Abstract
Background
Recent immunological data demonstrated that dendritic cells preferentially recognize advanced glycation end product (AGE) modified proteins, upregulate expression of the receptor for AGE (RAGE), and consequently bias the immune response towards allergy.
Methods
Peanut extract was characterized by mass spectrometry (MS) to elucidate the specific residues and specific AGE modifications found in raw and roasted peanuts and on rAra h 1 that was artificially glycated by incubation with glucose or xylose. The binding of the RAGE-V1C1 domain to peanut allergens was assessed by PAGE and Western analysis with anti-Ara h 1, 2, and 3 antibodies. IgE binding to rAra h 1 was also assessed using the same methods.
Results
AGE modifications were found on Ara h 1 and Ara h 3 in both raw and roasted peanut extract. No AGE modifications were found on Ara h 2. MS and Western blot analysis demonstrated RAGE binds selectively to Ara h 1 and Ara h 3 derived from peanut extract whereas the analysis failed to demonstrate Ara h 2 binding to RAGE. rAra h 1 with no AGE modifications did not bind RAGE, however after AGE modification with xylose, rAra h 1 bound to RAGE.
Conclusions
AGE modifications to Ara h 1 and Ara h 3 can be found in both raw and roasted peanuts. RAGE was demonstrated to selectively interact with AGE modified rAra h 1. If sensitization to peanut allergens occurs in dendritic cells via RAGE interactions, these cells are likely interacting with modified Ara h 1, and Ara h 3, and not Ara h 2.
Introduction
In the United States, the estimated prevalence of food allergy is 2.7%, where 1.3% is specifically sensitive to peanuts (1). Adverse reactions to peanuts can be triggered by extremely small doses and are the most frequent type of fatal anaphylaxis to food allergens (2, 3). However the reasons for this hypersensitivity are poorly understood. It has been suggested that food processing may contribute modifications, such as advanced glycation end products (AGEs), that are inappropriately recognized by the immune system and contribute to allergic sensitization (4). Studies with antibodies raised against common AGEs and advanced lipoxidation end products (ALEs) concluded that the major peanut allergens Ara h 1 and Ara h 3 were commonly modified with carboxymethylysine, malondialdehyde, and hydroxynonenal, while such modifications were less common in Ara h 2 (5). In addition, IgE derived from allergic patients more strongly recognizes roasted peanut extracts than raw (4). It should be noted that patients only raise antibodies against antigens to which they are exposed, and in the United States it is uncommon to encounter raw peanuts. So that while patient IgE recognizes roasted peanuts it does not demonstrate that AGEs necessarily contribute to sensitization.
However, there are several new lines of evidence that AGE modifications do contribute to allergic sensitization. Buttari, et al., demonstrated that dendritic cells stimulated with AGE-modified protein activated more IL-4 producing T-cells than IFN-γ producing T cells indicating a possible Th2 bias (6). Similar results were found by Hilmenyuk, et al., in comparing dendritic cells pulsed with AGE-modified OVA and regular OVA. The CD4+ T cells co-cultured with the dendritic cells produced more of IL-5 and reduced production of IFN-γ in response to the AGE modified OVA, again implying a Th2 bias (7). Another study also found enhanced CD4+ T cell activation in response to AGE-modified OVA, however the uptake by dendritic cells was mediated by scavenger receptor class A type I and II (SR-AI/II) and not RAGE (receptor for AGEs) or galectin-3 (8). These experiments provide good evidence that AGE modification of proteins can bias the immune response towards allergic sensitization.
The most logical receptors to recognize proteins with AGE modifications would be RAGE or other scavenger type receptors with promiscuous affinity for a variety of structurally different ligands. Previous studies in Caco-2 cells, which are a model for intestinal epithelia, showed that RAGE activation by AGEs stimulated MAP-kinases (9), which were shown to influence cellular proliferation in a cancer model (10). Very recently, AGE-modified Ara h 1 was demonstrated to influence the proliferation of Caco-2 cells (11). The proliferation depended on the incubation time and temperature used to induce the formation of AGEs, indicating that specific chemical structures were important in influencing the pro-inflammatory network (11).
The previously mentioned study is among the few to characterize what chemical modifications occur to the peanut allergens (5). The antibody technology utilized was not capable of identifying which specific residues were modified. In this paper we attempted to characterize, at the atomic level, the modifications that occur in roasted peanuts and in controlled reactions with sugars using mass spectrometry. We tested the modified and unmodified proteins for binding to RAGE and allergic patient-derived IgE. The results are the first to directly demonstrate RAGE recognition of AGE-modified peanut allergens and should contribute to a better understanding of the innate immune system recognition of peanut allergens.
Materials and Methods
Allergens and Peanut Extracts
Raw peanuts were oven-roasted at 177 °C (350 °F) for 15 minutes. Raw and roasted peanuts were crushed with a mortar and pestle. Two grams of ground peanut were extracted at 60 °C with 30 ml of phosphate–buffered saline (PBS), 1M NaCl, pH 7.4 for 15 minutes, by manually shaking and vortexing the extraction mix. Peanut extracts were allowed to stand for 15 min in a 60 °C bath, and the supernatant was centrifuged at 6,000 g for 5 min at room temperature. The supernatants were further centrifuged at 16,000 g for 5 min, and frozen at -20 °C. Recombinant Ara h 1 was expressed and purified as previously described (12).
