Abstract
Background
Leishmania major and an uncharacterized species have been reported from human patients in a cutaneous leishmaniasis (CL) outbreak area in Ghana. Reports from the area indicate the presence of anthropophilic Sergentomyia species that were found with Leishmania DNA.
Methodology/Principal Findings
In this study, we analyzed the Leishmania DNA positive sand fly pools by PCR-RFLP and ITS1 gene sequencing. The trypanosome was determined using the SSU rRNA gene sequence. We observed DNA of L. major, L. tropica and Trypanosoma species to be associated with the sand fly infections. This study provides the first detection of L. tropica DNA and Trypanosoma species as well as the confirmation of L. major DNA within Sergentomyia sand flies in Ghana and suggests that S. ingrami and S. hamoni are possible vectors of CL in the study area.
Conclusions/Significance
The detection of L. tropica DNA in this CL focus is a novel finding in Ghana as well as West Africa. In addition, the unexpected infection of Trypanosoma DNA within S. africana africana indicates that more attention is necessary when identifying parasitic organisms by PCR within sand fly vectors in Ghana and other areas where leishmaniasis is endemic.
Author Summary
Cutaneous leishmaniasis (CL) is one of the world's most neglected diseases transmitted by female sand flies and affecting mostly developing countries with about 1.2 million cases every year. In most African countries, the disease is typically caused by one of two species of Leishmania parasite: L. major or L. tropica. Clinical symptoms of both infections are similar, producing ulcerative and nodular lesions. Notwithstanding their similarity, lesions caused by L. major self-heal and bestow immunity to re-infection and therapy, if applied, is often by antimonials administered intra-lesionally, whereas the treatment of CL caused by L. tropica is difficult. Differentiating between these agents in any CL focus is important. Following the outbreak of CL in Ghana, we focused on the sand fly species present in the area in order to detect Leishmania DNA in potential vectors. Our study provides evidence on the occurrence of L. tropica and L. major DNA, and the detection of Trypanosoma DNA in Sergentomyia sand flies in Ghana. These findings have considerable implications in determining the epidemiology and dynamics of the disease. Significantly, our study supports the possibility of Sergentomyia sand flies as the vectors of CL in Ghana other than Phlebotomus, which contains all currently known vectors for Leishmania in the Old World.
Introduction
Cutaneous leishmaniasis (CL) is a vector-borne parasitic disease of humans and other mammals caused by cell-infecting flagellate protozoa of the genus Leishmania, transmitted by female phlebotomine sand flies. The disease occurs in 98 tropical, subtropical and temperate countries worldwide and it is estimated that 1.2 million new cases of CL occur per year [1]. CL of the Old World is caused by five species of Leishmania: L. tropica, L. major, L. aethiopica, L. infantum and L. donovani [2]. Clinically, cutaneous lesions due to L. tropica last much longer and are more difficult to treat than those due to L. major [3]. To date, there is no information available on the distribution of L. tropica in West Africa.
Sand fly species of the genus Phlebotomus are the putative vectors of Leishmania in the Old World [4]; however, few studies have recently suggested the possible involvement of some species of the genus Sergentomyia in the transmission of Leishmania in the Old World. Studies conducted in cutaneous leishmaniasis foci in Iran, Mali and Portugal have shown the detection of L. major DNA in S. sintoni [5], S. darlingi [6] and S. minuta [7], respectively. Earlier, L. major was also isolated from S. garnhami in Kenya [8]. Other reports have also detected L. donovani DNA in S. babu in India [9] and more recently, L. siamensis DNA in S. gemmea in Thailand [10].
In Ghana, L. major [11] and uncharacterized species [12] have been identified as the causative agents of human cutaneous leishmaniasis in an area of CL outbreak. In order to control the transmission of Leishmania spp. in an endemic area, information on potential sand fly vectors as well as their associated Leishmania species is of paramount importance since vector dispersion is one of the major factors that determine the potential rate of pathogen dissemination. Thus, this study was designed to determine and detect Leishmania DNA within sand flies collected in the outbreak area of CL in Ghana by molecular analysis in order to ascertain if they were infected by either L. major [11] or the yet to be characterized species [12]. This information is necessary to implicate the potential vectors in the area for a better understanding of the epidemiology of leishmaniasis in Ghana. Here, we report the confirmation of L. major DNA and first detection of L. tropica DNA in Sergentomyia species using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and DNA sequencing analyses, as well as Trypanosoma species associated with sand fly infections in Ghana.
Materials and Methods
Sand fly capture and taxonomic identification
Previously collected indoor resting sand flies from three CL outbreak communities; Klefe, Hlefi and Taviefe in Ho District of Ghana were used in this study. Verbal informed consent was obtained from residents in each of the community. The collections were done in August, September and November 2007 from human habitats using manual aspirator. Each unfed and blood-fed female fly was dissected under sterile conditions by cutting off the head and last three abdominal segments with a pair of sterilized entomological needles and forceps. The head and last three abdominal segments of each fly was mounted on microscope slides in Puri's medium and identified using taxonomic keys [13], [14]. For each blood-fed fly, the remainder of the body was processed under sterile condition and subjected to blood-meal analysis (Boakye et al, unpublished results). Afterwards, the dissected thorax and attached anterior abdomen of unfed and the processed specimens known to have fed on human-blood (Boakye et al, unpublished results) were separately pooled according to species and locality (Table 1) for DNA extraction and infection molecular analysis.
