Hox protein function. PLoS Genet. 9, e1003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A., Technau G. M. (2008). Multiple roles for Hox genes in segment-specific shaping of CNS lineages. Fly (Austin) 2, 316–319 [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A., Renner S., Technau G. M. (2008). Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development 135, 3435–3445 [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E. (1991). Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111, 437–449 [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Crosby M. A. (1988). The Abdominal-B gene of Drosophila melanogaster: overlapping transcripts exhibit two different spatial distributions. EMBO J. 7, 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Vernós I., Marco R., Morata G. (1985). Genetic organization of Drosophila bithorax complex. Nature 313, 108–113 [DOI] [PubMed] [Google Scholar]
- Schmid A., Chiba A., Doe C. Q. (1999). Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development 126, 4653–4689 [DOI] [PubMed] [Google Scholar]
- Schmidt H., Rickert C., Bossing T., Vef O., Urban J., Technau G. M. (1997). The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev. Biol. 189, 186–204 [DOI] [PubMed] [Google Scholar]
- Shen C. P., Jan L. Y., Jan Y. N. (1997). Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90, 449–458 [DOI] [PubMed] [Google Scholar]
- Skeath J. B. (1999). At the nexus between pattern formation and cell-type specification: the generation of individual neuroblast fates in the Drosophila embryonic central nervous system. BioEssays 21, 922–931 [DOI] [PubMed] [Google Scholar]
- Skeath J. B., Carroll S. B. (1994). The achaete-scute complex: generation of cellular pattern and fate within the Drosophila nervous system. FASEB J. 8, 714–721 [DOI] [PubMed] [Google Scholar]
- Struhl G., White R. A. (1985). Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell 43, 507–519 [DOI] [PubMed] [Google Scholar]
- Subramanian V., Meyer B. I., Gruss P. (1995). Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83, 641–653 [DOI] [PubMed] [Google Scholar]
- Suska A., Miguel-Aliaga I., Thor S. (2011). Segment-specific generation of Drosophila Capability neuropeptide neurons by multi-faceted Hox cues. Dev. Biol. 353, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Pfeifle C. (1989). A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 [DOI] [PubMed] [Google Scholar]
- Technau G., Campos-Ortega J. A. (1985). Fate-mapping in wildtype Drosophila melanogaster: II. Injection of horseradish peroxidase in cells of the early gastrula stage. Rouxs Arch. Dev. Biol. 194, 196–212 [Google Scholar]
- Technau G. M., Berger C., Urbach R. (2006). Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev. Dyn. 235, 861–869 [DOI] [PubMed] [Google Scholar]
- Thali M., Müller M. M., DeLorenzi M., Matthias P., Bienz M. (1988). Drosophila homoeotic genes encode transcriptional activators similar to mammalian OTF-2. Nature 336, 598–601 [DOI] [PubMed] [Google Scholar]
- Thor S. (1995). The genetics of brain development: conserved programs in flies and mice. Neuron 15, 975–977 [DOI] [PubMed] [Google Scholar]
- Tiong S., Bone L. M., Whittle J. R. (1985). Recessive lethal mutations within the bithorax-complex in Drosophila. Mol. Gen. Genet. 200, 335–342 [DOI] [PubMed] [Google Scholar]
- Tsuji T., Hasegawa E., Isshiki T. (2008). Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development 135, 3859–3869 [DOI] [PubMed] [Google Scholar]
- Urbach R., Technau G. M. (2003). Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development 130, 3621–3637 [DOI] [PubMed] [Google Scholar]
- van den Akker E., Forlani S., Chawengsaksophak K., de Graaff W., Beck F., Meyer B. I., Deschamps J. (2002). Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development 129, 2181–2193 [DOI] [PubMed] [Google Scholar]
- van Nes J., de Graaff W., Lebrin F., Gerhard M., Beck F., Deschamps J. (2006). The Cdx4 mutation affects axial development and reveals an essential role of Cdx genes in the ontogenesis of the placental labyrinth in mice. Development 133, 419–428 [DOI] [PubMed] [Google Scholar]
- von Hilchen C. M., Bustos A. E., Giangrande A., Technau G. M., Altenhein B. (2013). Predetermined embryonic glial cells form the distinct glial sheaths of the Drosophila peripheral nervous system. Development 140, 3657–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. (1985). opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell 40, 55–62 [DOI] [PubMed] [Google Scholar]
- White R. A., Wilcox M. (1984). Protein products of the bithorax complex in Drosophila. Cell 39, 163–171 [DOI] [PubMed] [Google Scholar]
- White K., Grether M. E., Abrams J. M., Young L., Farrell K., Steller H. (1994). Genetic control of programmed cell death in Drosophila. Science 264, 677–683 [DOI] [PubMed] [Google Scholar]
- Zavortink M., Sakonju S. (1989). The morphogenetic and regulatory functions of the Drosophila Abdominal-B gene are encoded in overlapping RNAs transcribed from separate promoters. Genes Dev. 3 12A, 1969-1981 [DOI] [PubMed] [Google Scholar]
- Zhai Z., Fuchs A. L., Lohmann I. (2010). Cellular analysis of newly identified Hox downstream genes in Drosophila. Eur. J. Cell Biol. 89, 273–278 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ungar A., Fresquez C., Holmgren R. (1994). Ectopic expression of either the Drosophila gooseberry-distal or proximal gene causes alterations of cell fate in the epidermis and central nervous system. Development 120, 1151–1161 [DOI] [PubMed] [Google Scholar]