SDS-PAGE and Western Blot Analyses
Each of the samples were mixed with 3x sample loading buffer and incubated for 10 minutes at 65 °C prior to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 4-20 % Novex Tris-HCl gel (Invitrogen). The gel was placed into Gel-Code Blue stain (Pierce, Rockford, IL) solution for 1 hour, then destained in distilled water for 1 hour and photographed. Detailed procedures of the Western blotting are given in the supplemental material. Immunocap values for patients 1-5 are, respectively: >100, 23.1, 14.5, 21.5, and 37.1 kU/L.
Glycation of recombinant Ara h 1
Roasting-induced Maillard reaction or glycation was performed as described in a previously established simulated roasting model with slight modifications (4). Purified rAra h 1 (0.5 mg/ml) was solubilized in PBS and incubated at 55 °C in the presence of 0.25 M glucose or xylose, for 0, 1, 2, 4, 7, or 10 days. Reactions were stopped by either addition of sample buffer for SDS-PAGE and western blot analysis or frozen at -20 °C for use in immunoprecipitation and mass spectrometry analysis.
RAGE pull down experiments
Human RAGE V1C1 domain was expressed as a fusion with maltose binding protein (MBP) and purified as described previously (13). Fresh amylose resin (50-200 ul of slurry) was washed 3-5x with PBS buffer by repeated centrifugation in small PCR tubes. The MBP-RAGE was incubated for 1 hour while gently rocking with the amylose resin at 3 mg/ml of slurry, which was reported as the maximum capacity of the resin (New England Biolabs, Ispswich, MA). Excess MBP-RAGE was removed with 3-5 washes of PBS. The resin and MBP-RAGE were incubated with either the peanut extract, rAra h 1, or the negative control Bos d 6 for 1 hour with gentle rocking. Again, excess protein was removed by repeated washing. The MBP-RAGE was eluted by adding PBS with 10 mM maltose. Xylose and glucose treated allergens rAra h 1 and Bos d 6 were obtained by incubating the allergens with 250 mM sugar for the times indicated, up to 10 days. The samples were dialyzed into PBS prior to the binding experiment to remove excess sugars that might interfere with the MBP-RAGE (maltose binding protein) construct’s binding to the amylose resin. Peanut extract concentrations were determined by Bradford assay to be 8-15 mg/ml. The amount of extract added to the resin was designed to not be more than 3 mg/ml.
Mass Spectrometry Sample Preparation
Raw and roasted peanut extracts were analyzed by gel-enhanced liquid chromatography mass spectrometry (GeLC-MS) or off-line/on-line two-dimensional liquid chromatography mass spectrometry in efforts to enhance the detection of AGE modifications on the peanut allergens. For specific details please see the supplementary material. Recombinant Ara h 1 was analyzed by standard nano LC-ESI-MS/MS.
Results
RAGE binding to recombinant Ara h 1
RAGE is commonly known to promiscuously bind a variety of substrates. In order to insure that there were no endogenous AGE modifications or post-translational modifications on Ara h 1 that may interact with RAGE, the first binding experiments utilized rAra h 1 from bacteria. As described in the methods, MBP-tagged RAGE (V1C1 domain) was bound to an amylose resin and utilized to ‘pull down’ potential ligands. Figure 1 shows a Western blot using an antibody selective for Ara h 1 to probe the samples obtained from pull down experiment. The results in panel A show the important negative control first, in which rAra h 1 that was not incubated with sugars is not retained by RAGE (lanes 4 and 5), and instead remains in the supernatant (lanes 2 and 3). Secondly, Figure 1B shows that rAra h 1 incubated with glucose and xylose for 10 days forms multimers due to crosslinking of the subunits from AGE modifications. Clearly the xylose is more efficient than glucose in creating these multimeric modifications. Thirdly, the xylose-modified rAra h 1 is retained by RAGE in this pull-down experiment (panel B lanes 4 versus 5), while the glucose-modified rAra h 1 is not (panel B lanes 6 versus 7).
Figure 1. The binding of rAra h 1 to RAGE before and after the Maillard reaction in the presence of glucose or xylose.
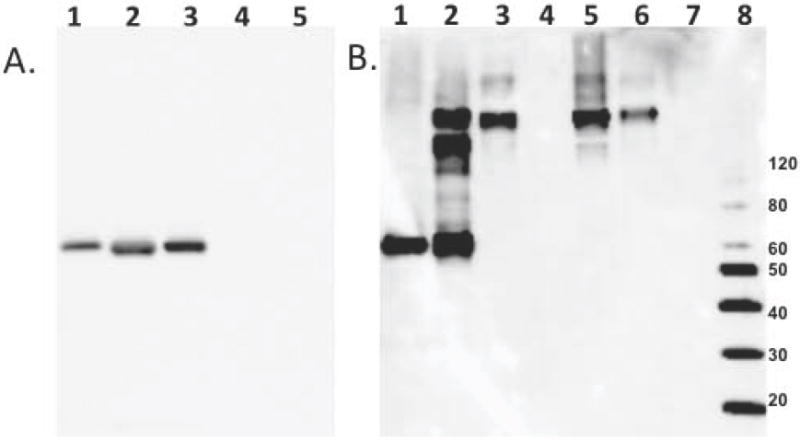
Panel A is a Western blot with anti-Ara h 1 antibody binding to 1) untreated rAra h 1 control, 2) untreated rAra h 1 supernatant from pull-down with RAGE-bound amylose resin or 3) amylose resin alone and 4) co-elution with MBP-RAGE from amylose resin or 5) amylose resin alone. Panel B is a anti-Ara h 1 Western blot of 1) untreated rAra h 1, 2) heated with glucose or 3) xylose for 10 days, 4) xylose treated rAra h 1 supernatant from pull-down with RAGE, or 5) co-elution with MBP-RAGE from amylose resin , and 6) glucose treated rAra h 1 supernatant from pull-down with RAGE, or 7) co-elution with MBP-RAGE from amylose resin , 8) Molecular weight marker.