Table 1. Summary of Sergentomyia sand flies screened by molecular biological method in this study.
| Location | Sand fly species | Number of unfed pools* (positive pools) | Number of blood-fed pools* ¶ (positive pools) | Total |
| Klefe | S. africana africana | 5 | 1 | 6 |
| S. ingrami | 10 | 2 (1)c | 12 | |
| S. simillima | 12 | 2 | 14 | |
| S. dissimillima | 13 | 0 | 13 | |
| S. hamoni | 5 (1)a | 0 | 5 | |
| Total | 45 | 5 | 50 | |
| Hlefi | S. africana africana | 19 (1)b | 1 | 20 |
| S. ingrami | 7 (2)c | 1 | 8 | |
| S. simillima | 4 | 2 | 6 | |
| S. dissimillima | 5 | 0 | 5 | |
| Total | 35 | 4 | 39 | |
| Taviefe | S. africana africana | 4 | 0 | 4 |
| S. ingrami | 4 (1)a | 1 | 5 | |
| S. simillima | 3 | 0 | 3 | |
| S. dissimillima | 1 | 0 | 1 | |
| Total | 12 | 1 | 13 |
Ten female specimens in each pool.
Identified human-blood fed specimens (Boakye et al, unpublished results).
L. tropica DNA detection according to sequence analysis.
Trypanosoma sp. DNA detection according to sequence analysis.
L. major DNA detection according to sequence analysis.
DNA extraction, Leishmania ITS1 PCR-RFLP and trypanosomatid SSU rRNA gene amplification
Ten (10) unfed and blood-fed flies belonging to the same species were pooled and homogenized in animal tissue lysis buffer (Qiagen) containing proteinase K. DNA was extracted using the Qiagen DNA mini kit (Qiagen, Valencia, CA) according to the manufacturer instructions and 0.5 µl of the extract was used as PCR templates.
The leishmanial ribosomal internal transcribed spacer 1 (ITS1) region was amplified, using primers L5.8S and LITSR [15]. The reactions were carried out in volumes of 25 µl containing 200 µM of each dNTP, 1.5 mM MgCl2, 2 units Taq polymerase and 500 nM of each primer. Each PCR reaction included a positive control (DNA from a reference strain: L. major - IPAP/EG/1989/S1-177 and L. tropica - MHOM/SU/1974/K27) and a negative control (water). After initial denaturation at 95°C for 2 min, PCR amplification were performed with 34 cycles consisting of denaturation (95°C for 20 sec), annealing (53°C for 30 sec), and extension (72°C for 1 min) followed by a final extension cycle at 72°C for 6 min. An aliquot of 5 µl of each PCR reaction was separated by electrophoresis on 2% agarose gel and visualized. Each ITS1 amplicon was digested with the restriction endonuclease HaeIII (Invitrogen) for the species identification. The restriction fragments were run and visualised on 2% agarose gel and compared with those of reference strains of L. major (IPAP/EG/1989/S1-177) and L. tropica (MHOM/SU/1974/K27).
For the identification of Trypanosoma species, the small subunit ribosomal RNA (SSU rRNA) gene was amplified from the sand fly using primer specific for SSU rRNA gene of trypanosomatids (TRY927F: GAAACAAGAAACACGGGAG and TRY927R: CTACTGGGCAGCTTGGA) [16], [17], [18].
PCR amplification of sand fly COI and18S rRNA genes
Validation of morphologically identified sand fly specimens was done using mitochondrial cytochrome c oxidase gene subunit I (COI) and 18S rRNA genes. Amplification of the COI gene was accomplished using the primers LCO I490 (5′-GGTCAACAAATCATAAAGATATTGG3′) and HCO 2198 (5′TAAACTTCAGGGTGACCAAAAAATCA-3′) [19]. The reaction was carried out in a volume of 15 µl using a pair of primers (1 µM each) and 2× AmpliTaq Gold PCR Master Mix (Applied Biosystems, NJ, USA). After initial denaturation at 95°C for 5 min, amplification was performed with 37 cycles consisting of denaturation at 94°C for 30 sec, annealing at 55°C for 45 sec, extension at 72°C for 1 min 30 sec, followed by a final extension at 72°C for 10 min. For the 18S rRNA locus, the sequence of interest was amplified using the primers Lu. 18S rRNA-1S (5′-TGCCAGTAGTTATATGCTTG-3′) and Lu. 18S rRNA-1R (5′-TTACGCGCCTGCTGCCTTCC-3′) [20]. The reaction was carried out in a volume of 15 µl using a pair of primers (0.4 µM each) and 2× AmpliTaq Gold PCR Master Mix (Applied Biosystems, NJ, USA). An initial denaturation was done for 5 min at 95°C, followed by PCR amplification for 30 cycles of denaturation (95°C for 1 min), annealing (55°C for 1 min), and polymerization (72°C for 1 min), with a final extension of 10 min at 72°C. Amplified products were resolved on 2% agarose gels.
Molecular cloning and nucleotide sequencing
Infection and sand fly PCR products were directly cloned into plasmid using a pGEM-T Easy Vector System (Promega, Madison, WI). Escherichia coli DH5 α cells was transformed with a ligated product and plated onto Luria Bertani agar plates containing ampicillin (50 µg/ml), 5-bromo-4-chloro-3-indolyl β-D-galactoside (36 µg/ml), and isopropyl β-D-thiogalactoside (40 µg/ml). Plasmid DNAs were extracted with QIAprep Spin Miniprep Kit (Qiagen) and the insert of the plasmids were sequenced by the dideoxy chain termination method using BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
Phylogenetic analysis
Sequences from both strands were aligned using Clustal W software [21] and imported into MEGA (Molecular Evolutionary Genetics Analysis) version 4.0 [22]. Phylogenetic trees were constructed by the neighbor-joining (NJ) method with the algorithm of MEGA program. Bootstrap values were determined with 1,000 replicates of the datasets. ITS1 gene sequences used for the analysis were L. major (IPAP/EG/1989/SI-177, DQ295824; MHOM/GH/2004/HO-004, DQ295825; MHOM/EG/2006/RTC-64, FJ460456; MHOM/TN/1997/LPN162, FN677342; MHOM/IL/1967/JERICHO II, EU326229), L. tropica (MHOM/EG/2006/RTC-66, FJ460457; IROS/NA/1976/ROSSI-II, AJ000302; MHOM/KE/1984/NLB297, AJ000301; MHOM/TN/1988/TAT3, AJ300485) and Leishmania sp. (MHOM/GH/2006/TVE, EF524071). The trypanosomatid SSU rRNA gene sequence was analyzed with those of T. avium (AB566384, AF416559), T. brucei rhodesiense (AJ009142), T. corvi (AY461665), T. microti (AJ009158), T. kuseli (AB175626), T. cruzi (AJ009149, AJ009150), T. rangeli (AJ012417), T. congolense (AJ009145, AJ009146), T. mega (AJ009157), T. rotatorium (AJ009161), T. fallisi (AF119806) and Trypanosoma species isolated from toads (EU021231, EU021232), Lutzomyia sand flies (EU021241, EU021242, EU021243, EU021244, EU021245, EU021237), Phlebotomus sand fly (AB520638), mosquitoes (AF416561), and hippoboscid (AF416562).