Mass spectrometry results (Table 1) confirmed that under the conditions examined, glucose only weakly allows for the formation of AGEs on recombinant Ara h 1. Interestingly, the Schiff base is the species detected. The Schiff base is the first step in the formation of AGEs and suggests that perhaps higher glucose concentration, longer incubation times, or higher temperatures may induce additional AGEs. In light of these mass spectrometric results, it is not surprising that glucose-modified rAra h 1 did not interact with RAGE and is likely related to the reduced efficiency of glucose versus xylose in forming AGE modifications in the same amount of time.
Table 1.
AGE Modifications on rAra h 1
| Recombinant Ara h 1 + xylose | |||||
|---|---|---|---|---|---|
| Peptide Sequence | m/z | MW obs (M+H) | MW theor (M+H) | delta MW | Modification |
| GPLLSILKAFN | 616.2 | 1231.4 | 1230.7 | 0.7 | CML |
| IFLAGDKDNVIDQIEK | 626.3 | 1876.9 | 1875 | 1.9 | CML |
| VAKISMPVNTPGQFEDFFPASSR | 862.1 | 2584.3 | 2583.3 | 1 | CML |
| DLAFPGSGEQVEKLIK | 614 | 1840 | 1839 | 1 | Pyrraline |
| IFLAGDKDNVIDQIEK | 643.3 | 1927.9 | 1926 | 1.9 | Pyrraline |
| NTLEAAFNAEFNEI(RR) | 670.9 | 2010.7 | 2009.9 | 0.8 | Di-CML1 |
| Recombinant Ara h 1 + glucose | |||||
| AMVIVVVNKGTGNLELVAVR | 749.2 | 2245.6 | 2244.3 | 1.3 | Fructose Lysine |
extensive y-ion series and precursor mass confirm this peptide with the addition of 116 Da, but the position of the 116 Da addition is ambiguous
Table 1 compares the AGE modifications and the identity of the modified residues in the xylose and glucose treated samples. The most common modification identified was a carboxymethyl (CM) adduct, which occurred on both arginine and lysine residues. Some peptides, such as the IFLAG, were found modified with different AGEs. All of the peptides that were modified by glucose, were also found to be modified in the sample incubated with xylose. Only the DLAF peptide was uniquely modified by xylose. Figure 2 and supplementary material display mass spectrometry results with AGEs present.
Figure 2. Identification of a Maillard reaction modified Ara h 1 peptide using LC-MS/MS.
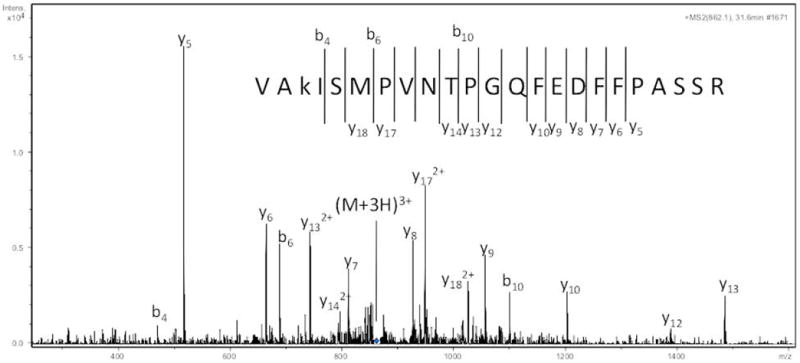
The fragment ion spectrum of an ion of m/z 862.1 is shown in the graph with the amino acid sequence shown in the figure corresponding to residues 260-283 of Ara h 1 with a carboxymethyllysine modification.
IgE binding to AGE modified proteins
Figure 3 shows the binding of IgE from five peanut allergic patients to the glucose and xylose treated rAra h 1. In panel A, the staining shows that the total protein content appears reduced at the longer incubation times with the sugars. The large number of free radicals that form during the progression of the Maillard reaction begin to attack and degrade the protein into smaller fragments that most likely run off of the bottom of the gel. What remains of the AGE modified protein (and fragments thereof) is less visible and appears as smears, due to extensive levels of modifications. In each case, the intensity of the Western blot (Figure 3B) closely matches the staining of the protein by coomassie. This shows that the patient IgE recognizes both the monomeric and covalently linked multimeric forms of rAra h 1 that form due to AGE modifications. The total IgE binding does not appear to be significantly affected by the incubation of rAra h 1 with sugars although there may be a slight reduction in IgE binding to the xylose modified samples at longer incubation times as the epitopes become extensively modified. Based on Figure 3 and the MS analysis in Table 1, the patient IgE extensively recognizes both the AGE modified and unmodified allergens. These results demonstrate that this in vitro system produces relevant modified allergens.
Figure 3. SDS-PAGE and IgE binding to rAra h 1 subjected to the Maillard reaction with glucose and xylose.
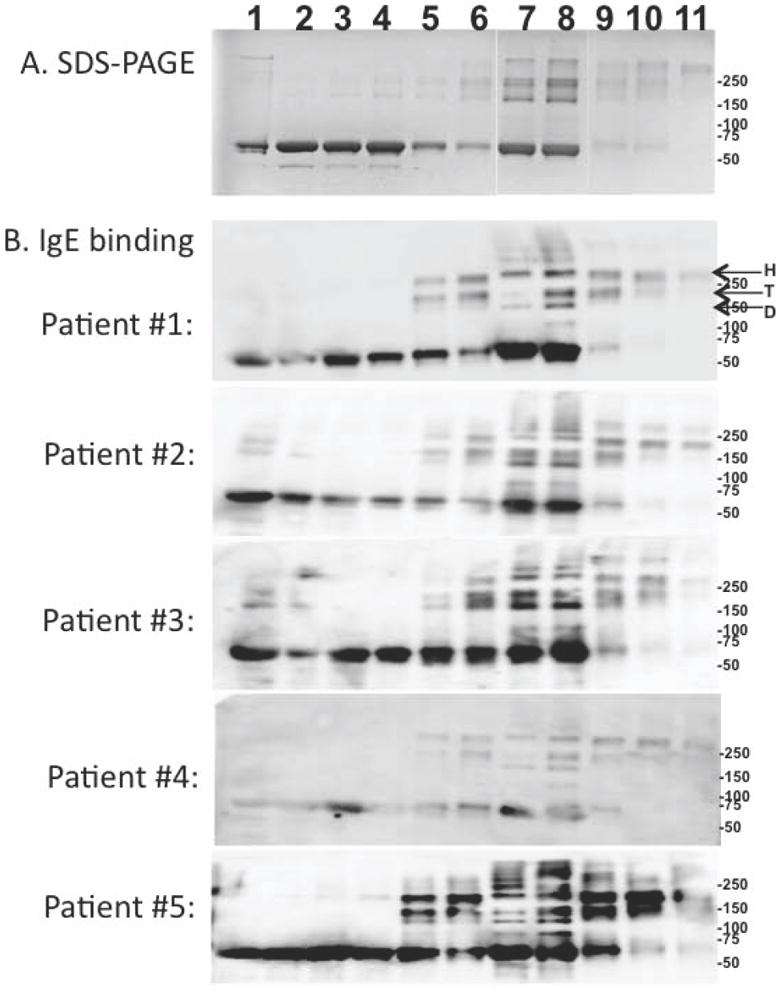
Panel A is an SDS-PAGE of rAra h 1: 1) untreated or heated in the presence of (lanes 2-6) glucose or (lanes 7-11) xylose for 1, 2, 4, 7 and 10 days, respectively. Panel B is an IgE Western blot analysis of 5 peanut allergic patient sera binding to the samples aligned with and described in panel A. Indicated by arrows are the location of the Ara h 1 hexamer (H), trimer (T) and dimer(D). Immunocap values for patients 1-5 are, respectively: >100, 23.1, 14.5, 21.5, and 37.1
To further test if the patient IgE recognized the AGE modifications exclusively, the milk allergen Bos d 6 was treated in the same in vitro system as was rAra h 1. Similar to the results in Figure 1, the xylose-treated Bos d 6 interacted most strongly with RAGE (data not shown). Supplementary Figure 1 demonstrates that the peanut-allergic patient sera did not recognize the AGE modified Bos d 6. In summary, RAGE is specific for AGE modifications while the patient IgE is allergen specific (See Supplementary Material).
RAGE interactions with allergens from roasted peanut extract
Figure 4 shows the results of a second RAGE pull-down experiment, however this time raw and roasted peanut extracts were used. Panel A shows a stained SDS gel for proper comparison of the amount of protein used in the lanes containing raw versus roasted peanut. Western blots are shown in panels B, C, and D utilizing mAbs specific for Ara h 1, Ara h 2, and Ara h 3 respectively. The results in panel B indicate that Ara h 1 is weakly pulled down by RAGE from peanut extract, from both the raw and roasted samples. Although, the bands are in lanes 4 and 6 are weak, Ara h 1 was readily identified with 12 MS/MS spectra corresponding to 3 unique peptides (See supplemental material), with a Distinct Summed MS/MS Search Score of 42.2, and 5% sequence coverage. In panel B, Ara h 2 was not detected in the RAGE pull down at the lower molecular weight (9 kDa) usually associated with monomeric Ara h 2. There are faint bands at higher molecular weight that could indicate the presence of aggregates with some affinity for RAGE. MS analysis never detected any Ara h 2 fragments with AGE modifications (see Table 2 and discussion below).