Sand fly COI gene sequences were analyzed with those of Sergentomyia babu babu (HQ585351, HQ585357), S. vadhanurensis (HQ585345, HQ585348), S. bailyi (HQ585381 and HQ585384), S. punjabenensis (HQ585375, HQ585379), Phlebotomus papatasi (JN172077) and the New World sand fly species Lutzomyia longiflocosa (FJ437273). The sand fly 18S rRNA gene sequences were analyzed with those of S. barraudi (JQ790518), S. queenslandi (HM775498), S. magna (AJ391741), S. babu (JN581685), S. minuta (AJ244419), S. schwetzi (AJ391739), S. buxtoni (AJ391737), S. dentata (AJ244423), S. dubia (AJ391738), S. clydei (AJ391742), S. ghesquierei (AJ391743), P. papatasi (AJ391726) and the New World sand fly species Lu. longipalpis (AJ244429).
Statistical analysis
The infection rate in sand flies was determined using the poolScreen2 program generously provided by Dr. Charles Katholi (Department of Biostatistics and Division of Geographic Medicine, University of Alabama at Birmingham, USA) [23]. The algorithm was used to calculate the maximum likelihood estimate (MLE) of infection in sand fly species at 95% confidence intervals.
Accession numbers
AB759711 508df2892113a5dbfe0001d5.clone1
AB759712 508df2892113a5dbfe0001d5.clone2
AB759713 508df2892113a5dbfe0001d5.clone3
AB759714 508f999a2113a5dfe60001a2.cloneIS1
AB759715 508f999a2113a5dfe60001a2.cloneIS2
AB759716 508f999a2113a5dfe60001a2.cloneIS3
AB759971 508fa9992113a5dbfe00028e.cloneCOI1
AB759972 508fa9992113a5dbfe00028e.cloneCOI2
AB759973 508fa9992113a5dbfe00028e.cloneCOI3
AB787189 511af92b12c2e8dff9003639.clone4
AB787190 511af92b12c2e8dff9003639.clone5
AB787191 511af92b12c2e8dff9003639.clone6
AB787192 511af92b12c2e8dff9003639.cloneCOI4
AB787193 511af92b12c2e8dff9003639.cloneCOI5
AB787194 511af92b12c2e8dff9003639.cloneCOI6
AB787195 511af92b12c2e8dff9003639.cloneIS4
AB787196 511af92b12c2e8dff9003639.cloneIS5
AB787197 511af92b12c2e8dff9003639.cloneIS6
Results
Detection of trypanosomatids within sand flies
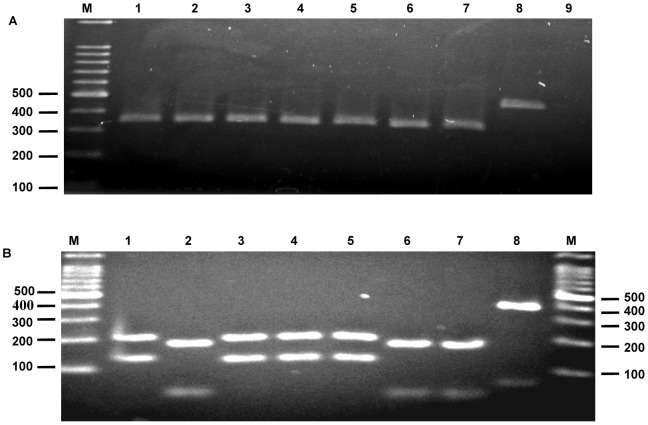
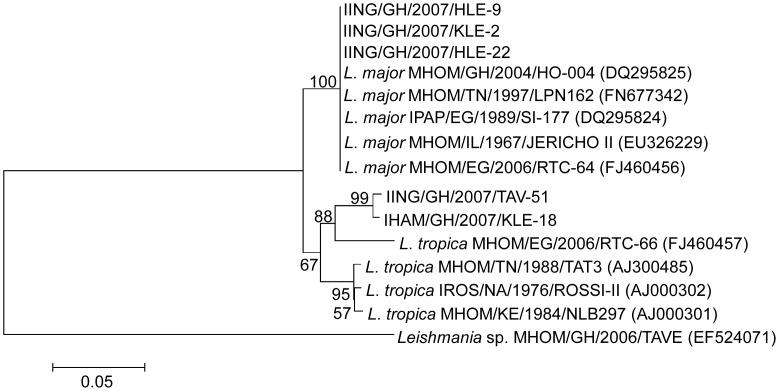
Sergentomyia sand flies of 102 pools were analyzed (Table 1), each containing ten females (in total 1,020 insects): S. africana africana (2 blood-fed, 28 unfed pools), S. ingrami (4 blood-fed, 21 unfed pools), S. simillima (4 blood-fed, 19 unfed pools), S. dissimillima (19 unfed pools), and S. hamoni (5 unfed pools). ITS1-PCR showed 6 positives, with a unique band of approximately 340 bp in 5 pools, namely, 4 S. ingrami and 1 S. hamoni as expected from the results obtained from the reference strains and the other yielding a fragment of approximately 500 bp in 1 S. africana africana pool (Fig. 1A). No Leishmania DNA was detected in S. simillima and S. dismillima pools from the three CL foci. The six positive samples were subjected to RFLP for species identification and their subsequent digestion with the endonuclease HaeIII produced fragments characteristic of L. major in samples 3–5 (reported in: Boakye et al, unpublished results) and the L. major reference DNA, and L. tropica in samples 6–7 and the L. tropica reference DNA, whereas sample 8 displayed an uncharacteristic RFLP pattern (Fig. 1B). To confirm authenticity of the RFLP analysis, both the 340 bp and 500 bp ITS1 PCR products obtained in the sand fly positive samples were subjected to sequencing analysis. The ITS1 DNA sequences obtained from the 3 (1 blood-fed from Klefe, 2 unfed from Hlefi) S. ingrami pools [IING/GH/2007/HLE-9 (GeneBank accession number: AB759711), IING/GH/2007/KLE-2 (AB759712), and IING/GH/2007/HLE-22 (AB759713)] were identical to each other and showed 99% identity