Figure 4. Detecting the binding of Ara h 1, Ara h 2 and Ara h 3 from raw and roasted peanut extracts to RAGE.
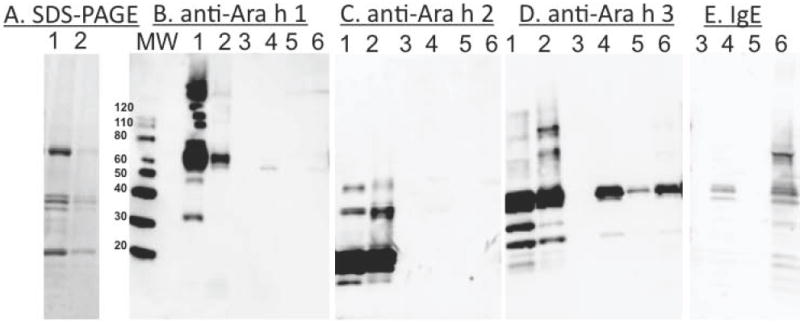
SDS-PAGE (Panel A) and Western blot of pull down experiments with (Panel B) anti-Ara h 1 , (Panel C) anti-Ara h 2, (Panel D) anti-Ara h 3 antibodies, and (Panel E) anti-IgE from pooled sera of patients #2 and #3 are shown. The lanes are: 1) Raw and 2) roasted peanut extracts that were subjected to MBP-RAGE amylose resin pull down experiments. Lanes 3 and 5 of each panel are supernatants from pull down experiments and lanes 4 and 6 contain material from raw and roasted peanut extracts, respectively, that co-elute with the RAGE from amylose resin. MW indicates the molecular weight marker. All panels are scaled to the molecular weight markers at the left of panel B.
Table 2.
AGE Modifications in Peanut Extract
| Ara h 1 (raw) | |||||
|---|---|---|---|---|---|
| Peptide Sequence | m/z | MW obs (M+H) | MW theor (M+H) | delta MW | Modification |
| EGALML(hydroxylP)(HFNSK) | 473.2 | 1417.6 | 1417.7 | 0.1 | CM1 |
| Ara h 3 (raw) | |||||
| NGIEETICSASVKK | 769.9 | 1538.8 | 1536.8 | 2 | CML |
| QLKNNNPFKFFVP(hydroxlP)FDHQSMR | 889.7 | 2667.1 | 2666.3 | 0.8 | CM |
| Ara h 1 (roasted) | |||||
| IFLAGDKDNVIDQIEKQAK | 754.6 | 2261.8 | 2261.2 | 0.6 | CML |
| VAKISMPVNTPGQFEDFFPASSR | 862.2 | 2584.6 | 2583.3 | 1.3 | CML |
| SSENNEGVIVKVSK | 517.2 | 1549.6 | 1547.8 | 1.8 | CML |
| DLAFPGSGEQVEKLIK | 597.1 | 1791.1 | 1788.9 | 1.2 | CML |
| Ara h 3 (roasted) | |||||
| NGIEETICSASVKK | 769.9 | 1538.8 | 1536.8 | 2 | CML |
| QLKNNNPFKFFVPPFDHQSMR | 889.7 | 2667.1 | 2666.3 | 0.8 | CM2 |
extensive y-ion series and precursor mass confirm this peptide with the addition of 58 Da ut the exact position of the 58 Da addition can only be localized to the 5 C-terminal residues
extensive y-ion series and precursor mass confirm this peptide with the addition of 58 Da ocalized on the extreme N-terminal residue
Figure 4 panel D shows that Ara h 3 is most efficiently pulled down by RAGE according to the Western blot analysis. In addition, the faint band in lane 5 may indicate either non-specific interactions of AGE modified Ara h 3 with the amylose resin or that the RAGE binding was saturated, but the bands are significantly stronger when RAGE is present (Lanes 5 vs 6). The MS results confirmed the presence of Ara h 3 and/or ten other variants or iso-allergens of Ara h 3 (Supplementary Table 1). In the NCBI database there are 15 different proteins from Arachis hypogaea with >68 % sequence identity to Ara h 3 (Supplementary Table 2). According to the World Health Organization and International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature (www.allergen.org), an allergen with >67 % sequence identity may be considered an iso-allergen. Because of the high sequence identity among the short peptides identified by MS it is difficult to uniquely identify from which protein the peptide was derived. We are confident that the Western and MS data demonstrate that RAGE pulls down Ara h 3, or one or more of these iso-allergens.
In addition, Figure 4 panel E shows IgE binding detected by Western blot using a pool of sera from patients 2 and 3 and the same samples used for the analyses in panels B-D. The patient’s IgE appears to detect the roasted extract more strongly than the raw after co-elution with RAGE. This is due to less total peanut protein that co-elutes with RAGE from the raw extract which contains fewer AGE modifications. The bands in panel E most closely match those of Ara h 3, and Ara h 1 although there are some additional bands. These additional bands may be other peanut allergens with AGE modifications besides Ara h 1, 2, or 3.
Table 2 describes the AGE modifications found in Ara h 1 and Ara h 3 in peanut extracts. Only a few peptides could be confidently identified as having carboxymethyl (CM) adducts in both the raw and roasted extracts. In the roasted extract we were only able to identify CM adducts on four peptides from Ara h 1 and two peptides from Ara h 3. Two of the peptides were similarly identified with CM adducts in the artificial glycation with rAra h 1. Mass spectra with these AGE modified peptides are shown in the supplemental material. Notably, no AGEs were found on Ara h 2 despite both Ara h 2.01 and Ara h 2.02 being readily detected in both raw and roasted peanut extracts. Ara h 2.01 was identified with a Distinct Summed MS/MS Score of 275.8, which corresponded to 334 spectra of 18 unique peptides and 78 % sequence coverage. Ara h 2.02 was identified with a Distinct Summed MS/MS Score of 158.4, which corresponded to 50 spectra of 10 unique peptides and 48 % sequence coverage. The fact that no glycation events could be detected was surprising, given the number of lysine and arginine residues on the surface of Ara h 2 in the recently described structure (14). Note that Tables 1 and 2 do not purport to be a complete catalogue of all possible AGE modifications. These are the modifications that were most confidently identified with mass spectrometry.
Figure 5 shows the location of the identified AGE modifications on the structure of Ara h 1 (12). The residues with modifications are distributed apparently randomly over the protein surface. Most of the lysine modifications occur on the “front face”, which is likely a consequence of the higher concentration of lysines on this surface.
Figure 5. Identification of AGE-modified residues identified on the surface of Ara h 1.
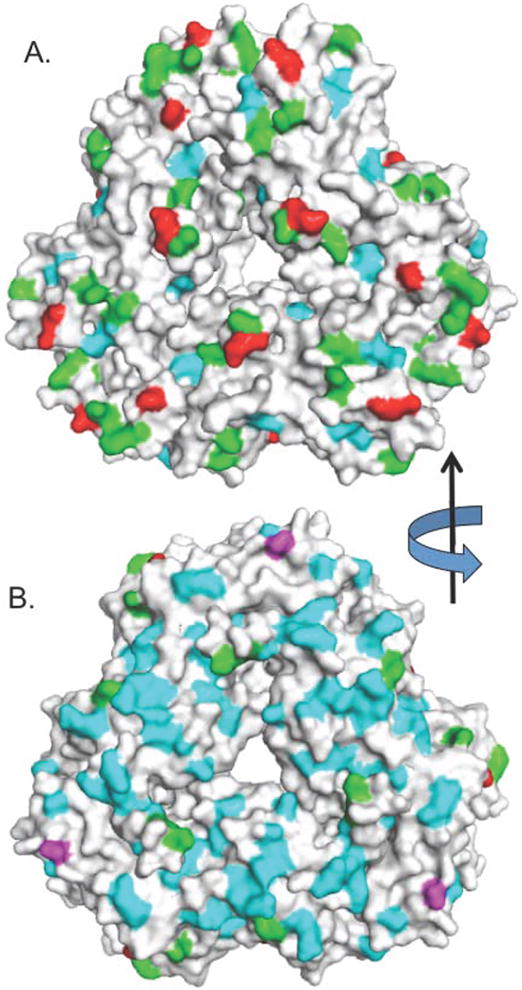
A surface rendering of Ara h 1 showing two views of the protein, rotated 180° about the y axis. Panel A is the “front” and panel B is the “back”. Unmodified lysines are colored green, modified lysines are red, umodified arginines are cyan, and modified arginines are magenta. The data are a composite of Tables 1 and 2.
Discussion
The peanut has been identified as one of the most potent allergenic foods (15). Extracts from roasted peanuts were shown to bind higher IgE levels than extracts from raw peanuts (4) and other studies support a role for AGE modifications in this phenomenon (17). Recent studies have suggested that AGE-RAGE interactions may contribute to allergic sensitization (6, 7) so we assessed the recognition and binding of peanut allergens to RAGE.
This paper describes evidence supporting RAGE binding to peanut allergens that have been specifically modified by AGEs. These studies establish that RAGE does not interact with unmodified rAra h 1, but does interact with AGE-modified rAra h 1 (Figure 1). Secondly, we find that RAGE can bind AGE-modified Ara h 1 and Ara h 3 (Figure 4). Mass spectrometry identified explicit AGE modifications associated with specific residues on Ara h 1 and Ara h 3 (Tables 1 and 2) and largely confirmed the analysis of the Western blots. We detected similar AGE modifications on the same peptides in Ara h 1 as were described by Hebling et al (18). However, in contrast with our results, that study did not mention any AGE modifications in the raw peanut extract. The lack of modifications to Ara h 2 was similar to that described with mAb detection of AGEs by Chung et al (5), and no modifications to Ara h 2 are mentioned in the MS analysis of Hebling et al (18). The reported large number of modifications in Chassaigne et al. may seem to be in conflict with this result, however the list of modifications to Ara h 2 in that study include primarily N-terminal modifications and not AGEs (19). An important contribution of this study is that AGE modifications were found on both raw and roasted peanut allergens. These results suggest that future studies on the effect of AGE modifications from roasted peanut extract should utilize a recombinant protein instead of raw peanut extract as a control.