to several L. major ITS1 sequences in GenBank, including isolates from Ghana, Egypt, Kenya, Sudan and Tunisia. The ITS1 sequences obtained from 1 unfed S. ingrami pool from Taviefe [IING/GH/2007/TAV-51 (AB787189)] and 1 unfed S. hamoni pool from Klefe [IHAM/GH/2007/KLE-18 (AB787190)] were different at 4 positions of the nucleotides, which include the deletion of 2 nucleotides in the former. These samples showed high similarity with several L. tropica ITS1 sequences in GenBank at 99% identity. In the phylogenetic tree, the 3 L. major sequences determined in this study were grouped into L. major cluster with 100% bootstrap support value, and the 2 L. tropica sequences determined were located in L. tropica cluster (Fig. 2).
Figure 1. Detection of Leishmania DNA in sand flies.
A. PCR of Leishmania internal transcribed spacer 1 (ITS1) region amplified within pools of female Sergentomyia sand flies captured indoors and Leishmania spp. controls. M: 100 bp size marker; Lanes 1 and 2 (L. major and L. tropica reference strains, respectively); Lanes 3 to 6, S. ingrami pools, Lane 7, S. hamoni pool; Lane 8, S. africana africana pool (∼500 bp) and Lane 9, negative control. B. HaeIII digestion of restriction fragment length polymorphisms of ITS1 PCR products shown in A. M: 100 bp size marker; Lane 1, L. major (IPAP/EG/1989/S1-177); Lane 2, L. tropica (MHOM/SU/1974/K27); Lanes 3 to 6, S. ingrami pools; Lane 7, S. hamoni pool and Lane 8, S. africana africana pool.
Figure 2. Phylogenetic tree of ITS1 gene sequences among species.
Leishmania ITS1 gene was amplified within Leishmania DNA positive S. ingrami pools (IING/GH/2007/HLE-9, IING/GH/2007/KLE-2, IING/GH/2007/HLE-22 and IING/GH/2007/TAV-51) and S. hamoni pool (IHAM/GH/2007/KLE-18) and sequenced. Analysis was performed by the neighbor-joining method on the sequences together with those from 10 Leishmania species including the 2 human isolates from Ghana, L. major (MHOM/GH/2004/HO-004) and an uncharacterized Leishmania sp. (MHOM/GH/2006/TAVE). The scale bar represents 0.05% divergence. Bootstrap values are reported at nodes.
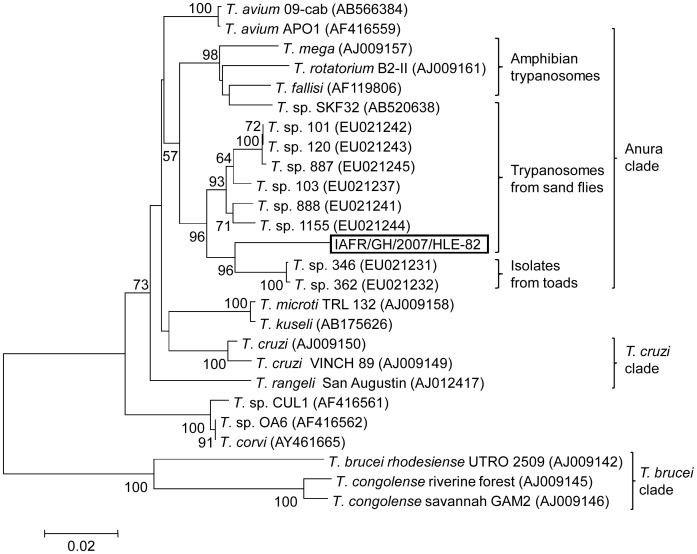
The ITS1 gene sequence of the 500 bp amplicon was determined and analyzed with the NCBI BLAST. Unexpectedly, the sequence did not match with those of Leishmania species but rather showed similarity to Trypanosoma spp., suggesting that the DNA sequence obtained belongs to the genus Trypanosoma. For further identification, the sample was analyzed with the SSU rRNA gene because this gene has been well studied in trypanosomatids [16], [17], [18]. The SSU rRNA gene sequence obtained [IAFR/GH/2007/HLE-82 (AB787191)] had 93% identity with those of trypanosomes isolated from toads. A phylogenetic tree constructed with this sequence and SSU rRNA sequences of Trypanosoma species showed that the Trypanosoma DNA sequence determined from S. africana africana pool collected from Hlefi was located in the clade comprising trypanosomes isolated from Brazilian toads (Trypanosoma sp. 362 and 364) [24], but the sequence did not completely match those from any reported species (Fig. 3). In addition, the Trypanosoma SSU rRNA sequence determined was separated from Trypanosoma species isolated from Amazon sand flies (T. sp. 101, 120, 887, 103, 888, and 1155) [24] and in a desert area of Pakistan (T. sp. SKF32) [18], which have closer relationships with anuran trypanosomes (Fig. 3). This result suggests that the Trypanosoma DNA detected within S. africana africana in this study is a novel or genetically uncharacterized Trypanosoma species possibly of amphibians.