A comparison of the amount of Ara h 1 detected by Western blot analysis in raw versus roasted peanut extracts indicated a reduced signal intensity for the allergen from roasted peanut 1 (Figure 4A, lanes 2 and 3). This result is apparently contradictory to previous work that demonstrated an increase in the amount of Ara h 1 in solution in peanut extracts that is dependent on the roasting time (20). This increase occurred in spite of a reduction, upon roasting, of the binding capacity of one of the two anti-Ara h 1 monoclonal antibodies from the ELISA used to measure the allergen. Similarly, the epitope recognized by the antibody used in the immunoblot analysis could have been affected by roasting, resulting in a reduced binding to Ara h 1 from roasted peanut. We emphasize that AGE-modified Ara h 1 is recognized by RAGE, as demonstrated in Figure 1. The detection of Ara h 1 in the extract may also be weak because Ara h 3 in roasted extract is more effectively competing for RAGE binding.
It is interesting to discuss why the patient IgE does not appear to significantly differentiate between the AGE modified and unmodified rAra h 1. There are several conclusions to draw. First, consumption of raw peanuts is rare, hence the patients in question were likely primarily exposed to roasted peanuts. However, we have demonstrated here that even ‘raw’ peanuts may contain a substantial number of AGE modifications. Therefore, the IgE binding data here suggests that the AGE modifications do not substantially modify patient epitopes, because the IgE binds well to the control rAra h 1 that is untreated and contains no AGE modifications. The data on AGE modified Bos d 6 further demonstrate that the patient IgE is not specific for the AGE modifications. In contrast, RAGE is specific for AGE modifications, and is not protein specific. To be clear on the separate issues of RAGE and IgE binding in the allergic response: RAGE is hypothesized to be important in the initial innate immune response that biases sensitization towards an allergic response. IgE binding is part of the later adaptive immune response from B-cells.
In addition to AGE modifications, other protein modifications, including gylcosylation, phosphorylation, and cysteinylation, have been studied for their ability to modify the immunological response (21). In the case of the birch pollen allergen Bet v 1, nitrosylation of the protein was demonstrated to alter the presentation of Bet v 1 peptides on dendritic cells and increase proliferation, and IL-5 and IFN-γ production in splenocytes (22, 23). While cataloging the AGE modifications to the peanut allergens, numerous other enzymatic and non-enzymatic modifications were identified such as malondialdehyde and hydroxy-prolines (data not shown). For this paper we have focused on reporting the AGE modifications and RAGE interactions. An important conclusion of this paper is that RAGE does bind to AGE-modified Ara h 1 and Ara h 3. If allergic sensitization is enhanced by RAGE-dependent interactions, it is apparently feasible that AGE modified Ara h 1 and Ara 3 are involved.
Supplementary Material
Acknowledgments
This research was supported in part by Research Project Number Z01- ES102885-01 to REL, Z01-ES50161 to KBT, and Z01-ES102488-05 to JGW in the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health. This work was supported in part by the Agricultural Research Service, U.S. Department of Agriculture.
Footnotes
Author Contributions
GAM conceived of the study, performed RAGE binding experiments, and wrote the manuscript. LLE and HP provided RAGE and contributed to the writing. KJ, AS, LJD, and JGW performed mass spectrometry experiments and analysis. REL and KBT analyzed data and contributed to the writing. SJM, BKH, HC, SR, JBN performed Western analysis, provided allergens, and contributed to the writing. AP contributed peanut extracts, analysis, and worked on the writing.
Conflicts of Interest
There are no conflicts of interest
References
- 1.Liu AH, Jaramillo R, Sicherer SH, Wood RA, Bock SA, Burks AW, et al. National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005-2006. The Journal of Allergy and Clinical Immunology. 2010;126(4):798–806. doi: 10.1016/j.jaci.2010.07.026. e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock SA, Munoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001-2006. Journal of Allergy and Clinical Immunology. 2007;119(4):1016–1018. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
- 3.Wensing M, Penninks AH, Hefle SL, Koppelman SJ, Bruijnzeel-Koomen CAFM, Knulst AC. The distribution of individual threshold doses eliciting allergic reactions in a population with peanut allergy. Journal of Allergy and Clinical Immunology. 2002;110(6):915–920. doi: 10.1067/mai.2002.129235. [DOI] [PubMed] [Google Scholar]
- 4.Maleki SJ, Chung SY, Champagne ET, Raufman JP. The effects of roasting on the allergenic properties of peanut proteins. Journal of Allergy and Clinical Immunology. 2000;106(4):763–768. doi: 10.1067/mai.2000.109620. [DOI] [PubMed] [Google Scholar]
- 5.Chung SY, Champagne ET. Association of end-product adducts with increased IgE binding of roasted peanuts. Journal of Agricultural and Food Chemistry. 2001;49(8):3911–3916. doi: 10.1021/jf001186o. [DOI] [PubMed] [Google Scholar]
- 6.Buttari B, Profumo E, Capozzi A, Facchiano F, Saso L, Sorice M, et al. Advanced glycation end products of human beta(2) glycoprotein I modulate the maturation and function of DCs. Blood. 2011;117(23):6152–6161. doi: 10.1182/blood-2010-12-325514. [DOI] [PubMed] [Google Scholar]
- 7.Hilmenyuk T, Bellinghausen I, Heydenreich B, Ilchmann A, Toda M, Grabbe S, et al. Effects of glycation of the model food allergen ovalbumin on antigen uptake and presentation by human dendritic cells. Immunology. 2010;129(3):437–445. doi: 10.1111/j.1365-2567.2009.03199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilchmann A, Burgdorf S, Scheurer S, Waibler Z, Nagai R, Wellner A, et al. Glycation of a food allergen by the Maillard reaction enhances its T-cell immunogenicity: role of macrophage scavenger receptor class A type I and II. The Journal of Allergy and Clinical Immunology. 2010;125(1):175–183. doi: 10.1016/j.jaci.2009.08.013. e171-111. [DOI] [PubMed] [Google Scholar]
- 9.Zill H, Gunther R, Erbersdobler HF, Folsch UR, Faist V. RAGE expression and AGE-induced MAP kinase activation in Caco-2 cells. Biochemical and Biophysical Research Communications. 2001;288(5):1108–1111. doi: 10.1006/bbrc.2001.5901. [DOI] [PubMed] [Google Scholar]
- 10.Buzzi N, Colicheo A, Boland R, de Boland AR. MAP kinases in proliferating human colon cancer Caco-2 cells. Molecular and Cellular Biochemistry. 2009;328(1-2):201–208. doi: 10.1007/s11010-009-0090-9. [DOI] [PubMed] [Google Scholar]
- 11.Teodorowicz M, Fiedorowicz E, Kostyra H, Wichers H, Kostyra E. Effect of Maillard reaction on biochemical properties of peanut 7S globulin (Ara h 1) and its interaction with a human colon cancer cell line (Caco-2) European Journal of Nutrition. 2013 doi: 10.1007/s00394-013-0494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chruszcz M, Maleki SJ, Majorek KA, Demas M, Bublin M, Solberg R, et al. Structural and immunologic characterization of Ara h 1, a major peanut allergen. The Journal of Biological Chemistry. 2011;286(45):39318–39327. doi: 10.1074/jbc.M111.270132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H, Boyington JC. The 1.5 angstrom Crystal Structure of Human Receptor for Advanced Glycation Endproducts (RAGE) Ectodomains Reveals Unique Features Determining Ligand Binding. Journal of Biological Chemistry. 2010;285(52):40762–40770. doi: 10.1074/jbc.M110.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller GA, Gosavi RA, Pomes A, Wunschmann S, Moon AF, London RE, et al. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011;66(7):878–885. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer K, Burks W. Current developments in peanut allergy. Current Opinion in Allergy and Clinical Immunology. 2006;6(3):202–206. doi: 10.1097/01.all.0000225161.60274.31. [DOI] [PubMed] [Google Scholar]
- 16.Sicherer SH, Munoz-Furlong A, Sampson HA. Prevalence of peanut and tree nut allergy in the United States determined by means of a random digit dial telephone survey: A 5-year follow-up study. Journal of Allergy and Clinical Immunology. 2003;112(6):1203–1207. doi: 10.1016/s0091-6749(03)02026-8. [DOI] [PubMed] [Google Scholar]
- 17.Nesbit JB, Hurlburt BK, Schein CH, Cheng HP, Wei H, Maleki SJ. Ara h 1 structure is retained after roasting and is important for enhanced binding to IgE. Molecular Nutrition & Food Research. 2012;56(11):1739–1747. doi: 10.1002/mnfr.201100815. [DOI] [PubMed] [Google Scholar]
- 18.Hebling CM, McFarland MA, Callahan JH, Ross MM. Global Proteomic Screening of Protein Allergens and Advanced Glycation Endproducts in Thermally Processed Peanuts. Journal of Agricultural and Food Chemistry. 2013;61(24):5638–5648. doi: 10.1021/jf303554t. [DOI] [PubMed] [Google Scholar]
- 19.Chassaigne H, Norgaard JV, van Hengel AJ. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS) Journal of Agricultural and Food Chemistry. 2007;55(11):4461–4473. doi: 10.1021/jf063630e. [DOI] [PubMed] [Google Scholar]
- 20.Pomes A, Butts CL, Chapman MD. Quantification of Ara h 1 in peanuts: why roasting makes a difference. Clinical and Experimental Allergy. 2006;36(6):824–830. doi: 10.1111/j.1365-2222.2006.02490.x. [DOI] [PubMed] [Google Scholar]
- 21.Doyle HA, Mamula MJ. Posttranslational modifications of self-antigens. Annals of the New York Academy of Sciences. 2005;1050:1–9. doi: 10.1196/annals.1313.001. [DOI] [PubMed] [Google Scholar]
- 22.Karle AC, Oostingh GJ, Mutschlechner S, Ferreira F, Lackner P, Bohle B, et al. Nitration of the Pollen Allergen Bet v 1.0101 Enhances the Presentation of Bet v 1-Derived Peptides by HLA-DR on Human Dendritic Cells. Plos One. 2012;7(2) doi: 10.1371/journal.pone.0031483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruijthuijsen YK, Grieshuber I, Stocklinger A, Tischler U, Fehrenbach T, Weller MG, et al. Nitration enhances the allergenic potential of proteins. International Archives of Allergy and Immunology. 2006;141(3):265–275. doi: 10.1159/000095296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