Figure 3. Phylogenetic tree of SSU rRNA gene sequences among species.
The SSU rRNA (IAFR/GH/2007/HLE-82) gene was amplified within S. africana africana pool and sequenced. Phylogenetic analysis of the SSU rRNA gene sequences was performed by the neighbor-joining method on the sequence together with those from 25 Trypanosoma species. The sequences from the database are represented by the name of the species, isolates and GeneBank (accession number). The scale bar represents 0.02% divergence. Bootstrap values are reported at nodes.
The minimum infection rate, assuming that one infected insect was present in each positive pool was 0.49% for Leishmania spp. and 0.10% for Trypanosoma infection among the total population of pooled female sand flies. Using poolScreen2 algorithm, the maximum Leishmania DNA infection rate was estimated as follows: L. tropica infection rate in S. ingrami (1 out of 25 pools) and S. hamoni (1 out of 5 pools) were 4.07% (95% CI 1.259–2.080) and 2.21% (95% CI 6.864–1.094), respectively. Trypanosoma DNA infection in S. africana africana (1 out of 30 pools) was estimated at 3.38% (95% CI 1.045–1.731).
Validation of sand fly species by molecular biological method
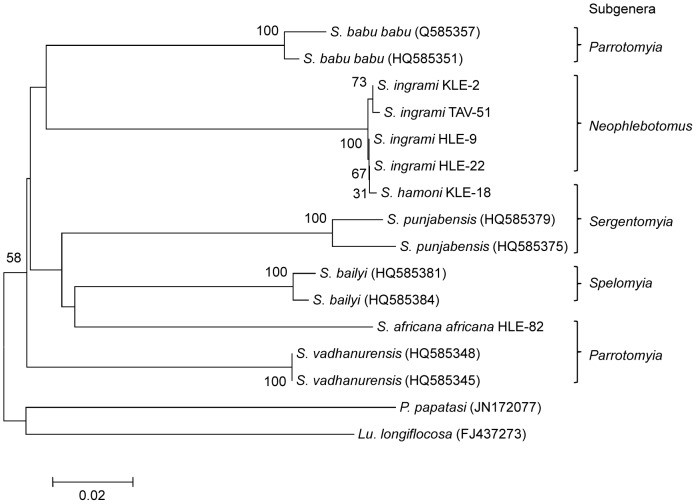
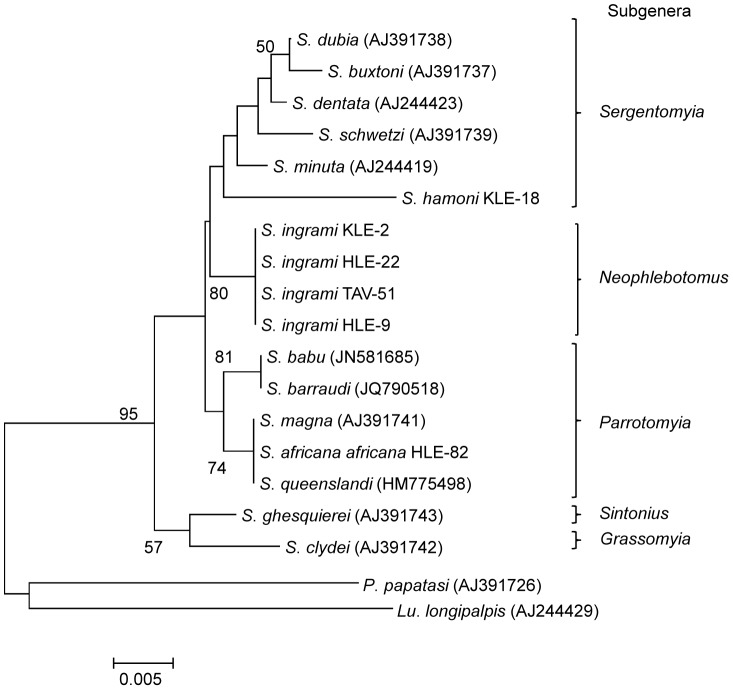
The COI gene sequences from infected flies (accession numbers: AB759971–AB759973, AB787192–AB787194) showed greater homology with those of Sergentomyia species registered in GeneBank than any other genus. A phylogenetic analysis was performed to observe the phylogenetic relationships of COI among Sergentomyia species. Among the three different infected Sergentomyia species included in phylogenetic analysis, only S. africana africana of the subgenus Parrotomyia was monophyletic (Fig. 4). All the four S. ingrami and one S. hamoni samples of the subgenera Neophlebotomus and Sergentomyia, respectively, were clustered in the same clade (Fig. 4). Similarly, 18S rRNA gene sequences from infected flies (accession numbers: AB759714–AB759716, AB787195–AB787197) had a greater degree of homology with those of Sergentomyia species (98–99%) than any Phlebotomus and Lutzomyia species (92–97%). In the phylogenetic tree, all species characterized in this study and those of other Sergentomyia species were located in their respective subgenus cluster (Fig. 5). These results confirmed that sand flies infected by Leishmania DNA belong to the genus Sergentomyia.
Figure 4. Phylogenetic tree of COI gene sequences among sand fly species.
The COI (S. ingrami HLE-9, S. ingrami HLE-22, S. ingrami KLE-2, S. ingrami TAV-51, S. hamoni KLE-18 and S. africana africana HLE-82) gene was amplified from infected sand flies and sequenced. Analysis of the sequences together with those from Sergentomyia species, Phlebotomus species and Lutzomyia species registered in GeneBank were performed. The bar scale represents 0.02% divergences. Bootstrap values are shown above or below branches. HLE, Hlefi community; KLE, Klefe community; TAV, Taviefe community.
Figure 5. Phylogenetic tree of 18S rRNA gene sequences among sand fly species.
The 18S rRNA (S. ingrami HLE-9, S. ingrami HLE-22, S. ingrami KLE-2, S. ingrami TAV-51, S. hamoni KLE-18 and S. africana africana HLE-82) gene was amplified from infected sand flies and sequenced. Analysis of the sequences together with those from Sergentomyia species, Phlebotomus species and Lutzomyia species registered in GeneBank were performed. The bar scale represents 0.005% divergences. Bootstrap values are shown above or below branches.
Discussion
In this study, we report the detection of Leishmania DNA within Sergentomyia species and their comparison with Leishmania species previously isolated from humans in Ho District. The correct identification and comparison of Leishmania species from sand flies and humans is of epidemiological importance for predictions of the risk and expansion of the disease in endemic and surrounding areas. Different PCR-based techniques are applied either in individual sand flies [6], [20] or in sand fly pools [25].
Three cutaneous leishmaniasis foci Klefe, Hlefi and Taviefe were identified in Ho, where both L. major [11] and uncharacterized species [12] were found as the etiologic agents. In the present study, only sand fly species of the genus Sergentomyia were identified from the CL focal area, consistent with previous reports from the same area [26], [27] and suggests the possible involvement of this genus in the transmission of CL. Our study revealed infection of L. major DNA in S. ingrami pools, and L. tropica DNA in one S. ingrami and one S. hamoni pools by ITS1 PCR-RFLP and sequencing analyses. These observations represent the first detection of L. tropica DNA and confirmation of L. major DNA in sand flies by gene sequencing in Ghana and support the earlier hypothesis that species of the genus Sergentomyia might be the vectors of CL in this area [26], [27]. The detection of L. tropica DNA in Ho focus is a novel finding and the first report in West Africa. In addition, DNA of Trypanosoma species was detected within one S. africana africana pool by the SSU rRNA gene analysis, which also represents the first report on infection of sand fly by trypanosomes other than Leishmania in West Africa. Furthermore, the blood-meal analysis of the blood-fed flies used in this study, revealed 3 anthropophilic Sergentomyia species; S. ingrami, S. africana africana and S. simillima (Boakye et al, unpublished results), [26].
Vector incrimination is an important part of any epidemiological studies on leishmaniasis. The vector role of a sand fly species in leishmaniasis focus is epidemiologically suspected when the species is predominant and proved anthropophilic behavior. This suspicion is strengthened when the same sand fly is found infected with the same leishmanial parasite as that found in man in the same place [28]. Phylogenetic analysis of the leishmanial ribosomal internal transcribed spacer 1 region indicates that the sequences from L. major DNA determined from S. ingrami align most closely with a human isolate from Ghana (MHOM/GH/2004/HO-004) [11]. Thus, based on the documented anthropophilic nature of S. ingrami (Boakye et al, unpublished results), [26], its abundance and the detection of L. major DNA within this sand fly as the same Leishmania species found in man from the same CL focus, suggests that S. ingrami is a possible vector of L. major transmission in the study area.
The amplification of Leishmania DNA in S. ingrami and S. hamoni, collected in Taviefe and Klefe communities, respectively, which clustered together with other ITS1 sequences of L. tropica, suggests that L. tropica might also be associated with the cases of CL in the outbreak area. Although phylogenetic analysis indicates that the ITS1 sequences from L. tropica DNA in Ghana have close affinity to an African isolate of L. tropica from Egypt, they were divergent from reported African strains. L. tropica is recognized as a very heterogeneous species of Leishmania and intra-specific microheterogeneity has been readily demonstrated [29]. The lack of information on the existence of L. tropica in human in this focus might be largely due to the small numbers of human specimens investigated [11], [12] and difficulties in distinguishing between L. major and L. tropica. The emergence of human CL due to L. tropica as an increasingly important public health problem is now being reported in many foci in Africa such as Libya, Kenya, Egypt and Morocco [30]–[33]. To our knowledge, this study presents the first detection of L. tropica DNA in Sergentomyia species.
The detection of L. tropica DNA in S. ingrami and S. hamoni in this area indicates the possibility of the two species participating in the parasite transmission cycle. The conventional wisdom was that there is specificity of Leishmania species and the vector hosts. However, it has been demonstrated that most sand fly species can support the development of multiple Leishmania species [34]. The mechanism of Leishmania attachment in permissive sand fly vectors as seen in Lutzomyia longipalpis and Phlebotomus arabicus was shown to be independent of midgut lipophosphoglycan (LPG) [35], whether such reported LPG-independent mechanism will apply to all permissive sand flies remains to be determined.
Another important finding is the infection of Trypanosoma DNA within S. africana africana. Phylogenetic analysis indicates that the SSU rRNA sequence from Trypanosoma DNA determined from S. africana africana in Ghana has a closer relationship with anuran trypanosomes, but was significantly divergent from all the reported strains [18], [24]. These observations strongly suggest that S. africana africana was infected by a novel or an uncharacterized Trypanosoma species. Several sand fly species have been reported to transmit Trypanosoma species in tropical and subtropical areas and most parasites were anuran trypanosomes [18], [36], [37]. To date, there is no data in the literature about the natural infections of S. africana africana by trypanosomes. The result presented in this study reinforces the importance of correct identification of parasitic organisms within insect vectors.
We have characterized for the first time DNA barcodes and 18S rRNA gene of S. ingrami, S. hamoni and S. africana africana. However, the sand fly COI phylogenetic analysis places both S. ingrami and S. hamoni in the same clade while the 18S rRNA gene tree separates them, several factors may explain species non-monophyly [38], but the most likely explanation, therefore, is incomplete lineage sorting. Notwithstanding this, both genes confirmed that all the infected sand fly species belonged to the genus Sergentomyia and support our evidence for species of this genus as the possible vectors of Leishmania in Ghana. Our findings question the dogma that Leishmania is exclusively transmitted by species of the genus Phlebotomus in the Old World. This observation warrants further investigation by microscopical examination of these sand fly species to obtain information on the localization of infection and presence of metacyclics, as well as initiating experimental studies in order to demonstrate their capacity to transmit the parasites. Additionally, surveillance would also be intensified aiming to identify the natural reservoir hosts in order to improve our understanding of the epidemiology of CL in the outbreak area.
Although sand flies of the genus Sergentomyia are considered as vectors of reptile Leishmania species, non-pathogenic to human [39], and S. schwetzi reported to be refractory to human Leishmania species [40], our data adds to few reported studies on the detection of human pathogenic Leishmania species in some Sergentomyia species: S. sintoni [5], S. darlingi [6], S. minuta [7], S. garnhami [8], S. babu [9] and S. gemmea [10], incriminating them as potential vectors of mammalian Leishmania species. Definitive conclusion that these Sergentomyia species are vectors of human Leishmania species awaits confirmation by demonstrating experimentally their capacity to transmit Leishmania parasite by biting to mammals. Though it is technically difficult to establish laboratory colonies of some species of sand flies and infection studies therein, it remains highly desirable for vector incrimination.
Taken together, the presence of multiple of Leishmania spp. in this CL focus suggests a more complex epidemiology for this outbreak than anticipated. Therefore, a proper understanding of the different parasites' life cycles and parasite-vector-reservoir interplays is needed for predicting the risk and expansion of the disease and applying effective prevention and control strategies in Ghana.
Acknowledgments
We thank Dr. Jeffrey T. Villinski, Vector Biology Program, U.S. Naval Medical Research Unit No. 3, Cairo, Egypt, for providing DNA of L. major (IPAP/EG/1989/S1-177) and L. tropica (MHOM/SU/1974/K27).
Funding Statement
This study was partly supported by funds from the Parasitology Department, Noguchi Memorial Institute for Medical Research, University of Ghana, Legon-Ghana, the US NAMRU 3, the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (Grant No. 23580424) and the Program for Leading Graduate Schools “Fostering Global Leaders in Veterinary Science for Contributing to One Health” (FO1), MEXT, Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Alvar J, Ve'lez ID, Bern C, Herrero M, Desjeux P, et al. (2012) Leishmaniasis Worldwide and Global Estimates of Its Incidence. PLoS ONE 7: e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO (2010) Control of the leishmaniasis: report of a meeting of the WHO Expert Committee on the control of leishmaniasis. World Health Organ Tech Rep Ser 949, Geneva.
- 3. Klaus S, Frankenburg S (1999) Cutaneous leishmaniasis in the Middle East. Clin Dermatol 17: 137–147. [DOI] [PubMed] [Google Scholar]
- 4.Munstermann LE (2004) Phlebotomine sand flies, the Psychodidae. In: Marquardt WC, Black WC, Freier JE, Hagedorn HH, Hemingway J, et al.., editor. Biology of Disease Vectors. Second edition. San Diego, CA: Elsevier. pp 141–151. [Google Scholar]
- 5. Parvizi P, Amirkhani A (2008) Mitochondrial DNA characterization of Sergentomyia sintoni populations and finding mammalian Leishmania infections in this sand fly by using ITS-rDNA. Iranian J Vet Res 22: 9–18. [Google Scholar]
- 6. Berdjane-Brouk Z, Koné AK, Djimde AA, Charrel RN, Ravel C, et al. (2012) First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous leishmaniasis foci in Mali. PloS one 7: e28266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Campino L, Cortes S, Dionísio L, Neto L, Afonso MO, et al. (2013) The first detection of Leishmania major in naturally infected Sergentomyia minuta in Portugal. Mem Inst Oswaldo Cruz Rio de Janeiro 108: 516–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mutinga MJ, Massamba NN, Basimike M, Kamau CC, Amimo FA, et al. (1994) Cutaneous leishmaniasis in Kenya: Sergentomyia garnhami (Diptera: Psychodidae), a possible vector of Leishmania major in Kitui District: a new focus of the disease. East Afr Med J 71: 424–428. [PubMed] [Google Scholar]
- 9. Mukherjee S, Hassan MQ, Ghosh A, Ghosh KN, Bhattacharya A, et al. (1997) Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. Am J Trop Med Hyg 57: 423–425. [DOI] [PubMed] [Google Scholar]
- 10. Kanjanopas K, Siripattanapipong S, Ninsaeng U, Hitakarun A, Jitkaew S, et al. (2013) Sergentomyia (Neophlebotomus) gemmea, a potential vector of Leishmania siamensis in southern Thailand. BMC Inf Dis 13: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fryauff DJ, Hanafi HA, Klena JD, Hoel DF, Appawu M, et al. (2006) ITS-I DNA sequence confirmation of Leishmania major as a cause of cutaneous leishmaniasis from an outbreak focus in the Ho District, southeastern Ghana. Am J Trop Hyg 75: 502–504. [PubMed] [Google Scholar]
- 12. Villinski JF, Klena JD, Abbassy M, Hoel DF, Puplampu N, et al. (2008) Evidence for a new species of Leishmania associated with a focal disease outbreak in Ghana. Diagn Microbiol Infect Dis 60: 323–327. [DOI] [PubMed] [Google Scholar]
- 13.Abonnenc E (1972). Les Phlebotomus de la Region Ethiopienne (Diptera, Psychodidae). Paris, ORSTOM. [Google Scholar]
- 14.Niang AA, Geoffory B, Angel G, Trouillet J, Killick-Kendrick R, et al. (2000) Les Phlebotomus de la d' Afrique de l' Quest: Institute de recherché pour le development, Institut fundamental d' Afrique noir l' Université Cheikh Anta Diop de Dakar.
- 15. El Tai NO, Osman OF, El Fari M, Presber W, Schönian G (2000) Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans R Soc Trop Med Hyg 94: 575–579. [DOI] [PubMed] [Google Scholar]
- 16. Noyes HA, Stevens JR, Teixeira M, Phelan J, Holz P (1999) A nested PCR for ssrRNA gene detects Trypanosoma binneyi in the platypus and Trypanosoma sp. in wombats and kangaroos in Australia. Int J Parasitol 29: 331–339. [DOI] [PubMed] [Google Scholar]
- 17. Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC (2004) Trypanosomes are monophyletic: evidence from genes for glyceraldehydes phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol 29: 1393–1404. [DOI] [PubMed] [Google Scholar]
- 18. Kato H, Uezato H, Sato H, Bhutto AM (2010) Natural infection of the sand fly Phlebotomus kazeruni by Trypanosoma species in Pakistan. Parasit Vectors 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato H, Uezato H, Katakura K, Calvopiña M, Marco JD, et al. (2005) Detection and identification of Leishmania species within naturally infected sand flies in the Andean areas of Ecuador by a polymerase chain reaction. Am J Trop Med Hyg 72: 87–93. [PubMed] [Google Scholar]
- 21. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 23. Katholi CR, Toé L, Merriweather A, Unnasch TR (1995) Determining the prevalence of Onchocerca volvulus infection in vector populations by PCR screening of pools of black flies. J Infect Dis 172: 1414–1417. [DOI] [PubMed] [Google Scholar]
- 24. Ferreira RC, De Souza AA, Freitas RA, Campaner M, Takata CS, et al. (2008) A phylogenetic lineage of closely related trypanosomes (Trypanosomatide, Kinetoplastida) of anurans and sand flies (Psychodidae, Diptera) sharing the same ecotopes in Brazilian Amazonia. J Eukaryot Microbiol 55: 427–435. [DOI] [PubMed] [Google Scholar]
- 25. Martín-Sánchez J, Gállego M, Barón S, Castillejo S, Morillas-Marquez F (2006) Pool screen PCR for estimating the prevalence of Leishmania infantum infection in sand flies (Diptera: Nematocera, Phlebotomidae). Trans R Soc Trop Med Hyg 100: 527–532. [DOI] [PubMed] [Google Scholar]
- 26. Kweku MK, Odoom S, Puplampu N, Desewu K, Nuako GK, et al. (2011) An outbreak of suspected cutaneous leishmaniasis in Ghana: lessons learnt and preparation for future outbreaks. Global Health Action 4: 5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boakye DA, Wilson MD, Kweku M (2005) A Review of leishmaniasis in West Africa. Ghana Med J 39: 94–97. [PMC free article] [PubMed] [Google Scholar]
- 28. Killick-Kendrick R (1990) Phlebotomine vectors of the leishmaniasis; a review. Med Vet Ent 4: 1–24. [DOI] [PubMed] [Google Scholar]
- 29. Schönian G, Schnur LF, El Fari M, Oskam L, Kolesnikov AA, et al. (2001) Genetic heterogeneity in the species Leishmania tropica revealed by PCR-based methods. Trans R Soc Trop Med Hyg 95: 217–224. [DOI] [PubMed] [Google Scholar]
- 30. Amro A, Gashout A, Al-Dwibe H, Alam MZ, Annajar B, et al. (2012) First molecular epidemiological study of cutaneous leishmaniasis in Libya. PLoS Negl Trop Dis 6: e1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sang DK, Njeru WK, Ashford RW (1992) A possible animal reservoir for Leishmania tropica s.l. in Kenya. Ann Trop Med Parasitol 86: 311–312. [DOI] [PubMed] [Google Scholar]
- 32. Shehata MG, Samy AM, Doha SA, Fahmy AR, Kaldas MR, et al. (2009) First report of Leishmania tropica from a classical focus of L. major in North-Sinai, Egypt. Am. J Trop Med Hyg 81: 213–218. [PubMed] [Google Scholar]
- 33. Rhajaoui M, Sebti F, Fellah H, Alam MZ, Nasereddin A, et al. (2012) Identification of the causative agent of cutaneous leishmaniasis in Chichaoua Province, Morocco. Parasite 19: 91-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Volf P, Myskova J (2007) Sand flies and Leishmania: specific versus permissive vectors. Trends Parasitol 23: 91–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Myskova J, Svobodova M, Stephen M, Beverley SM, Volf P (2007) A lipophosphoglycan-independent development of Leishmania in permissive sand flies. Microbes Infect 9: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lemos M, Morais DH, Carvalho VT, Ď Agosto M (2008) First record of Trypanosoma chattoni in Brazil and occurrence of other Trypanososma species in Brazilian frogs (Anura, Leptodactylidae). J Parasitol 94: 148–151. [DOI] [PubMed] [Google Scholar]
- 37. Viola LB, Campaner M, Takata CSA, Ferreira RC, Rodrigues AC, et al. (2008) Phylogeny of snake trypanosomes inferred by SSU rDNA sequences their possible transmission by phlebotomines, and taxonomic appraisal by molecular, cross-infection and morphological analysis. Parasitology 135: 595–605. [DOI] [PubMed] [Google Scholar]
- 38. Jinbo U, Kato T, Ito M (2011) Current progress in DNA barcoding and future implications for entomology. Entomol Sci 14: 107–124. [Google Scholar]
- 39. Bates P (2007) Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol 37: 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sadlova J, Dvorak V, Seblova V, Warburg A, Votypka J, et al. (2013) Sergentomyia schwetzi is not a competent vector for Leishmania donovani and other Leishmania species pathogenic to humans. Parasit Vectors 6: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]